Abstract
Nonalcoholic fatty liver disease (NAFLD) is a major health problem and a leading cause of chronic liver disease in the United States and developed countries. In humans, genetic factors greatly influence individual susceptibility to NAFLD. The goals of this study were to compare the magnitude of interindividual differences in the severity of liver injury induced by methyl-donor deficiency among individual inbred strains of mice and to investigate the underlying mechanisms associated with the variability. Feeding mice a choline- and folate-deficient diet for 12 wk caused liver injury similar to NAFLD. The magnitude of liver injury varied among the strains, with the order of sensitivity being A/J ≈ C57BL/6J ≈ C3H/HeJ < 129S1/SvImJ ≈ CAST/EiJ < PWK/PhJ < WSB/EiJ. The interstrain variability in severity of NAFLD liver damage was associated with dysregulation of genes involved in lipid metabolism, primarily with a down-regulation of the peroxisome proliferator receptor α (PPARα)-regulated lipid catabolic pathway genes. Markers of oxidative stress and oxidative stress-induced DNA damage were also elevated in the livers but were not correlated with severity of liver damage. These findings suggest that the PPARα-regulated metabolism network is one of the key mechanisms determining interstrain susceptibility and severity of NAFLD in mice.—Tryndyak, V., de Conti, A., Kobets, T., Kutanzi, K., Koturbash, I., Han, T., Fuscoe, J. C., Latendresse, J. R., Melnyk, S., Shymonyak, S., Collins, L., Ross, S. A., Rusyn, I., Beland, F. A., Pogribny, I. P. Interstrain differences in the severity of liver injury induced by a choline- and folate-deficient diet in mice are associated with dysregulation of genes involved in lipid metabolism.
Keywords: nonalcoholic fatty liver, interindividual, oxidative stress
Nonalcoholic fatty liver disease (NAFLD), which represents several related liver disorders, ranging from simple hepatic steatosis to nonalcoholic steatohepatitis (NASH) and cirrhosis, is a major health problem and a leading cause of chronic liver disease in the United States and developed countries (1). Emerging evidence has indicated clearly that interindividual genetic factors may greatly influence individual susceptibility to NAFLD in humans (2–7). For instance, recent genome-wide association studies of subjects with biopsy-confirmed NAFLD have identified several single-nucleotide polymorphisms as potential genetic modifiers influencing risk and features of NAFLD (2, 4, 6).
Elucidating the underlying mechanisms of the individual differences in susceptibility to NAFLD is critical not only for identifying of subpopulations sensitive to NAFLD, but also for its treatment and prevention. However, investigating the molecular basis of how genetic factors influencing the susceptibility to NAFLD in humans is frequently impractical and always very complex, whereas using relevant animal models may substantially overcome the many limitations of human-only studies. Several different animal models (8, 9), including dietary manipulations, have been used to replicate human fatty liver disease. Each of these models has some advantages and limitations. For instance, feeding high-energy diets compromises the metabolic status and induces obesity but does not uniformly cause liver injury (9). In contrast, feeding either a methionine- and choline-deficient or choline-deficient diet to rats or mice causes liver injury that resembles morphological features of human NAFLD (9) but does not attain the compromised metabolic status observed in patients.
Although most of the current studies that have used relevant animal models of human NAFLD have focused on identifying the underlying molecular mechanisms of the pathogenesis and progression of NAFLD (10), less attention has been given to intraspecies determinants for the predisposition to NAFLD that are applicable in the genetically diverse human population (11–14). In this respect, population-based mouse models that capture the broad genetic diversity similar to that found in humans (15) offer substantial advantages in understanding specific molecular pathways that determine individual susceptibility to numerous pathological states (16). More important, the results of these population-based studies may help identify determinants of the predisposition to NAFLD.
Based on these considerations, the goal of this study was to investigate the underlying molecular mechanisms associated with interstrain differences in sensitivity to liver injury induced by methyl-donor deficiency among individual inbred strains of mice. We demonstrate that interstrain variability in the severity of NAFLD induced by a choline- and folate-deficient (CFD) diet associated with dysregulated lipid metabolism, primarily with a diminished expression of peroxisome proliferator receptor α (PPARα)-regulated fatty acid oxidation pathway genes, including the acyl-coenzyme A oxidase 1 (Acox1) and medium-chain acyl-coenzyme A dehydrogenase (Mcad). This is a driving force that triggers lipid accumulation in hepatocytes and the development of NAFLD and determines the extent of NAFLD-associated liver injury. Markers of oxidative stress and oxidative stress-induced DNA damage were also elevated in the livers of mice fed the CFD diet but were not correlated with severity of liver damage.
MATERIALS AND METHODS
Animals and experimental design
Male A/J, C57BL/6J, C3H/HeJ, 129S1/SvImJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ mice (6 wk of age) were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). These strains were selected because they provide an excellent representation of genetic diversity and their genomes have been fully sequenced (15). The mice were housed in sterilized cages in a temperature-controlled room (24°C) with a 12-h light/dark cycle, and given ad libitum access to water and NIH-41 irradiated pelleted diet. At 8 wk of age, mice from each strain were allocated randomly to control and experimental groups. Mice in the methyl-donor-deficient experimental groups were maintained on a diet low in methionine (0.17% w/w) and lacking choline and folic acid (diet no. 519541, choline- and folate-deficient, iron-supplemented, and l-amino acid-defined diet; Dyets, Bethlehem, PA, USA) for 12 wk. Mice in the methyl-adequate control groups received the same diet supplemented with 0.4% methionine, 0.3% choline bitartrate, and 2 mg/kg folic acid. Diets were stored at 4°C before use and given ad libitum with replacement 2×/wk. Body weights of the mice were recorded weekly. Five experimental and 5 control mice from each strain were euthanized by exsanguination following deep isoflurane anesthesia 12 wk after diet initiation. The livers were excised, and a slice of the median lobe was fixed in neutral buffered formalin for 48 h for histopathological examination. The remaining liver was snap-frozen immediately in liquid nitrogen and stored at −80°C for subsequent analyses. All experimental procedures were reviewed and approved by the National Center for Toxicological Research Animal Care and Use Committee.
