Abstract
Endemic areas of human T-lymphotropic virus type 1 (HTLV-1) have been reported in Japan as well as tropical Africa, Central and South America and Melanesia. The existence of two subgroups, i.e., the transcontinental and Japanese subgroups, was reported in Japan. In the present study, we provide data on the ratio of the two subgroups in each endemic area and infection foci and examine the distribution of HTLV-1 in Japan and neighboring areas. A 657 bp fragment of env region of HTLV-1 proviral genome was successfully amplified for 183 HTLV-1 positive DNA samples. The subgroup determination was done by RFLP reactions using endonucleases HpaI and HinfI. The northern part of mainland Kyushu, represented by Hirado and Kumamoto, was monopolized by the Japanese subgroup, while the transcontinental subgroup ranged from 20 to 35% in the Pacific coast areas of Shikoku (Kochi), the Ryukyu Archipelago (Kakeroma and Okinawa) and Taiwan. An interesting finding in the present study is the presence of the transcontinental subgroup in Kochi, suggesting the endemicity of the transcontinental subgroup along the Kuroshio Current.
Keywords: Japanese subgroup, transcontinental subgroup, human migration, Kuroshio Current
Introduction
Human T-lymphotropic virus type 1 (HTLV-1) was first isolated in 1980 [1] and has been identified as a causative agent of adult T cell leukemia (ATL) and HTLV-1 associated myelopathy/tropical spastic paraparesis (HAM/TSP). HTLV-1 has three major transmission routes: from mother to infant through breast milk, from male to female through semen, and to blood recipients through the lymphocytes of HTLV-1 carriers. These transmission routes, especially mother-to-child transmission, allow HTLV-1 to pass from generation to generation and localize within family, community and ethnic groups. Thus, the elucidation of the geographical distribution of HTLV-1 has important ethno-epidemiological implications [2].
In view of this unique fact, a large number of phylogeographycal and epidemiological studies have been conducted within and beyond the borders of Japan, and valuable results have been obtained. Firstly, endemic areas were reported in tropical Africa, the Caribbean basin, Central and South America, Papua New Guinea and other islands of Melanesia, as well as Japan [3, 4]. Secondly, there are three major lineages existing worldwide: the Melanesian subtype, the Central African subtype, and the cosmopolitan subtype, ubiquitous in endemic areas around the world [5, 6]. Thirdly, the cosmopolitan subtype is further divided into three major subgroups: A, B, and C, which correspond to the transcontinental subgroup, the Japanese subgroup, and the West African subgroup, respectively [7, 8]. Fourthly, within Japan, endemicity is found in Kyushu and Okinawa, and small infection foci are seen in coastal islands of the Japan Sea and the Pacific side of Shikoku, Kii and Tohoku, while most of Honshu is HTLV-1-free [3]. Furthermore, a few endemic areas have been found in areas neighboring Japan: Nogliki of Sakhalin, Kinmen, Fujian and Taiwan [7, 9–11]. Fifthly, the existence of two different subgroups of HTLV-1, i.e., the transcontinental and Japanese subgroups, in Japan and clusters of the former subgroup in Kyushu and the Ryukyu islands were reported [12].
In the present study, we provide data on the ratio of the two subgroups in each endemic area and infection foci within Japan and use that data to elucidate the distribution of HTLV-1 in Japan and neighboring area.
Materials and Methods
DNA samples from a total of 197 anonymous HTLV-1 positive donors were obtained from the Joint Study on Predisposing Factors of ATL Development (JSPFAD) and used in the present study. Of the 197 samples, 40 were gathered in Hokkaido (Hokkaido University Hospital), four in Iwate (Iwate Medical University), 30 in Kochi (Kochi Medical School Hospital), 50 in Hirado (Nagasaki University Hospital), 23 in Kumamoto (Kumamoto University Hospital) and 50 in Okinawa (Okinawa Kyodo Hospital).
