Abstract
Sucrose nonfermenting 1 (Snf1)-related kinase (SNRK) is a serine/threonine kinase with sequence similarity to AMP-activated protein kinase (AMPK); however, its function is not well characterized. We conducted a gene array to determine which genes are regulated by SNRK. The array demonstrated that SNRK overexpression increased the levels of genes involved in cell proliferation, including calcyclin-binding protein (CacyBP), a member of the ubiquitin ligase complex that targets nonphosphorylated β-catenin for degradation. We confirmed that SNRK increased CacyBP mRNA and protein, and decreased β-catenin protein in HCT116 and RKO colon cancer cells. Furthermore, SNRK inhibited colon cancer cell proliferation, and CacyBP down-regulation reversed the SNRK-mediated decrease in proliferation and β-catenin. SNRK overexpression also decreased β-catenin nuclear localization and target gene transcription, and β-catenin down-regulation reversed the effects of SNRK knockdown on proliferation. SNRK transcript levels were reduced in human colon tumors compared to normal tissue by 35.82%, and stable knockdown of SNRK increased colon cancer cell tumorigenicity. Our results demonstrate that SNRK is down-regulated in colon cancer and inhibits colon cancer cell proliferation through CacyBP up-regulation and β-catenin degradation, resulting in reduced proliferation signaling. These findings reveal a novel function for SNRK in the regulation of colon cancer cell proliferation and β-catenin signaling.—Rines, A. K., Burke, M. A., Fernandez, R. P., Volpert, O. V., Ardehali, H. Snf1-related kinase inhibits colon cancer cell proliferation through calcyclin binding protein-dependent reduction of β-catenin.
Keywords: SNRK, CacyBP
Sucrose nonfermenting 1 (Snf1)-related kinase (SNRK) is a novel serine/threonine kinase and a member of the AMP-activated protein kinase (AMPK)-related family. The AMPK proteins are serine/threonine kinases that play a central role in cellular energy regulation. They share sequence similarity in their catalytic domains with the Saccharomyces cerevisiae protein Snf1, which is activated by low glucose and enables survival on alternative carbon sources (1). AMPK is the most widely studied member of this family and is activated by increases in the cellular AMP:ATP or ADP:ATP ratio (2). Active AMPK restricts ATP-consuming anabolic pathways, such as cell proliferation, cell growth, and metabolic substrate synthesis, and increases ATP-generating processes, such as glucose and fatty acid oxidation. These effects of AMPK are potentially therapeutically useful for restricting cancer cell proliferation and growth, treating metabolic disorders, and enhancing heart function during stress (3, 4). Other AMPK family members are also involved in an array of cellular processes related to metabolism, proliferation, and cell polarity (5, 6).
Although AMPK has been widely studied, little is known about the function of SNRK. The mRNA of SNRK is broadly expressed in the tissues of adult mice, rats, and humans, as indicated by Northern blot and in situ analyses (7–9). SNRK is a monomeric enzyme that contains a nuclear localization signal (NLS), an ATP-binding domain, and an active serine/threonine kinase domain with a conserved T-loop threonine residue (T173). Current knowledge about the function of SNRK suggests that it may be involved in neuronal apoptosis and blood vessel development. SNRK mRNA is up-regulated in the nucleus during apoptosis induced by low potassium in rat cerebellar granule neurons (7), and the zebrafish ortholog Snrk-1 is essential for angioblast arterial and vein specification (10). SNRK is also activated by liver kinase B1 (LKB1), which phosphorylates the T-loop threonine residue of SNRK (11), AMPK, and 11 other AMPK-related kinases (12). Unlike AMPK, SNRK does not require an additional stimulus, such as increased AMP:ATP, for activation by LKB1.
The purpose of our study was to identify novel functions and downstream targets of SNRK. We performed a gene array analysis and showed that SNRK alters several genes involved in cell proliferation and DNA synthesis, including calcyclin-binding protein (CacyBP; refs. 13, 14). CacyBP is a member of the E3 ubiquitin ligase SCFTBL1 complex that enhances proteasomal degradation of nonphosphorylated β-catenin (15), which translocates to the nucleus and increases expression of proliferation-associated genes (16, 17). β-catenin phosphorylation sites are commonly mutated in colon cancers, leading to accumulation of nonphosphorylated β-catenin that can engage in enhanced proliferation signaling (18). We verified that SNRK up-regulates CacyBP and reduces β-catenin protein levels in colon cancer cells. SNRK reduces colon cancer cell proliferation and DNA synthesis, and the mechanism is through CacyBP up-regulation and subsequently decreased β-catenin and downstream proliferation signaling. SNRK levels are decreased in human colon cancer tissue, and stable knockdown of SNRK increases in vitro colon cancer cell tumorigenicity. These results indicate that SNRK is a regulator of cell proliferation, and identify a SNRK-CacyBP-β-catenin signaling axis. Furthermore, the data suggest a novel mechanism by which nonphosphorylated β-catenin levels and proliferation can be modified in colon cancer cells.
MATERIALS AND METHODS
Cells and reagents
Cell lines were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured at 37°C in 5% CO2. HEK 293 cells were cultured in modified essential medium (MEM; Mediatech, Manassas, VA, USA) supplemented with 1 mM sodium pyruvate, penicillin/streptomycin, and 10% FBS. HCT116 cells were cultured in McCoy's 5a modified medium (ATCC) supplemented with penicillin/streptomycin and 10% FBS. RKO cells were cultured in Eagle's minimum essential medium (EMEM; ATCC, Manassas, VA, USA) supplemented with penicillin/streptomycin and 10% FBS.