Tissue processing, histological analysis, and criteria for pathology assessment
After 48 h, a slice of the median lobe of the liver that had been fixed in 10% neutral buffered formalin was trimmed, processed, and embedded in infiltrating medium (Surgipath Formula R; Leica Biosystems, Richmond, IL, USA), sectioned at ∼5 μm, mounted on a glass slide, and stained with hematoxylin and eosin. The hematoxylin- and eosin-stained sections were evaluated for steatosis, inflammation, necrosis, hepatocellular karyocytomegaly, and oval cell proliferation and graded using a severity score system for each of the morphological parameters as follows: grade 0, absent; grade 1, minimal; grade 2, mild; grade 3, moderate; and grade 4, severe changes. The data are presented as total liver pathology scores, which represent the combined mean severity for all of the lesions detected in the livers of the mouse strains.
RNA extraction and gene expression analysis using microarray technology
Total RNA was extracted from liver tissue using RNeasy Mini kits (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Gene expression profiles in the livers of A/J, C3H/HeJ, and WSB/EiJ mice fed the control or the CFD diet were determined utilizing Agilent whole-genome 4 × 44K mouse microarrays (Agilent Technologies, Santa Clara, CA, USA). Sample labeling and microarray processing were performed as detailed in the One-Color Microarray-Based Gene Expression Analysis 5.5 protocol (Agilent Technologies). The hybridized slides were scanned with an Agilent DNA Microarray scanner (Agilent Technologies) at 5-μm resolution. The resulting images were analyzed by determining the Cy3 fluorescence intensity of all gene spots (features) on each array using the Agilent Feature Extraction 10.7 software. The raw data were then uploaded into the ArrayTrack database (17). The median fluorescence intensity of all the pixels within one feature was taken as the intensity value for that feature. The raw intensity values were then normalized using 75 percentile channel scaling normalization using ArrayTrack. A list of differentially expressed genes was generated with ArrayTrack using a t test at P < 0.05 and a fold change at >2.0.
Functional analysis of significant genes
Ingenuity Pathway Analysis (IPA) 9.0 software (Ingenuity Systems, Redwood City, CA, USA) was used to determine canonical pathways that were enriched for significant mRNA transcripts identified from the t test analysis. Significance values were calculated based on a right-tailed Fisher's exact test that determined whether a pathway was overrepresented by calculating whether the genes in a given pathway were enriched within the data set compared to all genes on the array; P < 0.05 was selected as the cutoff for significance based on IPA threshold recommendations. Only those pathways with a P value below the threshold and having >3 representative genes in the data set were considered significant.
Quantitative reverse transcription–polymerase chain reaction (qRT-PCR)
Total RNA (2 μg) was reverse transcribed using random primers and the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's protocol, and cDNA was analyzed in a 96-well plate assay format using the 7900HT Fast Real-Time PCR System (Applied Biosystems). Each plate contained one experimental gene and a housekeeping gene. All primers were obtained from Applied Biosystems. The cycle threshold (Ct) for each sample was determined from the linear region of the amplification plot. The ΔCt values for all genes were determined relative to the endogenous control β-actin (Actb). The ΔΔCt values were calculated using treated group means relative to strain-matched control group means (18). The fold change data were calculated from the ΔΔCt values. All qRT-PCR reactions were conducted in triplicate and repeated twice.
Western blot analysis of protein levels
Liver tissue lysates were prepared by homogenization of 50 mg of tissue in 500 μl of lysis buffer (50 mM Tris-HCl, pH 7.4; 1% Nonidet P-40; 0.25% sodium deoxycholate; 150 mM NaCl; 1 mM EDTA; 1 mM PMSF; 1 μg/ml each of aprotinin, leupeptin, and pepstatin; 1 mM Na3VO4; and 1 mM NaF), sonication, and incubation at 4°C for 30 min, followed by centrifugation at 10,000 g at 4°C for 20 min. Extracts containing equal quantities of proteins were separated by SDS-PAGE on 8–15% polyacrylamide gels and transferred to PVDF membranes. Membranes were probed with primary antibodies against PPARα (1:1000; Abcam, Cambridge, MA, USA), PPARγ (1:200; Santa Cruz Biotechnology, Santa Cruz, CA, USA), and CCAAT/enhancer binding protein α (CEBPα; 1:200; Santa Cruz Biotechnology). Horseradish peroxidase-coupled donkey anti-goat secondary antibody (Santa Cruz Biotechnology) was used for visualization. Chemiluminescence detection was performed with Immobilon Western Chemiluminescent HRP Substrate (Millipore Corp., Billerica, MA, USA) and measured directly by a UVP BioSpectrum Imaging System (UVP, Upland, CA, USA). Equal protein loading was confirmed by immunostaining against glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:5000; Sigma-Aldrich, St. Louis, MO, USA).
Determination of hepatic glutathione content
Levels of reduced glutathione (GSH) and oxidized glutathione (GSSG) in the livers of control mice and mice fed the CFD diet were measured by high-performance liquid chromatography coupled with coulometric electrochemical detection, as described earlier (19).
Liquid chromatography combined with electrospray tandem mass spectrometry analysis of 8-oxodeoxyguanosine (8-oxodG) in hepatic DNA
Genomic DNA from the livers of control mice and mice fed the CFD diet was isolated by a slight modification of the method reported by Nakamura et al. (20). The levels of 8-oxodG in hepatic DNA were measured by liquid chromatography combined with electrospray tandem mass spectrometry, as described previously (21).
