Abstract
Microbial fuel cells (MFCs) are often inoculated from a single wastewater source. The extent that the inoculum affects community development or power production is unknown. The stable anodic microbial communities in MFCs were examined using three inocula: a wastewater treatment plant sample known to produce consistent power densities, a second wastewater treatment plant sample, and an anaerobic bog sediment. The bog-inoculated MFCs initially produced higher power densities than the wastewater-inoculated MFCs, but after 20 cycles all MFCs on average converged to similar voltages (470±20 mV) and maximum power densities (590±170 mW m−2). The power output from replicate bog-inoculated MFCs was not significantly different, but one wastewater-inoculated MFC (UAJA3 (UAJA, University Area Joint Authority Wastewater Treatment Plant)) produced substantially less power. Denaturing gradient gel electrophoresis profiling showed a stable exoelectrogenic biofilm community in all samples after 11 cycles. After 16 cycles the predominance of Geobacter spp. in anode communities was identified using 16S rRNA gene clone libraries (58±10%), fluorescent in-situ hybridization (FISH) (63±6%) and pyrosequencing (81±4%). While the clone library analysis for the underperforming UAJA3 had a significantly lower percentage of Geobacter spp. sequences (36%), suggesting that a predominance of this microbe was needed for convergent power densities, the lower percentage of this species was not verified by FISH or pyrosequencing analyses. These results show that the predominance of Geobacter spp. in acetate-fed systems was consistent with good MFC performance and independent of the inoculum source.
Keywords: community analysis, convergence, microbial fuel cells
Introduction
In a microbial fuel cell (MFC), exoelectrogenic bacteria on the anode degrade organic matter and produce electrical current. Exoelectrogens occur widely in nature (Logan, 2009), and MFCs have been successfully inoculated using pure cultures (Nevin et al., 2008; Ishii et al., 2008b; Watson and Logan, 2010), aerobic and anaerobic wastewater streams (Min et al., 2005; Ki et al., 2008), as well as environmental samples such as seawater, rivers and soil (Bond et al., 2002; Ieropoulos et al., 2010; Jiang et al., 2010).
The role of different inoculum sources on the communities that develop and the associated MFC performances under otherwise identical experimental conditions has not been well studied. Holmes et al. (2004) examined anodic communities and compared reactor performance of sediment-type MFCs inoculated with samples from salt-water marshes, freshwater and marine sediments. They found that all anodic communities were dominated by different orders of Deltaproteobacteria, indicating that this class of bacteria was linked to exoelectrogenic activity. MFCs inoculated with wastewater have produced more power than a natural environment inoculum in some cases, but not in others. Anaerobic and aerobic wastewater effluent inocula were compared with a river water inoculum in a continuous-flow, acetate-fed MFC (Ieropoulos et al., 2010). The aerobic effluent (1.16 mW m−2) and river water (1.14 mW m−2) inocula produced a higher maximum power than the anaerobic effluent inoculum (0.89 mW m−2). In contrast, a wastewater inoculum produced a higher open circuit voltage (785 mV) than a soil inoculum (684 mV) (Jiang et al., 2010) when fed acetate, but power densities were not reported. The communities that developed in these wastewater studies were not reported.
Another issue that arises in many MFC studies is a lack of replicate reactors. It has been noted that while replication is important to achieve statistical confidence in molecular community analyses, ∼60% of microbial diversity studies do not include replicate analyses (Prosser, 2010). Several MFC studies have been conducted using only a single reactor (Bond et al., 2002; Aelterman et al., 2006; Jong et al., 2006; Reimers et al., 2007; Xing et al., 2008b). The lack of replicates makes it difficult to determine if the reported microbial communities and reactor performances would be reproducible. Fortunately, there seems to be more of a trend in recent studies to use replicates, with most studies reporting on duplicate (Holmes et al., 2004; Rismani-Yazdi et al., 2011; Sasaki et al., 2011) or triplicate reactors (Faimali et al., 2010; Nelson et al., 2010; Jung and Regan, 2011). It is particularly important to establish better linkages between reactor performance and microbial communities because other factors affect reactor performance, such as differing reactor internal resistances and architectures.
Comparisons of microbial communities are also complicated by the fact that many studies use only one molecular technique to analyze the anodic community. The two most commonly used community analysis techniques in MFC studies have been denaturing gradient gel electrophoresis (DGGE) (Rismani-Yazdi et al., 2007; Erable et al., 2009; Zang et al., 2010; Jung and Regan, 2011) and clone libraries (Ishii et al., 2008a; Ki et al., 2008; Kiely et al., 2011a; Kiely et al., 2011b; Sun et al., 2010) using PCR amplicons of 16S rRNA genes. Pyrosequencing can be used to increase the sensitivity of community analysis compared with DGGE or clone libraries, allowing identification of less-abundant (but possibly still important) members of the community, while eliminating cloning bias (Lee et al., 2010). However, all of these post-PCR methods are potentially subject to quantitative bias associated with differential amplification efficiency and rRNA operon copy number. The use of multiple analysis techniques may provide greater confidence on the composition of the community. For example, fluorescent in-situ hybridization (FISH) is a useful non-PCR based technique that can be used to corroborate the findings of other molecular techniques (Kiely et al., 2011c), but it has rarely been used in MFC studies.
In this study we used four different molecular analysis techniques (16S rRNA gene-targeted DGGE, clone libraries and pyrosequencing and FISH) to study the anodic communities that develop in MFCs. Three different inocula were studied in triplicate reactors over time. Through these analyses, we were able to link reactor performance to shifts in community composition and predominance of important exoelectrogenic species in MFCs that were otherwise identical in architecture and chemical solutions.
Materials and methods
Reactor configuration, inoculation and electrochemical monitoring
Single-chamber air-cathode MFCs (28 ml) were constructed from Lexan cubes to form a 4-cm long cylindrical chamber 3 cm in diameter (Cheng and Logan, 2007). Anodes were made from a graphite fiber brush (2.5 cm diameter, 2.5 cm long) (Logan et al., 2007) (PANEX33 160 K, ZOLTEK, St Louis, MO, USA) and cathodes were carbon cloth (type B-1B, E-TEK) containing 0.5 mg cm−2 Pt (Cheng et al., 2006). Reactors were kept in a constant 30 °C chamber and fed a medium consisting of 1 g l−1 sodium acetate and Wolfe's vitamins (5 ml l−1) and minerals (12.5 ml l−1) (Balch et al., 1979) in 50 mℳ phosphate buffer (Na2HPO4, 4.56 g l−1; NaH2PO4 monohydrate, 2.45 g l−1; NH4Cl, 0.31 g l−1; KCl, 0.13 g l−1) (Xing et al., 2009; Kiely et al., 2011b). There was no attempt made to prevent light intrusion into the reactors.
Reactors (triplicate) were inoculated using samples from three different sources: the PSU (Pennsylvania State University) Wastewater Treatment Plant primary clarifier effluent, which has been used in a number of MFC studies with relatively good and consistent performance (Liu et al., 2005; Kiely et al., 2011b); University Area Joint Authority Wastewater Treatment Plant (UAJA) primary clarifier effluent; and sediment from a stagnant bog in the Black Moshannon Lake State Park, PA, USA (Bog). The UAJA plant is much larger, and serves more rural communities, compared with the PSU plant that treats ‘city' wastewater. Wastewater reactors were inoculated in a 1:1 ratio of wastewater to medium. The Bog reactors were inoculated using a 1:3 ratio of sediment to medium to maintain similar viscosity to the wastewater inocula. All reactors were then fed every 3 days (with 3 days being equivalent to one cycle), with complete medium replacement.
Voltage (E) was measured every 20 min across an external resistor (R=1 kΩ) using a data collection system (Keithley, Cleveland, OH, USA). Power (P=E2/R) and current (I=E/R) were normalized by the cathode projected surface area (7.1 cm2). Polarization curves were generated using the single-cycle method (Watson and Logan, 2011) by varying external resistances every 20 min from 10 kΩ to 50 Ω. All statistical calculations using a t-test were based on α=0.05. The Coulombic efficiency in similar fixed-resistance systems has been well studied (∼20% for an external resistance of 1000 Ω) and was not measured in these experiments (Zhang et al., 2009).
