Abstract
Methanococcus maripaludis grown syntrophically with Desulfovibrio vulgaris was compared with M. maripaludis monocultures grown under hydrogen limitation using transcriptional, proteomic and metabolite analyses. These measurements indicate a decrease in transcript abundance for energy-consuming biosynthetic functions in syntrophically grown M. maripaludis, with an increase in transcript abundance for genes involved in the energy-generating central pathway for methanogenesis. Compared with growth in monoculture under hydrogen limitation, the response of paralogous genes, such as those coding for hydrogenases, often diverged, with transcripts of one variant increasing in relative abundance, whereas the other was little changed or significantly decreased in abundance. A common theme was an apparent increase in transcripts for functions using H2 directly as reductant, versus those using the reduced deazaflavin (coenzyme F420). The greater importance of direct reduction by H2 was supported by improved syntrophic growth of a deletion mutant in an F420-dependent dehydrogenase of M. maripaludis. These data suggest that paralogous genes enable the methanogen to adapt to changing substrate availability, sustaining it under environmental conditions that are often near the thermodynamic threshold for growth. Additionally, the discovery of interspecies alanine transfer adds another metabolic dimension to this environmentally relevant mutualism.
Keywords: methanogen, syntrophy, coculture, alanine utilization, sulfate-reducing bacteria
Introduction
Virtually, all methane released to the biosphere, estimated to be approximately 1 billion tons per year, are produced by methanogenic archaea living in a close association with other anaerobic microorganisms (Thauer and Shima, 2008). When electron acceptors, such as sulfate, nitrate, Mn(IV) or Fe(III), are absent or present at low concentrations, organic carbon in anoxic environments is converted to CO2 and methane via a microbial food web consisting of anaerobic bacteria, microeukaryotes, syntrophic bacteria and methanogens. Anaerobic bacteria are principally responsible for the hydrolysis of biopolymers and, in association with syntrophic bacteria, ferment organic carbon to acetic acid, CO2 and hydrogen. At the low hydrogen partial pressures, typical of most anoxic habitats (<10 Pa), these are the major fermentation products and the principle substrates for methanogens. Thus, methanogens rely on microorganisms that occupy the lower tiers of this anaerobic food web for their primary growth substrates. In turn, this food-web functions optimally only when the hydrogen concentrations are maintained at a low level by methanogenic activity. At higher hydrogen concentrations, many fermentation reactions do not yield sufficient energy to support growth. Transfer of the reduced metabolic byproducts, here mainly in the form of hydrogen, from lower trophic levels to the metabolically specialized methanogens is generically termed ‘interspecies electron/hydrogen transfer'. Thus, there is a close cooperativity and interdependency between organisms producing hydrogen (sometimes referred to as proton-reducing syntrophs) and hydrogenotrophic methanogens.
We have examined the genetic and metabolic basis of this common interdependency using a model system composed of Desulfovibrio vulgaris growing in syntrophic association with the hydrogenotrophic methanogen Methanococcus maripaludis (Stolyar et al., 2007; Walker et al., 2009; Hillesland and Stahl, 2010). The pairing with D. vulgaris, a representative sulfate-reducing bacterium, provides a model system well suited for the study of this type of microbial mutualism, as both organisms have sequenced genomes (Heidelberg et al., 2004; Hendrickson et al., 2004) and established genetic systems (Moore and Leigh, 2005; Keller et al., 2009). We previously used this pairing to identify an alternative electron-transfer system used by D. vulgaris when growing syntrophically (Walker et al., 2009). In the current study, we sought to characterize M. maripaludis grown under both syntrophic and hydrogen-limited conditions.
M. maripaludis utilizes a very narrow range of substrates for carbon and energy (primarily CO2, hydrogen and formate). However, genomic and biochemical studies of this well-characterized methanogen reveal a large number of isofunctional genes, suggesting the potential for adaptive responses to conditions not found in the standard laboratory cultures. Previous studies (Hendrickson et al., 2007) examined the transcriptional response of M. maripaludis grown under hydrogen limitation, a proxy condition for its natural environment, where hydrogen concentrations are generally <10 Pa (Thauer et al., 2008). That study revealed hydrogen limitation greatly increased the expression of genes encoding enzymes of the methanogenic pathway that reduce or oxidize the electron-carrying deazaflavin coenzyme F420, primarily in response to hydrogen limitation, not growth rate. Although the low hydrogen concentration used in that study more closely reflected environmental conditions, it did not capture the syntrophic lifestyle that sustains most environmental populations of methanogens. In order to better approximate the natural condition, we characterized M. maripaludis growing in syntrophic association with a Desulfovibrio in chemostats on a lactate-based medium. In the absence of sulfate, the two organisms are obligately coupled: the Desulfovibrio is dependent upon M. maripaludis to maintain the low H2 concentration required for lactate fermentation and M. maripaludis is dependent upon the Desulfovibrio as a source of hydrogen. Here we present a system-level study of M. maripaludis physiology as compared between syntrophic growth versus growth under hydrogen limitation.