Determination of genomic DNA damage
The extent of genomic DNA damage was determined by measuring the levels of apurinic/apyrimidinic (AP) sites by using DNA Damage Quantification AP Site Counting Kit (Dojindo Molecular Technologies, Rockville, MD, USA) according to the manufacturer's protocol. Briefly, the levels of biotin-tagged AP sites in genomic DNA were measured quantitatively by an avidin-biotin-peroxidase assay at 650 nm in a Termomax Microplate Reader (Molecular Devices, Sunnyvale, CA, USA), using the linear calibration curve with the standards supplied with the kit. The data were calculated as number of AP sites per 105 nt and presented as percentage change from control values. Each sample was analyzed in triplicate.
Statistical analyses
Results are presented as means ± sd. Data were analyzed by 1-way analysis of variance (ANOVA), with pairwise comparisons being made by the Student-Newman-Keuls method. When necessary, the data were natural log transformed before conducting the analyses to maintain a more equal variance or normal data distribution. Pearson product-moment correlation coefficients were used to determine the relationship between levels of gene expression, GSH, GSSG, 8-oxodG, and AP sites, and GSSG/GSH ratio and histopathology scores. Values of P < 0.05 were considered significant.
RESULTS
Histopathological evaluation of liver injury induced by the CFD diet
Feeding mice the CFD diet caused pathological changes in the liver that share key features with NAFLD, including cytoplasmic alteration, steatosis, necrosis, inflammation, oval cell hyperplasia, and karyomegaly. However, the magnitude of NAFLD-associated liver injury differed among the individual mouse strains, with the following order based on total liver pathology scores: A/J ≈ C57BL/6J ≈ C3H/HeJ < 129S1/SvImJ ≈ CAST/EiJ < PWK/PhJ < WSB/EiJ (Fig. 1A and Supplemental Table S1).
Figure 1.
Histopathological lesions in the livers of mice fed a CFD diet for 12 wk. A) Total liver pathology scores of hepatic lesions in mice fed the CFD diet. Total liver pathology scores represent the mean severity for all of the lesions detected in the livers of mice fed the CFD diet. Bars with different letters differ significantly between strains. B) Representative hematoxylin and eosin staining of liver tissues from A/J (a, d), C3H/HeJ (b, e), and WSB/EiJ (c, f) mice fed the control diet (a–c) or the CFD diet (d–f). Arrows indicate inflammatory foci; arrowheads indicate single-cell necrosis.
Figure 1B shows representative hematoxylin and eosin staining of liver tissues from A/J, C3H/HeJ, and WSB/EiJ mice fed control diet and the CFD diet. Lipid accumulation in hepatocytes was characterized by microvesicular, macrovesicular, and, most commonly, mixed (microvesicular and macrovesicular) patterns of fat deposition. Hepatocytes with microvesicular fat accumulation appeared foamy, with the cytoplasm partially or completely filled with numerous small lipid vacuoles, which did not displace the nucleus to the periphery. This pattern of steatosis was most pronounced in A/J mice. Macrovesicular fatty change was morphologically characterized by hepatocytes mostly containing a moderately large to large and well-defined single rounded vacuole within each cell. The nucleus and cytoplasm were displaced to the periphery in such cells. This pattern was commonly observed in PWK/PhJ and WSB/EiJ mice, strains exhibiting the greatest degree of liver injury. Inflammatory foci (Fig. 1B, marked by arrows) were composed of a mixed population of polymorphonuclear and mononuclear cells, mainly neutrophils, macrophages, and lymphocytes. Single-cell necrosis (Fig. 1B, marked by arrowheads) was one of the most common histopathological observations in both of the PWK/PhJ and WSB/EiJ mouse strains, but was most severe in the WSB/EiJ mice.
Microarray analysis of gene expression in the livers of mice fed the CFD diet
To determine whether or not gene expression profiles mirrored the pathological changes found in the livers of mice fed the CFD diet, hepatic gene expression profiles were examined in control and methyl-donor-deficient A/J, C3H/HeJ, and WSB/EiJ mice using high-throughput Agilent whole genome 4 × 44K mouse microarrays. These mice exhibited strain-specific differences in the magnitude of liver injury induced by the CFD diet, with A/J and C3H/HeJ mice being among the least susceptible and WSB/EiJ mice being the most sensitive (Fig. 1 and Supplemental Table S1).
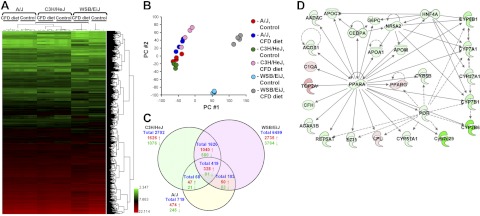
Unsupervised hierarchical clustering of the gene expression data showed that each strain could be distinguished by its hepatic gene expression profile (Fig. 2A), with the A/J and C3H/HeJ mice more closely related to each other than WSB/EiJ mice. Furthermore, mice fed the CFD diet could be distinguished from mice fed the control diet within each strain, with the difference within the WSB/EiJ strain being the greatest. The tight clustering of samples within each strain/diet group indicated high quality data that would allow subtle differences in gene expression to be identified.
Figure 2.
Whole-genome microarray analysis of gene expression in the livers of A/J, C3H/HeJ, and WSB/EiJ mice fed control diet or a CFD diet. A) Heat map illustrating significant differences in global gene expression among A/J, C3H/HeJ, and WSB/EiJ mice fed control or CFD diets. Color bar identifies high-expressed (red) and low-expressed (green) genes. B) PC analysis illustrating similarities within groups, as evidenced by tight clustering of samples within groups, and differences among control A/J, C3H/HeJ, and WSB/EiJ mice as well as between A/J, C3H/HeJ, and WSB/EiJ mice fed the CFD diet. C) Venn diagram showing genes that were significantly different among A/J, C3H/HeJ, and WSB/EiJ mice fed the CFD diet. D) Molecular network interactions among genes in the livers of WSB/EiJ mice significantly differ between control and deficient groups. The IPA 9.0 database was used to determine and visualize molecular pathways enriched by the significant mRNA transcripts. Up-regulated genes are shown in red, down-regulated genes in green.