DNA extraction
Anode samples were taken during cycles 1, 3, 4, 5, 11 and 16 by cutting ∼0.2 g (wet weight; corresponding to ∼2% of the anode) of the anode fibers using flame-sterilized scissors. The samples were taken mid-cycle during peak electrochemical activity. Anode removal would not affect performance as recent tests with identical reactors have shown that up to 75% of the anode brush can be removed without adversely affecting voltage or power generation (Hutchinson et al., 2011). DNA was extracted using the PowerSoil DNA Isolation Kit (MO-BIO Laboratories, Carlsbad, CA, USA) according to the manufacturer's instructions. DNA samples from all cycles were used for cluster analysis of DGGE profiles via Principal Component Analysis to screen for convergence of the anodic communities from the different inocula. The convergent anode samples obtained during cycle 16 were then further analyzed by FISH and used for the clone library and pyrosequencing analyses. DNA samples were stored at −20 °C before analysis.
16S rRNA gene clone libraries
16S rRNA gene clone libraries were generated for the anode communities present during cycle 16 as previously described (Kiely et al., 2011b), but with different primers. Extracted DNA was amplified via PCR (Zuo et al., 2008) with a static annealing temperature of 55 °C to obtain fragment lengths of 960 bp using bacterial primers 530F (5′-GTCCCAGCMGCCGCGG-3′) and 1490R (5′-GGTTACCTTGTTACGACTT-3′) (Wani et al., 2006). NCBI BLAST (blastn algorithm) was then used to analyze the DNA sequences using the NCBI Genomes database (Altschul et al., 1990).
Shannon diversity indices, Good's coverage values and rarefaction curves were calculated using the Mothur computer software (Schloss et al., 2009). Sequences were first aligned in MEGA 4 (Tamura et al., 2007). Rarefaction curves are an average composed of 1000 randomizations. Approximately 90 clones were analyzed for each sample and deposited into GenBank under accession numbers JF817397–JF818119.
Pyrosequencing and analysis
Nine additional bacterial phylogenetic libraries were produced from the cycle 16 anode samples by pyrosequencing utilizing the GS FLX sequencer and Titanium series chemistry (454 Life Sciences, Branford, CT, USA). Universal bacterial PCR primers 343F 5′-TACGGRAGGCAGCAG-3′ and 926R 5′-CCGTCAATTYYTTTRAGTTT-3′ (Liu et al., 2007; Wang and Qian, 2009) were used and also included pyrosequencing adapters, keys and multiplex identifiers. PCR was conducted in 50 μl volumes containing 1 × PCR master mix (Roche Applied Science, Indianapolis, IN, USA), 0.4 μℳ of each primer and 2 μl of DNA template (5 ng μl−1). PCR was performed at the following cycling conditions: initial denaturation at 94 oC for 5 min, and 25 cycles of 94 °C denaturation for 15 s, annealing at 56 °C for 45 s and a 1-min extension at 72 °C, followed by a final extension at 72 °C for 8 min. Four reactions were conducted for each sample and amplicons were combined before removing salts and unincorporated primers using a Qiagen MinElute PCR purification kit (Qiagen Inc., Valencia, CA, USA) (Bibby et al., 2010). Amplicons were visualized on a 1.2% agarose gel and extracted if smearing was observed using a Qiagen MinElute gel extraction kit (Qiagen Inc.). Each purified DNA amplicon was pooled at equal concentrations for sequencing. Before sequencing, DNA concentration and purity were confirmed on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Sequencing was performed using an emPCR Lib-A Kit (Roche Applied Science) at the Yale Center for Genome Analysis. Reads were subjected to quality control at the machine based on run-produced Phred scores. Keypass, dots and mixed filters assessed the quality of the whole read while the quality of read ends was checked by signal intensity and primer filters.
Quantitative sequence analysis was performed using programs in the Quantitative Insights Into Microbial Ecology (QIIME) tool box (Caporaso et al., 2010). Within QIIME, sequences were sorted, and primer and multiplex identifier sequences were trimmed. Denoising was conducted using Titanium Pyronoise software (Quince et al., 2009; Caporaso et al., 2010; Quince et al., 2011). Similar sequences were clustered into operational taxonomic units based on 97% identity, aligned with the Greengenes core set (DeSantis et al., 2006), and taxonomy was assigned using the RDP (ribosomal database project) classifier (Cole et al., 2009). Within QIIME, rarefaction curves using the denoised data were generated using the alpha_rarefaction.py workflow script with OTUs clustered at 97% identity. The unprocessed DNA sequences obtained in this study have been deposited in the MG-RAST (Meyer et al., 2009) archive under accession number 4465374.3. Multiplex identifier information is included in Supplementary Table S1.
DGGE analysis
Fragments of 16S rRNA genes from cycles 1 and 16 were amplified via PCR using universal primers 968F (5′-AACGCGAAGAACCTTAC-3′) and 1401R (5′-CGGTGTGTACAAGACCC-3′). A GC clamp (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCC-3′) was attached to 968F (Ren et al., 2007). Samples were amplified with a G-Storm 1 Thermocycler (GRI Laboratories, Braintree, Essex, UK) using a previously described temperature profile (Xing et al., 2010) and stored at 4 °C. DGGE gels were prepared as previously described (Xing et al., 2008a). Electrophoresis was conducted for 12 h at 60 °C and 70 V in a 0.5 × TAE buffer solution to achieve effective band separation. Gels were then silver stained as previously described (Bassam et al., 1991).
The most visually prominent bands from each gel were excised using sterilized pipette tips, crushed in 50 μl of TE buffer (10 mℳ Tris-HCl, 1 mℳ EDTA (pH 8.0)) and eluted overnight at 4 °C. The eluted DNA was used as the template for PCR as described above excluding the GC clamp. The PCR products were purified using a PCR cleanup kit (Qiagen) followed by ligation of the DNA fragments into a TOPO 2.1 vector (Invitrogen, Carlsbad, CA, USA) and transformation into chemically competent E. coli cells. Plasmids were extracted (96-well Plasmid Extraction Kit, Omega Bio-Tek, Norcross, GA, USA) from eight random colonies per band and the insert DNA was sequenced (ABI 3730XL, Applied Biosystems, Carlsbad, CA, USA) to phylogenetically identify the bands and inspect for comigration of bands derived from different templates.
PCA of the DGGE profiles was conducted using two methods in order to determine the convergence of the anode communities. Both methods used presence/absence of bands and scoring band intensities (scale from 0–5). One method used a computer software program (GelCompar II, Applied-Maths, Austin, TX, USA), while the other used visual observation (Fry et al., 2006). PCA plots were generated using Minitab 16 (Minitab, State College, PA, USA) to determine the similarity of the replicates at different times and to evaluate temporal community shifts in the reactors.
FISH
The final anode sample taken during cycle 16 was analyzed using FISH for comparison with clone library and pyrosequencing results. Samples were immediately fixed using 4% paraformaldehyde for 3 h at 4 °C, washed in sterile phosphate buffer (FISH-PBS; 10 mℳ NaH2PO4, 130 mℳ NaCl, pH 7.2–7.4), and stored at –20 °C in 1:1 PBS to ethanol solution. A G. sulfurreducens specific probe 5′-labeled with Alexa Fluor 594 (GEO2; 5′-GAAGACAGGAGGCCCGAAA-3′) (Invitrogen) along with 2 unlabeled helper probes (HGEO2-2; 5′-CTAATGGTACGCGGACTCATCC-3′ and HGEO2-1; 5′-GTCCCCCCCTTTTCCCGCAAG-3′) (Richter et al., 2007) were each added to a final concentration of 10 ng μl−1 and hybridized in 1 ℳ NaCl solution containing 0% formamide for 1.5 h at 46 °C (Kiely et al., 2010). Samples were then washed in a 1-ℳ NaCl solution at 48 °C and stained with Slow Fade Gold containing DAPI (Invitrogen) to identify all bacterial and archaeal cells. Visualization of DAPI and probe-conferred fluorescence was then performed on an Olympus BX61 epifluorescent microscope. The percentage of G. sulfurreducens in each reactor sample was determined by counting at least 1000 total cells from 10 different fields. No probe and nonsense probe controls were included to examine for autofluorescence and non-specific binding of the dye, respectively. Verification of the FISH probe specificity was performed using probeCheck (Loy et al., 2008) and through negative hybridizations with two closely related bacteria previously found in acetate-fed MFCs (Pelobacter propionicus, G. metallireducens) (data not shown). Each species tested had six mismatches from the probe used in this study.
