Abstract
β-catenin is a central component of the adaptor complex that links cadherins to the actin cytoskeleton in adherens junctions and thus, it is a good candidate to sense and transmit mechanical forces to trigger specific changes inside the cell. To fully understand its molecular physiology, we must first investigate its mechanical role in mechanotransduction within the cadherin system. We have studied the mechanical response of β-catenin to stretching using single-molecule force spectroscopy and molecular dynamics. Unlike most proteins analyzed to date, which have a fixed mechanical unfolding pathway, the β-catenin armadillo repeat region (ARM) displays low mechanostability and multiple alternative unfolding pathways that seem to be modulated by its unstructured termini. These results are supported by steered molecular dynamics simulations, which also predict its mechanical stabilization and unfolding pathway restrictions when the contiguous α-helix of the C-terminal unstructured region is included. Furthermore, simulations of the ARM/E-cadherin cytosolic tail complex emulating the most probable stress geometry occurring in vivo show a mechanical stabilization of the interaction whose magnitude correlates with the length of the stretch of the cadherin cytosolic tail that is in contact with the ARM region.
Introduction
The importance of mechanical force in regulating intracellular signaling and gene expression during embryonic development, tissue morphogenesis, and cell differentiation in normal physiology as well as in pathological conditions like cancer is well established (1). However, the molecular mechanisms by which cells sense and respond to mechanical stress through intracellular biochemical changes remain unclear. Cell adhesion receptors like cadherins and integrins are key components of this mechanotransduction machinery because they fulfill three different functions: cell-to-cell and cell-to-extracellular matrix adhesion; connecting the membrane to the cytoskeleton inside the cell; and the regulation of intracellular signaling pathways, including gene expression (1). The nanomechanical properties of the talin rod, a key component of the adaptor complex that links the extracellular matrix to the cytoskeleton through integrins, have been recently characterized (2). This structure was found to display complex mechanical features, including the exposure of cryptic binding sites to vinculin upon stretching, advancing our understanding of the mechanisms of force transduction within this pathway.
Initiation of cell-cell contacts requires interactions through cadherins, an important class of cell-cell adhesion receptors (3) linked to the cortical actin cytoskeleton through connecting proteins. β-catenin is a key component of the plasma membrane-cytoskeleton chain of the cadherin adhesion system (Fig. 1 A), being a multifunctional protein that acts both as a structural adaptor protein and as a transcription cofactor translocating to the nucleus in the Wnt signaling pathway (4). Thus, β-catenin has the potential to coordinate changes in gene expression with dynamic changes in cell adhesion and migration. Indeed, its deregulation can lead to alterations in cell fate, adhesion, and migration, provoking the development of different types of cancer (4). Furthermore, this protein has been recently related to neurodegeneration (5).
Figure 1.
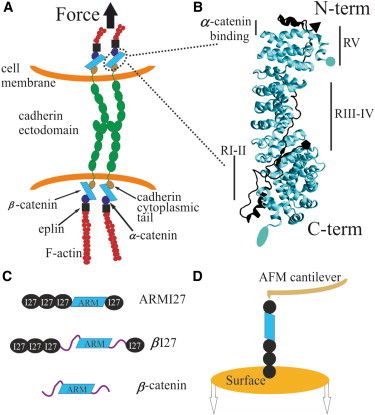
Tertiary and quaternary structure of β-catenin in the adherens junction and the experimental setup for its nanomechanical analysis. (A) Schematic representation of the cadherin adhesion system and possible force applied through the cell-cell adhesion contact. β-catenin is an adaptor protein involved in cell-cell adhesion that can also translocate to the nucleus and act as a transcriptional cofactor when the Wnt signaling pathway is active (4). (B) Crystal structure of the ARM region from β-catenin (lighter structure) complexed with E-cadherin cytoplasmic tail (darker structure). The ARM region is in the middle of the protein and consists of 12 ARM repeats stacked onto one another in a superhelical tertiary structure. β-catenin binds both α-catenin (close to its N-terminal region) and the cadherin cytoplasmic domain (through the ARM region). Circle and pentagon mark the N-termini of β-catenin and cadherin respectively, which are oriented in opposite directions. Ellipse and triangle mark the C-termini of β-catenin and cadherin respectively. (C) We analyzed several fusion proteins comprising four repeats of the titin I27 marker, used for single-molecule identification, flanking either the region of 12 ARM repeats from β-catenin (ARMI27) or the full-length β-catenin (βI27). We also analyzed the full-length β-catenin without the markers. (D) Cartoon representation of the experimental setup: proteins were immobilized on top of a substrate and after their attachment to the cantilever tip of an AFM, these molecules were axially stretched (in the N-C direction).
Mechanotransduction often involves conformational changes in the protein domains that are subjected to tension (1,2). The cadherin-catenin contact is known to be subjected to mechanical stress (3,6), and a molecular clutch model has been proposed as the mechanism to connect the system to the cytoskeleton (7). Thus, it seems likely that catenins, the main component in the adaptor complex of these contacts, are good candidates to sense and/or transmit mechanical forces to the cell interior to trigger biochemical changes. Significantly, it has been proposed that tension may regulate the fraction of β-catenin that is either bound to cell-cell junctions or free in the cytosol and, in doing so, it could stabilize the cell-cell contacts and control downstream signaling (8). Because catenins connect cadherins to the actin cytoskeleton, knowing how these central proteins respond to force is of paramount importance to understand the basis of these mechanotransduction processes.
β-catenin binds to the cytoplasmic domain of cadherins (9) and α-catenin in a ternary complex (10), and the exact chain of components that physically links them to the actin cytoskeleton has been solved (11). Like many proteins with a putative mechanical function (12), β-catenin is modular, containing 12 characteristic armadillo (ARM) repeats in its central region (R1 to R12, from N- to C-termini) that mediate protein-protein interactions (13,14) and that are flanked by two long disordered terminal regions, which have been postulated to modulate their binding to ligands (15,16). Each ARM repeat is formed by ∼42 aa residues arranged in three α-helices so that the region has an overall solenoid structure ((13,14); Fig. 1 B). This ARM region (aa 138 to 664 of the full-length β-catenin (13)) binds the cytoplasmic domain of cadherin ((9,17); Fig. 1 B), whereas the N-terminal disordered region including ARM R1 (aa 120–151) binds to α-catenin ((10); Fig. 1 B). The ARM region contains three sites for tyrosine phosphorylation that could downregulate the fraction of β-catenin bound to α-catenin (Tyr-142) and cadherins (Tyr-489 or Tyr-654) upon phosphorylation (18–20).