Furthermore, DNA was extracted from peripheral blood donated by five anonymous HTLV-1 carriers on Ishigaki Island, Japan (Yaeyama County, Ishigaki City, Okinawa Prefecture). The analysis of samples donated by the Yaeyama residents was approved by the ethics committee of the Institute of Tropical Medicine, Nagasaki University, Japan (Approval No. 10012147).
A 657 bp fragment of env region was amplified by nested PCR. The first reactions were performed in 20 µl volumes containing 1 µl (ca. 50 ng) of the extracted DNA, 200 µM (final conc.) of dNTP mixture, 0.25 µM (final conc.) of the primer sets, 2 µl of 10 × Ex Taq Buffer and 0.5U TaKaRa Ex Taq HS (TAKARA BIO Inc., Shiga, Japan). The external primers were TAATAGCCGCCAGTGGAAAG (nucleotide positions according to the J02029 sequence: 5027–5046) and AGTCCTTGGAGGCTGAACG (6786–6768). The thermal conditions were as follows: 5-min denature at 94°C, 40 cycles of 40 sec at 94°C, 30 sec at 61°C and 40 sec at 72°C, and 10-min final extention at 72°C. The second reactions were performed in 40 µl volumes containing 2 µl of the first PCR product, 200 µM (final conc.) of dNTP mixture, 0.25 µM (final conc.) of the primer sets, 4 µl of 10 × Ex Taq Buffer and 1U TaKaRa Ex Taq HS. The internal primers were CTCCCTTCTAGTCGACGCTCCAGG (5685–5708) and CGTCTGTTCTGGGCAGCATA (6341–6322). The thermal conditions were as follows: 2-min denature at 95°C, 35 cycles of 20 sec at 95°C, 20 sec at 58°C and 30 sec at 72°C, and 2-min final extention at 72°C.
All of the 35 samples from Hokkaido, all of the four from Iwate, 28 of 30 from Kochi, 44 of 50 from Hirado, 21 of 23 from Kumamoto, 46 of 50 from Okinawa and all of the five from Yaeyama were well amplified. RFLP reactions were performed using endonucleases HpaI and HinfI as designed by Yang et al. [7]. The digested DNA fragments were electrophoresed on 2% agarose gel pre-stained with ethidium bromide and visualized.
Results and Discussion
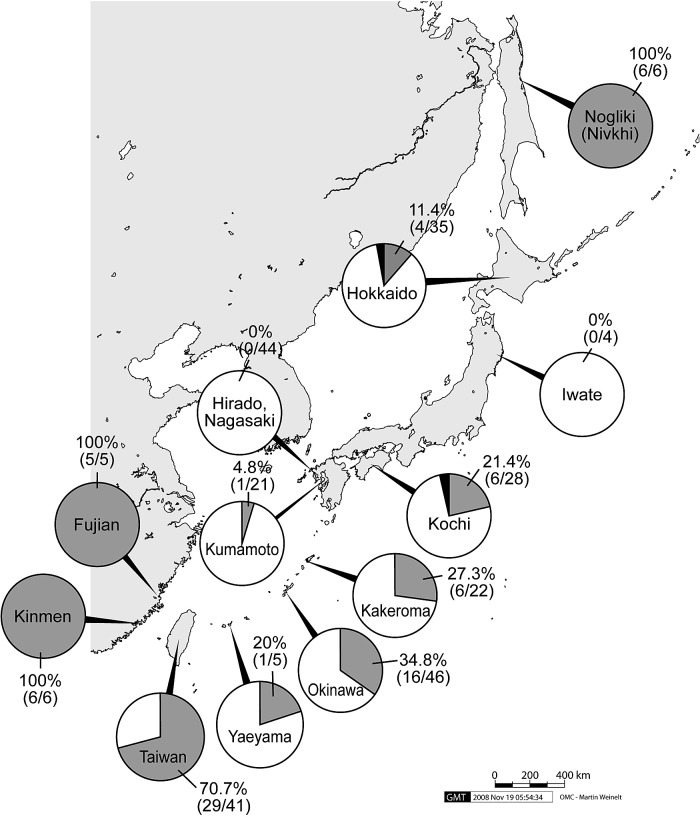
All except one of the HTLV-1 isolates from Iwate, Hirado and Kumamoto were determined as the Japanese subgroup, while 20–35% of the isolates from Hokkaido, Kochi, Okinawa and Yaeyama were determined as the transcontinental subgroup (Fig. 1). The electrophoresis profile of two isolates (Hokkaido and Kochi) was consistent with neither the Japanese nor the transcontinental subgroup but similar to the West African/Caribbean subgroup shown by Yang et al. [7]. Thus, these were tentatively treated as “undetermined” in the present paper.