Materials and methods
Strains
Transcriptional and proteomic analyses were performed using M. maripaludis S2 and D. vulgaris Hildenborough. Additionally, seven mutant strains (all described in previous investigations) of M. maripaludis were used during phenotypic growth comparisons. Details for all strains are shown in Table 1.
Table 1. Summary of strains used during investigation.
| Strain | Mutant | Gene(s) | Description | Reference |
|---|---|---|---|---|
| Methanococcus maripaludis strains | ||||
| S2 | NA | NA | Wild-type strain | Jones et al, 1983 |
| Mm1145 | ΔfruA | Mmp1382 | Selenocysteine-containing F420-reducing hydrogenase deletion mutant | Hendrickson and Leigh, 2008 |
| Mm1183 | ΔfrcA | Mmp0820 | Cysteine-containing F420-reducing hydrogenase deletion mutant | Hendrickson and Leigh, 2008 |
| Mm1184 | ΔfrcAΔfruA | Mmp1382Mmp0820 | Double mutant of fruA and frcA | Hendrickson and Leigh, 2008 |
| Mm1020 | Δmtd | Mmp0372 | F420-dependent methylenetetrahydromethanopterin dehydrogenase deletion mutant | Hendrickson and Leigh, 2008 |
| Mm1097 | Δhmd | Mmp0127 | Hydrogen-dependent methylenetetrahydromethanopterin dehydrogenase deletion mutant | Hendrickson and Leigh, 2008 |
| Mm1002 | Δald | Mmp1513 | Alanine dehydrogenase deletion mutant | Moore and Leigh, 2005 |
| Mm1018 | ΔagcS | Mmp1511 | Sodium:alanine symporter deletion mutant | Moore and Leigh, 2005 |
| Desulfovibrio vulgaris strain | ||||
| Hildenborough (=ATCC 29 579) | NA | NA | Wild-type strain | ATCC |
Abbreviations: NA, not applicable.
Biomass production
Three biological replicates of cocultures and hydrogen-limited M. maripaludis monocultures were grown in chemostats as previously described (Haydock et al., 2004; Walker et al., 2009). Briefly, cocultures were grown on 30 mℳ lactate in previously described coculture medium (CCM) (Walker et al., 2009). M. maripaludis monocultures were grown in Bioflo 110 bioreactors (1.3 l vessel capacity; New Brunswick Scientific Co., Edison, NJ, USA) on modified CCM containing 30 mℳ acetate instead of lactate. Sodium sulfide was replaced in the medium with sparging (approximately 13 ml min−1 of a 1% H2S:N2 mixture) of hydrogen sulfide gas. Chemostat setup and the medium and gas delivery systems were identical to a previously described system (Haydock et al., 2004).
A 1-ml glycerol stock of previously grown coculture or monoculture was used to inoculate 100 ml of CCM (amended with sulfate for monocultures) in a 200 ml serum vial. Cultures were incubated in the dark at 37 °C with a shaking speed of 250 r.p.m. When the cultures reached an OD600 of 0.25–0.30, they were transferred to a 3-l FairMenTec chemostat (FairMenTec GmbH, Göttingen, Germany) filled with 2 l of CCM. The chemostat was run in batch mode at 37 °C with a stirring speed of 250 r.p.m. N2:CO2 (90:10) was flushed through a sterile cotton plug before entering the headspace of the reactor. Flow rate was maintained at 0.25 ml min−1 using an Alicat Scientific mass controller (MC-20SCCM-D; Alicat Scientific, Tucson, AZ, USA). Headspace concentrations of CH4, CO2, H2, H2S, O2 and N2 were monitored at 30 min intervals using a Hiden Analytical QIC-20 mass spectrometer (Hiden Analytical, Warrington, UK). For coculture experiments, a culture medium flow rate of 1.3 ml min−1 (25.6 h retention time) was initiated when the OD600 reached 0.325 –0.350. The chemostat was assumed to be at steady state when the variance of OD600 readings was <10% over three retention periods. Cultures were harvested through ice-chilled sterile stainless steel tubing connected to the chemostat culture medium exhaust line. Falcon tubes (50 ml) were stored in an anoxic chamber and pre-chilled on ice prior to harvesting. The tubes were centrifuged at 4 °C for 15 min at 3220 g, after which the supernatant was poured off and the tubes were immediately frozen at –80 °C. Samples were shipped overnight on dry ice.