Here, we have characterized the nanomechanics of β-catenin both experimentally and in silico. First, by using single-molecule force spectroscopy (SMFS) to stretch the protein axially (N-C direction), we have analyzed its ARM region and its full-length form including the N- and C-terminal regions. Furthermore, we have performed molecular dynamics (MD) simulations of the process of stretching of the ARM region (21), the ARM region with an extra α-helix from the unstructured C-terminal region. Finally, we analyzed the mechanical response of the complex formed by the ARM region with different stretches of the E-cadherin cytoplasmic tail ((17), Fig. 1 B and Fig. S1 in the Supporting Material) emulating the likely pulling geometry that is expected to exist in vivo (Fig. 1 A) by stretching both structures from the N-terminus.
Material and Methods
SMFS
SMFS was carried out using a custom-made atomic force microscope (AFM) described elsewhere (22). Before each experiment, the cantilever tip was cleaned for 1 min with an UV/Ozone ProCleaner Plus lamp (Bioforce Nanosciences, Ames, IA). The chip was then immersed in buffer and the spring constant of the cantilever was calibrated by the equipartition theorem (cantilevers were MLCT-AUHW, Veeco Probes, Camarillo, CA; with a spring constant of ∼40 pN/nm; or BL-RC, Olympus, Tokyo, Japan; with a spring constant of ∼30 pN/nm). To perform the AFM experiments we used gold-coated coverslips, either home-made (see the Supporting Material) or commercial ones (Arrandee, Werther, Germany), as well as Nitrilotriacetic acid (NTA)-Ni2+ functionalized glass coverslips (see the Supporting Material) with similar results. We placed 10–40 μl of either full-length β-catenin or the fusion heteropolyproteins (Fig. 1 C) onto the surface and allowed 15–60 min for protein binding. We then washed the surface several times with the working buffer, containing phosphate buffered saline/0.2 mM EDTA/5mM dithiothreitol at pH 7.4. The force versus extension traces were recorded using the length-clamp mode (12) at a constant pulling speed of 400 nm/s (Fig. 1 D). The force noise at this pulling speed (∼9 pN) was estimated as the width of a Gaussian fitting to the force noise of the noncontact region of the recordings (Fig. S2 A). To rule out the possibility of overlooking force data that could lie below this noise, we increased the sensitivity (noise/signal ratio) by lowering the pulling speed (noise ∼4 pN at 20 nm/s, Fig. S2 A) and we found similar types of recordings (Fig. S2 B). Furthermore, the lack of a significant overlapping between the histograms of the noise and that of the unfolding forces at 400 nm/s strongly suggests that the shape of the force distribution reflects the intrinsic mechanical behavior of the constructs studied (Fig. S2 C).
SMFS analysis
Constructs termed βI27 and ARMI27 (Fig. 1 C) had a built-in internal control for single-molecule identification: flanking repeats of the I27 marker (a model system in the field (12)) that allows the identification of unfolding events from the ARM structure (23,24). The magnitudes of interest in these experiments are the unfolding force (FU, related to the barrier that resists the mechanical unfolding of the protein) and the increase in contour length (ΔLC, related to the length of the protein that is force hidden). The built-in internal control allowed us to be confident that we had stretched the complete ARM region (plus the disordered terminals in the case of βI27) only when a recording contained four I27 unfolding events (FU ∼200 pN and ΔLC ∼28 nm (25)). The β-catenin region always unfolded before the I27 modules and therefore, we adopted a further criterion for the selection of recordings: the contour length at which the first I27 force peak appeared (termed LCβ and obtained by fitting the force peaks to the worm-like chain model of protein elasticity ((26), Fig. 2 A) should correspond to the expected length of the flanked region under study. This criterion also enabled us to sort out populations originated from the ARM region from those corresponding to the I27 markers (Fig. S3 and Fig. S4). The βI27 construct yielded comparatively less diagnostic recordings (i.e., carrying four I27 force peaks). In the case of β-catenin alone, for the analysis we used the total contour length as the only criterion to select single-molecule recordings (781 aa ∼312 nm (27)). We also analyzed the length at which the first unfolding peak appeared (LC0) to determine if some regions of the protein were unfolded before the pulling process started.
Figure 2.
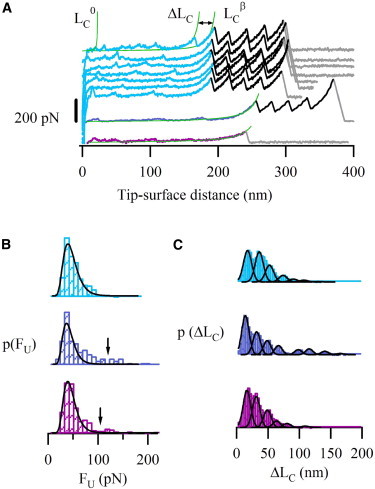
Single-molecule force spectroscopy of the ARM region and full-length β-catenin. (A) Force-extension recordings from the mechanical unfolding of the three constructs from Fig. 1C: ARMI27 (top, from LC0 to LCβ: ARM region; from LCβ to the last force peak: I27 modules), the βI27 (middle, from LC0 to LCβ: full-length β-catenin region; from LCβ to the last force peak: I27 modules) and β-catenin (bottom). Peaks were fitted using the worm-like chain model of polymer elasticity (smooth lines (26)), which allows to calculate contour lengths (LC0, ΔLC, and LCβ). The region under study in the three constructs unfolded in a similar way, with weak forces and variable ΔLC values (several traces from ARMI27 construct are shown as a representative example, Fig. S6). The I27 marker shows an ΔLC of 28 nm and a FU of around 200 pN, in accordance with previous reports (25). (B) FU histograms. Lines correspond to log-normal fittings (χ2 are 1.5 × 10−5, 8.5 × 10−5, and 2.3 × 10−5 for ARMI27, βI27 and β-catenin, respectively). Only 0.4% of the data points lie above 100 pN for ARMI27 (top), being 10% for βI27 (middle), and β-catenin (bottom) (containing unstructured terminal regions). This suggests that the ARM region is somehow stabilized by the outer regions. (C) The ΔLC histograms for the proteins show similar (though slightly shifted) multimodal distributions with the mean values close to the length of one or multiple ARM repeats (see details in the main text). ARMI27 (top), βI27 (middle), and β-catenin (bottom).