Results
Electrochemical performance
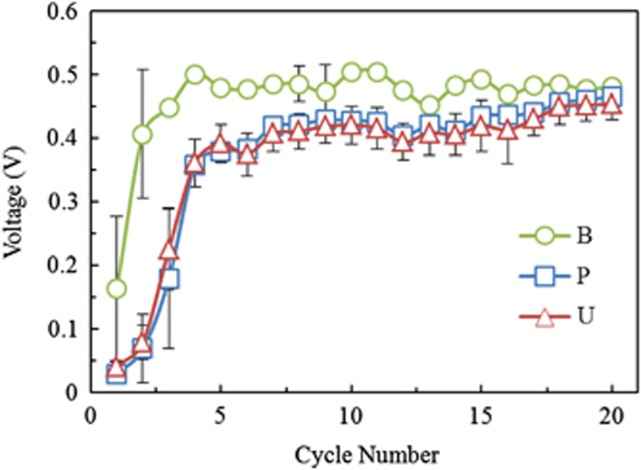
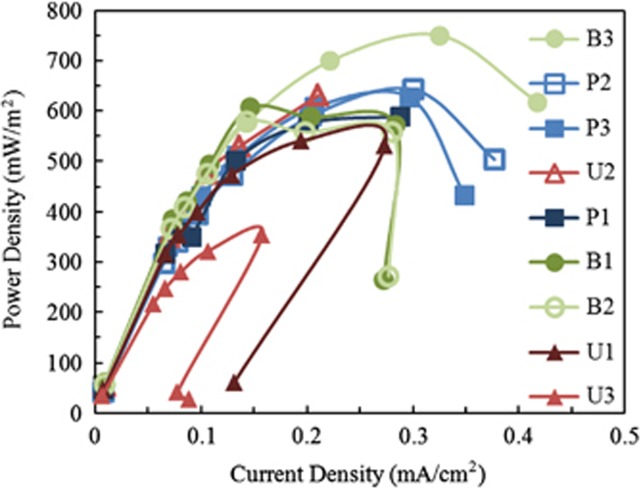
The bog-inoculated reactors initially produced higher maximum voltages on average than the wastewater-inoculated MFCs, but after 20 cycles, all reactors converged to a maximum voltage output of 470±20 mV (Figure 1). Based on polarization tests, maximum power densities on cycle 16 were not significantly different for eight of the nine reactors (P>0.05), but the UAJA3 replicate produced significantly (P=0.005, t-test) less voltage (data not shown) and power (354 mW m−2; Figure 2) than the other samples. The maximum power density was 590±110 mW m−2 based on all nine reactors, and 620±60 mWm−2 excluding the UAJA3 replicate (n=8).
Figure 1.
Average maximum voltage (n=3) in each cycle for each inoculum source (B=bog, P=PSU and U=UAJA).
Figure 2.
Power density curves generated from cycle 16 showing that UAJA replicate 3 produced far less power than all other reactors, including other UAJA replicates. Legend order mimics the order of the curves based on max power density.
16S rRNA gene clone library and pyrosequencing analysis
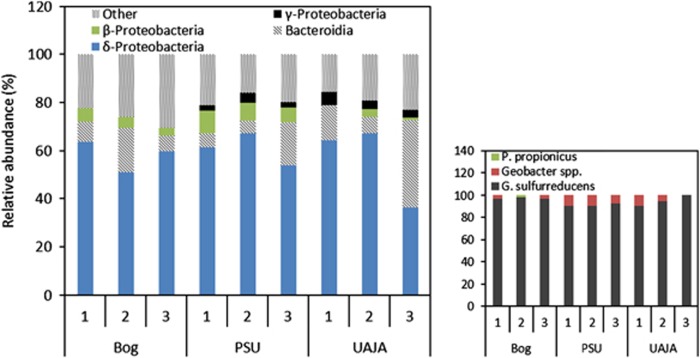
Based on the 16S rRNA gene clone libraries, during cycle 16 all reactors were found to be predominated by clones most similar to Geobacter spp. (>95% identity to Geobacter spp. with most sequences having >97% identity to G. sulfurreducens). On average, 61±6% (n=8, excluding UAJA3) of the clones were similar to Geobacter spp., except for replicate UAJA3, which had 36%. Clones distantly related to Bacteroidetes (89% identity or lower) were the second most abundant taxonomic group in all of the anode communities (11±6%, n=8), but were 36% of the clone library for the community in the UAJA3 replicate. Other sequences identified in the clone analysis were <95% similar to members of Betaproteobacteria (5±3%, n=9) and Alphaproteobacteria (2±2%, n=9) (Figure 3).
Figure 3.
Abundance of clones with sequence identity to dominant classes in each reactor from cycle 16. UAJA3 had the lowest number of clones showing similarity to Deltaproteobacteria. The group labeled as other consists of sequences with <95% identity. Insert: the majority of the Deltaproteobacteria were Geobacter sulfurreducens.
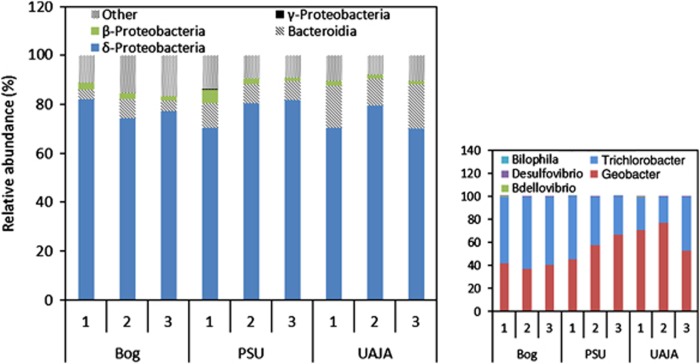
A higher relative abundance of sequences with similarity to Deltaproteobacteria (81±4%) (Figure 4) was identified by pyrosequencing compared with 16S rRNA gene clone libraries. Bacteroidetes (10±5%) and Firmicutes (8±3%) were the other two most prominent phyla present. The Deltaproteobacteria sequences obtained by pyrosequencing were dominated by the closely related genera Geobacter and Trichlorobacter, as identified by the RDP II classifier (Cole et al., 2005). It has been proposed that Trichlorobacter be reclassified as Geobacter (Nevin et al., 2007), so the two genera are grouped together in this analysis. The main difference between the two results is that pyrosequencing analysis did not confirm the clone library result that showed a lower abundance of clones with sequence similarity to G. sulfurreducens in the UAJA3 replicate.
Figure 4.
Community analysis results obtained from pyrosequencing. No significant difference in the amount of Proteobacteria was seen in any reactor. Insert: the Deltaproteobacteria was made up almost entirely of Geobacter and Trichlorobacter (95%). Other minor genera were detected by pyrosequencing that were not detected in the clone libraries.
There was no difference in diversity among the anode communities derived from the different inocula at cycle 16 based on the Shannon Diversity Index of each reactor generated from clone library data (1.78±0.22, n=9, P>0.05). Good's Coverage values indicated that on average 82% of the total species richness was accounted for in each community (Pommier et al., 2007) (Table 1). Rarefaction curves generated using both Mothur and QIIME (454 sequences) also suggest that there is little difference between the diversity of each of the reactors. The rarefaction curves using both data sets approached, but did not achieve, a plateau despite using more than 1500 sequences for the pyrosequencing data set compared with ∼100 sequences using the 16S rRNA gene clone libraries. While this might suggest that the diversity of the bacterial communities was not fully captured here, it is clear that the predominance of Geobacter was shown using both approaches.