Transcriptional analysis
Whole-transcriptome microarrays containing 70-mer probes for each of the 1722 M. maripaludis S2 open reading frames were spotted on UltraGAPS glass slides (Corning Life Sciences, Corning, NY, USA) using a BioRobotics Microgrid II arrayer (Genomic Solutions, Ann Arbor, MI, USA). Each slide also contained 70-mer probes for 3531 D. vulgaris Hildenborough open reading frames. Each slide had duplicate spots for each open reading frame. Each biological replicate was hybridized to at least three slides. Thus, each log2 expression level descried here was obtained using triplicate biological replicates/slides, for each of which there were duplicate on-chip technical replicates.
RNA isolation, quantification and transcription were performed as previously described (Walker et al., 2009) and fluorescently labeled using Cy5-dUTPs. Labeled RNA was compared against Cy3-dUTP-labeled genomic DNA and computational analyses were performed (Walker et al., 2009). For log2 R calculations, R=signal intensity ratio of coculture/monoculture. Z-values were calculated as described in Mukhopadhyay et al. ( 2006). In this study, genes with absolute Z-score values >1 were considered significantly changed. Gene-expression data are available at Microbes Online (www.microbesonline.org) and under GEO reference GSE30831, GSM764979 and GSM764978.
Protein preparation, labeling and analysis
Cell pellets from biological triplicates were pooled into 1 ml of lysis buffer (500 mℳ triethylammonium bicarbonate with 4 ℳ urea, pH=7). The samples were lysed by sonication on ice for 3 min of active time with pulses of 5 s on and 10 s off. Because of high amounts of DNA associated with the M. maripaludis samples, 40 μl of RQ-1 DNAse (1U μl−1; Promega, Madison, WI, USA) was added to each sample and 1 ℳ MgCl2 was added to a final concentration of 10 mℳ. The samples were set at 37 °C for 1.5 h, after which the samples were clarified by centrifugation at 10 000 g for 30 min at 4 °C. Protein concentration was determined using the Biorad Assay (Biorad, Hercules, CA, USA). To provide a control for the coculture, equal amounts of protein were mixed from M. maripaludis and D. vulgaris, referred to hereafter as synthetic blend. The iTRAQ (isobaric tags for relative and absolute quantitation) labeling was carried out as previously described (Redding et al., 2006) and samples were labeled as follows: tag114–coculture; tag115–synthetic blend; tag116–synthetic blend; and tag117–coculture. This strategy allowed two technical replicates for each of the samples, increasing confidence for proteins that show differential expression. All the collected data were processed using Protein Pilot (AB SCIEX, Framingham, MA, USA). The data is computed as ratios, so as to provide relative change. Because the actual ratio of D. vulgaris to M. maripaludis in the coculture was not 1:1, normalization of the fold change (coculture versus synthetic blend) was done based on the cellular ratio data obtained using DAPI (4'6-diamidino-2-phenylindole)-stained cell counts, which indicated that the true ratio was 80:20 (Walker et al., 2009). For D. vulgaris proteins, the fold change was normalized by taking the log2 ratio of the coculture to synthetic blend and subtracting the log2 value of 80/50, whereas the M. maripaludis fold change was normalized by subtracting the log2 value of 20/50. A detailed explanation of this normalization strategy and the complete proteomics data set are provided in the Supplementary Information and Supplementary Table S1. Normalized protein log2 ratios >|2| were considered to be significantly changed.
Metabolite analysis
Cell pellets and supernatants from 50 ml cultures were collected (in triplicate) for cocultures and monocultures at OD600 ∼0.3. Metabolites were extracted from both pellets and supernatant using methanol extraction and lyophilized as described previously (Baidoo et al., 2008). Briefly, 2 ml pre-chilled methanol was used to extract metabolites from pellet, whereas 1 ml supernatant was extracted with 1 ml methanol. A total of 30 ml water (deionized high-performance liquid chromatography grade) was added to extracts and frozen in liquid N2 and lyophilized. All reagents used were high-performance liquid chromatography grade. Capillary electrophoresis (CE) separation and mass spectrometry (MS) analysis were conducted as previously described (Baidoo et al., 2008). The standard concentration curves using commercially available standards of pyruvate and alanine (Sigma, St Louis, MO, USA) were used to obtain absolute levels of target metabolites in pmol mg−1 dry cell weight.