Human SNRK cDNA was cloned into the pEGFP-N3 vector to create the SNRK-GFP plasmid. Site-directed mutagenesis was performed on SNRK-GFP with the Stratagene QuikChange site mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) to create SNRK-NLS-GFP (mutation K456A) and SNRK-T173A-GFP. To obtain untagged SNRK and SNRK-T173A, cDNA from pEGFP-N3 vectors was excised and ligated into pCMV-Script. Human CacyBP-pCMV6-XL5 was purchased from Origene (Rockville, MD, USA). shRNA lentivirus against SNRK was obtained from the GIPZ Lentiviral shRNAmir library (Open Biosystems, Lafayette, CO, USA; oligo ID V2LHS_173445), as well as a nonsilencing control shRNA lentivirus (oligo ID RHS4346). siRNA for CacyBP was obtained from Thermo Scientific (Lafayette, CO, USA; siGenome SMARTpool siRNA D-012521) and siRNA for β-catenin was from Qiagen (Valencia, CA, USA; SI02662478). SNRK antibody was obtained from Millipore; GFP, actin, and GAPDH antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA); Lamin A/C, CacyBP, β-catenin, and tubulin antibodies were from Cell Signaling (Danvers, MA, USA). (S)-MG132 was purchased from Cayman Chemical (Ann Arbor, MI, USA).
Cell transfection and transduction
Transfection was performed with Lipofectamine with Plus reagent (Invitrogen, Carlsbad, CA, USA) at a 2:1 Lipofectamine:DNA ratio in HEK 293 cells and with 2:1 Lipofectamine 2000:DNA in HCT116 cells for 48 h before analysis. Viruses for shRNA were amplified using CaCl2 transfection in HEK 293T cells for 3 d, followed by collection of supernatant and filtration. For treatment, virus was added in equal amounts to cells for 3 d with puromycin (1 μg/ml). siRNA was transfected into cells using HiPerfect Transfection reagent (Qiagen) for 72 h. For combined treatments, cells were transfected with plasmid 24 h after siRNA treatment or were transfected with plasmid or siRNA 48 h after initial shRNA treatment.
Cell lysis and Western blot analysis
For total protein lysates, cell lysis buffer consisted of 50 mM Tris (pH 7.5), 1 mM EGTA, 1 mM EDTA, 1% v/v Triton X-100, 1 mM sodium orthovanadate, 10 mM sodium β-glycerophosphate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 270 mM sucrose, 0.1% v/v β-mercaptoethanol, and 1× Protease Arrest (G Biosciences, St. Louis, MO, USA). Cells were washed in cold PBS, incubated with cold lysis buffer for 15 min on ice, and spun down at 13,200 rpm for 15 min, and the supernatant was collected and used in downstream applications. Protein concentration was determined using the Bradford assay (Thermo Scientific). Equal amounts of protein were loaded into a precast NuPAGE gel, transferred to a nitrocellulose membrane (Invitrogen), and blotted with the indicated primary antibodies and appropriate horseradish peroxidase-conjugated secondary antibodies. Chemiluminescent signal was produced using ECL Plus Western blotting substrate (Pierce, Lafayette, CO, USA). Densitometry was performed using ImageJ software (U.S. National Institutes of Health, Bethesda, MD, USA).
Nuclear and cytoplasmic separation of lysates was conducted using the NE-PER nuclear and cytoplasmic extraction kit (Thermo Scientific) according to the manufacturer's protocol. Protein concentration was determined by the BCA assay (Thermo Scientific).
Gene array studies
Biotin-labeled cRNA was generated from high-quality total RNA using the Illumina TotalPrep RNA amplification kit (Ambion Biosystems, Carlsbad, CA, USA). The gene array studies and analysis were performed at the Northwestern University Genomics Core Facility. Briefly, 250 ng of total RNA was reverse transcribed with an oligo (dT) primer bearing the T7 promoter. The first-strand cDNA produced in the reaction was used to make the second-strand cDNA. Purified second-strand cDNA along with biotin UTPs were used to generate biotinylated, antisense RNA of each mRNA in an in vitro transcription reaction. Size distribution profiles for the labeled cRNA samples were evaluated by a bioanalyzer. Approximately 1.5 μg of purified labeled cRNAs were hybridized at 55°C overnight with the Illumina Human-6 v2 expression Beadchip and washed the following day. Signal was developed with Streptavidin-Cy3 and scanned with an Illumina BeadArray Reader. Full gene array data have been deposited in the U.S. National Center for Biotechnology Information (NCBI) Gene Expression Omnibus GEO (19) and are accessible through GEO Series accession number GSE30185 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30185).
Kinase assay
To measure kinase activity, 50 μg of transfected cell lysate was added to 10 mM MgAC and 100 μM ATP with or without 2.5 μM H3.3 substrate (New England Biolabs, Ipswich, MA, USA) for 90 min at room temperature, then added to an equal volume of kinase Glo Plus reagent (Promega, Fitchburg, WI, USA) for 1 h before being read for endpoint luminescence, as previously reported (10). Activity without H3.3 substrate was subtracted as background from activity measured with H3.3 substrate.
qRT-PCR
Total RNA was isolated from cells using RNA-Stat (Tel-test, Friendswood, TX, USA), treatment with RNase-free DNase I (Invitrogen, Carlsbad, CA, USA) to digest genomic DNA, and ethanol precipitation of RNA. cDNA was obtained using the Taqman Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA). Quantitative real-time PCR was conducted with Fast SYBR Green Master Mix (Applied Biosystems) on the 7500 Fast real-time PCR System (Applied Biosystems). Specificity of the primers was determined by melting curve analysis. Expression was normalized to 18s rRNA. Primer sequences used were CacyBP, 5′-CAGAAATCACAGAAGAAAGCAGAAC-3′ and 5′-TTTCACGCTATAGCCCGTTGT-3′; c-Myc, 5′-CGTCTCCACACATCAGCACAA-3′ and 5′-TCTTGGCAGCAGGATAGTCCTT-3′; Cyclin D1, 5′-GCATGTTCGTGGCCTCTAAGA-3′ and 5′-CGGTGTAGATGCACAGCTTCTC-3′; 18s rRNA, 5′-CGGGTCGGGAGTGGGT-3′ and 5′-GAAACGGCTACCACATCCAAG-3′; and SNRK, 5′-TGTCCCTCTCAACACAAGTGGTT-3′ and 5′-GAGGACGGGCGCACTCT-3′.