Table 1. Shannon Diversity Indices and Good's coverage values calculated from 16S rRNA gene clone library and pyrosequencing data taken during cycle 16.
|
16S rRNA clone libraries |
Pyrosequencing |
||||
|---|---|---|---|---|---|
| No. of clones | Shannon diversity | Good's coverage (%) | No. of sequences | Shannon diversity | |
| Bog 1 | 93 | 1.57 | 86 | 3413 | 3.82 |
| Bog 2 | 92 | 1.94 | 80 | 1757 | 4.04 |
| Bog 3 | 92 | 1.45 | 89 | 1698 | 4.07 |
| Bog-average | 1.65 | 85 | 3.98 | ||
| PSU 1 | 86 | 1.86 | 81 | 3073 | 5.07 |
| PSU 2 | 94 | 1.53 | 84 | 2675 | 4.36 |
| PSU 3 | 95 | 1.99 | 82 | 2745 | 4.32 |
| PSU-average | 1.79 | 82 | 4.58 | ||
| UAJA 1 | 95 | 1.90 | 79 | 1022 | 4.39 |
| UAJA 2 | 88 | 1.72 | 79 | 1043 | 4.25 |
| UAJA 3 | 91 | 2.07 | 81 | 3595 | 4.69 |
| UAJA-average | 1.90 | 80 | 4.44 | ||
Abbreviations: PSU, Pennsylvania State University; UAJA, University Area Joint Authority Wastewater Treatment Plant.
Community shifts with DGGE
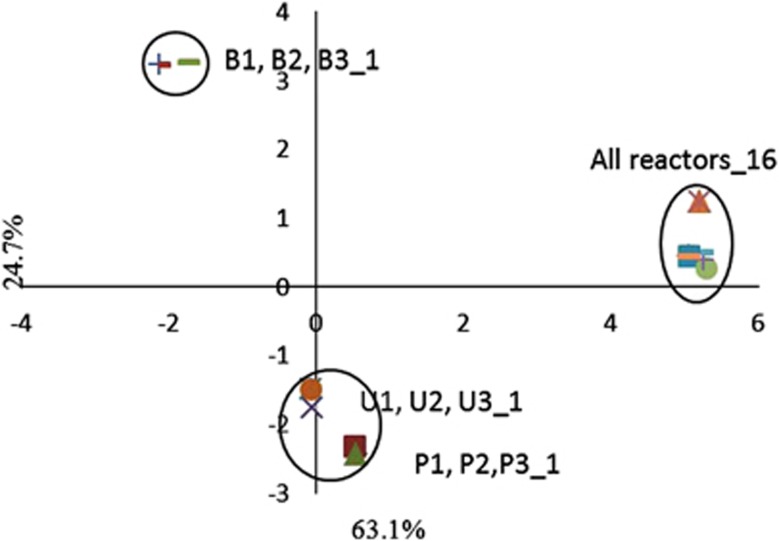
Analysis of predominant bands in DGGE gels using PCA showed convergence of the final microbial communities at cycle 16 (Supplementary Figure S1). During the first cycle, the Bog results clustered separately (quadrant 1) from the two wastewater inoculum sources (quadrant 4) (Figure 5). By the end of the study, all of the data clustered in quadrant 2. The computer-generated PCA (Warmink and van Elsas, 2008) produced the same results as manual scoring, indicating no bias in the manual scoring method (Supplementary Figure S2).
Figure 5.
PCA plot of the dominant (darkest) bands from the DGGE gels representing the communities at cycle 1 and cycle 16. This plot contrasts the initial community of wastewater-inoculated reactors (P=PSU and U=UAJA) with the bog-inoculated reactors (B). Over the duration of the experiment all communities converged irrespective of inoculum source.
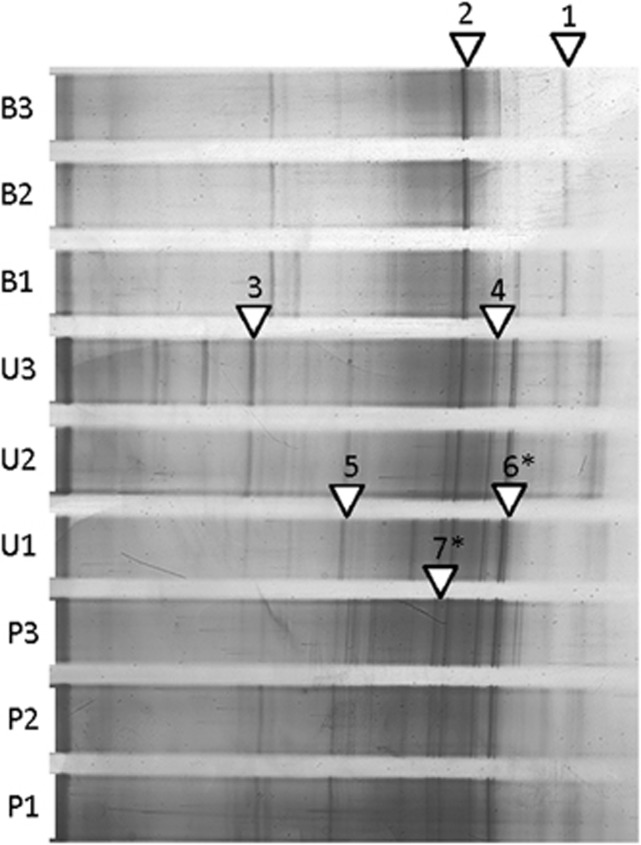
Seven of the visually darkest bands in the DGGE gels during cycle 16 were excised and sequenced, assuming that these bands would represent the predominant species in the communities (Figure 6). None of the dark bands analyzed (bands 2, 3 and 4) showed sequence identity to Geobacter spp., which was shown to be dominant in the 16S rRNA gene clone library and pyrosequencing analysis. Analysis of additional bands (1 and 5) revealed the presence of bacteria that were most closely related to the known isolates Geobacter spp. (95% identity) and ‘Candidatus Protochlamydia sp.' (89%). Other bands sequenced were most closely related to the known Firmicutes isolates Clostridium sp. (89%) and Alkaliphilus sp. (89%), and the Gammaproteobacterium Pseudomonas sp. (99%) (Table 2). Cloning and sequencing of these prominent DGGE bands did not show evidence of DNA comigration. Other sequenced bands (6 and 7) did exhibit comigration, but did not include Geobacter.
Figure 6.
DDGE gel from cycle 16 showing bands that were excised and identified. Gels were used in the PCA to monitor shifts in overall microbial community throughout the experiment. (B=bog, P=PSU and U=UAJA). Asterisks denote bands that exhibited comigration during analysis.
Table 2. DGGE band identification.
| Closest known isolate | Identity (%) | Found in 16S rRNA gene clone library | |
|---|---|---|---|
| Band 1 | Geobacter sulfurreducens | 99 | Yes |
| Band 2 | Alkaliphilus oremlandii | 89 | Yes |
| Band 3 | Pseudomonas fluorescens | 99 | Yes |
| Band 4 | Clostridium sticklandii | 89 | Yes |
| Band 5 | Candidatus Protochlamydia amoebophila | 89 | Yes |
| Band 6 | Rhodothermus marinus | 83 | No |
| Bartonella quintana | 81 | Yes | |
| Band 7 | Exiguobacterium sp. | 88 | No |
| Clostridium tetani | 84 | Yes |
Abbreviation: DGGE, denaturing gradient gel electrophoresis
FISH
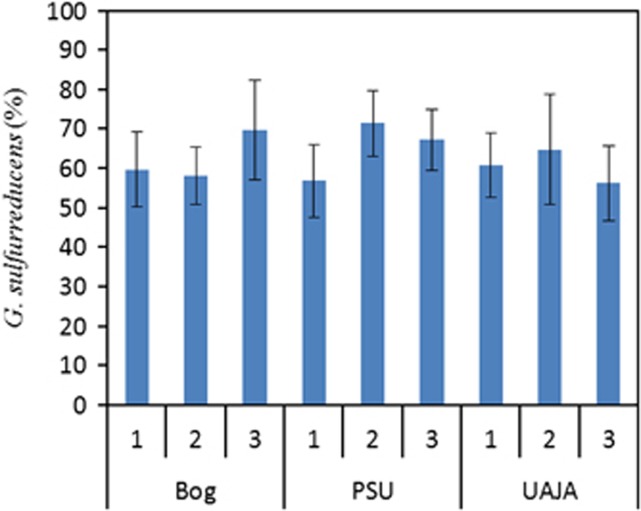
FISH confirmed that a large portion of the biofilm communities was G. sulfurreducens. On the basis of probe-positive cell counts, 63±6% of DAPI-stained cells were G. sulfurreducens (n=30 per sample) (Figure 7). FISH analysis did not confirm the clone library results that the UAJA3 sample had a significantly smaller population of G. sulfurreducens.