Phenotypic growth analyses
All the phenotypic growth assays were carried out in 17 ml Hungate tubes (BellCo Glass, Vineland, NJ, USA) equipped with rubber stoppers and crimp-tops. Cultures were incubated at 37 °C in the dark with a 300 r.p.m. shaking speed. Each tube contained 8 ml of CCM amended with 30 mℳ of electron donor (lactate or pyruvate). The headspace contained an overpressure of 18 kPa of N2:CO2 (80:20). Dilution series out to 10−8 were initiated from 1 ml glycerol stocks of each Methanococcus mutant and D. vulgaris. Methanococcus cultures were grown in CCM lacking lactate and amended with 5 mℳ acetate and 250 kPa overpressure of H2:CO2 (80:20). Desulfovibrio cultures were grown in CCM amended with 30 mℳ sulfate. Cocultures were established by combining 0.5 ml of exponentially growing Methanococcus and 1 ml of exponentially growing Desulfovibrio from the highest dilutions. Cocultures were transferred (1% v/v) three times to ensure dilution of any residual sulfate/acetate or H2 before inoculating triplicate tubes for growth experiments. Tubes were monitored for growth using OD600 readings blanked against uninoculated medium.
Results
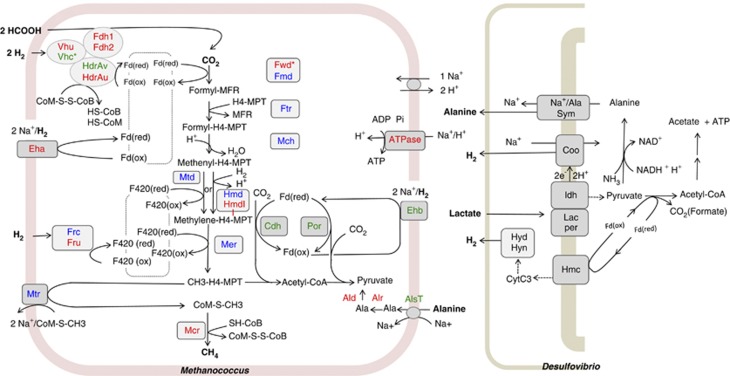
The most general change associated with syntrophic growth was a decrease in transcripts for energy-consuming biosynthetic functions (for example, pyruvate oxidoreductase (Por), acetyl-CoA decarbonylase/synthase and energy-conserving hydrogenase B (Ehb), see Figure 1 for enzymatic reaction depictions) and an increase in transcripts in the energy-generating methanogenesis pathway (see Table 2 and Supplementary Tables S2 and S4; Supplementary Figure S1) plots transcriptional changes according to function as defined by clusters of orthologous groups). Figure 1 illustrates the differential expression of transcripts observed in coculture compared with hydrogen-limited M. maripaludis monocultures. However, compared with growth in monoculture under hydrogen limitation, transcript levels for isofunctional genes often diverged, with transcripts of one variant significantly increasing, whereas the other was little changed or significantly decreased in their relative abundances. A common feature was an apparent increase in transcripts for functions using H2 directly as reductant, versus those using the reduced deazaflavin (coenzyme F420). In several cases, the proteins were confidently identified (Table 3 and Figure 2) and the corresponding changes generally corroborated the observations at the transcript level. The greater importance of direct reduction by H2 was supported by improved syntrophic growth of a deletion mutant in an F420-dependent dehydrogenase of M. maripaludis (described in more detail below). Metabolite, transcript and proteomic analyses also pointed to a unique, although undefined, role for alanine utilization within this syntrophic coupling.
Figure 1.