Measurement of cellular proliferation
To measure proliferation by cell counting, treated cells were counted using trypan blue staining and a hemocytometer, and an equal number of viable cells for each sample was plated in triplicate. Cells were subsequently collected by trypsinization after 24, 48, and 72 h, resuspended in PBS and trypan blue, and counted on a hemacytometer. To measure proliferation by MTS reduction, equal numbers of viable treated cells were plated into a 96-well plate, grown for 72 h, and mixed with MTS/PMS reagent from the CellTiter 96 AQueous nonradioactivity cell proliferation assay (Promega) for 1 h before reading absorbance at 490 nm to measure MTS reduction.
DNA synthesis
Treated cells were serum starved for 24 h, incubated with 10 μM 5-bromodeoxyuridine (BrdU) in serum-containing medium for 6–24 h, then fixed and stained with anti-BrdU-adenomatous polyposis coli (APC) antibody using the APC BrdU Flow Kit (BD Pharmingen, Franklin Lakes, NJ, USA), and analyzed by flow cytometry.
Luciferase reporter assay
HCT116 cells were transfected with GFP or SNRK plasmids, and RKO cells were transduced with shRNA, then cotransfected 24 h later with Super 8x TOPflash or FOPflash plasmids (courtesy of Randall T. Moon, Howard Hughes Research Institute, Seattle, WA, USA; ref. 20; Addgene plasmid 12456) along with pRL-TK Renilla plasmid at a 4:1 TOPflash:pRL-TK ratio using Lipofectamine 2000. After 48 h, luciferase activity was measured in a luminometer with the Dual Glo Luciferase Assay (Promega, Fitchburg, WI, USA). Luciferase activity was normalized to transfection efficiency by measuring Renilla activity with the same assay, and activity measured with FOPflash was subtracted as background from activity measured with TOPflash.
TissueScan cDNA array
mRNA expression of SNRK in matched normal and tumor human tissue was determined using the TissueScan real-time colon cancer cDNA array III (HCRT103) from Origene, according to the manufacturer's protocol.
Colony formation assay
RKO cells were stably transduced with nonsilencing control or SNRK shRNA with puromycin selection, then clonally expanded. For the colony formation assay, viable cells from independent cultures of stably transduced RKO cells were counted, 250 cells were plated in 6-well plates, grown for 7 d, fixed with 6% glutaraldehyde, stained with 3% crystal violet, and then washed in water.
Soft agar colony growth assay
For the soft agar colony growth assay, viable cells from independent cultures of stably transduced RKO cells were counted, then 2.4 × 104 cells in 2 ml EMEM with 0.4% low-melting-point agarose and 10% serum were plated on 2 ml EMEM with 1% low-melting-point agarose and 10% serum in a 6-well plate. Cells were grown for 3 wk, and then fluorescent colonies >50 cells were scored in a microscope.
Statistical analysis
Data are presented as means ± se of measurements. Statistical significance for analysis with 2 groups was determined by an unpaired 2-tailed Student's t test. Statistical significance for ≥3 groups was determined by ANOVA followed by a post hoc analysis using the Tukey-Kramer test. Values of P < 0.05 were considered statistically significant.
RESULTS
Gene array analysis of SNRK overexpression
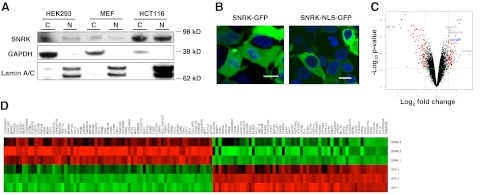
SNRK contains an NLS at its C terminus, and a previous study showed that SNRK protein is present in the nuclei of rat granular neurons (7). To verify that SNRK localizes to the nucleus, we isolated cytoplasmic and nuclear fractions from HEK 293 cells, mouse embryonic fibroblasts (MEFs), and colon cancer HCT116 cells and measured SNRK levels with Western blot analysis. SNRK was detected in both the nucleus and cytoplasm (Fig. 1A) of all three cell types. We also overexpressed GFP-fusion proteins of wild-type SNRK (SNRK-GFP) and SNRK with a point mutation in its NLS (SNRK-NLS-GFP) in HEK 293 cells. While wild-type SNRK localized to both the cytoplasm and the nucleus, SNRK-NLS was almost exclusively restricted to the cytoplasm (Fig. 1B). These results show that SNRK is present in both the cytoplasm and nucleus and that its nuclear localization can be abrogated by mutating the NLS.
Figure 1.
SNRK is present in the nucleus and cytoplasm and alters the expression of genes associated with DNA synthesis and proliferation. A) SNRK protein levels in cytoplasmic (C) and nuclear (N) fractions from HEK 293, MEF, and HCT116 cells. GAPDH was used as a cytoplasmic loading control, and Lamin A/C was used as a nuclear loading control. B) Confocal microscopy images of HEK 293 cells transfected with wild-type SNRK (SNRK-GFP) and SNRK with a mutation in its NLS (SNRK-NLS-GFP). Scale bars = 10 μm. C) Volcano plot of the gene array. The volcano plot indicates the size of the biological effect (fold change) vs. the reproducibility of the result (statistical P value). Each gene is represented as a dot. Red dots represent the genes with P < 0.01, false discovery rate < 0.05, and fold change > 1.5. See Table 1 for gene ontology analysis based on biological processes. D) Heat map of the gene array. Each row is one sample; each column is one probe. Red represents higher expression than average, green represents lower expression than average; n = 3.
Since SNRK is present in the nucleus, we hypothesized that SNRK plays a role in the regulation of gene expression. To test this hypothesis, we overexpressed SNRK-GFP or GFP in HEK 293 cells, and then performed a gene array analysis (Fig. 1C, D, Table 1, and Supplemental Tables S1–S3) with sample quality control (Supplemental Fig. S1). The expression of several genes was altered in response to SNRK overexpression. When the altered genes were categorized by function, processes related to cell proliferation (such as DNA replication and cell cycle progression) and metabolism were among the most significantly altered by SNRK overexpression (Table 1). On the basis of this gene ontology analysis, we focused on the role of SNRK in proliferation in this study.
Table 1.