The unfolding force histograms were fitted to a log normal function (Fig. 2 B, solid curves), whereas the ΔLC and LC0 histograms were fitted to several Gaussian curves (Figs. 2 C and Fig. S5, respectively). Igor Pro 6 software (Wavemetrics, Oswego, OR) was used for this analysis.
Although the force distributions were fitted only to one log normal function (due to the low number of high force events, Fig. 2 B), for statistical purposes the force values were sorted out into two groups based on whether they laid above or below the force value that minimizes the probability density function (arrows in Fig. 2 B, middle and bottom). These values were 100 pN for ARMI27, 110 pN for βI27, and 95 pN for β-catenin. Statistical results are expressed as mean ± SE.
MD simulations
The crystal structures of the ARM region and the ARM region carrying a stacked α-helix at the C-terminus (ARMHC) of human β-catenin are 533 and 644 aa long, respectively (PDB codes 1qz7 and 2z6h (21,14)). The long size of this structure makes their modeling by MD difficult if an explicit solvent is used, because the water box needed to hydrate the whole molecule would be extremely large and would exceed the regular computing capacity. The generalized Born solvation model (generalized Born/surface area (28)) overcomes this limitation by using a force field that emulates the properties of an explicit solvent, and predicts the mechanical properties as accurately as explicit water models (29).
The initial atomic coordinates were taken from the crystal structure of the ARM region of human β-catenin (1qz7) because it contains the complete backbone chain (including the R10 repeat (21)). To complete the R10 loop of 2z6h (which is incomplete in this deposited structure), we performed a structural alignment with 1qz7, taking the backbone of the R10 loop from the latter structure as initial guessing coordinates. Moreover, the structures with PDB codes 1i7x and 1i7w were combined to obtain the ARM region complexed with different stretches of a murine E-cadherin cytoplasmic tail (termed ARM/E-cadhcyt, Fig. S1 (17)). We removed the phosphorous groups from 1i7w (Ser-840, Ser-842, and Ser-848) and generated four different structures comprising residues 784–840, 784–850, 784–840/854–879, and 784–850/854–879 from the E-cadherin cytoplasmic tail (hyphen indicates an interval, whereas the slash bar indicates a gap). These residues correspond to the interacting regions RI-III, RI-IV, RI-III/V and RI-IV/V from the E-cadherin tail, respectively (Fig. S1 (17)). Because in the solved atomic structure of the E-cadherin tail a few residues are missing, we emulated a continuous chain between solved fragments by imposing a restraint (400 kcal mol−1 Å−2 and 4.6 or 1.6 nm for the gaps RIII/V and RIV/V, respectively) between the boundaries of the gap during the pulling of the generated complexes. Parameters were built using AMBER 10 (30) in combination with the parm99 force field and the atomic coordinates from the 1qz7 PDB entry and the completed 2z6h. A previously reported generalized Born/surface area model was used (31,32) in conjunction with the parameters described elsewhere (28). The surface area was computed with the LCPO model (31). The cutoff distance for the potential was 12 Å with a switching distance of 10 Å. The salt concentration of monovalent counterions was set to 0.1 M, which was implemented by a modified generalized Born model that takes account of mobile monovalent counterions through the Debye-Hückel limiting law for ionic interactions (33). We first ran a minimization of 5 ps. Subsequently, with 1 fs as the time step, we increased the temperature from 0 to 298 K over 20 ps with restrained positions for Cα atoms. Finally, before performing free MD, we removed the restraints and stabilized the system for 8 ps. Free MD simulations were performed for 4 ns with a time step of 2 fs. We selected the coordinates of the protein at 1, 2, and 3 ns of the free MD as starting points for the steered molecular dynamics (SMD). A distance restraint (Xrestr) to the Cα at the N- and C-termini was then applied (krestr = 5 kcal mol−1 Å−2). In the case of ARM/E-cadhcyt complex, the distance restraint was imposed to the N-termini of both structures, emulating the most probable geometry of the possible physiological stress (Fig. 1, A and B). The initial value of these restraints was equal to the distance between the terminal Cα atoms, and this distance was increased at a rate of 1 Å/ps. Trajectories visualization, extraction of Cα root mean-square deviation (RMSD), and the distance between the terminal Cα atoms (X) were obtained with the VMD software package (34). These data were then loaded into Igor Pro 6 software (Wavemetrics) to calculate the forces as follows: F = −krestr × (X–Xrestr).
Results and Discussion
Nanomechanics of β-catenin ARM region
AFM-based SMFS (Fig. 1 D) enables individual protein molecules to be axially pulled (i.e., in the N-C direction), so that the magnitude of the resistance barriers involved in the stretching process can be measured in the so-called length-clamp mode (12). The initial recordings obtained showed that the whole ARM region was stretched before the I27 modules (Fig. 2 A), indicating that it has lower mechanostability than that of the I27 markers. The ARM region unfolding events showed a highly variable pattern in ΔLC values (Figs. 2, A and C, and Fig. S6) with an average unfolding force FU = 44 ± 1 pN (n = 278, Fig. 2 B, top) and a single event with higher unfolding force FU = 113 ± 9 pN (here the error is the measurement error). The best fitting of the unfolding force histogram was obtained by a log normal function (solid curves in Fig. 2 B; see Materials and Methods).
In the force-extension traces, the ΔLC values between consecutive unfolding events correspond to the region of the protein that is force-hidden behind a mechanical barrier. As a result, repeat-containing proteins typically display sawtooth pattern recordings (12). Interestingly, the ARM region unfolding traces showed force events variable in number, position, and increase in contour length (Figs. 2 A and Fig. S6), as opposed to a typical sawtooth pattern, which indicates the possibility of unfolding through different pathways. Moreover, we examined the FU vs. ΔLC plots because any clustering of the data would point to the existence of a preferential unfolding pathway (Fig. S3 D). Similarly, the plot of ΔLC vs. the tip-surface distance shows a complete scatter (Fig. S3 C). The lack of correlation between FU and ΔLC, and ΔLC vs. the tip-surface supports the idea of multiple alternative unfolding pathways as opposed to a single one.
