Abstract
The past 50 years has witnessed the emergence of new viral and bacterial pathogens with global effect on human health. The hyperinvasive group A Streptococcus (GAS) M1T1 clone, first detected in the mid-1980s in the United States, has since disseminated worldwide and remains a major cause of severe invasive human infections. Although much is understood regarding the capacity of this pathogen to cause disease, much less is known of the precise evolutionary events selecting for its emergence. We used high-throughput technologies to sequence a World Health Organization strain collection of serotype M1 GAS and reconstructed its phylogeny based on the analysis of core genome single-nucleotide polymorphisms. We demonstrate that acquisition of a 36-kb genome segment from serotype M12 GAS and the bacteriophage-encoded DNase Sda1 led to increased virulence of the M1T1 precursor and occurred relatively early in the molecular evolutionary history of this strain. The more recent acquisition of the phage-encoded superantigen SpeA is likely to have provided selection advantage for the global dissemination of the M1T1 clone. This study provides an exemplar for the evolution and emergence of virulent clones from microbial populations existing commensally or causing only superficial infection.—Maamary, P. G., Ben Zakour, N. L., Cole, J. N., Hollands, A., Aziz, R. K., Barnett, T. C., Cork, A. J., Henningham, A., Sanderson-Smith, M., McArthur, J. D., Venturini, C., Gillen, C. M., Kirk, J. K., Johnson, D. R., Taylor, W. L., Kaplan, E. L., Kotb, M., Nizet, V., Beatson, S. A., Walker, M. J. Tracing the evolutionary history of the pandemic group A streptococcal M1T1 clone.
Keywords: Streptococcus pyogenes, virulence factors, reemergent pathogens
The discovery and broad application of antimicrobials since the mid-20th century has reduced morbidity and mortality associated with numerous infectious agents. Nonetheless, the past 50 years has witnessed the emergence and resurgence of microbial pathogens, such as HIV, SARS, H5N1 (bird flu), H1N1 (swine flu), and multiply drug-resistant enterocococci, staphylococci, Escherichia coli, and Mycobacterium tuberculosis (1, 2). While infection with group A Streptococcus (GAS) has remained endemic in many indigenous populations (3), the incidence of severe GAS disease in Western countries had, until recently, steadily declined beginning in the early 20th century (4, 5). In the mid-1980s, a resurgence in serious streptococcal infections in the United States was attributed to a unique M1T1 GAS clone (6). This M1T1 clone has been globally disseminated and is frequently associated with severe life-threatening conditions, such as “flesh-eating” necrotizing fasciitis and streptococcal toxic shock-like syndrome (STSS) (4, 7, 8). The overrepresentation of the GAS M1T1 clone in these severe disease episodes may correspond to the high prevalence of this lineage in uncomplicated cases of pharyngitis. Nevertheless, in comparison with other GAS lineages, the predominance of this M1T1 clone in the GAS population is striking, regardless of disease topology (9).
The emergence of the M1T1 clone has been attributed to the acquisition of novel phages encoding the superantigen SpeA2 and the DNase Sda1/SdaD2 (10, 11) and a horizontally acquired 36-kb M12 GAS chromosomal region encoding the virulence factors streptolysin O and NAD glycohydrolase (11). The superantigen SpeA, which is resistant to degradation by SpeB, has been implicated in enhancing the severity of invasive M1T1 disease (12), while the bacteriophage-encoded Sda1 is a potent virulence factor (13) that protects M1T1 GAS from neutrophil killing by degrading the DNA framework of neutrophil extracellular traps (NETs; refs. 14, 15, 16).
Initial evolutionary studies used a comparative genomic approach to determine differences between M1T1 clonal isolates and the M1 isolate SF370 (10, 11). On the basis of these findings, the emergence of the globally disseminated M1T1 clone was proposed to comprise two major horizontal gene transfer events: first the acquisition of bacteriophages encoding SpeA2 and the DNase Sda1, then the recombinatorial replacement of the 36-kb chromosomal segment encoding enhanced expression of streptolysin O and NAD glycohydrolase from a serotype M12 parental strain (11). A separate PCR screening study for the speA2 and sda1 genes generated evidence suggesting that introduction of the speA2-encoding phage may have preceded the sda1-encoding phage in the M1 background (17).
With access to an extensive World Health Organization collection of North American serotype M1 isolates and comprehensive genomic data generated by high-throughput, Illumina technology, we hypothesize that acquisition of the 36-kb genome segment from serotype M12 GAS and the bacteriophage-encoded DNase Sda1 results in increased virulence of the M1T1 precursor and occurred relatively early in the molecular evolutionary sequence. The more recent acquisition of the phage-encoded superantigen SpeA may then have provided selection advantage for the global dissemination of the M1T1 clone.
MATERIALS AND METHODS
GAS strains and culture
GAS serotype M1 isolates examined in this study were selected to ensure coverage of the restriction enzyme analysis (REA) types described by Johnson et al. (9) and also include representatives of the globally disseminated M1T1 clone (18) and the genome sequenced M1 isolate SF370 (19) (Table 1). Strains were routinely cultured on commercial horse-blood agar or in static liquid medium at 37°C in Todd-Hewitt broth supplemented with 1% (w/v) yeast extract (THY).
Table 1.