Figure 7.
Percentage of G. sulfurreducens in each reactor determined by FISH analysis, based on 10 images taken from different locations of each anode sample.
Discussion
Voltage and power comparisons
After a long acclimation time (20 cycles), the voltages of all the reactors converged to a similar voltage (470±20 mV) and power density (590±110 mW m−2) regardless of inoculum source. This conclusion would not be different based on any combination of duplicates. The average voltages and maximum power calculated using the two most disparate replicates from the UAJA-inoculated reactors produced 450±34 mV and 490±190 mW m−2, compared with 453±25 mV and 510±140 mW m−2 based on triplicate reactors. These values for voltage (P=0.90) and power (P=0.92) are not significantly different when based on duplicates or triplicates. Power production in MFCs with flat or brush anodes is limited by the cathode (Fan et al., 2008; Rismani-Yazdi et al., 2008). MFCs with brush anodes have been shown to produce lower internal resistances and larger power densities than reactors with flat anode materials (Logan et al., 2007), and therefore the systems used here were all cathode-limited in power generation. Even under the conditions where the anode did not limit power generation, the average performance of the reactors was independent of the initial inoculum once all reactors reached steady conditions. Given that UAJA3 produced significantly less power than the other replicates by the end of the study, our conclusions could have been different based on the analysis of a single reactor. Although the Bog sample initially produced a significantly higher voltage than the wastewater-inoculated reactors, and these MFCs reached their maximum voltage after only three cycles, they eventually became similar in performance to the other inocula after 17 more cycles. This more rapid acclimation was likely due to consistently anaerobic conditions for microbial communities in the bog compared with a wastewater effluent.
Wastewater from the PSU treatment plant has previously been used in a large number of studies (Liu et al., 2005; Cheng and Logan, 2007; Logan et al., 2007; Xing et al., 2010; Kiely et al., 2011b) with results generally consistent with those reported here. Maximum voltages produced here with this and the other inocula (470±20 mV) were generally consistent with those in several previous studies (500±40 mV, n=5) where 50 mℳ PBS, 1 g l−1 sodium acetate and single-chamber air-cathode MFCs were also used (Liu et al., 2005; Cheng and Logan, 2007; Logan et al., 2007; Xing et al., 2010; Kiely et al., 2011b). Maximum power densities have been slightly larger in some cases (704±30 mW m−2, after the cathodic biofilm was removed) (Kiely et al., 2011b) and smaller in others (506 mW m−2) (Liu et al., 2005) than obtained here (590±110 mW m−2) with the same buffer. In some studies, power densities were only reported at higher buffer concentrations, precluding direct comparisons (Cheng and Logan, 2007; Logan et al., 2007; Xing et al., 2010). Differences in maximum power for otherwise identical conditions could be due to methods used to obtain polarization data, which can result in higher power densities owing to Type M power overshoot (Watson and Logan, 2011), the use of different resistors or acclimation times before recording voltages; biofilms on the cathode, which can reduce performance; or better intrinsic performance of the cathodes leading to overall higher power densities. Some power density curves examined here also exhibited Type D power overshoot, where the power density curve doubles back instead of reaching higher current densities. We now know that this type of overshoot results in an underestimation of maximum power densities (Hong et al., 2011; Watson and Logan, 2011). Type D overshoot arises from the inability of the bacterial community to transfer electrons fast enough to the anode when the external resistance is lowered (Watson and Logan, 2011). Power overshoot implies that the capacity for these reactors to function at higher current densities would have required additional acclimation to low resistances, which was not done here (Watson and Logan, 2011).
Comparison of the community composition using different molecular techniques
Analysis of communities from the different inocula showed similar amounts of G. sulfurreducens based on 16S rRNA gene clone library analysis (61±6%) and FISH (63±6%), but a higher amount based on pyrosequencing data (81±4%). A similar conclusion of the predominance of Geobacter in acetate-fed MFC anode communities has been previously reported (Chae et al., 2009; Kan et al., 2011). In this study, acetate positively selected for Geobacter, which had a key role in its dominance of the communities in this study. Although Geobacter is seen to dominate in acetate-fed reactors, it is also able to grow using lactate (Call and Logan, 2011) and syntrophically using formic acid (Sun et al., 2012) showing that Geobacter is prevalent in systems using several different substrates. PCA of DGGE band patterns indicated a convergence of the communities to a similar composition after 16 cycles, but sequencing of the darkest bands did not reveal a predominance of G. sulfurreducens or any other specific genus in the community. One possible cause for the differences among some of these techniques is that different primer sequences were used for each technique because of methodological constraints such as amplicon length for full denaturation to give effective separation via DGGE (Wu et al., 1998) and sequencing length for pyrosequencing. The primer sets used here were chosen based on their common use in previous studies (Table 3). The possible effect these primers had on the resulting communities was analyzed with the RDP probe match function for sequences >1200 bp with no mismatches (Cole et al., 2009). In this analysis, the primers used here were compared with other bacterial primers reported in the literature for their identity to Deltaproteobacteria, Geobacter and Bacteroidia, three of the most prevalent taxa in the clone library and pyrosequencing analyses (Table 3). It should be noted that low values for reported primers that target the extreme ends of the 16S rRNA gene could result from relatively fewer sequences covering these regions in the RDP database. There also could be an inflation of matches for a particular primer if some sequences in the database were originally derived from the use of that primer and the primer sequence was not trimmed from the deposited sequence. The match scores for the DGGE primers used here are lower than those obtained for other common DGGE, clone library and pyrosequencing primers for all three taxa. This could in part explain the absence of Geobacter as a predominant community member in the DGGE analyses. Additionally, the greater specificity of the pyrosequencing primers to Geobacter compared with the clone libraries primers could partially explain the difference in Geobacter abundance. However, both primer sets have differences in targeting Bacteroidia, yet clone libraries (11±6%) and pyrosequencing (10±5%) gave similar results for sequences in this class. Despite these differences, in the final analysis, relatively similar responses were obtained for the different wastewater inocula based on both community analysis and time to reach maximum power densities. This indicates that these wastewater sources were equivalent as inocula. However, they were less effective in terms of achieving rapid acclimation of the reactors compared with the bog inoculum.