The analysis of the ΔLC histogram (Fig. 2 C, top) showed a distribution that could be fitted to seven Gaussian functions (ΔLC maxima: 16.0, 34.6, 50.9, 72.4, 89, 106, and 130 nm; σ = 9.2 nm; χ2 = 1.5 × 10−5). This multimodal distribution can be interpreted as the result of unfolding either single or multiple ARM repeats, given the number of aa contained in each ARM repeat (42 aa on average) and considering 0.4 nm as the length of a stretched aa ((27); i.e., ∼17 nm/ARM repeat). Indeed, evidence for the existence of distinct conformers (i.e., molecular plasticity) in the ARM region has been already reported (35). Furthermore, the occurrence of the unfolding events was variable: we can find from two to nine force peaks in different force extension traces (with an average value of 4.5 β-catenin peaks/trace). Considering that the FU value (∼44 pN) is larger than the noise (Fig. S2 C) one can assume that the results obtained are due to the intrinsic behavior of the molecule under our experimental conditions (i.e., variable number of force peaks per recording with different ΔLC values). Thus, this variability suggests that some ARM repeats could be unfolded with pulling forces below 9 pN or, alternatively, some repeats unfold almost simultaneously with the similar unfolding force, indicating the likely existence of cooperativity in the mechanical unfolding of the ARM repeats. To rule out the possibility that some force events were overlooked due to the force noise, we also performed pulling experiments with a higher signal/noise ratio (by decreasing the pulling speed), which depicted a similar behavior (Fig. S2 B).
Moreover, as some unfolding recordings showed an initial region lacking force peaks, we have also analyzed the distance at which the first force peak (LC0) appears, to examine the extent the molecule was previously unfolded (before stretching, Fig. 2 and Fig. S5). This analysis allows extracting important information: first, peaks that appear at an extension shorter than the length of the folded molecule (11 nm coming from the ARM region + 5 nm from each I27 × 4 = 31 nm; solid red line in Fig. S5) could in principle be originated either from unbinding events (when detaching the ARM region from the surface or from artifactual interactions between some ARM repeats and the flanking I27 modules) or from the rupture of the forces that maintained the tertiary structure (ARM superhelix); second, most of the events lay above this length, suggesting that some ARM repeats unfold below our detection limit or are in equilibrium between structured and unstructured conformations, similar to what has been recently shown for ankyrin (ANK) repeats 5–6 of IκBα (36).
The nanomechanics of full-length β-catenin
To analyze the possible mechanical effects of the unstructured terminal regions, we examined the nanomechanics of the full-length form. We started analyzing a protein construct consisting of the full-length β-catenin flanked by I27 markers (βI27, Fig. 1 C). The force traces were very similar to those obtained during the unfolding of the ARMI27 construct (Fig. 2 A, middle). βI27 FU histogram (Fig. 2 B, middle) showed similar low force events to those obtained from the ARM region, with FU (47 ± 2 pN, n = 86), but also a few events with an FU of 140 ± 8 pN (n = 12).
The corresponding ΔLC histogram (Fig. 2 C, middle) also showed a distribution that could be fitted to several Gaussian functions (ΔLC maxima:12.5, 30.4, 47.2, 65.2, 97, 114, 139, and 163 nm; σ = 8.2 nm; χ2= 3.7 × 10−5). Similar to ARMI27, the plot of ΔLC vs. the tip-surface distance shows a complete scatter (Fig. S3 C and Fig. S4 C). Furthermore, as shown in Fig. S4 D, βI27 also showed no correlation between FU and ΔLC, supporting the idea of multiple, alternative unfolding pathways.
The flanking terminal regions of β-catenin seem to be unstructured (and thus one could expect them to be stretched without mechanical resistance (23)) and are thought to interact weakly with the ARM region (14,15). We therefore examined the LC0 values (data not shown), observing that some events appear at an extension shorter than the expected length if the N- and C-terminal segments were not interacting (and thus could be stretched showing no mechanical resistance): 11 nm (ARM) + 5 nm × 4 (I27) + 0.4 nm × 250 aa (N- and C-termini) = 131 nm. These data suggest that, before the expected length, we are detecting unbinding or unfolding events, although the exact origin of these force peaks is unknown.
To rule out the theoretical possibility that the high forces observed in βI27 could also be originated from artifactual interactions with the I27 marker (Fig. 2 B, middle), we analyzed β-catenin alone (without the markers). As this approach lacks a single-molecule marker, the selection of recordings had to be done based solely on the full-length criterion, being particularly careful in selecting only recordings that were clean in the proximal region (a region prone to noise originated from nonspecific interactions, see Methods (37)). These force-extension recordings were essentially similar to those obtained from the other two constructs (Fig. 2 A, bottom). The force histogram had two populations (Fig. 2 B, bottom) comparable to those observed in the other two constructs, one of them with an FU of 43 ± 1 pN (n = 273) and the other with FU of 125 ± 4 pN (n = 33).
The corresponding ΔLC histogram also had a multimodal distribution that could be fitted to six Gaussian functions (ΔLC maxima: 14.9, 29.7, 47.4, 62.3, 79, and 108 nm; σ = 8.2 nm; χ2 = 1.3 × 10−5) (Fig. 2 C, bottom). As with βI27 and ARMI27, both the plots of ΔLC vs. tip surface distance and FU vs. ΔLC show a complete scattering (Fig. S7, A and B, respectively) supporting, again, the idea of multiple alternative unfolding pathways. The analysis of the first force event (not shown) shows that we can find peaks at shorter lengths than the one expected for the native protein (11 nm (ARM) + 0.4 nm × 250 aa (N and C unstructured termini) = 111 nm). This behavior is similar to that observed for βI27, meaning that we cannot distinguish if the first force peak arises from an unbinding event (terminals/surface, terminals/ARM, surface/ARM) or from ARM repeat unfolding.