Streptococcus pyogenes isolates and isogenic mutant strains used in this study
| Isolate | Infection | emm type | REA type | PFGE type | covRS allele | SpeB phenotype | Ref. |
|---|---|---|---|---|---|---|---|
| 89158 | I | 1.46 | b | A | WT | + | 9 |
| L485 | U | 1.0 | f | B | WT | + | 9 |
| SF370 | U | 1.6 | ND | C | WT | + | 19 |
| NS696 | U | 1.0 | ND | D | WT | + | 23 |
| AB1551 | U | 1.0 | e | D | WT | + | 9 |
| 90223 | I | 1.0 | g | E | WT | + | 9 |
| GT94052 | U | 1.0 | h | E | WT | + | 9 |
| GT94966 | I | 1.0 | d | E | WT | + | 9 |
| 5636 | I | 1.0 | c | E | WT | + | This study |
| 5448 | I | 1.0 | c | E | WT | + | 18 |
| 5448AP | AP | 1.0 | ND | ND | Mutant | − | 36 |
| 5636ΔspeA | NA | 1.0 | ND | ND | WT | + | This study |
| 5636ΔspeA-comp | NA | 1.0 | ND | ND | WT | + | This study |
AP, animal passage; I, invasive; NA, not applicable; ND, not determined; PFGE, pulsed-field gel electrophoresis; REA, restriction enzyme analysis; U, uncomplicated; WT, wild type.
SpeB activity assays and Western blot analysis
SpeB cysteine protease activity in cell-free overnight GAS supernatants was determined using a modified azocaseinolytic assay (20) or by culture on Columbia skim milk agar (21). Previously described methods were used to determine the GAS SpeB phenotype following subcutaneous murine passage (22). Detection of SpeB in stationary phase supernatants by Western blot was performed using standard protocols (22).
emm Sequence typing
The emm sequence type of isolates used in this study was determined as described previously (23), and it was designated in accordance with established criteria (http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm).
DNA-DNA microarray
The oligonucleotide microarrays and methodology used for DNA-DNA microarray experiments were described previously (24). Under established criteria (24), all median-normalized fluorescence values <40 were designated absent.
Pulsed-field gel electrophoresis (PFGE)
Pulsed-field gel electrophoresis (PFGE) was performed using the method of Ramachandran et al. (25).
Genome sequencing
Genomic DNA was isolated from overnight liquid cultures using the DNeasy blood and tissue kit (Qiagen, Valencia, CA, USA), according to the manufacturer's guidelines. Genomic fragment libraries for whole-genome sequencing were prepared at the Australian Genome Research Facility (University of Queensland, St. Lucia, QLD, Australia) using the Illumina TruSeq DNA library preparation protocol (Illumina, Inc., San Diego, CA, USA). Libraries were pooled for sequencing on the Illumina Genome Analyzer II instrument. Of note, the strain 5448 (18), as well as the previously sequenced strain S. pyogenes MGAS5005 (11), used here as an internal reference, were both run on the Illumina HiSeq 2000 instrument. Paired-end 76-bp sequence reads were generated for all strains, with the exception of 5448 and MGAS5005, for which paired-end 100-bp sequence reads were generated. For each GAS isolate, between 759,056 and 8,879,651 read pairs were obtained, corresponding to an estimated average coverage of 62× to 964×. FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/) was used to assess the quality of the sequence reads. All reads for the 10 different M1 strains have been deposited in the European Nucleotide Archive (ENA) under the study accession no. ERP001330.
Single-nucleotide polymorphism (SNP) detection
Reads were mapped to the MGAS5005 reference genome (GenBank CP000017) using two different tools, MAQ (26) and BWA (27). Following read mapping with BWA, a consensus pileup was produced using Samtools (28), and putative SNPs were called using bcftools (28). Aligned reads containing SNPs predicted by both approaches were also manually inspected before being annotated.
Phylogenetic analysis
Whole-genome SNP-based phylogenetic reconstruction using the maximum-likelihood model was carried out with 505 high-quality SNPs. Of note, SNPs occurring in prophage-related regions, as well as those occurring in the horizontally acquired 36-kb M12 chromosomal segment, were excluded to minimize interferences of SNPs associated with recombination events. The maximum-likelihood estimation was implemented using PhyML (29) with the nucleotide substitution model T92, as recommended by jModelTest (30), and without substitution rate heterogeneity correction or invariant estimation. Clade support was evaluated by analyzing 1000 bootstrap pseudoreplicates.
Transcriptional microarray
The glass slide DNA microarrays used for transcriptional analysis in this study were obtained from the Pathogen Functional Genomics Research Center (sponsored by the U.S. National Institute of Allergy and Infectious Diseases) at the J. Craig Venter Institute (Rockville, MD, USA). These arrays (S. pyogenes version 2) comprised 70-mer oligonucleotide probes representing various GAS core and prophage open reading frames (ORFs) in addition to 500 Arabidopsis thaliana control 70-mers. All probes representing MGAS5005 and SF370 ORFs were used for analysis; however, all prophage sequences and those within the 36-kb slo- and nga-encoding region were excluded to facilitate examination of the core M1 genome. Single-color hybridizations were performed essentially as outlined in the hybridization of labeled DNA and cDNA probes protocol (available online at http://pfgrc.jcvi.org/index.php/microarray/protocols.html) employed by the Pathogen Functional Genomics Resource Center and the Institute for Genomic Research (J. Craig Venter Institute). Slides were scanned using a GenePix 4000B scanner (Axon Instruments, Sunnyvale, CA, USA) and processed with GenePixPro 4.0 (Axon Instruments). Transcriptional analyses were undertaken using GeneSpring GX 11.5 (Agilent Technologies, Columbia, MD, USA), and microarray data submitted to the U.S. National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) in accordance with MIAME standards (GEO accession no. GSE35568).
Neutrophil killing assays
Assays for GAS survival following incubation with human neutrophils in vitro were performed as described previously (24, 31). These assays were conducted using midlogarithmic phase bacteria at a multiplicity of infection (MOI) of 10:1 (GAS:neutrophils).