Gene ontology (GO) analysis based on biological processes
| GO term | P | Significant gene number | Increase ∼ decrease |
|---|---|---|---|
| DNA replication initiation | 1.28E-06 | 9 | 7∼2 |
| Biopolymer metabolic process | 4.55E-06 | 243 | 132∼113 |
| Regulation of transcription, DNA-dependent | 8.80E-06 | 127 | 74∼55 |
| Proline biosynthetic process | 5.08E-05 | 4 | 3∼1 |
| Organelle organization and biogenesis | 6.16E-05 | 69 | 42∼28 |
| Negative regulation of cellular metabolic process | 8.91E-05 | 30 | 17∼14 |
| DNA-dependent DNA replication | 0.00012472 | 13 | 9∼4 |
| Negative regulation of transcription, DNA-dependent | 0.0002571 | 19 | 14∼6 |
| DNA replication | 0.00027656 | 19 | 14∼5 |
| Actin filament organization | 0.00038397 | 7 | 3∼4 |
| Chromosome organization and biogenesis | 0.00055253 | 30 | 21∼10 |
| DNA metabolic process | 0.00058124 | 48 | 33∼16 |
| Chromosome organization and biogenesis | 0.00067976 | 29 | 20∼10 |
| Regulation of cell cycle | 0.00068108 | 38 | 20∼18 |
See Fig. 1.
SNRK regulates the expression of CacyBP and β-catenin
We examined the gene array results to identify potential targets of SNRK that are involved in cellular proliferation. One of the altered genes was CacyBP, whose mRNA levels were 1.6-fold higher with SNRK overexpression than with GFP overexpression (P=3.97×10−6, false discovery rate of 0.0015). CacyBP is involved in the ubiquitination and subsequent proteosomal degradation of nonphosphorylated β-catenin (21, 22). Mutations in β-catenin phosphorylation sites are commonly found in colorectal cancers (18), resulting in increased oncogenic β-catenin signaling. Thus, we investigated whether SNRK regulates CacyBP and β-catenin levels in two colon cancer cell lines with different levels of β-catenin. We used HCT116 cells, which have a high level of β-catenin, to study SNRK overexpression, and RKO cells, which have a very low basal level of β-catenin caused by the absence of its plasma membrane-associated pool (23), to study SNRK down-regulation. Both HCT116 and RKO cells have intact wild-type APC (18, 24), which is required for the CacyBP-associated ubiquitination of β-catenin.
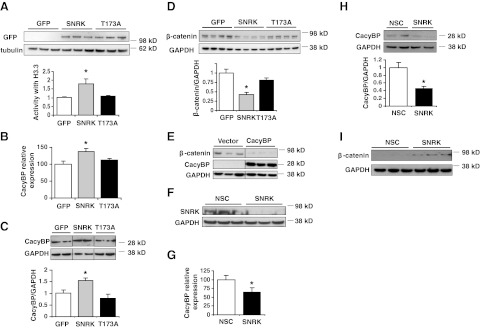
We first assessed whether SNRK overexpression in colon cancer cells results in the production of the active enzyme by using a luminescence-based kinase assay with histone H3.3 as a SNRK substrate, as previously reported (10, 11). A SNRK-T173A mutant, in which the conserved T-loop residue is mutated to inhibit SNRK activation, and GFP were used as controls. H3.3 substrate-specific kinase activity was significantly higher in lysates from cells that overexpress SNRK than in those expressing GFP or the SNRK-T173A mutant enzyme (Fig. 2A). Lysates from GFP- and SNRK-T173A-overexpressing cells still had some background activity, which was most likely caused by other cellular kinases that phosphorylate the substrate used in our assay. We also measured activity of the GFP-tagged constructs compared to untagged constructs, and the activity was similar regardless of the presence of the tag, demonstrating that the GFP tag does not interfere with the activity of SNRK (Supplemental Fig. S2A).
Figure 2.
SNRK increases CacyBP levels and decreases β-catenin protein levels in colon cancer cells. A) Expression and activity of SNRK-GFP and SNRK-T173A-GFP in HCT116 cells; n = 3. B) Levels of CacyBP mRNA in HCT116 cells with overexpression of SNRK or controls; n = 9. C) Levels of CacyBP protein in HCT116 cells with overexpression of SNRK or controls. Top panel: representative Western blots (lanes have been rearranged from the same blot for clarity). Bottom panel: summary of densitometry; n ≥ 3. D) Levels of β-catenin protein in HCT116 cells with overexpression of SNRK or controls. Top panel: representative Western blots. Bottom panel: summary of densitometry; n = 7. E) Levels of β-catenin protein in HCT116 cells with overexpression of CacyBP or vector control (lanes shown are rearranged from the same blot). F) Western blot of SNRK protein in RKO cells treated with SNRK or nonsilencing control (NSC) shRNA. G) Levels of CacyBP mRNA with SNRK down-regulation in RKO cells; n = 10. H) Levels of CacyBP protein with SNRK down-regulation in RKO cells. Top panel: representative Western blots. Bottom panel: summary of densitometry; n = 3. I) Levels of β-catenin protein with SNRK down-regulation in RKO cells. *P < 0.05 vs. GFP and T173A or vs. NSC.
To confirm that SNRK up-regulates CacyBP, we overexpressed SNRK, GFP, or the SNRK-T173A control in HCT116 cells, and saw a significant increase in the levels of CacyBP mRNA and protein with SNRK relative to the controls (Fig. 2B, C). Since CacyBP increases the proteolysis of β-catenin, we then measured β-catenin protein levels in response to SNRK overexpression. β-catenin levels decreased significantly with SNRK overexpression compared to the controls (Fig. 2D). We also verified that SNRK overexpression similarly increased CacyBP and decreased β-catenin in HEK 293 cells (Supplemental Fig. S2B, C). In addition, to independently confirm that CacyBP increases β-catenin degradation in HCT116 cells, we found that overexpression of CacyBP alone also reduces β-catenin levels (Fig. 2E).
In RKO cells, SNRK shRNA treatment significantly decreased SNRK protein levels (Fig. 2F). SNRK knockdown reduced CacyBP mRNA (Fig. 2G) and protein levels (Fig. 2H) and increased β-catenin protein levels (Fig. 2I). Together, these findings demonstrate that SNRK regulates CacyBP and β-catenin expression in colon cancer cells.