Taken altogether, our experimental results indicate that the unfolding process of the ARM region and the full-length β-catenin proceeds in a very similar manner, showing similar average unfolding forces, similar ΔLC profiles, and multiple unfolding pathways with several intermediates. Furthermore, the N- and C-termini of β-catenin could be modulating the mechanical stability of the protein, as suggested by the increased frequency of high force peaks (above 100 pN) in the FU distributions of the two constructs carrying the N/C termini (0.4% for ARMI27 and ∼11% for βI27 and β-catenin, Fig. 2 B, middle and bottom).
Stretching β-catenin in silico
Finally, to study the atomic details of the unraveling of the ARM repeat region, we performed MD simulations of the ARM region using the reported crystal structures for the human β-catenin, one carrying exclusively the ARM region (PDB code 1qz7 (21)) and the other with an extra α-helix at the C-terminus (ARMHC; PDB code 2z6h (14)). In the case of the ARM region, repeats R1, R10, and R12 displayed larger deviations than the other repeats during 4 ns of free MD (Fig. S8 A, top). As expected, repeats located at the free ends of the ARM region (repeats R1 and R12) showed more mobility than those repeats located in the inner part of the structure. The larger fluctuations of R10 could be attributed to the extended length of this repeat, which carries a long loop with a fluctuating secondary structure (data not shown). Similar to what was observed experimentally, the unfolding force traces from the ARM region (Fig. 3 A, top) show low mechanical stability, compared with I27 unfolding under the same conditions (29). To follow the unfolding sequence during the simulations of the stretching of the ARM region, we plotted the backbone RMSD of each ARM repeat (Fig. S8 B, left), showing that those repeats located at the N-terminal region (R1, R2, and R3) unfolded first in all the simulations of this structure (although we were pulling from both termini). Some of the repeats showed unfolding intermediates and a significant freedom in the unfolding pathway, i.e., a nonordered pathway, as the unfolding of several repeats was observed at different N-C distances in the three trajectories (see R4 or R11 in Fig. S8 B, left). These results support the idea that the unfolding pathway of the ARM repeats is not fixed.
Figure 3.
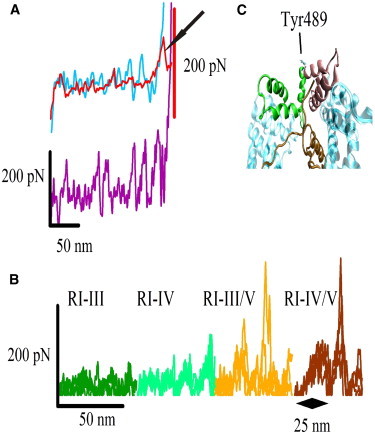
SMD simulations of β-catenin and β-catenin/E-cadherin complex. (A) Force traces for the stretching of the ARM (top) and the ARMHC structures (bottom). An experimental unfolding curve (top, arrow) of the ARM region is superimposed to the in silico curve, showing the similarity between both curves. The structure carrying helix C (ARMHC, bottom) shows slightly higher unfolding force peaks. This evidence goes in line with the increase of high force events observed experimentally when the unstructured termini are present in the structures (Fig. 2B). Thus, β-catenin termini could somehow stabilize the ARM repeats. (B) Superimposition of the four force traces for each β-catenin/E-cadherin complex. The magnitude of the force peaks is higher as longer stretches of E-cadherin are present. A high force peak appears upon breaking the interaction between R1 and RV. (C) Detail of the conformational changes experienced by R8 and R9 ARM repeats resulting in the exposure of the buried Tyr-489.
In the case of the free MD of ARMHC, the RMSD of the protein backbone for R1, R10, and R12 was comparable to those of the other repeats (Fig. S8 A, bottom), whereas the extra helix at the C-terminus had a larger RMSD due to its terminal residues. It seems that interactions between helix C and R12 (three hydrophobic residues and two hydrogen bonds (14)) place the whole structure into a lower energy state, and R1, R10, and R12 do not deviate too much. The SMD simulations showed the same unfolding hierarchy for R1, R2, and R3 as that shown in the ARM structure, although the number of force peaks and their magnitude were slightly higher (Fig. 3 A, bottom), indicating that this structure is in a deeper energy state and shows almost the same unfolding pathway for all the trajectories (Fig. S8 B, right). Thus, little changes in the conformation could place the protein in different energy states, deriving in different unfolding pathways (38).
Although the time window in simulations is much shorter than in AFM experiments, these results qualitatively reproduce, at least in the case of the ARM structure, the variability of the mechanical unfolding pathway and the low FU values observed in the experiments. Nevertheless, we have no experimental evidence for the tendency observed in the simulations toward a directional unfolding from N- to C-terminus. Furthermore, the results obtained with ARMHC indicate a certain stabilization of the ARM repeats by interactions with helix C (for example, see R11 for ARM and ARMHC, Fig. S8 B), an effect that could also occur in the presence of the entire N- and C-termini, considering our experimental results.
Stretching ARM/E-cadhcyt complex in silico
In addition to studying the response of the ARM region to mechanical force, we also simulated the mechanics of the whole ARM/E-cadhcyt complex. Because it has been shown that cadherins are subjected to force (1) and can even apply force from inside the cell to the outside (3), it is important to know how do the proteins that integrate this system respond to force, and how this response is altered when the proteins interact with their corresponding partners. In this context, it is critical to assess how β-catenin transmits force as such in its natural environment, in complex with the cadherin cytoplasmic tail (along the whole ARM region) and with α-catenin (between residues 118–149). To this end, we setup four structures carrying different extensions of the E-cadhcyt interacting region (Fig. S1). α-catenin structure was not included in the complex because it interacts with only six residues at the N-terminus of the ARM structure. Because cadherin ectodomain corresponds to the N-terminal region of the protein and α-catenin binds to the N-terminal region of ARM repeats, we emulated the possible mechanical stress by the distance restraints imposed on both N-termini (ARM and E-cadhcyt). Interestingly, during free MD, ARM repeats R1 and R10 (and R12, to a less extent) show differential RMSD values, depending on the length of E-cadhcyt structure in complex with the ARM region (Fig. S9 B). In particular, R1 shows the largest deviation when the whole E-cadhcyt region is present in the complex, whereas R10 shows larger deviations only when RIV of E-cadhcyt is absent of the structure.