Human epithelial cell adherence assays
Assays for the GAS adherence to HEp-2 human epithelial cells in vitro were performed as described previously (31).
In vivo SpeB-switching assays
To monitor the phase-switch in SpeB phenotype during local infection, C57BL/J6 mice were subcutaneously challenged with 107 cfu/100 μl GAS, then SpeB-switching assays undertaken as described previously (14, 24). Representative SpeB-negative colonies were selected for sequence analysis to confirm loss of SpeB expression through covRS mutation using standard protocols (14).
Transgenic murine infection model
Subcutaneous GAS infection of the humanized plasminogen transgenic AlbPLG1 mouse model utilized in this study has been previously described (14, 24, 32).
Construction of a recombinant speA-knockout GAS strain
A ligation-independent cloning (LIC) strategy was employed for the generation of the speA knockout pHY304-LIC vector used in this study (33). Briefly, the 400 and 450 bases immediately upstream and downstream of the speA ORF, respectively, and the cat gene were amplified by PCR, while the pHY304-LIC vector was prepared by PmeI digestion (oligonucleotide primer sequences are given in Supplemental Table S1). Following treatment with T4 polymerase, all fragments were combined and transformed into DH5α E. coli by heat shock. Electrotransformation and allelic replacement mutagenesis were undertaken using standard protocols (15, 34). Successful allelic replacement was confirmed by PCR and sequence analysis.
RESULTS
Genomic characterization and phylogenetic analysis of M1 GAS
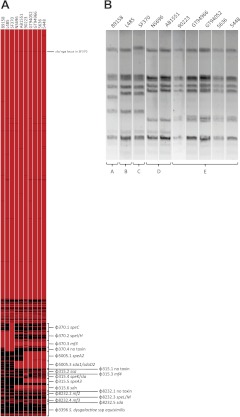
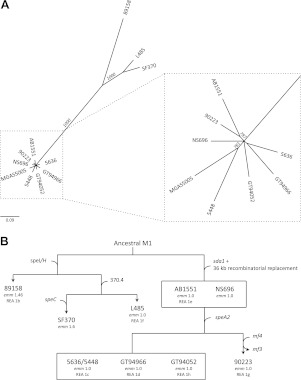
The GAS serotype M1 isolates examined in this study were selected to ensure coverage of the REA types described by Johnson et al. (ref. 9 and Table 1). Both the M1T1 clonal strain 5636 and the previously characterized M1T1 clonal strain 5448 were included in this cohort to represent the globally disseminated M1T1 clone (14, 18, 24, 35). Preliminary DNA microarray analysis revealed that carriage of core GAS genes was conserved across the M1 isolates examined, with prophage sequences comprising the major source of variation in gene complement (Fig. 1A). As documented for an M1T1 subpopulation (10), isolate 90223 had undergone a toxin exchange, losing the otherwise ubiquitous mf3 and acquiring the homologous DNase-encoding mf4. PFGE analysis revealed the 8 REA types represented in this strain set could be categorized into 5 PFGE types (Fig. 1B). The observation of 4 REA types sharing the same PFGE profile as the M1T1 clone indicates conserved phage carriage across these M1T1-like isolates. GAS M1 strains 89158, L485, SF370, NS696, AB1551, 90223, GT94052, GT94966, 5636, and 5448 were subjected to Illumina genome sequencing to reveal fine-scale genomic features that distinguish M1T1 GAS. A list of core genome SNPs compared to MGAS5005 was generated to provide insight into the evolutionary relatedness of these isolates. All SNPs detected within phage boundaries, as well as those occurring in the horizontally acquired 36-kb M12 chromosomal segment, were excluded. All isolates positive for carriage of the sda1-encoding prophage and the horizontally acquired 36-kb M12 chromosomal segment displayed highly conserved SNP-profiles (Fig. 2A). We propose a model for the evolution of the globally disseminated M1T1 clone (Fig. 2B) in which the majority of genetic variation in serotype M1 GAS accumulated prior to the acquisition of the speA-encoding prophage, an event that occurred relatively recently in evolutionary history and is likely to have triggered the subsequent global spread of the M1T1 clone.
Figure 1.
Preliminary genotyping of S. pyogenes serotype M1 isolates. A) DNA-DNA microarray heat map of M1 isolates generated using an oligonucleotide glass slide array described previously and comprised probes representing the M1 core ORFeome (24). Additional probes representing prophage ORFs from GAS serotypes M1, M3, M18, and S. dysgalactiae subsp. equisimilis were included on the array and have been labeled on the heat map. Heat map was generated from background-subtracted median-normalized fluorescence units. Values equal to or exceeding threshold were designated present (red), while those below were considered absent (black). B) PFGE of M1 isolates digested with SmaI. Strains under examination are given at the top of the gel, and isolates are grouped into PFGE types A–E.
Figure 2.
Genomic characterization of S. pyogenes serotype M1 isolates. A) Maximum likelihood tree with 1000 bootstrap replicates generated using a list of core genome SNPs. Bootstrap values >500 are given in italics. All SNPs occurring in prophage regions, and the 36-kb SLO- and NADase-encoding recombinatorial replacement region, were excluded for analysis. Scale bar represents the number of base substitutions per site. B) Proposed model for the evolution of the globally disseminated M1T1 clone. Acquisition of the phage-encoded genes speI/H, speC, sda1, speA, mf4, and the toxin-negative phage 370.4 are indicated. The recombination event resulting in the transfer of a 36-kb chromosomal segment from a parental M12 strain is also shown. The emm sequence type of each isolate and REA pattern according to Johnson et al. (9) are given. emm subtypes have been designated according to established criteria (http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm). Isolates contained within boxes are equivalent for gene carriage, emm sequence type, and REA type (where known). The globally disseminated M1T1 clone is represented by the isolates 5636 and 5448.