Fig. 1.
Ratio of the transcontinental subgroup (grey) to the Japanese subgroup (white) of HTLV-1 cosmopolitan subtype in various localities of East Asia. The data of Nogliki, Kakeroma, Taiwan, Kinmen and Fujian were cited from Syrtsev et al. [10], Eguchi et al. [13], Yang et al. [7], Chen et al. [9] and Wang et al. [11], respectively.
The uneven distribution of the transcontinental and Japanese subgroups in the endemic areas of Japan was clarified in the present study, whereas only the transcontinental subgroup was reported from neighboring areas such as Nogliki of Sakhalin, Kinmen, and Fujian [9–11].
The northern part of mainland Kyushu, represented by Hirado and Kumamoto, seems to be monopolized by the Japanese subgroup. On the other hand, the presence of the transcontinental subgroup ranges from 20 to 35% in the Pacific coast areas of Shikoku (Kochi), the Ryukyu Archipelago (Kakeroma [13] and Okinawa) and Taiwan [7]. An interesting finding in the present study is the presence of the transcontinental subgroup in Kochi, suggesting the endemicity of the transcontinental subgroup along the Kuroshio Current.
A north-flowing ocean current on the west side of the Pacific Ocean, the Kuroshio Current has played the role of an aorta for migration and transportation along the Pacific coast of southwestern Japan since prehistoric times. The endemicity of the transcontinental subgroup along the Kuroshio Current might reflect this human movement. If so, we need to pay more attention to the date and mode of local human movements which may have implications in the epidemiology of HTLV-1 and other infectious agents such as hepatitis B virus [14].
Acknowledgements
We gratefully thank Dr. Osamu Ikehara (Okinawa Prefectural Yaeyama Hospital, Japan) who gathered samples on Ishigaki I., and Drs. Junko Okumura, Masahiro Hashizume, Toshihiko Sunahara and Hidefumi Fujii for their important suggestions. The authors thank the staff members in all the collaborating institutions and Mr. Makoto Nakashima, Ms. Takako Akashi, and other staff members in the central office of the JSPFAD for their efforts in sample processing and biologic assays.
Role of Funding Source
This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (20390186, 221S0001, 23659354, 23590800), Cooperative Research Grant (2009-E-1) of the Institute of Tropical Medicine, Nagasaki University and by the Global Center of Excellence Program at Nagasaki University. No sponsor, however, participated in the study design, in the collection, analysis, and interpretation of data, in the writing of the report, or in the decision to submit the paper for publication.
Author Disclosure Statement
Drs. Otani, Yamamoto and Eguchi have full access to all the data in the study and hold final responsibility for the decision to submit for publication. All authors declare that they have no conflict of interest.