The SMD results show a clear mechanical stabilization of the complex when longer stretches of E-cadhcyt are complexed with the ARM region (Fig. 3 B). The mechanical resistance observed when regions RI-III and RI-IV are complexed with the ARM region is close to zero, but some force peaks appear when RV is included in the structure. Remarkably, only when the E-cadherin regions RI-V are present, the ARM region undergoes a conformational change after the first 25 nm of pulling leading to separation of repeats R8 and R9 without being unfolded (showing small deviation in the RMSD plot) (Figs. 3 C and Fig. S10) and Tyr-489 seems to get more exposed and accessible to the solvent (Fig. 3 C and Fig. 4 C) thus being more accessible. This conformational change could, hence, provide a mechanism to increase β-catenin turnover and the dynamics of cadherin adhesion by modulating the Tyr-489 accessibility to protein kinases like Abl (20). Moreover, at the end of the stretching process, when the RI-III/V and RI-IV/V regions are in complex with the ARM repeats and E-cadhcyt is completely elongated, a high force peak results from the unbinding of repeat R1 from E-cadherin region RV (Fig. 3 B).
Figure 4.
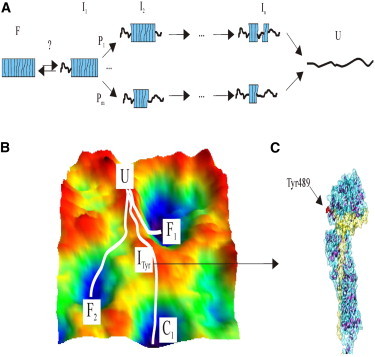
Cartoon representation of the possible scenario for the multiple mechanical unfolding pathways of β-catenin. (A) For simplicity, this model is restricted to the ARM region, which is represented as a box. Based on LC0 analysis and considering our MD simulations results, we hypothesize that several ARM repeats would be unfolded beforehand. Afterward, depending on the initial state, many unfolding pathways (P) with a variety of intermediate states (I) with different ARM repeats composing the mechanical blocks would be possible. (B) Illustration showing the possible roughness in the energy landscape of β-catenin, containing several minima with similar energy levels, and different unfolding trajectories. Fi illustrates the different initial folded states, whereas C1 represents the minima corresponding to the cadherin-complexed state. ITyr is the intermediate where Tyr-489 is greatly exposed. U is the unfolded state. (C) Cadherin cytoplasmic tail binding can canalize β-catenin unfolding pathway (43), allowing the ARM region to open at Tyr-489, which is susceptible to Abl-mediated phosphorylation.
General discussion on β-catenin nanomechanics
Taken together, our data suggest that the mechanical properties of the ARM region of β-catenin are very sensitive to small changes in its structure. This region seems to follow multiple unfolding pathways upon stretching, which suggests that the process takes place through a rough energy landscape with shallow energy minima (Fig. 4, A and B). It is interesting to compare our results with those obtained from other solenoid repeat proteins, specifically those proteins containing ANK repeats, although both types of solenoid proteins may not be strictly comparable. Thus, the mechanical unfolding of gankyrin, a 7-ANK repeat protein, was shown to proceed by multiple pathways compatible with a rugged and complex energy landscape (39). In addition, SMFS experiments of ankyrin B, using 24 and 12 repeats fragments, showed that ANK repeats can unfold either individually or two at a time (40). Furthermore, the chemically determined energy landscape of a fragment of ankyrin R composed of 12 ANK repeats is predicted to be very rough with several possible configurations (38), whereas the chemical unfolding of Notch appears as a two-state cooperative process (41,42). These results on ANK repeats cannot be easily reconciled, and it looks like the actual unfolding pathway could highly depend on the energy state of the protein before the denaturation and on the unfolding agent (chemical, thermal, or mechanical), with local perturbations being critical for the energetics of the unfolding process.
Comparing the results obtained with βI27 and β-catenin, we can conclude that the increase in the frequency of high force events originates exclusively from the presence of the β-catenin termini (0.4% for ARMI27 and ∼11% for βI27 and β-catenin). Although we cannot discard the possibility that these force peaks might originate from interactions between the termini and other elements (surface or ARM region), if we consider that the experiments were performed on surfaces of different chemical nature, we can speculate that these high force events could originate from the unbinding of β-catenin termini from the ARM region. Nevertheless, they could also be originated by some stabilization of the mechanical unfolding of the ARM region mediated by the terminal regions, as predicted by MD simulations of the ARM region with the α-helix at C termini.
The mechanostability of β-catenin is in the range of that of talin (2), and more interestingly, in the range of the forces sustaining cadherin-cadherin adhesions (43) and much lower than the mechanostability of C-cadherin ectodomain (44) and other adhesion receptors (37). Furthermore, the mechanical stability we measured is comparable to that recently reported for the full-length β-catenin (45). However, as the data from the latter study were obtained without the use of single-molecule markers, they are not strictly comparable to our results. That study showed that full-length β-catenin most often unfolds in a rather discrete manner, although it was still possible to observe at least two ARM repeats unfolding simultaneously. It must be noted that only less than one-third of the molecule was stretched in those experiments, i.e., ∼100 nm (the full length, 718 aa, would correspond to ∼312 nm, considering 0.4 nm/aa (27)), and therefore, in this setup it is not possible to discard an interaction or modulation of the unfolding/refolding process by the unstructured termini. Furthermore, in this kind of experiment the cyclic collapse/extension trajectories have been proposed to stem from the hopping over an entropic barrier that emerges upon application of force to an extended polypeptide and that does not correlate with the final folding transition (46). From our results, the fact that the ΔLC histograms are multimodal, unlike the force histogram, indicates that similar force values are needed to unravel a stack or a single ARM repeat, as if the protein was constituted by mechanical blocks (each one consisting of one or several ARM repeats, Fig. 4 A), all of them with similar mechanical stability. This observation suggests the existence of cooperative effects among repeats in the mechanical blocks comprising more than one ARM repeat.