Expression profiling of M1 GAS
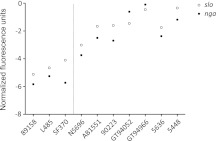
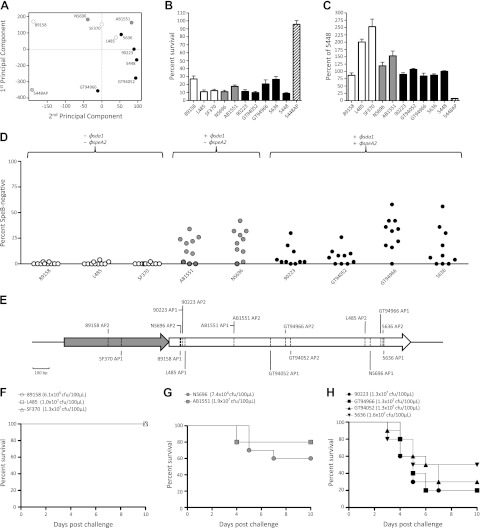
The GAS isolates included in this study were selected to ensure that the covRS wild-type form of each REA type was represented. All isolates were found to produce the cysteine protease SpeB (Table 1), and alignment of the covRS operon with the previously described reference 5448 (14) confirmed the integrity of this global regulator. Transcriptomic microarray analysis revealed that the relative expression of core GAS genes was conserved across the isolates studied, with major differences confined to horizontal gene transfer regions. As has been previously documented (11), virulence genes slo and nga located within the recombinatorial 36-kb chromosomal segment were up-regulated in M1 isolates harboring the horizontally acquired 36-kb M12 chromosomal region (Fig. 3). To compare the similarity of these transcriptomes, the animal-passaged isogenic covS mutant form of 5448 (5448AP) was included as a transcriptionally distinct reference. This strain has been previously characterized (36) and represents the hyperinvasive form of M1T1 GAS that is frequently isolated from cases of invasive disease and is consistently recovered following passage of the wild-type M1T1 clone in vivo (14, 36, 37). Relative to 5448AP and the divergent 89158, the remaining strains clustered in a principle component analysis (PCA) revealing a conserved expression profile among historic and contemporary M1 isolates (Fig. 4A). In accordance with previous work (8, 11, 38), transcription of prominent GAS virulence genes, including the antiphagocytic factors SLO, NAD glycohydrolase, and capsule, in addition to the secreted toxins SIC (streptococcal inhibitor of complement) and SpyA (C3 family ADP-ribosyltransferase), was up-regulated in the covS mutant 5448AP compared to all M1 isolates studied (results not shown).
Figure 3.
Midlogarithmic phase transcriptomic analysis of M1 isolates in vitro. Expression of the streptolysin O (slo)- and NAD glycohydrolase (nga)-encoding genes in M1 isolates containing intact covRS loci. Dotted line separates strains containing historic slo and nga genes (left) from those containing the contemporary allele (right) acquired from a serotype M12 donor strain. Data are presented as normalized fluorescence units; larger fluorescence values correspond with increased expression.
Figure 4.
Phenotypic analysis and interactions of GAS with the innate immune response in vitro and in vivo. Shading indicates carriage of the sda1 and speA prophages as follows: sda1−/speA−, open symbols and bars; sda1+/speA−, shaded symbols and bars; sda1+/speA+, solid symbols and bars. The animal-passaged covS mutant 5448AP is indicated by striped symbols and bars. A) Principal component analysis at the midlogarithmic phase of growth in vitro. B) Percentage survival of S. pyogenes following coincubation with human neutrophils in vitro (mean±se). C) Adherence of S. pyogenes to the human epithelial HEp-2 cell line in vitro (mean±se). D) Percentage of SpeB− cfu recovered following a 3-d subcutaneous passage of SpeB+ S. pyogenes in C57BL/J6 mice. Each data point represents a single inoculated mouse. Data for isolate SF370 reproduced from previous work (14). E) Map of inactivating covRS mutations in representative SpeB− cfu recovered following subcutaneous passage. For each strain, two representative animal-passaged variants were selected from independent mice (n=1 for SF370, as loss of SpeB expression was detected in only a single mouse). F–H) Percentage survival of humanized plasminogen transgenic AlbPLG1 mice (n=10) following subcutaneous infection with the sda1−/speA− isolates 89158, L485, and SF370 (F), the sda1+/speA− isolates NS696 and AB1551 (G), and the sda1+/speA+ isolates 90223, GT94052, GT94966, and 5636 (H).
Host-pathogen interactions, invasive capacity and murine virulence of M1 GAS
Each of the isolates in this study was susceptible to killing by human neutrophils (P<0.001; Fig. 4B) and displayed significantly increased adherence to the HEp-2 human epithelial cell line in comparison to the covS mutant 5448AP (P<0.001; Fig. 4C). While increased adherence compared to wild-type 5448 was observed in the divergent isolates L485 and SF370 (P<0.001), the relative adherence of the remaining isolates was equivalent to that of the M1T1 strain 5448. We next examined the capacity of the M1 isolates in this study to switch to the invasive SpeB-negative covRS mutant form during subcutaneous murine passage. In accordance with previous studies using 5448 (14, 35), the M1T1 strain 5636 was also found to SpeB-switch during replication in vivo (Fig. 4D). This observation was consistent for all sda1+ isolates (Fig. 4D), corroborating the hypothesis that, under pressure from the immune response, Sda1 serves as a selective force for covRS mutation (14). The limited capacity of the M1 isolates 89158, L485, and SF370 to SpeB-switch is consistent with our previous findings that selection for this switch is removed in the absence of Sda1 (14). Representative SpeB-negative colonies recovered from murine passage were chosen and covRS sequence analysis confirmed that loss of SpeB expression could be mapped to mutations in this operon (Fig. 4E; Supplemental Table S2). Compared to the M1T1 strain 5636, the reduced frequency of switching in the M1 isolates 89158, L485, and SF370 correlated with attenuated virulence in the humanized plasminogen transgenic AlbPLG1 mouse model of subcutaneous infection (P<0.05; Fig. 4F). The capacity of all sda1+ isolates to switch in vivo correlated with increased virulence in this model (Fig. 4G, H).