References
- 1.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci U S A 1980; 77(12): 7415–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonoda S, Li HC, Tajima K. Ethnoepidemiology of HTLV-1 related diseases: ethnic determinants of HTLV-1 susceptibility and its worldwide dispersal. Cancer Sci 2011; 102(2): 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tajima K. Ethnic distribution of HTLV-1-associated diseases. Clinical Virology 1992; 20(5): 366–373 [Google Scholar]
- 4.Verdonck K, Gonzalez E, Van Dooren S, Vandamme AM, Vanham G, Gotuzzo E. Human T-lymphotropic virus 1: Recent knowledge about an ancient infection. Lancet Infect Dis 2007; 7: 266–281 [DOI] [PubMed] [Google Scholar]
- 5.Liu HF, Goubau P, Van Brussel M, Van Laethem K, Chen YC, Desmyter J, Vandamme AM. The three human T-lymphotropic virus type I subtypes arose from three geographically distinct simian reservoirs. J Gen Virol 1996; 77: 359–368 [DOI] [PubMed] [Google Scholar]
- 6.Van Dooren S, Verschoor EJ, Fagrouch Z, Vandamme AM. Phylogeny of primate T lymphotropic virus type 1 (PTLV-1) including various new Asian and African nonhuman primate strains. Infect Genet Evol 2007; 7: 374–381 [DOI] [PubMed] [Google Scholar]
- 7.Yang YC, Hsu TY, Liu MY, Lin MT, Chen JY, Yang CS. Molecular subtyping of human T-lymphotropic virus type I (HTLV-I) by a nested polymerase chain reaction-restriction fragment length polymorphism analysis of the envelope gene: two distinct lineages of HTLV-I in Taiwan. J Med Virol 1997; 51: 25–31 [PubMed] [Google Scholar]
- 8.Yamashita M, Ishida T, Ohkura S, Miura T, Hayami M. Phylogenetic characterization of a new HTLV type 1 from the Ainu in Japan. AIDS Res Hum Retroviruses 2001; 17(8): 783–787 [DOI] [PubMed] [Google Scholar]
- 9.Chen YM, Ting ST, Lee CM, Liu WT, Pan WH, Cheng ATA, Chou P. Community-based molecular epidemiology of HTLV type I in Taiwan and Kinmen: Implication of the origin of the cosmopolitan subtype in northeast Asia. AIDS Res Hum Retroviruses 1999; 15(3): 229–237 [DOI] [PubMed] [Google Scholar]
- 10.Syrtsev AV, Yamashita M, Senyuta NB, Susova OY, Hayami M, Gurtsevitch VE. HTLV-I infection among Nivkhi people in Sakhalin: Comparative serologic and phylogenetic analyses for 9 years. Int J Cancer 2000; 87: 379–381 [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Li X, Song A, Zhang C, Chen Y, Chen C, Lin Y, Shun L, Li L, Liu Y, Yang J, Yang B, Tang Q, Harrison TJ. Prevalence and partial sequence analysis of human T cell lymphotropic virus type I in China. J Med Virol 2005; 76(4): 613–618 [DOI] [PubMed] [Google Scholar]
- 12.Vidal AU, Gessain A, Yoshida M, Mahieux R, Nishioka K, Tekaia F, Rosen L, De The G. Molecular epidemiology of HTLV type I in Japan: Evidence for two distinct ancestral lineages with a particular geographical distribution. AIDS Res Hum Retroviruses 1994; 10(11): 1557–1566 [DOI] [PubMed] [Google Scholar]
- 13.Eguchi K, Fujii H, Oshima K, Otani M, Matsuo T, Yamamoto T. Human T-lymphotropic virus type 1 (HTLV-1) genetic typing in Kakeroma island, an island at the crossroads of the Ryukyuans and Wajin in Japan, providing further insights into the origin of the virus in Japan. J Med Virol 2009; 81: 1450–1456 [DOI] [PubMed] [Google Scholar]
- 14.Orito E, Ichida T, Sakugawa H, Sata M, Horiike N, Hino K, Okita K, Okanoue T, Iino S, Tanaka E, Suzuki K, Watanabe H, Hige S, Mizokami M. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology 2001; 34(3): 590–594 [DOI] [PubMed] [Google Scholar]