Conclusion
Our results suggest that the β-catenin ARM region is very sensitive to small changes in its structure and follows multiple unfolding pathways upon stretching, which could be modulated by its unstructured termini and ligand binding. We can speculate that if β-catenin is subjected to tension in vivo, the axial forces that this structure is expected to experience or to transfer are on the range of the known mechanostabilities of the other components of the mechanical circuit: the force borne by cadherin/cadherin adhesion (43) and by cadherin ectodomains (44). Given that β-catenin shows low mechanical stability and even several ARM repeats are unfolded with low mechanical resistance (as the LC0 analysis shows), and considering the high chemical affinity for its ligands (40 and 100 nM for the cadherin tail and α-catenin, respectively (16,47)) we suggest that β-catenin structure could serve as a mechanical buffer under high forces (F > FU ∼50 pN) being able to unfold a significant portion of its structure to preserve the links between the cadherin ectodomain and the actin cytoskeleton (through α-catenin and EPLIN, 11). Moreover, due to the similar mechanical stabilities of cadherin-cadherin bonds and β-catenin, when the latter and the cadherin tail interact by their five contact regions, forming a mechanical stable complex, β-catenin could also serve as a force transmitter.
To fully understand the mechanical role of β-catenin in mechanotransduction through the cadherin-catenin system, future experiments should explore the kinetics of this adaptor platform under different forces (below β-catenin mechanical stability, above the cadherin-cadherin rupture forces, and values in between) upon the binding of different specific ligands.
Acknowledgments
We thank J. Clarke for kindly providing the pRSETA-(I27)8 vector and A. Cano for the full-length β-catenin clone in pQE32 vector.
This work was funded by grants from the Ministerio de Ciencia e Innovación (BIO2007-67116), the Consejería de Educación de la Comunidad de Madrid (S-0505/MAT/0283), and the Consejo Superior de Investigaciones Científicas (200620F00) to M.C.-V., and from the Ministerio de Ciencia e Innovación (BFU2009-10052) and the Consejería de Educación de la Comunidad de Madrid (P2010/BMD-2457) to M.M. J.O. and A.M.V. are recipients of PhD fellowships from the Consejería de Educación de la Comunidad de Madrid and the Fundación Ramón Areces, respectively.
The authors declare no conflict of interest.
Supporting Material
References
- 1.Schwartz M.A., DeSimone D.W. Cell adhesion receptors in mechanotransduction. Curr. Opin. Cell Biol. 2008;20:551–556. doi: 10.1016/j.ceb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.del Rio A., Perez-Jimenez R., Sheetz M.P. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganz A., Lambert M., Ladoux B. Traction forces exerted through N-cadherin contacts. Biol. Cell. 2006;98:721–730. doi: 10.1042/BC20060039. [DOI] [PubMed] [Google Scholar]
- 4.Nelson W.J., Nusse R. Convergence of Wnt, β-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godin J.D., Poizat G., Humbert S. Mutant huntingtin-impaired degradation of β-catenin causes neurotoxicity in Huntington’s disease. EMBO J. 2010;29:2433–2445. doi: 10.1038/emboj.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dzamba B.J., Jakab K.R., DeSimone D.W. Cadherin adhesion, tissue tension, and noncanonical Wnt signaling regulate fibronectin matrix organization. Dev. Cell. 2009;16:421–432. doi: 10.1016/j.devcel.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giannone G., Mège R.M., Thoumine O. Multi-level molecular clutches in motile cell processes. Trends Cell Biol. 2009;19:475–486. doi: 10.1016/j.tcb.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Chen C.S., Tan J., Tien J. Mechanotransduction at cell-matrix and cell-cell contacts. Annu. Rev. Biomed. Eng. 2004;6:275–302. doi: 10.1146/annurev.bioeng.6.040803.140040. [DOI] [PubMed] [Google Scholar]
- 9.Perez-Moreno M., Fuchs E. Catenins: keeping cells from getting their signals crossed. Dev. Cell. 2006;11:601–612. doi: 10.1016/j.devcel.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aberle H., Butz S., Hoschuetzky H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J. Cell Sci. 1994;107:3655–3663. doi: 10.1242/jcs.107.12.3655. [DOI] [PubMed] [Google Scholar]
- 11.Abe K., Takeichi M. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc. Natl. Acad. Sci. USA. 2008;105:13–19. doi: 10.1073/pnas.0710504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberhauser A.F., Carrión-Vázquez M. Mechanical biochemistry of proteins one molecule at a time. J. Biol. Chem. 2008;283:6617–6621. doi: 10.1074/jbc.R700050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber A.H., Nelson W.J., Weis W.I. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 14.Xing Y., Takemaru K., Xu W. Crystal structure of a full-length β-catenin. Structure. 2008;16:478–487. doi: 10.1016/j.str.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castaño J., Raurell I., García de Herreros A. β-catenin N- and C-terminal tails modulate the coordinated binding of adherens junction proteins to β-catenin. J. Biol. Chem. 2002;277:31541–31550. doi: 10.1074/jbc.M204376200. [DOI] [PubMed] [Google Scholar]
- 16.Choi H.J., Huber A.H., Weis W.I. Thermodynamics of β-catenin-ligand interactions: the roles of the N- and C-terminal tails in modulating binding affinity. J. Biol. Chem. 2006;281:1027–1038. doi: 10.1074/jbc.M511338200. [DOI] [PubMed] [Google Scholar]
- 17.Huber A.H., Weis W.I. The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 18.Roura S., Miravet S., Duñach M. Regulation of E-cadherin/Catenin association by tyrosine phosphorylation. J. Biol. Chem. 1999;274:36734–36740. doi: 10.1074/jbc.274.51.36734. [DOI] [PubMed] [Google Scholar]
- 19.Murase S., Mosser E., Schuman E.M. Depolarization drives β-catenin into neuronal spines promoting changes in synaptic structure and function. Neuron. 2002;35:91–105. doi: 10.1016/s0896-6273(02)00764-x. [DOI] [PubMed] [Google Scholar]
- 20.Lilien J., Balsamo J. The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of β-catenin. Curr. Opin. Cell Biol. 2005;17:459–465. doi: 10.1016/j.ceb.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Xing Y., Clements W.K., Xu W. Crystal structure of a β-catenin/axin complex suggests a mechanism for the β-catenin destruction complex. Genes Dev. 2003;17:2753–2764. doi: 10.1101/gad.1142603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valbuena A., Oroz J., Carrión-Vázquez M. Quasi-simultaneous imaging/pulling analysis of single polyprotein molecules by atomic force microscopy. Rev. Sci. Instrum. 2007;78:113707. doi: 10.1063/1.2794732. [DOI] [PubMed] [Google Scholar]