Role of SpeA in M1T1 clone pathogenesis
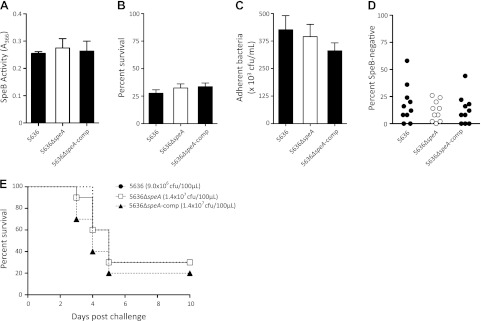
SpeA is a major virulence determinant known to contribute to immune dysregulation and generation of an exaggerated inflammatory response, marked by a distinct cytokine profile (12, 39, 40, 41, 42). We set out to examine whether SpeA contributes to GAS virulence, in ways apart from dysregulation of immune cells. Thus, we generated an isogenic speA− knockout mutant in the M1T1 strain 5636, designated 5636ΔspeA. This mutation was restored to the original speA+ genotype (5636ΔspeA-comp) in accordance with Koch's molecular postulates (43). As expected, this covRS intact speA− mutant and complemented mutant were positive for SpeB production (Fig. 5A). We subjected 5636ΔspeA and 5636ΔspeA-comp to neutrophil killing and HEp-2 adherence assays in vitro; abrogation of speA neither affected resistance to neutrophil killing, adherence to epithelial cells (P<0.01; Fig. 5B, C), the capacity for a switch to the hyperinvasive covRS mutant form during subcutaneous murine passage (Fig. 5D), nor the virulence of 5636ΔspeA in the transgenic AlbPLG1 mouse model (P<0.05; Fig. 5E).
Figure 5.
Characterization of the speA+ M1T1 strains 5636 and 5636ΔspeA-comp (solid symbols and bars) and the isogenic speA− knockout mutant 5636ΔspeA (open symbols and bars). A) SpeB activity in cell-free stationary phase supernatants (mean±se). B) Percentage survival during coculture with human neutrophils in vitro (mean±se). C) Relative adherence to the human epithelial HEp-2 cell line in vitro (mean±se). D) Capacity to switch to an invasive SpeB− phenotype at the site of local infection was determined during a 3-d subcutaneous passage in C57BL/J6 mice. Each data point represents the percentage of SpeB− cfu recovered from a single mouse (n=10/strain). E) Relative virulence of 5636 (9.0×106 cfu/dose), 5636ΔspeA (1.4×107 cfu/dose), and 5636ΔspeA-comp (1.4×107 cfu/dose) was assessed over a 10-d period following subcutaneous infection of humanized plasminogen transgenic AlbPLG1 mice (n=10 mice/strain).
DISCUSSION
Horizontal gene transfer has been central to the emergence and reemergence of contemporary microbial pathogens (1). The recrudescence of severe GAS infection has been largely attributed to the horizontal exchange and diversification of toxin-encoding elements (10, 44).
The streptodornase Sda1 is known to contribute to virulence of the GAS M1T1 clone (14, 15). In this study, we observe that the capacity of M1 GAS to switch to the invasive covRS mutant form during local infection occurs in all M1 GAS carrying the sda1 gene, including the globally disseminated M1T1 clone and the speA− M1 strains NS696 and AB1551. This observation suggests that increased virulence associated with carriage of the sda1-encoding bacteriophage is unlikely, per se, to have resulted in global dissemination of the M1T1 clone. The most closely related streptodornase to Sda1 has been described in M12 serotype GAS (44). The horizontally acquired 36-kb M12 GAS chromosomal region and sda1-encoding bacteriophage both appear to have been acquired from a GAS M12 background (11, 13). In combination with the findings presented in this study, these observations suggest that the incorporation of these elements into the M1 genome may have possibly occurred simultaneously.
The incidence of infection with GAS expressing SpeA superantigen markedly declined throughout the early 1900s (45). The speA2 allele of M1T1 GAS was first described following the resurgence of STSS episodes in the mid-1980s and, unlike the speA1 allele, was confined to this single clonal lineage (12). SpeA2 contains a single amino acid substitution in a region that may play a role in superantigen mitogenicity, which is otherwise highly conserved among staphylococcal and streptococcal superantigens (12). This superantigen preferentially binds human leukocyte antigen (HLA)-DQ (46), and disruption of speA in M1 GAS has been shown to reduce promitogenic activity 2-fold (40). We observe that an M1T1 speA-knockout mutant is not attenuated for virulence in a humanized murine model of skin infection, which is consistent with previous studies (40, 41). This observation is likely due to the reduced capacity of murine MHC class II molecules to bind SpeA, as infection of mice carrying the HLA-DQ transgene with speA+ M1T1 GAS has been shown to increase immune activation and lymphoblastic tissue infiltration (42).