SNRK decreases cellular proliferation in colon cancer cells
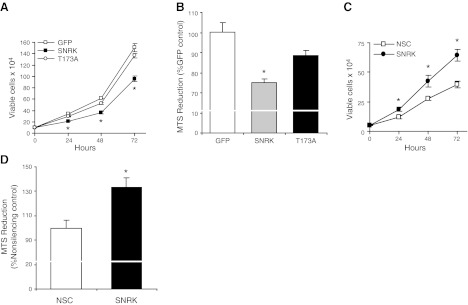
Since SNRK alters the expression of CacyBP and β-catenin, we examined whether SNRK affects cell proliferation in colon cancer cells. Cellular proliferation was significantly lower in SNRK-transfected HCT116 cells than in cells transfected with GFP or SNRK-T173A, as measured by viable cell number (Fig. 3A) and MTS reduction (Fig. 3B). We also down-regulated SNRK in RKO cells and measured cell proliferation. Proliferation, as measured by viable cell number (Fig. 3C) and MTS reduction (Fig. 3D), was increased in response to SNRK down-regulation. These results demonstrate that SNRK decreases cell proliferation in colon cancer cells. We also found the same effects of SNRK on cell proliferation in HEK 293 cells (Supplemental Fig. S2D–G).
Figure 3.
SNRK decreases colon cancer cell proliferation. A) Proliferation in HCT116 cells transfected with SNRK-GFP, SNRK-T173A-GFP, or GFP, as determined by viable cell counting with trypan blue exclusion at 24, 48, and 72 h; n ≥ 3. B) Proliferation in HCT116 cells after 72 h, as measured by MTS reduction; n ≥ 3. C) Proliferation in RKO cells treated with nonsilencing control (NSC) or SNRK shRNA, as measured by viable cell number at 24, 48, and 72 h; n = 3. D) Proliferation in RKO cells with down-regulation of SNRK at 72 h, as measured by MTS reduction; n = 3. *P < 0.05 vs. GFP and T173A or vs. NSC.
SNRK reduces the nuclear localization of β-catenin and the expression of β-catenin-regulated genes in colon cancer cells
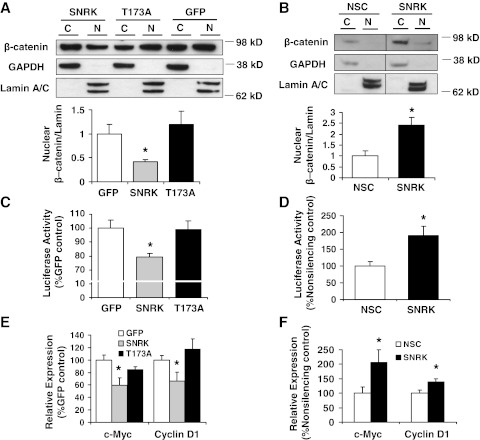
SNRK decreases CacyBP expression, and CacyBP has been found to reduce levels of nuclear β-catenin (15, 21). Nuclear β-catenin signaling is clinically relevant in colon cancer, as β-catenin enables transcription of oncogenic target genes when it translocates to the nucleus and coactivates T-cell factor (TCF)/lymphoid enhancer factor (LEF) transcription factors, leading to increased expression of genes such as c-Myc and cyclin D1 (16, 17). Thus, we next studied the regulation of β-catenin nuclear accumulation by SNRK in colon cancer cells. Overexpression of SNRK in HCT116 cells decreased levels of nuclear β-catenin compared to the GFP and SNRK-T173A controls (Fig. 4A). Conversely, down-regulation of SNRK in RKO cells led to increased levels of nuclear β-catenin (Fig. 4B).
Figure 4.
SNRK regulates β-catenin nuclear accumulation and β-catenin target expression in colon cancer cells. A, B) Top panels: Western blots of β-catenin in cytoplasmic (C) and nuclear (N) fractions in transfected HCT116 cells (A) and RKO cells treated with nonsilencing control (NSC) or SNRK shRNA (B). Bottom panels: summary of densitometry for nuclear β-catenin; n = 3. C) TOPflash luciferase reporter assay in transfected HCT116 cells; n ≥ 4. D) TOPflash luciferase reporter assay in RKO cells with SNRK down-regulation; n = 3. E) c-Myc and cyclin D1 mRNA expression in transfected HCT116 cells; n ≥ 10. F) c-myc and cyclin D1 mRNA expression in RKO cells with down-regulated SNRK; n ≥ 10. *P < 0.05 vs. GFP and T173A or vs. NSC.
We then studied whether the transcription of β-catenin target genes is regulated by SNRK by using the TOPflash luciferase reporter, which contains 7 copies of the consensus TCF/LEF transcription binding site. We cotransfected HCT116 cells with SNRK or controls and with the Super8x TOPflash luciferase reporter construct, or with Super 8x FOPflash (which contains mutated consensus sites) as a measure of background luminescence. Compared with controls, SNRK overexpression resulted in reduced TOPflash luciferase expression, indicating decreased β-catenin target expression (Fig. 4C). We also transfected SNRK shRNA-infected RKO cells with TOPflash, and found increased TCF/LEF expression with SNRK knockdown (Fig. 4D). To further demonstrate that SNRK regulates β-catenin target expression, we performed quantitative real-time PCR for the β-catenin targets c-Myc and cyclin D1. SNRK overexpression in HCT116 cells reduced the expression of both c-Myc and cyclin D1 (Fig. 4E), while SNRK down-regulation in RKO cells increased their expression (Fig. 4F). These results verify that SNRK not only regulates the expression and nuclear accumulation of β-catenin, but also its downstream signaling.