- 23.Li H., Oberhauser A.F., Fernandez J.M. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc. Natl. Acad. Sci. USA. 2001;98:10682–10686. doi: 10.1073/pnas.191189098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steward A., Toca-Herrera J.L., Clarke J. Versatile cloning system for construction of multimeric proteins for use in atomic force microscopy. Protein Sci. 2002;11:2179–2183. doi: 10.1110/ps.0212702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrión-Vazquez M., Oberhauser A.F., Fernandez J.M. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bustamante C., Marko J.F., Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 27.Ainavarapu S.R., Brujic J., Fernandez J.M. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys. J. 2007;92:225–233. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsui V., Case D.A. Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers. 2000-2001;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 29.Valbuena A., Oroz J., Carrión-Vázquez M. On the remarkable mechanostability of scaffoldins and the mechanical clamp motif. Proc. Natl. Acad. Sci. USA. 2009;106:13791–13796. doi: 10.1073/pnas.0813093106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Case D.A., Darden T.A., Kollman P.A. University of California; San Francisco: 2008. AMBER 10. [Google Scholar]
- 31.Hawkins G.D., Cramer C.J., Truhlar D.G. Pairwise solute descreening of solute charges from a dielectric medium. Chem. Phys. Lett. 1995;246:122–129. [Google Scholar]
- 32.Hawkins G.D., Cramer C.J., Truhlar D.G. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J. Phys. Chem. 1996;100:19824–19839. [Google Scholar]
- 33.Srinivasan J., Trevathan M.W., Case D.A. Application of a pairwise generalized Born model to proteins and nucleic acids: inclusion of salt effects. Theor. Chem. Acc. 1999;101:426–434. [Google Scholar]
- 34.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 35.Ritco-Vonsovici M., Ababou A., Horton M. Molecular plasticity of β-catenin: new insights from single-molecule measurements and MD simulation. Protein Sci. 2007;16:1984–1998. doi: 10.1110/ps.072773007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamboy J.A., Kim H., Komives E.A. Visualization of the nanospring dynamics of the IkappaBalpha ankyrin repeat domain in real time. Proc. Natl. Acad. Sci. USA. 2011;108:10178–10183. doi: 10.1073/pnas.1102226108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrión-Vázquez M., Oberhauser A.F., Martínez-Martín D. Protein nanomechanics, as studied by AFM single-molecule force spectroscopy. In: Arrondo J.L.R., Alonso A., editors. Advanced Techniques in Biophysics. Springer-Verlag; Berlin, Heidelberg: 2006. pp. 163–245. [Google Scholar]
- 38.Werbeck N.D., Itzhaki L.S. Probing a moving target with a plastic unfolding intermediate of an ankyrin-repeat protein. Proc. Natl. Acad. Sci. USA. 2007;104:7863–7868. doi: 10.1073/pnas.0610315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serquera D., Lee W., Itzhaki L.S. Mechanical unfolding of an ankyrin repeat protein. Biophys. J. 2010;98:1294–1301. doi: 10.1016/j.bpj.2009.12.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee G., Abdi K., Marszalek P.E. Nanospring behaviour of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 41.Bradley C.M., Barrick D. Limits of cooperativity in a structurally modular protein: response of the Notch ankyrin domain to analogous alanine substitutions in each repeat. J. Mol. Biol. 2002;324:373–386. doi: 10.1016/s0022-2836(02)00945-2. [DOI] [PubMed] [Google Scholar]
- 42.Tripp K.W., Barrick D. The tolerance of a modular protein to duplication and deletion of internal repeats. J. Mol. Biol. 2004;344:169–178. doi: 10.1016/j.jmb.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 43.Baumgartner W., Hinterdorfer P., Drenckhahn D. Cadherin interaction probed by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 2000;97:4005–4010. doi: 10.1073/pnas.070052697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oroz J., Valbuena A., Carrión-Vázquez M. Nanomechanics of the cadherin ectodomain: “canalization” by Ca2+ binding results in a new mechanical element. J. Biol. Chem. 2011;286:9405–9418. doi: 10.1074/jbc.M110.170399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim M., Abdi K., Marszalek P.E. Fast and forceful refolding of stretched α-helical solenoid proteins. Biophys. J. 2010;98:3086–3092. doi: 10.1016/j.bpj.2010.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berkovich R., Garcia-Manyes S., Fernández J.M. Hopping around an entropic barrier created by force. Biochem. Biophys. Res. Commun. 2010;403:133–137. doi: 10.1016/j.bbrc.2010.10.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koslov E.R., Maupin P., Rimm D.L. α-catenin can form asymmetric homodimeric complexes and/or heterodimeric complexes with β-catenin. J. Biol. Chem. 1997;272:27301–27306. doi: 10.1074/jbc.272.43.27301. [DOI] [PubMed] [Google Scholar]
- 48.Behrens J., von Kries J.P., Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 49.Miroux B., Walker J.E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 50.Rief M., Gautel M., Gaub H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 51.Hossain M.D., Furuike S., Kinosita K., Jr. The rotor tip inside a bearing of a thermophilic F1-ATPase is dispensable for torque generation. Biophys. J. 2006;90:4195–4203. doi: 10.1529/biophysj.105.079087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varea J., Monterroso B., Menéndez M. Structural and thermodynamic characterization of Pal, a phage natural chimeric lysin active against pneumococci. J. Biol. Chem. 2004;279:43697–43707. doi: 10.1074/jbc.M407067200. [DOI] [PubMed] [Google Scholar]
- 53.Böhm G., Muhr R., Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- 54.Improta S., Politou A.S., Pastore A. Immunoglobulin-like modules from titin I-band: extensible components of muscle elasticity. Structure. 1996;4:323–337. doi: 10.1016/s0969-2126(96)00036-6. [DOI] [PubMed] [Google Scholar]
- 55.Zinober R.C., Brockwell D.J., Smith D.A. Mechanically unfolding proteins: the effect of unfolding history and the supramolecular scaffold. Protein Sci. 2002;11:2759–2765. doi: 10.1110/ps.0224602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans E., Ritchie K. Strength of a weak bond connecting flexible polymer chains. Biophys. J. 1999;76:2439–2447. doi: 10.1016/S0006-3495(99)77399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