Whole-genome phylogenetic analysis suggests that acquisition of speA in the 1980s provided the M1T1 index strain with a powerful selection advantage allowing for the global dissemination of the M1T1 clone. This improved fitness may result in enhanced colonization, immune defense, in vivo persistence, and/or host-host transmission. As a superantigen, SpeA is capable of activating a range of T-cell subsets via cross-linking conserved sequences of the T-cell receptor Vβ chain with constant regions of major histocompatibility complex class II molecules (47). It is possible that the superantigenic activity of SpeA nonspecifically deflects the host immune response, diminishes targeted immune clearance, and simulates an immunocompromised-type state, thereby providing an opportunity for microbial colonization and persistence in vivo. It has also been suggested that production of the SpeA2 allelic variant by M1T1 GAS enabled this strain to overcome herd immunity due to the absence of SpeA2-neutralizing antibodies in the host population prior to the upsurge of severe GAS disease episodes in the mid-1980s (4).
In a recent study, SLO has been shown to rapidly kill superficial mucosal cells, promoting the penetration of SpeA into lower epithelial layers and providing a gateway for GAS entry into deeper tissue with a minimal proinflammatory response (48). In deeper epithelial layers, SpeA can interact with active cells and subsequently elicit a cytokine cascade (48). These observations are particularly striking as the M1T1 clone carries the M12-derived slo allele that is associated with increased SLO production (11).
It is possible that the decline in GAS infections in Western countries throughout the 20th century concomitantly increased the reservoir of potential hosts for colonization, thus creating an opportunity for a founder effect. As such, a subpopulation of bacteria that is exposed to this increased host pool could, under the right conditions, become significantly overrepresented in subsequent generations for reasons unrelated to the fitness of “selected” clones (49). However, the coemergence of dominant clones in serotype M3, M12, and M28 GAS (9, 50), favors a hypothesis for enhanced pathogenic fitness through phage-driven dissemination of novel virulence factors.
More recent diversification due to phage exchange in M1T1 GAS has been described (10) and as exemplified in the case of GAS strain 90223, wherein the otherwise ubiquitous mf3 gene encoded on Φ5005.2 was replaced with the mf4-encoding module, likely derived from a related serotype M3 prophage (51). The subsequent success of this mf4-positive subpopulation, in the context of the parental mf3-positive M1T1 pandemic clone, will be defined by either genetic drift (relative neutrality of mf4 acquisition), natural selection (relative benefit/deficit of mf4 acquisition), or a combination of both factors (49).
This study has resolved the sequence of evolutionary events leading to the emergence of the GAS M1T1 pandemic clone. Acquisition of speA correlates with the global dissemination of this highly conserved clone. In this genetic background, we suggest that SpeA functions primarily in immune cell dysregulation and inflammatory modulation and, thereby, enhances bacterial fitness, and does not contribute to other virulence mechanisms. We further suggest that the acquisition of SpeA into the M1T1 virulome triggered GAS M1T1 global dissemination.
Supplementary Material
Acknowledgments
P.M. is the recipient of an Australian postgraduate award. J.C. and A.H. are recipients of Australian National Health and Medical Research Council (NHMRC) Overseas Biomedical Training Fellowships. M.S.S. is the recipient of an NHMRC Career Development Fellowship.
This work was funded by Medical Research Service, Department of Veterans Affairs (M.K.), U.S. National Institutes of Health grant AI077780 (V.N., M.W.), NHMRC of Australia grant 573401 (M.W.), and Australian International Science Linkages grant CG110095 (M.W., M.K., V.N.).
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- GAS
- group A Streptococcus
- HLA
- human leukocyte antigen
- LIC
- ligation-independent cloning
- ORF
- open reading frame
- PFGE
- pulsed-field gel electrophoresis
- REA
- restriction enzyme analysis
- SNP
- single-nucleotide polymorphism
- STSS
- streptococcal toxic shock-like syndrome
REFERENCES
- 1. Morens D. M., Folkers G. K., Fauci A. S. (2004) The challenge of emerging and re-emerging infectious diseases. Nature 430, 242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johnson J. R., Johnston B., Clabots C., Kuskowski M. A., Castanheira M. (2010) Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51, 286–294 [DOI] [PubMed] [Google Scholar]
- 3. Carapetis J. R., Walker A. M., Hibble M., Sriprakash K. S., Currie B. J. (1999) Clinical and epidemio logical features of group A streptococcal bacteraemia in a region with hyperendemic superficial streptococcal infection. Epidemiol. Infect. 122, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aziz R. K., Kotb M. (2008) Rise and persistence of global M1T1 clone of Streptococcus pyogenes. Emerg. Infect. Dis. 14, 1511–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Katz A. R., Morens D. M. (1992) Severe streptococcal infections in historical perspective. Clin. Infect. Dis. 14, 298–307 [DOI] [PubMed] [Google Scholar]
- 6. Cleary P. P., Kaplan E. L., Handley J. P., Wlazlo A., Kim M. H., Hauser A. R., Schlievert P. M. (1992) Clonal basis for resurgence of serious Streptococcus pyogenes disease in the 1980s. Lancet 339, 518–521 [DOI] [PubMed] [Google Scholar]