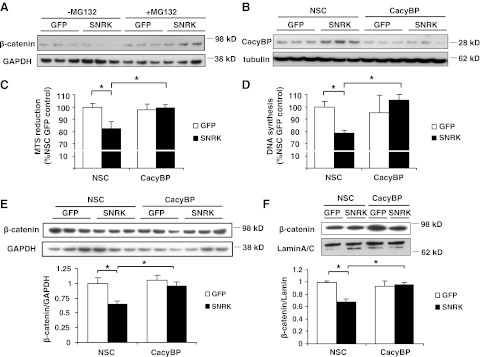
CacyBP is necessary for the SNRK-induced reduction in colon cancer cell proliferation
Because CacyBP is regulated by SNRK, we hypothesized that the mechanism for the antiproliferative effects of SNRK is through increased CacyBP and reduced β-catenin. First, because CacyBP contributes to APC-mediated proteasomal degradation of β-catenin, we determined whether proteasomal activity is necessary for the SNRK-mediated decrease in β-catenin. We found that the proteasomal inhibitor MG132 reversed the reduction in β-catenin caused by SNRK overexpression (Fig. 5A). Next, we measured proliferation and DNA synthesis with SNRK overexpression combined with CacyBP knockdown. Compared with nonsilencing control siRNA, CacyBP siRNA resulted in a significant decrease in CacyBP protein in HCT116 cells (Fig. 5B). SNRK overexpression significantly decreased MTS reduction (Fig. 5C) and DNA synthesis (Fig. 5D) compared to GFP overexpression with nonsilencing control siRNA, as expected. However, there was no significant difference between SNRK and GFP in MTS reduction or DNA synthesis with CacyBP knockdown (Fig. 5C, D). Thus, CacyBP expression is necessary for SNRK to reduce proliferation and DNA synthesis. In addition, CacyBP knockdown abolished the decrease in β-catenin protein levels in response to SNRK overexpression in HCT116 cells (Fig. 5E), as well as the decrease in nuclear β-catenin levels (Fig. 5F).
Figure 5.
CacyBP down-regulation reverses the decrease in proliferation caused by SNRK. A) Western blots of β-catenin protein in HCT116 cells transfected with GFP or SNRK and with or without 24 h of 5 μM MG132 treatment. B) Western blots of CacyBP protein in HCT116 cells transfected with GFP or SNRK and treated with CacyBP or nonsilencing control (NSC) siRNA. C, D) Proliferation measured by MTS reduction (C) and DNA synthesis measured by BrdU incorporation (D) in HCT116 cells transfected with SNRK or GFP and treated with CacyBP or NSC siRNA; n = 6. E, F) Top panels: Western blots of total β-catenin (E) and nuclear β-catenin (F) in HCT116 cells transfected with GFP or SNRK and treated with CacyBP or NSC siRNA. Bottom panels: summary of densitometry; n = 3. Legends indicate plasmids used; axis labels indicate siRNA used. CacyBP, CacyBP siRNA. *P < 0.05.
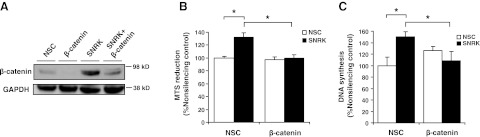
β-Catenin is necessary for increased colon cancer cell proliferation caused by SNRK knockdown
Next, we determined whether changes in β-catenin expression are necessary for the regulation of proliferation by SNRK. We down-regulated SNRK and β-catenin in RKO cells (Fig. 6A) and then measured proliferation and DNA synthesis. As expected, compared to nonsilencing control, SNRK knockdown alone increased proliferation, as measured by MTS reduction (Fig. 6B) and DNA synthesis (Fig. 6C). However, β-catenin knockdown reversed this effect of SNRK knockdown, reducing proliferation (Fig. 6B) and DNA synthesis (Fig. 6C) to control levels. These findings demonstrate that the regulation of β-catenin is a necessary mechanism by which SNRK reduces proliferation of colon cancer cells.
Figure 6.
β-Catenin is necessary for the increase in proliferation caused by SNRK knockdown. A) Western blots of β-catenin protein in RKO cells treated with NSC or SNRK shRNA and NSC or β-catenin siRNA. NSC, treatment with NSC shRNA and siRNA; β-catenin, treatment with NSC shRNA and β-catenin siRNA; SNRK, treatment with SNRK shRNA and NSC siRNA; SNRK+β-catenin, treatment with SNRK shRNA and β-catenin siRNA. B) Proliferation as measured by MTS reduction in RKO cells treated with NSC or SNRK shRNA and NSC or β-catenin siRNA. Legend indicates shRNA used; axis labels indicate siRNA used; n ≥ 3. C) DNA synthesis as measured by BrdU incorporation in RKO cells treated with NSC or SNRK shRNA and NSC or β-catenin siRNA. Legend indicates shRNA used; axis labels indicate siRNA used; n ≥ 7. *P < 0.05.
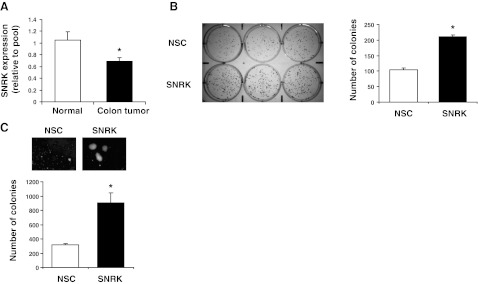
SNRK expression is decreased in human colon cancer tissue, and stable knockdown of SNRK increases the tumorigenicity of colon cancer cells
We then measured levels of SNRK transcript expression in human colon cancer and matched normal tissue using a colon cancer cDNA array. SNRK levels were down-regulated in colon cancer compared with normal tissue (Fig. 7A). Next, we determined whether stable SNRK knockdown increases colon cancer cell tumorigenicity. We stably transfected RKO cells with nonsilencing or SNRK shRNA and verified that SNRK and CacyBP protein levels were reduced, and that β-catenin levels and proliferation were increased (Supplemental Fig. S3). Stable SNRK knockdown increased adherent colony formation (Fig. 7B) and increased colony formation in an anchorage-independent soft agar assay (Fig. 7C), indicating increased in vitro tumorigenesis. These results demonstrate that SNRK is down-regulated in human colon cancer and that stable SNRK knockdown increases the tumorigenicity of colon cancer cells.
Figure 7.
SNRK is reduced in human colon cancer tissue, and stable SNRK knockdown increases the tumorigenicity of colon cancer cells in vitro. A) Quantitative real-time PCR using primers for human SNRK was conducted on samples from matched normal and tumor human colon tissue samples using an Origene TissueScan cDNA array; n = 22–23. B) Colony formation assay in RKO cells with stable knockdown of SNRK; n = 6. NSC, stable nonsilencing control shRNA treatment. C) Soft agar colony assay in RKO cells with stable knockdown of SNRK; n = 3. *P < 0.05.