Principal component (PC) analysis utilizing the entire gene expression dataset also showed the relatively tight clustering of each strain/diet group and the clear difference between methyl-donor-deficient and control diet liver gene expression profiles (Fig. 2B). The profiles of the 2 relatively resistant strains, A/J and C3H/HeJ, were clustered away from the susceptible WSB/EiJ strain along PC 1. The effect of the CFD diet within each strain was greatest for WSB/EiJ mice, followed by C3H/HeJ, and then A/J mice, and appeared to be mainly along PC 2. The amount of variability captured was 44.3 and 23.7% for PCs 1 and 2, respectively.
To identify genes that were differentially expressed between the control and the CFD diet groups within each strain, a t test, P < 0.05, coupled with a fold-change cutoff > 2 was applied. A total of 719, 2702, and 6499 genes were found to be differentially expressed in the livers of A/J, C3H/HeJ, and WSB/EiJ mice, respectively, fed the CFD diet (Fig. 2C). Even though the A/J and C3H/HeJ mice had a similar magnitude of NAFLD-associated liver injury (Fig. 1), there were nearly 4 times as many genes changing expression in response to the CFD diet in the C3H/HeJ strain as in the A/J strain. This suggests that gene expression profiles provide greater sensitivity than histopathological evaluation. The pattern of gene expression changes was also different, depending on the extent of liver injury. In the livers of A/J and C3H/HeJ mice, strains relatively resistant to NAFLD-associated liver injury, 60–66% of the differentially expressed genes were up-regulated by the CFD diet, whereas in the susceptible WSB/EiJ strain only 42% of the genes were up-regulated. The 2 relatively resistant strains also shared many of the same CFD diet-induced gene expression changes. Approximately 68% of the differentially expressed genes found in A/J mice were also found in C3H/HeJ mice; of the up-regulated genes, 79% were in common. In addition to this shared set of differentially expressed genes, the C3H/HeJ strain had more than 2000 other differentially expressed genes. The susceptible WSB/EiJ strain also shared this core set of differentially expressed genes and, in addition, showed ∼6000 other differentially expressed genes (Fig. 2C). In total, there were 419 (328 up-regulated and 91 down-regulated) differentially expressed genes induced by the CFD diet in all 3 strains, and the magnitude of the expression of many of these genes progressively increased with the magnitude of liver injury.
Pathway enrichment analysis of the differentially up-regulated and down-regulated genes in the livers of control mice and mice fed the CFD diet demonstrated a strong enrichment in genes associated with deregulated lipid metabolism, small-molecule biochemistry, molecular transport, cell death and proliferation, and inflammation; key deregulated pathways in the pathogenesis of NAFLD. Interestingly, the greatest effect of the CFD diet in mice was on the metabolic PPAR-regulated network (Fig. 2D; Ingenuity score 27).
Effect of the CFD diet on lipid metabolism pathways
To evaluate further the effect of the CFD diet on lipid metabolism pathways, the expression of key differentially expressed genes involved in the regulation of cellular lipid metabolism was examined in the livers of A/J, C57BL/6J, C3H/HeJ, 129S1/SvImJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ mice by qRT-PCR.
Figure 3 shows that feeding the CFD diet caused strain-dependent changes in the expression of lipid metabolism genes. The most noticeable changes induced by the CFD diet were a distinct up-regulation of the fatty acid translocase (Fat; Cd36) and stearoyl-coenzyme A desaturase 2 (Scd2), and a marked down-regulation of carbohydrate response element-binding protein (Chrebp) and PPARα-regulated lipid catabolic pathway genes (Fig. 3). This was evident by significantly lower protein level of PPARα (Fig. 4) and a decreased expression of Acox1 and Mcad, two key genes involved in peroxisomal and mitochondrial fatty acid β-oxidation (Fig. 3). Interestingly, strain-dependent changes in the expression of Scd2, Chrebp, PPARα, Acox1, and Mcad in the livers of mice fed the CFD diet strongly correlated with the degree of liver injury (Supplemental Figs. S1 and S2).
Figure 3.
Expression of selective markers of lipid metabolism in the livers of mice fed a CFD diet. Expression of Cd36, Apob, Srebf1, Chrebp, Fasn, Scd2, Acox1, Mcad, and Cpt1 genes was determined by qRT-PCR as detailed in Materials and Methods. Results are presented as an average fold change in the expression of each gene in the livers of mice fed the CFD diet relative to that in control groups, which were assigned a value of 1. Bars represent means ± sd (n=5). Bars with different letters differ significantly between strains.*P < 0.05 vs. control mice fed a control diet containing 0.4% methionine, 0.3% choline bitartrate, and 2 mg/kg folic acid.
Figure 4.
Western blot analysis of PPARα, PPARγ, and CEBPα proteins in the livers of control mice and mice fed a CFD diet. Western blot images are representative. Results are presented as an average fold change in the level of each protein in the livers of mice fed the CFD diet relative to that in control groups, which were assigned a value of 1. Bars represent means ± sd (n=5). Bars with different letters differ significantly between strains.*P < 0.05 vs. control mice fed a control diet containing 0.4% methionine, 0.3% choline bitartrate, and 2 mg/kg folic acid.