- 7. Cole J. N., Barnett T. C., Nizet V., Walker M. J. (2011) Molecular insight into invasive group A streptococcal disease. Nat. Rev. Microbiol. 9, 724–736 [DOI] [PubMed] [Google Scholar]
- 8. Aziz R. K., Kansal R., Aronow B. J., Taylor W. L., Rowe S. L., Kubal M., Chhatwal G. S., Walker M. J., Kotb M. (2010) Microevolution of group A streptococci in vivo: capturing regulatory networks engaged in sociomicrobiology, niche adaptation, and hypervirulence. PLoS One 5, e9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johnson D. R., Wotton J. T., Shet A., Kaplan E. L. (2002) A comparison of group A streptococci from invasive and uncomplicated infections: are virulent clones responsible for serious streptococcal infections? J. Infect. Dis. 185, 1586–1595 [DOI] [PubMed] [Google Scholar]
- 10. Aziz R. K., Edwards R. A., Taylor W. W., Low D. E., McGeer A., Kotb M. (2005) Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J. Bacteriol. 187, 3311–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sumby P., Porcella S. F., Madrigal A. G., Barbian K. D., Virtaneva K., Ricklefs S. M., Sturdevant D. E., Graham M. R., Vuopio-Varkila J., Hoe N. P., Musser J. M. (2005) Evolutionary origin and emergence of a highly successful clone of serotype M1 group a Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192, 771–782 [DOI] [PubMed] [Google Scholar]
- 12. Nelson K., Schlievert P. M., Selander R. K., Musser J. M. (1991) Characterization and clonal distribution of four alleles of the speA gene encoding pyrogenic exotoxin A (scarlet fever toxin) in Streptococcus pyogenes. J. Exp. Med. 174, 1271–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aziz R. K., Ismail S. A., Park H. W., Kotb M. (2004) Post-proteomic identification of a novel phage-encoded streptodornase, Sda1, in invasive M1T1 Streptococcus pyogenes. Mol. Microbiol. 54, 184–197 [DOI] [PubMed] [Google Scholar]
- 14. Walker M. J., Hollands A., Sanderson-Smith M. L., Cole J. N., Kirk J. K., Henningham A., McArthur J. D., Dinkla K., Aziz R. K., Kansal R. G., Simpson A. J., Buchanan J. T., Chhatwal G. S., Kotb M., Nizet V. (2007) DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat. Med. 13, 981–985 [DOI] [PubMed] [Google Scholar]
- 15. Buchanan J. T., Simpson A. J., Aziz R. K., Liu G. Y., Kristian S. A., Kotb M., Feramisco J., Nizet V. (2006) DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16, 396–400 [DOI] [PubMed] [Google Scholar]
- 16. Sumby P., Barbian K. D., Gardner D. J., Whitney A. R., Welty D. M., Long R. D., Bailey J. R., Parnell M. J., Hoe N. P., Adams G. G., Deleo F. R., Musser J. M. (2005) Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc. Natl. Acad. Sci. U. S. A. 102, 1679–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vlaminckx B. J., Schuren F. H., Montijn R. C., Caspers M. P., Beitsma M. M., Wannet W. J., Schouls L. M., Verhoef J., Jansen W. T. (2007) Dynamics in prophage content of invasive and noninvasive M1 and M28 Streptococcus pyogenes isolates in the Netherlands from 1959 to 1996. Infect. Immun. 75, 3673–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chatellier S., Ihendyane N., Kansal R. G., Khambaty F., Basma H., Norrby-Teglund A., Low D. E., McGeer A., Kotb M. (2000) Genetic relatedness and superantigen expression in group A Streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect. Immun. 68, 3523–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferretti J. J., McShan W. M., Ajdic D., Savic D. J., Savic G., Lyon K., Primeaux C., Sezate S., Suvorov A. N., Kenton S., Lai H. S., Lin S. P., Qian Y., Jia H. G., Najar F. Z., Ren Q., Zhu H., Song L., White J., Yuan X., Clifton S. W., Roe B. A., McLaughlin R. (2001) Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98, 4658–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Collin M., Olsen A. (2000) Generation of a mature streptococcal cysteine proteinase is dependent on cell wall-anchored M1 protein. Mol. Microbiol. 36, 1306–1318 [DOI] [PubMed] [Google Scholar]
- 21. Ashbaugh C. D., Warren H. B., Carey V. J., Wessels M. R. (1998) Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J. Clin. Invest. 102, 550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cole J. N., McArthur J. D., McKay F. C., Sanderson-Smith M. L., Cork A. J., Ranson M., Rohde M., Itzek A., Sun H., Ginsburg D., Kotb M., Nizet V., Chhatwal G. S., Walker M. J. (2006) Trigger for group A streptococcal M1T1 invasive disease. FASEB J. 20, 1745–1747 [DOI] [PubMed] [Google Scholar]
- 23. McKay F. C., McArthur J. D., Sanderson-Smith M. L., Gardam S., Currie B. J., Sriprakash K. S., Fagan P. K., Towers R. J., Batzloff M. R., Chhatwal G. S., Ranson M., Walker M. J. (2004) Plasminogen binding by group A streptococcal isolates from a region of hyperendemicity for streptococcal skin infection and a high incidence of invasive infection. Infect. Immun. 72, 364–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maamary P. G., Sanderson-Smith M. L., Aziz R. K., Hollands A., Cole J. N., McKay F. C., McArthur J. D., Kirk J. K., Cork A. J., Keefe R. J., Kansal R. G., Sun H., Taylor W. L., Chhatwal G. S., Ginsburg D., Nizet V., Kotb M., Walker M. J. (2010) Parameters governing invasive disease propensity of non-M1 serotype group A streptococci. J. Innate. Immun. 2, 596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramachandran V., McArthur J. D., Behm C. E., Gutzeit C., Dowton M., Fagan P. K., Towers R., Currie B., Sriprakash K. S., Walker M. J. (2004) Two distinct genotypes of prtF2, encoding a fibronectin binding protein, and evolution of the gene family in Streptococcus pyogenes. J. Bacteriol. 186, 7601–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H., Ruan J., Durbin R. (2008) Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 [DOI] [PubMed] [Google Scholar]