DISCUSSION
In this study, we investigated the role of SNRK in the proliferation of colon cancer cells. We showed that SNRK is present in both the cytoplasm and the nucleus and that SNRK regulates genes involved in DNA synthesis and cell cycle regulation. The overexpression of SNRK resulted in a significant decrease in proliferation, whereas the down-regulation of SNRK increased proliferation in colon cancer cell lines. Furthermore, CacyBP up-regulation and β-catenin down-regulation were necessary for these effects. SNRK expression was also reduced in human colon cancer tissue, and stable SNRK knockdown increased the tumorigenicity of colon cancer cells. Collectively, these studies indicate that SNRK regulates colon cancer cell proliferation through a CacyBP-β-catenin pathway.
We have demonstrated that SNRK increases CacyBP mRNA and protein expression. This is the first time that a kinase has been shown to regulate CacyBP, and our findings also introduce a novel mechanism by which the activity of the SCFTBL1 complex is regulated. Previously, the only SCFTBL1 complex member known to both be regulated and to induce changes in β-catenin levels was seven in absentia homologue-1 (Siah-1), which is increased by p53 in response to DNA damage (15). The regulation of CacyBP by SNRK suggests that SNRK may be a therapeutic target for the treatment of colon cancer, especially in cases where the phosphorylated residues of β-catenin are mutated, because the SCFTBL1 ubiquitination of β-catenin is phosphorylation-independent. An interesting next step will be to determine how SNRK regulates CacyBP levels and whether it interacts with transcription factors to alter CacyBP mRNA expression.
The role of CacyBP in cancer appears to be complex and cell type dependent. Previous studies have found that CacyBP decreases proliferation in gastric cancer (21) and renal carcinoma cells (25) and regulates the G1 checkpoint in MEF (26). CacyBP has also been found to reduce proliferation of breast cancer cells, and its expression is positively correlated with improved prognoses in breast cancer (27). However, CacyBP is also associated with increases in the proliferation of pancreatic cancer cells (28). There have also been conflicting findings about the relative expression of CacyBP in normal and cancerous tissues, with some reports finding elevated CacyBP expression in tumors and others finding reduced expression (21, 25, 29). The ubiquitination targets of CacyBP may vary depending on cell type, and this variation may determine whether CacyBP increases or decreases cancer cell proliferation. In addition, CacyBP may be involved in cellular functions other than proliferation that affect cancer progression. Our work shows that CacyBP reduces the proliferation of colon cancer cells and, consequently, suggests that SNRK and CacyBP up-regulation may be therapeutically desirable for targeting colon cancer.
Although some of the effects we have identified for SNRK may be considered relatively modest, they are proportional to the increase in the activity of the overexpressed SNRK protein in our studies (Fig. 2B). Furthermore, we have provided evidence that a modest change in SNRK levels is relevant, as its expression is moderately altered in human colon cancer tissue, and a conservative modulation of its levels markedly affects cancer cell tumorigenicity. Moreover, the overexpression of a kinase may sometimes lead to nonspecific cellular effects, but we believe that the overexpression experiments reported here accurately reflect the physiological function of SNRK for several reasons. First, we have used SNRK down-regulation to complement our findings from overexpression. Second, we have demonstrated that overexpressed SNRK activates a known SNRK substrate peptide. Finally, we have used a SNRK-T173A kinase mutant control, which has effects similar to those observed with the GFP control, thereby demonstrating that the effects of SNRK overexpression are dependent on its kinase activity and are not merely an artifact of kinase overexpression. Thus, the results from our overexpression studies provide crucial information about the physiological function of this poorly characterized kinase.
In summary, our results indicate that SNRK is down-regulated in human colon cancer, and that SNRK decreases cell proliferation by increasing CacyBP expression and reducing β-catenin levels and signaling. Thus, SNRK is a novel regulator of CacyBP and β-catenin protein levels, as well as colon cancer cell proliferation, and may be a potential therapeutic target for the treatment of colon cancer.
Supplementary Material
Acknowledgments
H.A. is supported by U.S. National Institutes of Health grants K02 HL107448, R01 HL087149, R01 HL104181, P01 HL108795, and the American Heart Association.