Table 3. Primer hits to the major phylogenetic taxa found in this study using RDP probe match.
| Molecular technique | Primer set | Sequence (5′ to 3′) | References | RDP probe match values (%) | ||
|---|---|---|---|---|---|---|
| |
|
|
|
Deltaproteobacteria |
Geobacter |
Bacteroidia |
| Clone libraries | a530F/1494R | GTCCCAGCMGCCGCGG GGTTACCTTGTTACCACTT | This study, Wani et al., 2006, | 0.1 0.3 | 0 0.2 | 0.1 0 |
| 530F/1494R | GTGCCAGCMGCCGCGG GGYTACCTTGTTACCACTT | Madrid et al., 2001 | 97 0.3 | 97 0.2 | 97 0 | |
| 27F/1541R | AGAGTTTGATCCTGGCTCAG AAGGAGGTGATCCAGCC | Löffler et al., 2000; Arturrson and Jansson, 2003; Kiely et al., 2011b | 26 3.8 | 28 6 | 24 0.3 | |
| 63/1389 | CAGGCCTAACACATGCAAGTC ACGGGCGGTGTGTACAAG | Lucas et al., 2003, Witt et al., 2011 | 0.8 78 | 0.2 82 | 6 31 | |
| 41F/1389R | GCTCAGATTGAACGCTGGCG ACGGGCGGTGTGTACAAG | Hongoh et al., 2003; Fisher et al., 2007 | 6.4 78 | 0.2 82 | 0 31 | |
| DGGE | 341F/534R | CCTACGGGAGGCAGCAG ATTACCGCGGCTGCTGG | Watanabe et al., 2001; Xing et al., 2006 | 96 86 | 98 98 | 98 98 |
| 341F/926R | CCTACGGGAGGCAGCAG CCGTCAATTCMTTTGAGTTT | Watanabe et al., 2001; Xing et al., 2006 | 96 80 | 98 96 | 98 96 | |
| 968F/1401R | AACGCGAAGAACCTTAC CGGTGTGTACAAGACCC | This study, Watanabe et al., 2001; Xing et al., 2006 | 59 1 | 15 1 | 0.5 0.3 | |
| 454 Sequencing | 343F/926R | TACGGRAGGCAGCAG CCGTCAATTYYTTTRAGTTT | This study, Liu et al., 2007 Wang and Qian, 2009 | 96 96 | 98 96 | 98 96 |
| 27F/342R | AGAGTTTGATCCTGGCTCAG CTGCTGCSYCCCGTAG | Kunin et al., 2010, | 27 96 | 28 98 | 24 98 | |
| 1114F/1392R | GCAACGAGCGCAACCC ACGGGCGGTGTGTRC | Engelbrektson et al., 2010; Kunin et al., 2010 | 94 80 | 97 86 | 0.7 37 | |
| FISH | GBS207 | GAAGACAGGAGGCCCGAAA | Richter et al., 2007 | 0.1 | 3 | 0 |
Abbreviations: DGGE, denaturing gradient gel electrophoresis; FISH, fluorescent in-situ hybridization, RDP, ribosomal database project.
This primer sequence was incorrectly written in the reference. The correct sequence is 5′-GTGCCAGCMGCCGCGG-3′ (Madrid et al., 2001; Rossetti et al., 2003). Deltaproteobacteria, Geobacter and Bacteroidia all had 97% probe match values when the correct sequence was used. The specificity of the PCR conditions would determine whether one mismatch would allow a primer to anneal to the template. This one mismatch did not appear to bias the targeting of any particular taxa as all probe match values for the incorrect primer gave ∼0% match.
Pyrosequencing analysis provided more depth of coverage to the community analysis than the 16S clone library analysis, which allowed a more detailed investigation of the bacteria present at low abundances in the anode community, indicating the presence of Acidobacteria, which includes the known exoelectrogen, Geothrix fermentans (Bond and Lovley, 2005) and Actinobacteria phyla, which were not identified using 16S rRNA gene clone libraries. The role that these members of the underrepresented phyla could have in electricity generation, however, is not known. Additionally, FISH was a useful tool in this study as G. sulfurreducens was highly enriched and could be easily targeted.
The clone library analysis was the only method that showed a possible change in the microbial community for the underperforming replicate UAJA3, indicating a reduced predominance of G. sulfurreducens and a greater predominance of clones (36%) most similar to Bacteroidetes, but at a very low (89%) identity. This change in the microbial community for the UAJA3 replicate does not necessarily indicate that the microbial community was the cause of the reduced performance of this replicate. It is possible that the performance reflects an undetectable physical difference of the reactor, such as a poor seal with a gasket allowing additional oxygen to leak into the reactor. Even this possible difference in the UAJA3 community cannot be certain because of the lack of agreement between the pyrosequencing and clone library approaches for this sample.
Conclusions
On the basis of performance and microbial community analyses of triplicate MFCs inoculated from three different sources, it was determined that electrochemical performance of the reactors converged after 2 months of operation (20 cycles). The use of at least two replicates was sufficient to characterize these systems in terms of electrochemical performance. When replicates produced similar voltages and power densities, community analysis of replicates produced similar results. Results of this study also suggest that at least one non-PCR based technique, such as FISH, should be used to verify findings of the community analysis and corroborate findings on microbial abundances in the biofilms.
Acknowledgments
This research was supported by Award KUS-I1-003-13 from the King Abdullah University of Science and Technology (KAUST) and by Award DGE-0750756 from the National Science Foundation (NSF) Graduate Student Fellowship Program.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aelterman P, Rabaey K, Pham TH, Boon N, Verstraete W. Continuous electricity generation at high voltages and currents using stacked microbial fuel cells. Environ Sci Technol. 2006;40:3388–3394. doi: 10.1021/es0525511. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller E, Myers EW, Lipman DJ. Basic logic alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Artursson V, Jansson JK. Use of Bromodeoxyuridine immunocapture to identify active bacteria associated with arbuscular mycorrhizal hyphae. Appl Environ Microbiol. 2003;69:6208–6215. doi: 10.1128/AEM.69.10.6208-6215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: reevaluation of a unique biological group. Microbiol Mol Biol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassam BJ, Caetano-Anollés G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- Bibby K, Viau E, Peccia J. Pyrosequencing of the 16S rRNA gene to reveal bacterial pathogen diversity in biosolids. Water Res. 2010;44:4252–4260. doi: 10.1016/j.watres.2010.05.039. [DOI] [PubMed] [Google Scholar]
- Bond DR, Holmes DE, Tender LM, Lovley DR. Electrode-reducing microorganisms that harvest energy from marine sediments. Science. 2002;295:483–485. doi: 10.1126/science.1066771. [DOI] [PubMed] [Google Scholar]
- Bond DR, Lovley DR. Evidence for involvement of an electron shuttle in electricity generation by Geothrix fermentans. Appl Environ Microbiol. 2005;71:2186–2189. doi: 10.1128/AEM.71.4.2186-2189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call DF, Logan BE. Lactate oxidation coupled to iron or electrode reduction by Geobacter sulfurreducens PCA. Appl Environ Microbiol. 2011;77:8791–8794. doi: 10.1128/AEM.06434-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K-J, Choi M-J, Lee J-W, Kim K-Y, Kim IS. Effect of different substrates on the performance, bacterial diversity, and bacterial viability in microbial fuel cells. Bioresource Technol. 2009;100:3518–3525. doi: 10.1016/j.biortech.2009.02.065. [DOI] [PubMed] [Google Scholar]
- Cheng S, Liu H, Logan BE. Increased performance of single-chamber microbial fuel cells using an improved cathode structure. Electrochem Commun. 2006;8:489–494. [Google Scholar]
- Cheng S, Logan BE. Ammonia treatment of carbon cloth anodes to enhance power generation of microbial fuel cells. Electrochem Commun. 2007;9:492–496. [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, et al. The ribosomal database project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, et al. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010;4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- Erable B, Roncato M-A, Achouak W, Bergel A. Sampling natural biofilms: A new route to build efficient microbial anodes. Environ Sci Technol. 2009;43:3194–3199. doi: 10.1021/es803549v. [DOI] [PubMed] [Google Scholar]