Effect of the CFD diet on the expression of cytochrome P450 (Cyp) genes
Figure 5 shows that feeding the CFD diet also caused substantial changes in the expression of PPARα-associated Cyp genes (22–26), characterized by a marked, but strain independent, down-regulation of Cyp2b10, Cyp2c29, and Cyp7b1 in the livers of all the strains. Feeding the CFD diet also reduced the expression of Cyp2e1 and Cyp27a1 in the livers of each of the mouse strains, with the exception of Cyp2e1 in A/J mice. Furthermore, the decrease of Cyp2e1 and Cyp27a1 was correlated strongly with the extent of liver injury (r=−0.747, P=2.5×10−13 for Cyp2e1 and r=−0.768, P=2.1×10−14 for Cyp27a1). Interestingly, the expression of these 2 genes in the livers of mice fed the CFD diet also correlated directly with the levels of PPARα (r=0.753, P=3.4×10−11 for Cyp2e1 and r=0.786, P=1.9×10−12 for Cyp27a1). The expression of Cyp4a14 was significantly increased in the livers of all but C57BL/6J-deficient mice.
Figure 5.
Expression of Cyp genes in the livers of mice fed a CFD diet. Results are presented as an average fold change in the expression of each gene in the livers of mice fed the CFD diet relative to that in control groups, which were assigned a value of 1. Bars represent means ± sd (n=5). Bars with different letters differ significantly between strains.*P < 0.05 vs. control mice fed a control diet containing 0.4% methionine, 0.3% choline bitartrate, and 2 mg/kg folic acid.
Effect of the CFD diet on oxidative stress
Oxidative stress and subsequent reactive oxygen species-induced damage to DNA and other biomolecules are well-established events in the pathogenesis of NAFLD (27, 28). In view of this, the hepatic content of GSH, a prevalent low-molecular-weight nonenzymatic antioxidant in mammalian cells (29), was evaluated in the livers of control mice and mice fed the CFD diet. Figure 6 shows that feeding the CFD diet caused a slight but significant decrease in the levels of hepatic GSH in A/J, C57BL/6J, CAST/EiJ, PWK/PhJ, and WSB/EiJ mice. The levels of GSSG and the GSSG/GSH ratio increased in the livers of all but C3H/HeJ mice. However, the changes in the levels of GSH, GSSG, and GSSG/GSH ratio were not highly correlated with the extent of liver injury (Fig. 6).
Figure 6.
Hepatic glutathione content in the livers of mice fed a CFD diet. A) Liver GSH and GSSG content and GSSG/GSH ratio in mice fed the CFD diet relative to that in control groups. Results are presented as an average fold change; control groups were assigned a value of 1. Bars represent means ± sd (n=5). Bars with different letters differ significantly between strains.*P < 0.05 vs. control mice fed a control diet containing 0.4% methionine, 0.3% choline bitartrate, and 2 mg/kg folic acid. B) Correlation plots of total liver pathology scores and the GSH and GSSG content and GSSG/GSH ratio in the livers of mice fed the CFD diet. Each symbol represents an individual animal in the CFD diet group of the respective mouse strain.
The levels of 8-oxodG in genomic DNA in A/J, C57BL/6J, C3H/HeJ, and PWK/PhJ mice fed the CFD diet did not differ from mice fed the control diet. In contrast, the levels of 8-oxodG in 129S1/SvImJ and CAST/EiJ mice were significantly decreased, while in WSB/EiJ mice were the levels of 8-oxodG slightly, but significantly elevated (Fig. 7). Feeding the CFD diet did not change the levels of AP sites in hepatic DNA, another indicator of oxidative damage to DNA, in any of the mouse strains (Fig. 7).
Figure 7.
Oxidative damage to genomic DNA in the livers of mice fed a CFD diet. A) Levels of 8-oxodG and AP sites in DNA isolated from the livers of mice fed the CFD diet relative to that in control groups. Bars represent means ± sd (n=5). Bars with different letters differ significantly between strains.*P < 0.05 vs. control mice fed a control diet containing 0.4% methionine, 0.3% choline bitartrate, and 2 mg/kg folic acid. B) Correlation plots of total liver pathology scores and the levels of 8-oxodG and AP sites in DNA in the livers of mice fed the CFD diet. Each symbol represents an individual animal in the MCFD diet group of the respective mouse strain.
The expression of base excision DNA repair genes, whose up-regulation is considered as a reliable biomarker of oxidative DNA damage (30), did not change, except for Ung and Lig1 genes, and, more important, did not correlate with the extent of liver injury (Supplemental Fig. S3).
DISCUSSION
In this study we demonstrate that feeding mice the CFD diet caused liver injury that resembled the liver pathology associated with NAFLD, including its key pathological features: steatosis, inflammation, apoptosis, and necrosis. The severity of liver injury was strain specific, with A/J, C57BL/6J, and C3H/HeJ mice being the least susceptible and PWK/PhJ and WSB/EiJ mice being the most sensitive.
The pathogenesis of NAFLD is often conceptualized as a 2-step process that consists of hepatic triglyceride accumulation leading to steatosis as a first step, followed by a second step that includes oxidative stress and inflammation. However, there is a lack of consensus on the significance of these events and the underlying mechanisms responsible for the development of NAFLD (31) and, more important, for differences in interindividual disease severity and sensitivity.
The results of our study suggest that dysregulated lipid metabolism determines the severity of NAFLD-associated liver injury. This was evidenced by strain-dependent prominent alterations in the hepatic lipid metabolism that correlated tightly with the extent of pathological changes in the livers of mice fed the CFD diet. In contrast, while oxidative stress and oxidative stress-induced DNA damage did increase in the livers of mice fed the CFD diet, the extent of the increase did not reflect the severity of liver damage. This was evidenced by strain-independent changes in the extent of oxidative stress and oxidative stress-induced DNA damage and a lack of correlation between them and the magnitude of liver injury in deficient mice.
Several different mechanisms may be responsible for fat accumulation in the livers during NAFLD, including increased fatty acid delivery, increased de novo fatty acid synthesis, decreased fatty acid oxidation, and/or impaired secretion of very low density lipoproteins (31). In the present study we found that all these mechanisms contribute to liver injury induced by feeding mice the CFD diet. However, among these processes, the inhibition of PPARα-mediated peroxisomal and mitochondrial fatty acid β-oxidation was the driving force that triggered lipid accumulation in the livers. This was evidenced by a marked down-regulation of PPARα, Acox1, and Mcad in the livers of deficient mice, which strongly correlated with the severity of NAFLD-related pathological changes in the livers.