The authors report no conflicts of interest.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- AMPK
- AMP-activated protein kinase
- APC
- adenomatous polyposis coli
- BrdU
- 5-bromodeoxyuridine
- CacyBP
- calcyclin-binding protein
- EMEM
- Eagle's minimum essential medium
- LEF
- lymphoid enhancer factor
- MEF
- mouse embryonic fibroblast
- NLS
- nuclear localization signal
- Snf1
- sucrose nonfermenting 1
- SNRK
- Snf1-related kinase
- TCF
- T-cell factor
REFERENCES
- 1. Celenza J. L., Carlson M. (1984) Cloning and genetic mapping of SNF1, a gene required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 4, 49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hardie D. G. (2007) AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell. Biol. 8, 774–785 [DOI] [PubMed] [Google Scholar]
- 3. Fogarty S., Hardie D. G. (2010) Development of protein kinase activators: AMPK as a target in metabolic disorders and cancer. Biochim. Biophys. Acta 1804, 581–591 [DOI] [PubMed] [Google Scholar]
- 4. Wong A. K., Howie J., Petrie J. R., Lang C. C. (2009) AMP-activated protein kinase pathway: a potential therapeutic target in cardiometabolic disease. Clin. Sci. (Lond.) 116, 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bright N. J., Thornton C., Carling D. (2009) The regulation and function of mammalian AMPK-related kinases. Acta Physiol. (Oxf.) 196, 15–26 [DOI] [PubMed] [Google Scholar]
- 6. Hurov J., Piwnica-Worms H. (2007) The Par-1/MARK family of protein kinases: from polarity to metabolism. Cell Cycle 6, 1966–1969 [DOI] [PubMed] [Google Scholar]
- 7. Yoshida K., Yamada M., Nishio C., Konishi A., Hatanaka H. (2000) SNRK, a member of the SNF1 family, is related to low K+-induced apoptosis of cultured rat cerebellar granule neurons. Brain Res. 873, 274–282 [DOI] [PubMed] [Google Scholar]
- 8. Becker W., Heukelbach J., Kentrup H., Joost H. G. (1996) Molecular cloning and characterization of a novel mammalian protein kinase harboring a homology domain that defines a subfamily of serine/threonine kinases. Eur. J. Biochem. 235, 736–743 [DOI] [PubMed] [Google Scholar]
- 9. Kertesz N., Samson J., Debacker C., Wu H., Labastie M. C. (2002) Cloning and characterization of human and mouse SNRK sucrose non-fermenting protein (SNF-1)-related kinases. Gene 294, 13–24 [DOI] [PubMed] [Google Scholar]
- 10. Chun C. Z., Kaur S., Samant G. V., Wang L., Pramanik K., Garnaas M. K., Li K., Field L., Mukhopadhyay D., Ramchandran R. (2009) Snrk-1 is involved in multiple steps of angioblast development and acts via notch signaling pathway in artery-vein specification in vertebrates. Blood 113, 1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jaleel M., McBride A., Lizcano J. M., Deak M., Toth R., Morrice N. A., Alessi D. R. (2005) Identification of the sucrose non-fermenting related kinase SNRK, as a novel LKB1 substrate. FEBS Lett. 579, 1417–1423 [DOI] [PubMed] [Google Scholar]
- 12. Lizcano J. M., Goransson O., Toth R., Deak M., Morrice N. A., Boudeau J., Hawley S. A., Udd L., Makela T. P., Hardie D. G., Alessi D. R. (2004) LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schneider G., Filipek A. (2011) S100A6 binding protein and Siah-1 interacting protein (CacyBP/SIP): spotlight on properties and cellular function. Amino Acids 41, 773–780 [DOI] [PubMed] [Google Scholar]
- 14. Filipek A., KuŸnicki J. (1998) Molecular cloning and expression of a mouse brain cDNA encoding a novel protein target of calcyclin. J. Neurochem. 70, 1793–1798 [DOI] [PubMed] [Google Scholar]
- 15. Matsuzawa S. I., Reed J. C. (2001) Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol. Cell. 7, 915–926 [DOI] [PubMed] [Google Scholar]
- 16. Tetsu O., McCormick F. (1999) Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398, 422–426 [DOI] [PubMed] [Google Scholar]
- 17. He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 18. Morin P. J., Sparks A. B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K. W. (1997) Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275, 1787–1790 [DOI] [PubMed] [Google Scholar]
- 19. Edgar R., Domrachev M., Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl. Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Veeman M. T., Slusarski D. C., Kaykas A., Louie S. H., Moon R. T. (2003) Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 13, 680–685 [DOI] [PubMed] [Google Scholar]
- 21. Ning X., Sun S., Hong L., Liang J., Liu L., Han S., Liu Z., Shi Y., Li Y., Gong W., Zhang S., Chen Y., Guo X., Cheng Y., Wu K., Fan D. (2007) Calcyclin-binding protein inhibits proliferation, tumorigenicity, and invasion of gastric cancer. Mol. Cancer Res. 5, 1254–1262 [DOI] [PubMed] [Google Scholar]
- 22. Santelli E., Leone M., Li C., Fukushima T., Preece N. E., Olson A. J., Ely K. R., Reed J. C., Pellecchia M., Liddington R. C., Matsuzawa S. (2005) Structural analysis of Siah1-Siah-interacting protein interactions and insights into the assembly of an E3 ligase multiprotein complex. J. Biol. Chem. 280, 34278–34287 [DOI] [PubMed] [Google Scholar]
- 23. Major M. B., Camp N. D., Berndt J. D., Yi X., Goldenberg S. J., Hubbert C., Biechele T. L., Gingras A. C., Zheng N., Maccoss M. J., Angers S., Moon R. T. (2007) Wilms tumor suppressor WTX negatively regulates WNT/β-catenin signaling. Science 316, 1043–1046 [DOI] [PubMed] [Google Scholar]
- 24. Da Costa L. T., He T. C., Yu J., Sparks A. B., Morin P. J., Polyak K., Laken S., Vogelstein B., Kinzler K. W. (1999) CDX2 is mutated in a colorectal cancer with normal APC/β-catenin signaling. Oncogene 18, 5010–5014 [DOI] [PubMed] [Google Scholar]
- 25. Sun S., Ning X., Liu J., Liu L., Chen Y., Han S., Zhang Y., Liang J., Wu K., Fan D. (2007) Overexpressed CacyBP/SIP leads to the suppression of growth in renal cell carcinoma. Biochem. Biophys. Res. Commun. 356, 864–871 [DOI] [PubMed] [Google Scholar]
- 26. Fukushima T., Zapata J., Singha N., Thomas M., Kress C., Krajewska M., Krajewski S., Ronai Z., Reed J., Matsuzawa S. (2006) Critical function for SIP, a ubiquitin E3 ligase component of the beta-catenin degradation pathway, for thymocyte development and G1 checkpoint. Immunity 24, 29–39 [DOI] [PubMed] [Google Scholar]
- 27. Nie F., Yu X. L., Wang X. G., Tang Y. F., Wang L. L., Ma L. (2010) Down-regulation of CacyBP is associated with poor prognosis and the effects on COX-2 expression in breast cancer. Int. J. Oncol. 37, 1261–1269 [DOI] [PubMed] [Google Scholar]
- 28. Chen X., Mo P., Li X., Zheng P., Zhao L., Xue Z., Ren G., Han G., Wang X., Fan D. (2011) CacyBP/SIP protein promotes proliferation and G1/S transition of human pancreatic cancer cells. Mol. Carcinog. 50, 804–810 [DOI] [PubMed] [Google Scholar]
- 29. Zhai H., Shi Y., Jin H., Li Y., Lu Y., Chen X., Wang J., Ding L., Wang X., Fan D. (2008) Expression of calcyclin-binding protein/Siah-1 interacting protein in normal and malignant human tissues: an immunohistochemical survey. J. Histochem. Cytochem. 56, 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.