- Faimali M, Chelossi E, Pavanello G, Benedetti A, Vandecandelaere I, De Vos P, et al. Electrochemical activity and bacterial diversity of natural marine biofilm in laboratory closed-systems. Bioelectrochemistry. 2010;78:30–38. doi: 10.1016/j.bioelechem.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Fan Y, Sharbrough E, Liu H. Quantification of the internal resistance distribution of microbial fuel cells. Environ SciTechnol. 2008;42:8101–8107. doi: 10.1021/es801229j. [DOI] [PubMed] [Google Scholar]
- Fisher M, Miller D, Brewster C, Husseneder C, Dickerman A. Diversity of Gut Bacteria of Reticulitermes flavipes as Examined by 16S rRNA Gene Sequencing and Amplified rDNA Restriction Analysis. Curr Microbiol. 2007;55:254–259. doi: 10.1007/s00284-007-0136-8. [DOI] [PubMed] [Google Scholar]
- Fry JC, Webster G, Cragg BA, Weightman AJ, Parkes RJ. Analysis of DGGE profiles to explore the relationship between prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol Ecol. 2006;58:86–98. doi: 10.1111/j.1574-6941.2006.00144.x. [DOI] [PubMed] [Google Scholar]
- Holmes DE, Bond DR, O'Neil RA, Reimers CE, Tender LR, Lovley DR. Microbial communities associated with electrodes harvesting electricity from a variety of aquatic sediments. Microb Ecol. 2004;48:178–190. doi: 10.1007/s00248-003-0004-4. [DOI] [PubMed] [Google Scholar]
- Hong Y, Call DF, Werner CM, Logan BE. Adaptation to high current using low external resistances eliminates power overshoot in microbial fuel cells. Biosens Bioelectron. 2011;28:71–76. doi: 10.1016/j.bios.2011.06.045. [DOI] [PubMed] [Google Scholar]
- Hongoh Y, Yuzawa H, Ohkuma M, Kudo T. Evaluation of primers and PCR conditions for the analysis of 16S rRNA genes from a natural environment. FEMS Microbiol Lett. 2003;221:299–304. doi: 10.1016/S0378-1097(03)00218-0. [DOI] [PubMed] [Google Scholar]
- Hutchinson A, Tokash J, Logan BE. Analysis of carbon fiber brush loading in anodes on startup and performance of microbial fuel cells. J Power Sources. 2011;196:9213–9219. [Google Scholar]
- Ieropoulos I, Winfield J, Greenman J. Effects of flow-rate, inoculum and time on the internal resistance of microbial fuel cells. Bioresource Technol. 2010;101:3520–3525. doi: 10.1016/j.biortech.2009.12.108. [DOI] [PubMed] [Google Scholar]
- Ishii S, Hotta Y, Watanabe K. Methanogenesis versus electrogenesis: morphological and phylogenetic comparisons of microbial communities. Biosci Biotech Bioch. 2008a;72:286–294. doi: 10.1271/bbb.70179. [DOI] [PubMed] [Google Scholar]
- Ishii S, Watanabe K, Yabuki S, Logan BE, Sekiguchi Y. Comparison of electrode reduction activities of Geobacter sulfurreducens and an enriched consortium in an air-cathode microbial fuel cell. Appl Environ Microb. 2008b;74:7348–7355. doi: 10.1128/AEM.01639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Li B, Jia W, Lei Y. Effect of inoculum types on bacterial adhesion and power production in microbial fuel cells. Appl Biochem Biotech. 2010;160:182–196. doi: 10.1007/s12010-009-8541-z. [DOI] [PubMed] [Google Scholar]
- Jong BC, Kim BH, Chang IS, Liew PWY, Choo YF, Kang GS. Enrichment, performance and microbial diversity of a thermophilic mediatorless microbial fuel cell. Environ Sci Technol. 2006;40:6449–6454. doi: 10.1021/es0613512. [DOI] [PubMed] [Google Scholar]
- Jung S, Regan JM. Influence of external resistance on electrogenesis, methanogenesis, and anode prokaryotic communities in microbial fuel cells. Appl Environ Microb. 2011;77:564–571. doi: 10.1128/AEM.01392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan J, Hsu L, Cheung ACM, Pirbazari M, Nealson KH. Current production by bacterial communities in microbial fuel cells enriched from wastewater sludge with different electron donors. Environ Sci Technol. 2011;45:1139–1146. doi: 10.1021/es102645v. [DOI] [PubMed] [Google Scholar]
- Ki D, Park J, Lee J, Yoo K. Microbial diversity and population dynamics of activated sludge microbial communities participating in electricity generation in microbial fuel cells. Water Sci Technol. 2008;58:2195–2201. doi: 10.2166/wst.2008.577. [DOI] [PubMed] [Google Scholar]
- Kiely P, Call D, Yates M, Regan J, Logan B. Anodic biofilms in microbial fuel cells harbor low numbers of higher-power-producing bacteria than abundant genera. Appl Microbiol Biot. 2010;88:371–380. doi: 10.1007/s00253-010-2757-2. [DOI] [PubMed] [Google Scholar]
- Kiely PD, Cusick R, Call DF, Selembo PA, Regan JM, Logan BE. Anode microbial communities produced by changing from microbial fuel cell to microbial electrolysis cell operation using two different wastewaters. Bioresource Technol. 2011a;102:388–394. doi: 10.1016/j.biortech.2010.05.019. [DOI] [PubMed] [Google Scholar]
- Kiely PD, Rader G, Regan JM, Logan BE. Long-term cathode performance and the microbial communities that develop in microbial fuel cells fed different fermentation endproducts. Bioresource Technol. 2011b;102:361–366. doi: 10.1016/j.biortech.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Kiely PD, Regan JM, Logan BE. The electric picnic: synergistic requirements for exoelectrogenic microbial communities. Curr Opin Biotech. 2011c;22:378–385. doi: 10.1016/j.copbio.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Lee T, Van Doan T, Yoo K, Choi S, Kim C, Park J. Discovery of commonly existing anode biofilm microbes in two different wastewater treatment MFCs using FLX Titanium pyrosequencing. Appl Microbiol Biot. 2010;87:2335–2343. doi: 10.1007/s00253-010-2680-6. [DOI] [PubMed] [Google Scholar]
- Liu H, Cheng S, Logan BE. Production of electricity from acetate or butyrate in a single chamber microbial fuel cell. Environ Sci Technol. 2005;39:658–662. doi: 10.1021/es048927c. [DOI] [PubMed] [Google Scholar]
- Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucl Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan BE. Exoelectrogenic bacteria that power microbial fuel cells. Nat Rev Microbiol. 2009;7:375–381. doi: 10.1038/nrmicro2113. [DOI] [PubMed] [Google Scholar]
- Logan BE, Cheng S, Watson V, Estadt G. Graphite fiber brush anodes for increased power production in air-cathode microbial fuel cells. Environ Sci Technol. 2007;41:3341–3346. doi: 10.1021/es062644y. [DOI] [PubMed] [Google Scholar]
- Loy A, Arnold R, Tischler P, Rattei T, Wagner M, Horn M. probeCheck—a central resource for evaluating oligonucleotide probe coverage and specificity. Environ Microbiol. 2008;10:2894–2898. doi: 10.1111/j.1462-2920.2008.01706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas FS, Broennimann O, Febbraro I, Heeb P. High diversity among feather-degrading bacteria from a dry meadow soil. Microb Ecol. 2003;45:282–290. doi: 10.1007/s00248-002-2032-x. [DOI] [PubMed] [Google Scholar]
- Löffler FE, Sun Q, Li J, Tiedje JM. 16S rRNA gene-based detection of tetrachloroethene-dechlorinating desulfuromonas and dehalococcoides species. Appl Environ Microbiol. 2000;66:1369–1374. doi: 10.1128/aem.66.4.1369-1374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid VM, Taylor GT, Scranton MI, Chistoserdov AY. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the cariaco basin. Appl Environ Microbiol. 2001;67:1663–1674. doi: 10.1128/AEM.67.4.1663-1674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Aglyamova G, Wang S, Buchanan-Carter J, Abrego D, Colbourne J, et al. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics. 2009;10:219. doi: 10.1186/1471-2164-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Cheng S, Logan BE. Electricity generation using membrane and salt bridge microbial fuel cells. Water Res. 2005;39:1675–1686. doi: 10.1016/j.watres.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Nelson KY, Razban B, McMartin DW, Cullimore DR, Ono T, Kiely PD. A rapid methodology using fatty acid methyl esters to profile bacterial community structures in microbial fuel cells. Bioelectrochemistry. 2010;78:80–86. doi: 10.1016/j.bioelechem.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Nevin KP, Holmes DE, Woodard TL, Covalla SF, Lovley DR. Reclassification of Trichlorobacter thiogenes as Geobacter thiogenes comb. nov. Int J Syst Evol Microbiol. 2007;57:463–466. doi: 10.1099/ijs.0.63408-0. [DOI] [PubMed] [Google Scholar]