ACOX1 is the first and rate-limiting enzyme that catalyzes peroxisomal β-oxidation of long-chain and very long chain fatty acids (32). It is well established that targeted disruption of the Acox1 gene in mice results in the development of severe microvesicular steatohepatitis (33). Several lines of evidence have established convincingly that PPARα, a member of the nuclear hormone receptor superfamily, is the main transcriptional regulator of a number of genes involved in fatty acid oxidation in liver, including Acox1 (34, 35). Specifically, a recent report by van der Meer et al. (33) demonstrated that the promoter region of the Acox1 gene contains a binding region for PPARα. In addition, it has been shown that sustained activation of PPARα increases hepatic fatty acid oxidation, reduces hepatic steatosis, and prevents obesity in Acox1−/−/ob/ob mice (35) and ethanol-fed C57BL/6J mice (36). In the present study,, we also demonstrated that lipid accumulation in the livers of CFD diet-fed mice may be associated with an impaired mitochondrial fatty acid β-oxidation caused by down-regulation of the Mcad gene. MCAD is one of the key enzymes involved in mitochondrial fatty acid β-oxidation (37). Previous reports have demonstrated that one of the main pathophysiological features of Mcad deficiency is fatty liver (37, 38). Hence, the observed concurrent down-regulation of PPARα, Acox1, and Mcad, which strongly correlated with the strain-dependent severity of liver injury, provides further support to the importance of the Pparα-regulated pathway in the pathogenesis of NAFLD. This also indicates that inhibition of Pparα function may increase susceptibility to NAFLD. This suggestion is further supported by previous observations showing the significance of the Pparα pathway for the proper liver functioning (39), including results of several studies demonstrating that Pparα−/− mice fed either a methionine- and choline-deficient diet (22), a diet containing alcohol (40), or a high-fat diet (25) develop more severe steatohepatitis than wild-type mice.
Another important finding is a substantially injury-dependent down-regulation of the Chrebp gene in the livers of mice fed the CFD diet. ChREBP is a major transcriptional regulator of hepatic lipogenic and glycolytic genes and a major determinant of systemic insulin sensitivity (41). It has been demonstrated that Chrebp−/− mice exhibited insulin resistance (42). Likewise, a decreased expression of ChREBP has been found in patients with NASH with severe insulin resistance (43). In view of this, our study suggests that PWK/PhJ, and WSB/EiJ mice exhibiting the greatest magnitude of liver injury and lowest degree of PPARα, Acox1, Mcad, and Chrebp expression in response to the CFD diet may be relevant mouse strains to study pathogenesis of NAFLD.
Previous reports have demonstrated an existence of an association between increased hepatic lipid accumulation and impaired functioning of Cyp450 enzymes in the pathogenesis of NAFLD (23, 44, 45). In view of this, an altered expression of PPAR-regulated Cyp450 genes, especially a strain-dependent down-regulation of Cyp2e1 and Cyp27a1 in the livers of mice fed the CFD diet, which correlated strongly with the levels of PPARα, is an additional and important finding of the present study.
In summary, the data presented herein point to dysregulated lipid metabolism, primarily a diminished functioning of the PPARα-regulated lipid catabolic pathway, as a driving force that triggers lipid accumulation in the liver, promotes development of NAFLD, and, more important, determines interstrain susceptibility and severity of NAFLD in mice. The strain-dependent alterations in the PPARα-regulated metabolism network may be associated with genetic variations between these strains. Nevertheless, further research is needed to find genetic and genomic markers that could identify individuals susceptible to NAFLD because the relatively small size of the mouse population used in this research limits exploration of the genomic variants that may explain observed differences between strains.
Supplementary Material
Acknowledgments
The authors declare no conflicts of interest. The views expressed in this paper do not necessarily represent those of the U.S. Food and Drug Administration.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- 8-oxodG
- 8-oxodeoxyguanosine
- Acox1
- acyl-coenzyme A oxidase 1
- AP
- apurinic/apyrimidinic
- CEBPα
- CCAAT/enhancer binding protein α
- Cd36
- fatty acid translocase (Fat)
- CFD
- choline- and folate-deficient
- Chrebp
- carbohydrate response element-binding protein
- Cyp
- cytochrome P450
- GSH
- reduced glutathione
- GSSG
- oxidized glutathione
- Mcad
- medium-chain acyl-coenzyme A dehydrogenase
- NAFLD
- nonalcoholic fatty liver disease
- NASH
- nonalcoholic steatohepatitis
- PC
- principal component
- PPARα
- peroxisome proliferator receptor α
- qRT-PCR
- quantitative reverse transcription-PCR
- Scd2
- stearoyl-coenzyme A desaturase 2
- qRT-PCR
- quantitative reverse transcription–polymerase chain reaction
REFERENCES