- 30. Posada D. (2009) Selection of models of DNA evolution with jModelTest. Methods Mol. Biol. 537, 93–112 [DOI] [PubMed] [Google Scholar]
- 31. Hollands A., Aziz R. K., Kansal R., Kotb M., Nizet V., Walker M. J. (2008) A naturally occurring mutation in ropB suppresses SpeB expression and reduces M1T1 group A streptococcal systemic virulence. PLoS One 3, e4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun H., Ringdahl U., Homeister J. W., Fay W. P., Engleberg N. C., Yang A. Y., Rozek L. S., Wang X., Sjobring U., Ginsburg D. (2004) Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science 305, 1283–1286 [DOI] [PubMed] [Google Scholar]
- 33. Eschenfeldt W. H., Lucy S., Millard C. S., Joachimiak A., Mark I. D. (2009) A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 498, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanderson-Smith M. L., Dinkla K., Cole J. N., Cork A. J., Maamary P. G., McArthur J. D., Chhatwal G. S., Walker M. J. (2008) M protein-mediated plasminogen binding is essential for the virulence of an invasive Streptococcus pyogenes isolate. FASEB J. 22, 2715–2722 [DOI] [PubMed] [Google Scholar]
- 35. Cole J. N., Pence M. A., von Kockritz-Blickwede M., Hollands A., Gallo R. L., Walker M. J., Nizet V. (2010) M protein and hyaluronic acid capsule are essential for in vivo selection of covRS mutations characteristic of invasive serotype M1T1 group A Streptococcus. MBio 1, e00191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aziz R. K., Pabst M. J., Jeng A., Kansal R., Low D. E., Nizet V., Kotb M. (2004) Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol. Microbiol. 51, 123–134 [DOI] [PubMed] [Google Scholar]
- 37. Sumby P., Whitney A. R., Graviss E. A., DeLeo F. R., Musser J. M. (2006) Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kansal R. G., Datta V., Aziz R. K., Abdeltawab N. F., Rowe S., Kotb M. (2010) Dissection of the molecular basis for hypervirulence of an in vivo-selected phenotype of the widely disseminated M1T1 strain of group A Streptococcus bacteria. J. Infect. Dis. 201, 855–865 [DOI] [PubMed] [Google Scholar]
- 39. Tomai M. A., Schlievert P. M., Kotb M. (1992) Distinct T-cell receptor V beta gene usage by human T lymphocytes stimulated with the streptococcal pyrogenic exotoxins and pep M5 protein. Infect. Immun. 60, 701–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sriskandan S., Unnikrishnan M., Krausz T., Cohen J. (1999) Molecular analysis of the role of streptococcal pyrogenic exotoxin A (SpeA) in invasive soft-tissue infection resulting from Streptococcus pyogenes. Mol. Microbiol. 33, 778–790 [DOI] [PubMed] [Google Scholar]
- 41. Unnikrishnan M., Cohen J., Sriskandan S. (2001) Complementation of a speA negative Streptococcus pyogenes with speA: effects on virulence and production of streptococcal pyrogenic exotoxin A. Microb. Pathog. 31, 109–114 [DOI] [PubMed] [Google Scholar]
- 42. Sriskandan S., Unnikrishnan M., Krausz T., Dewchand H., Van Noorden S., Cohen J., Altmann D. M. (2001) Enhanced susceptibility to superantigen-associated streptococcal sepsis in human leukocyte antigen-DQ transgenic mice. J. Infect. Dis. 184, 166–173 [DOI] [PubMed] [Google Scholar]
- 43. Falkow S. (1988) Molecular Koch's postulates applied to microbial pathogenicity. Rev. Infect. Dis. 10, S274–276 [DOI] [PubMed] [Google Scholar]
- 44. Beres S. B., Musser J. M. (2007) Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One 2, e800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cone L. A., Woodard D. R., Schlievert P. M., Tomory G. S. (1987) Clinical and bacteriologic observations of a toxic shock-like syndrome due to Streptococcus pyogenes. N. Engl. J. Med. 317, 146–149 [DOI] [PubMed] [Google Scholar]
- 46. Imanishi K., Igarashi H., Uchiyama T. (1992) Relative abilities of distinct isotypes of human major histocompatibility complex class II molecules to bind streptococcal pyrogenic exotoxin types A and B. Infect. Immun. 60, 5025–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kotb M. (1995) Bacterial pyrogenic exotoxins as superantigens. Clin. Microbiol. Rev. 8, 411–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brosnahan A. J., Mantz M. J., Squier C. A., Peterson M. L., Schlievert P. M. (2009) Cytolysins augment superantigen penetration of stratified mucosa. J. Immunol. 182, 2364–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baquero F. (2011) The 2010 Garrod lecture: the dimensions of evolution in antibiotic resistance: ex unibus plurum et ex pluribus unum. J Antimicrob. Chemother. 66, 1659–1672 [DOI] [PubMed] [Google Scholar]
- 50. Rogers S., Commons R., Danchin M. H., Selvaraj G., Kelpie L., Curtis N., Robins-Browne R., Carapetis J. R. (2007) Strain prevalence, rather than innate virulence potential, is the major factor responsible for an increase in serious group A streptococcus infections. J. Infect. Dis. 195, 1625–1633 [DOI] [PubMed] [Google Scholar]
- 51. Beres S. B., Sylva G. L., Sturdevant D. E., Granville C. N., Liu M., Ricklefs S. M., Whitney A. R., Parkins L. D., Hoe N. P., Adams G. J., Low D. E., DeLeo F. R., McGeer A., Musser J. M. (2004) Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infections. Proc. Natl. Acad. Sci. U. S. A. 101, 11833–11838 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.