- Nevin KP, Richter H, Covalla SF, Johnson JP, Woodard TL, Orloff AL, et al. Power output and columbic efficiencies from biofilms of Geobacter sulfurreducens comparable to mixed community microbial fuel cells. Environ Microbiol. 2008;10:2505–2514. doi: 10.1111/j.1462-2920.2008.01675.x. [DOI] [PubMed] [Google Scholar]
- Pommier T, CanbÄCk B, Riemann L, BostrÖM KH, Simu K, Lundberg P, et al. Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol. 2007;16:867–880. doi: 10.1111/j.1365-294X.2006.03189.x. [DOI] [PubMed] [Google Scholar]
- Prosser JI. Replicate or lie. Environ Microbiol. 2010;12:1806–1810. doi: 10.1111/j.1462-2920.2010.02201.x. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Meth. 2009;6:639. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport R, Turnbaugh P. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimers CE, Stecher HA, Westall JC, Alleau Y, Howell KA, Soule L, et al. Substrate degradation kinetics, microbial diversity, and current efficiency of microbial fuel cells supplied with marine plankton. Appl Environ Microbiol. 2007;73:7029–7040. doi: 10.1128/AEM.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren N, Xing D, Rittmann BE, Zhao L, Xie T, Zhao X. Microbial community structure of ethanol type fermentation in bio-hydrogen production. Environ Microbiol. 2007;9:1112–1125. doi: 10.1111/j.1462-2920.2006.01234.x. [DOI] [PubMed] [Google Scholar]
- Richter H, Lanthier M, Nevin KP, Lovley DR. Lack of electricity production by Pelobacter carbinolicus indicates that the capacity for Fe(III) oxide reduction does not necessarily confer electron transfer ability to fuel cell anodes. Appl Environ Microb. 2007;165:370–376. doi: 10.1128/AEM.00804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rismani-Yazdi H, Carver SM, Christy AD, Tuovinen OH. Cathodic limitations in microbial fuel cells: an overview. J Power Sources. 2008;180:683–694. [Google Scholar]
- Rismani-Yazdi H, Christy AD, Carver SM, Yu Z, Dehority BA, Tuovinen OH. Effect of external resistance on bacterial diversity and metabolism in cellulose-fed microbial fuel cells. Bioresource Technol. 2011;102:278–283. doi: 10.1016/j.biortech.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Rismani-Yazdi H, Christy AD, Dehority BA, Morrison M, Yu Z, Tuovinen OH. Electricity generation from cellulose by rumen microorganisms in microbial fuel cells. Biotechnol Bioeng. 2007;97:1398–1407. doi: 10.1002/bit.21366. [DOI] [PubMed] [Google Scholar]
- Rossetti S, Blackall LL, Majone M, Hugenholtz P, Plumb JJ, Tandoi V. Kinetic and phylogenetic characterization of an anaerobic dechlorinating microbial community. Microbiology. 2003;149:459–469. doi: 10.1099/mic.0.26018-0. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Morita M, Sasaki D, Hirano S-i, Matsumoto N, Ohmura N, et al. Methanogenic communities on the electrodes of bioelectrochemical reactors without membranes. J Biosci Bioeng. 2011;111:47–49. doi: 10.1016/j.jbiosc.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Call DF, Kiely PD, Wang A, Logan BE. Syntrophic interactions improve power production in formic acid fed MFCs operated with set anode potentials or fixed resistances. Biotechnol Bioeng. 2012;109:405–41. doi: 10.1002/bit.23348. [DOI] [PubMed] [Google Scholar]
- Sun Y, Zuo J, Cui L, Deng Q, Dang Y. Diversity of microbes and potential exoelectrogenic bacteria on anode surface in microbial fuel cells. J Gen Appl Microbiol. 2010;56:19–29. doi: 10.2323/jgam.56.19. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wang Y, Qian P-Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One. 2009;4:e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani AA, Surakasi VP, Siddharth J, Raghavan RG, Patole MS, Ranade D, et al. Molecular analyses of microbial diversity associated with the Lonar soda lake in India: an impact crater in a basalt area. Res Microbiol. 2006;157:928–937. doi: 10.1016/j.resmic.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Warmink JA, van Elsas JD. Selection of bacterial populations in the mycosphere of Laccaria proxima: is type III secretion involved[quest] ISME J. 2008;2:887–900. doi: 10.1038/ismej.2008.41. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Kodama Y, Harayama S. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J Microbiol Methods. 2001;44:253–262. doi: 10.1016/s0167-7012(01)00220-2. [DOI] [PubMed] [Google Scholar]
- Watson VJ, Logan BE. Power production in MFCs inoculated with Shewanella oneidensis MR-1 or mixed cultures. Biotechnol Bioeng. 2010;105:489–498. doi: 10.1002/bit.22556. [DOI] [PubMed] [Google Scholar]
- Watson VJ, Logan BE. Analysis of polarization methods for elimination of power overshoot in microbial fuel cells. Electrochem Commun. 2011;13:54–56. [Google Scholar]
- Witt V, Wild C, Anthony KRN, Diaz-Pulido G, Uthicke S. Effects of ocean acidification on microbial community composition of, and oxygen fluxes through, biofilms from the Great Barrier Reef. Environ Microbiol. 2011;13:2976–2989. doi: 10.1111/j.1462-2920.2011.02571.x. [DOI] [PubMed] [Google Scholar]
- Wu Y, Hayes VM, Osinga J, Mulder IM, Looman MWG, Buys CHCM, et al. Improvement of fragment and primer selection for mutation detection by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1998;26:5432–5440. doi: 10.1093/nar/26.23.5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D, Cheng S, Logan B, Regan J. Isolation of the exoelectrogenic denitrifying bacterium Comamonas denitrificans based on dilution to extinction. Appl Microbiol Biot. 2010;85:1575–1587. doi: 10.1007/s00253-009-2240-0. [DOI] [PubMed] [Google Scholar]
- Xing D, Cheng S, Regan JM, Logan BE. Change in microbial communities in acetate- and glucose-fed microbial fuel cells in the presence of light. Biosens Bioelectron. 2009;25:105–111. doi: 10.1016/j.bios.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Xing D, Ren N, Rittmann BE. Genetic diversity of hydrogen-producing bacteria in an acidophilic Ethanol-H2-Coproducing system, analyzed using the [Fe]-Hydrogenase gene. Appl Environ Microbiol. 2008a;74:1232–1239. doi: 10.1128/AEM.01946-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D, Zuo Y, Cheng S, Regan JM, Logan BE. Electricity generation by Rhodopseudomonas palustris DX-1. Environ Sci Technol. 2008b;42:4146–4151. doi: 10.1021/es800312v. [DOI] [PubMed] [Google Scholar]
- Xing DF, Nan Q, Song JX, Xu XL. Community of activated sludge based on different targeted sequence of 16S rDNA by denaturing gradient gel electrophoresis. Environ Sci. 2006;27:1424–1428. [PubMed] [Google Scholar]
- Zang G-L, Sheng G-P, Tong Z-H, Liu X-W, Teng S-X, Li W-W, et al. Direct electricity recovery from canna indica by an air-cathode microbial fuel cell inoculated with rumen microorganisms. Environ Sci Technol. 2010;44:2715–2720. doi: 10.1021/es902956e. [DOI] [PubMed] [Google Scholar]
- Zhang F, Cheng S, Pant D, Bogaert GV, Logan BE. Power generation using an activated carbon and metal mesh cathode in a microbial fuel cell. Electrochem Commun. 2009;11:2177–2179. [Google Scholar]
- Zuo Y, Xing D, Regan JM, Logan BE. Isolation of the exoelectrogenic bacterium Ochrobactrum anthropi YZ-1 by using a U-tube microbial fuel cell. Appl Environ Microbiol. 2008;74:3130–3137. doi: 10.1128/AEM.02732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.