- 1. Cohen J. C., Horton J. D., Hobbs H. H. (2011) Human fatty liver disease: old questions and new insights. Science 332, 1519–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chalasani N., Guo X., Loomba R., Goodarzi M. O., Haritunians T., Kwon S., Cui J., Taylor K. D., Wilson L., Cummings O. W., Chen Y. D., Rotter J. I. (2010) Genome-wide association study identifies variants associated with histologic features of nonalcoholic fatty liver disease. Gastroenterology 139, 1567–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hooper A., Adams L. A., Burnett J. R. (2011) Genetic determinants of hepatic steatosis in man. J. Lipid Res. 52, 593–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Speliotes E. K., Yerges-Armstrong. L. M., Wu J., Hernaez R., Kim L. J., Palmer C. D., Gudnason V., Eiriksdottir G., Garcia M. E., Launer L. J., Nalls M. A., Clark J. M., Mitchell B. D., Shuldiner A. R., Butler J. L., Tomas M., Hoffmann U., Hwang S. J., Massaro J. M., O'Donnell C. J., Sahani D. V., Salomaa V., Schadt E. E., Schwartz S. M., Siscovick D. S. NASH CRN, GIANT Consortium, MAGIC Investigators, Voight B. F., Carr J. J., Feitosa M. F., Harris T. B., Fox C. S., Smith A. V., Kao W. H., Hirschhorn J. N., Borecki I. B, and GOLD Consortium. (2011) Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 7, e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Di Rosa M., Malaguarnera L. (2012) Genetic variants in candidate genes influencing NAFLD progression. J. Mol. Med. (Berl.) 90, 105–118 [DOI] [PubMed] [Google Scholar]
- 6. Daly A. K., Ballestri S., Carulli L., Loria P., Day C. P. (2011) Genetic determinants of susceptibility and severity in nonalcoholic fatty liver disease. Expert Rev. Gastroenterol. Hepatol. 5, 253–263 [DOI] [PubMed] [Google Scholar]
- 7. Hernaez R. (2012) Genetic factors associated with the presence and progression of nonalcoholic fatty liver disease: a narrative review. Gastroenterol. Hepatol. 35, 32–41 [DOI] [PubMed] [Google Scholar]
- 8. Hebbard L., George J. (2011) Animal models of nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 8, 34–44 [DOI] [PubMed] [Google Scholar]
- 9. Maher J. J. (2011) New insights from rodent models of fatty liver disease. Antioxid. Redox Signal. 15, 535–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guillén N., Navarro M. A., Arnal, Noone E., Arbonés-Mainar J. M., Acín S., Surra J. C., Muniesa P., Roche H. M., Osada J. (2009) Microarray analysis of hepatic gene expression identifies new genes involved in steatotic liver. Physiol. Genomics 37, 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamazaki Y., Kakizaki S., Takizawa D., Ichikawa T., Sato K., Takagi H., Mori M. (2008) Interstrain differences in susceptibility to non-alcoholic steatohepatitis. J. Gastroenterol. Hepatol. 23, 276–282 [DOI] [PubMed] [Google Scholar]
- 12. Hill-Baskin A .E., Markiewski M. M., Buchner D. A., Shao H., DeSantis D., Hsiao G., Subramaniam S., Berger N. A., Croniger C., Lambris J. D., Nadeau J. H. (2009) Diet-induced hepatocellular carcinoma in genetically predisposed mice. Hum. Mol. Genet. 18, 2975–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pogribny I. P., Tryndyak V. P., Bagnyukova T. V., Melnyk S., Montgomery B., Ross S. A., Latendresse J. R., Rusyn I., Beland F. A. (2009) Hepatic epigenetic phenotype predetermines individual susceptibility to hepatic steatosis in mice fed a lipogenic methyl-deficient diet. J. Hepatol. 51, 176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nishikawa S., Sugimoto J., Okada M., Sakairi T., Takagi S. (2012) Gene expression in livers of BALB/C and C57BL/6J mice fed a high-fat diet. Toxicol. Pathol. 40, 71–82 [DOI] [PubMed] [Google Scholar]
- 15. Yang H., Wang J. R., Didion J. P., Buus R. J., Bell T. A., Welsh C. E., Bonhomme F., Yu A. H., Nachman M. W., Pialek J., Tucker P., Boursot P., McMillan L., Churchill G. A., de Villena F. P. (2011) Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 43, 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harrill A. H., Ross P. K., Gatti D. M., Threadgill D. W., Rusyn I. (2009) Population-based discovery of toxicogenomics biomarkers for hepatotoxicity using a laboratory strain diversity panel. Toxicol. Sci. 110, 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang H., Harris S. C., Su Z., Chen M., Qian F., Shi L., Perkins R., Tong W. (2009) ArrayTrack: an FDA and public genomic tool. Methods Mol. Biol. 563, 379–398 [DOI] [PubMed] [Google Scholar]
- 18. Schmittgen T. D., Livak K. J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 19. Melnyk S., Pogribna M., Pogribny I., Hine R. J., James S. J. (1999) A new HPLC method for the simultaneous determination of oxidized and reduced plasma aminothios using coulometric electrochemical detection. J. Nutr. Biochem. 10, 4900–4907 [DOI] [PubMed] [Google Scholar]
- 20. Nakamura J., La D. K., Swenberg J. A. (2000) 5′-nicked apurinic/apyrimidinic sites are resistant to beta-elimination by beta-polymerase and are persistent in human cultured cells after oxidative stress. J. Biol. Chem. 275, 5323–5328 [DOI] [PubMed] [Google Scholar]
- 21. Powell C. L., Kosyk O., Ross P. K., Schoonhoven R., Boysen G., Swenberg J. A., Heinloth A. N., Boorman G. A., Cunningham M. L., Paules R. S., Rusyn I. (2006) Phenotypic anchoring of acetaminophen-induced oxidative stress with gene expression profiles in rat liver. Toxicol. Sci. 93, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ip E., Farrell G. C., Robertson G., Hal P., Kirsch R., Leclercq I. (2003) Central role of PPARα-dependent hepatic lipid turnover in dietary steatohepatitis in mice. Hepatology 38, 123–132 [DOI] [PubMed] [Google Scholar]
- 23. Fisher C. D., Jackson J. P., Lickteig A. J., Augustine L. M., Cherrington N. J. (2008) Drug metabolizing enzyme induction pathways in experimental non-alcoholic steatohepatitis. Arch. Toxicol. 82, 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koh K. H., Xie H., Yu A. M., Jeong H. (2011) Altered cytochrome P450 expression in mice during pregnancy. Drug Metab. Dispos. 39, 165–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abdelmegeed M. A., Yoo S. H., Henderson L. E., Gonzalez F. J., Woodcroft K. J., Song B. J. (2011) PPARα expression protects male mice from high fat-induced nonalcoholic fatty liver. J. Nutr. 141, 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klein K., Thomas M., Winter S., Nussler A.K., Niemi M., Schwab M., Zanger U.M. (2012) PPARA: a novel genetic determinant of CYP3A4 in vitro and in vivo. [E-pub ahead of print] Clin. Pharmacol. Ther. doi: 10.1038/clpt.2011.336 [DOI] [PubMed] [Google Scholar]
- 27. Koek G. H., Liedor P. R., Bast A. (2011) The role of oxidative stress in non-alcoholic steatohepatitis. Clin. Chim. Acta 412, 1297–1305 [DOI] [PubMed] [Google Scholar]
- 28. Rolo A. P., Teodoro J. S., Palmeira C. M. (2012) Role of oxidative stress in the pathogenesis of nonalcoholic steatohepatitis. Free Radic. Biol. Med. 52, 59–69 [DOI] [PubMed] [Google Scholar]
- 29. Franco R., Schoneveld O. J., Pappa A., Panayiotidis M. I. (2007) The central role of glutathione in the pathophysiology of human diseases. Arch. Physiol. Biochem. 113, 234–258 [DOI] [PubMed] [Google Scholar]
- 30. Powell C. L., Swenberg J. A., Rusyn I. (2005) Expression of base excision DNA repair genes as biomarker of oxidative DNA damage. Cancer Lett. 229, 1–11 [DOI] [PubMed] [Google Scholar]
- 31. Marra F., Gastaldelli A., Svegliati Baroni G., Tell G., Tiribelli C. (2008) Molecular basis and mechanisms of progression of non-alcoholic steatohepatitis. Trends Mol. Med. 14, 72–81 [DOI] [PubMed] [Google Scholar]
- 32. Vluggens A., Andreoletti P., Viswakarma N., Jia Y., Matsumoto K., Kulik W., Khan M., Huang J., Guo D., Yu S., Sarkar J., Singh I., Rao M. S., Wanders R. J., Reddy J. K., Cherkaoui-Malki M. (2010) Reversal of mouse Acyl-CoA oxidase 1 (ACOX1) null phenotype by human ACOX1b isoform. Lab. Invest. 90, 696–705 [DOI] [PubMed] [Google Scholar]
- 33. Fan C. Y., Pan J., Chu R., Lee D., Kluckman K. D., Usuda N., Singh I., Yeldandi A. V., Rao M. S., Maeda N., Reddy J. K. (1996) Hepatocellular and hepatic peroxisomal alterations in mice with a disrupted peroxisomal fatty acyl-coenzyme A oxidase gene. J. Biol. Chem. 271, 24698–24710 [DOI] [PubMed] [Google Scholar]
- 34. Van der Meer D. L., Degenhardt T., Väisänen S., de Groot P. J., Heinäniemi M., de Vries S. C., Müller M., Carlberg C., Kersten S. (2010) Profiling of promoter occupancy by PPARalpha in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Res. 38, 2839–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang J., Jia Y., Fu T., Viswakarma N., Bai L., Rao M. S., Zhu Y., Borensztajn J., Reddy J. K. (2012) Sustained activation of PPAR{alpha} by endogenous ligands increases hepatic fatty acid oxidation and prevents obesity in ob/ob mice. FASEB J. 26, 628–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fischer M., You M., Matsumoto M., Crabb D. W. (2003) Peroxisome proliferator-receptor α (PPARα) agonist treatment reverses PPARα dysfunction and abnormalities in hepatic lipid metabolism in ethanol-fed mice. J. Biol. Chem. 278, 27997–28004 [DOI] [PubMed] [Google Scholar]
- 37. Spiekerkoetter U., Wood P. A. (2010) Mitochondrial fatty acid oxidation disorders: pathophysiological studies in mouse models. J. Inherit. Metab. Dis. 33, 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tolwani R. J., Hamm D. A., Tian L., Sharer J. D., Vockley J., Rinaldo P., Matern D., Schoeb T. R., Wood P. A. (2005) Medium-chain acyl-CoA dehydrogenase deficiency in gene-targeted mice. PLoS Genet. 1, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pyper S. R., Viswakarma N., Yu S., Reddy J. K. (2010) PPARalpha: energy combustion, hypolipidemia, inflammation and cancer. Nucl. Recept. Signal. 8, e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakajima T., Kamijo Y., Tanaka N., Sugiyama E., Tanaka E., Kiyosawa K., Fukushima Y., Peters J. M., Gonzalez F. J., Aoyama T. (2004) Peroxisome proliferator-activated receptor alpha protects against alcohol-induced liver damage. Hepatology 40, 972–980 [DOI] [PubMed] [Google Scholar]
- 41. Uyeda K., Repa J. J. (2006) Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 4, 107–110 [DOI] [PubMed] [Google Scholar]
- 42. Iizuka K., Bruick R. K., Liang G., Horton J. D., Uyeda K. (2004) Deficiency of carbohydrate response element binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. U. S. A. 101, 7281–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Benhamed F., Denechaud P. D., Lemoine M., Robichon C., Moldes M., Bertrand-Michel J., Ratziu V., Serfaty L., Housset C., Capeau J., Girard J., Guillou H., Postic C. (2012) The lipogenic transcription factor ChREBP dissociates hepatic steatosis from insulin resistance in mice and humans. J. Clin. Invest. 122, 2176–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gómez-Lechón M. J., Jover R., Donato M. T. (2009) Cytochrome p450 and steatosis. Curr. Drug Metab. 10, 692–699 [DOI] [PubMed] [Google Scholar]
- 45. Buechler C., Weiss T. S. (2011) Does hepatic steatosis affect drug metabolizing enzymes in the liver? Curr. Drug Metab. 12, 24–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.