Conceptual schematic of M. maripaludis and D. vulgaris syntrophic interaction, highlighting the central energy-generating and -consuming the pathways of the methanogen. Relative changes (Table 2) in transcript abundance during syntrophic growth are indicated by red (increase) and green (decrease) coloration. Blue coloration indicates no statistically significant change as specified in the Materials and methods section. Hydrogen-limited monocultures served as the control growth condition. Oxidation of formate to CO2 and H2 coupled with coenzyme F420-reduction (not depicted) is catalyzed by two alternative formate dehydrogenases, Fdh1 and Fdh2. One of two membrane-bound energy-conserving hydrogenases (Eha and Ehb) couple the chemiosmotic energy of ion gradients to H2 oxidation and ferredoxin reduction. Of these, Ehb generates the low potential electron carrier used for anabolism, whereas Eha is hypothesized to function primarily in the energy-generating methanogenesis pathway, generating low potential-reducing equivalents for the reduction of CO2 to formylmethanofuran (Major et al., 2010). Two different formylmethanofuran dehydrogenases catalyze this first step in methanogenesis, tungsten (Fwd) and molybdenum (Fmd) forms. Transfer of the formyl group from methanofuran to methanopterin by Ftr and subsequent elimination of H2O by Mch yields methenyl-H4-methanopterin. Two different enzymes can then reduce methenyl-H4-MPT to methylene-H4MPT, one (Mtd) using H2 as reductant and the other (Hmd) using reduced coenzyme F420. M. maripaludis has an Hmd paralog of unknown function (Mmp1716, HmdII) that may also function in this step (Hendrickson et al., 2004). Reduction of methylene-H4MPT by another F420-dependent reductase (Mer) yields methyl-H4MPT. The reduced coenzyme F420 required for the formation of methyl-H4MPT by these two steps is generated by one of two alternative F420-reducting hydrogenase (Fru and Frc). The final steps to methane production are catalyzed by a methyl transferase (Mtr) and a reductase (Mcr) coupled to two forms of a F420-nonreducing hydrogenase (Vhu and Vhc). The mixed disulfide (CoM-CoB) produced by reduction of methyl coenzyme M is then reduced by one of two forms of the heterodisulfide reductase determined by the composition of the HdrA subunit (HdrAU or HdrAV). Fdh/ Hdr/Vhu/ Fwd are reported to have protein–protein interactions (Costa et al., 2010). (Note: The interaction with Fwd could not be depicted here without compromising the clarity of the figure. Vhc is not part of this interaction). Other reactions include the transport of alanine (AlsT), and subsequent coversion to pyruvate via an alanine racemase (Alr) and dehydrogenase (Ald). Additional M. maripaludis proteins shown: pyruvate oxidoreductase, acetyl-CoA decarbonylase/synthase. The D. vulgaris metabolic pathway is based upon results as described in Walker et al., 2009 and is updated to include an unspecified sodium/alanine transporter (Na+/ala sym). Other D. vulgaris proteins shown: lactate permease (lac per), lactate deydrogenase (ldh), membrane-bound Coo hydrogenase (Coo), high-molecular weight cytochrome (Hmc), periplasmic hydrogenases (Hyd and Hyn), cytochrome c3 (Cyt c3) and oxidized and reduced ferredoxin (Fd).
Table 2. Expression ratios of selected genes in the central energy-generating and -consuming pathways of M. maripaludis.
| Gene number | ID |
Coculture versus H2-limited monoculture |
H2-limited versus Pi-limiteda |
||
|---|---|---|---|---|---|
| log2Rb | Z-score | log2Rb | P-value | ||
| F420-interacting proteins | |||||
| Mmp0058 | mer | n.a. | n.a. | 2.51 (5.7) | 4.80E-11 |
| Mmp0372 | mtd | 0.55 (1.5) | 0.79 | 4.14 (17.6) | 2.30E-10 |
| Mmp1298 | fdhAI | 1.70 (3.2) | 2.22 | 2.26 (4.8) | 1.50E-04 |
| Mmp0138 | fdhA2 | 1.54 (2.9) | 0.77 | 0.41 (1.3) | 7.60E-03 |
| Mmp1385 | fruB | 3.29 (9.8) | 2.49 | 1.83 (3.6) | 3.00E-09 |
| Mmp0818 | frcG | −1.50 (0.35) | 0.79 | 0.16 (1.1) | 7.30E-03 |
| Methanogenesis pathway proteins (energy-generating) | |||||
| Mmp0127 | hmd | −1.15 (0.45) | 0.75 | −0.20 (0.87) | 2.60E-01 |
| Mmp1716 | hmdII | 1.04 (2.1) | 1.43 | −0.09 (0.93) | 7.40E-03 |
| Mmp1155 | hdrB1 | 0.55 (1.5) | 0.94 | 0.22 (1.2) | 1.50E-02 |
| Mmp1053 | hdrB2 | 1.10 (2.1) | 1.5 | 0.46 (1.4) | 9.60E-04 |
| Mmp1697 | hdrAu | 0.81 (1.7) | 1.12 | 0.40 (1.3) | 3.30E-03 |
| Mmp0825 | hdrAv | −1.09 (0.47) | 1.05 | 0.23 (1.2) | 8.60E-04 |
| Mmp1559 | mcrA | 1.31c (2.5) | 1.95 | −0.30 (0.81) | 6.70E-03 |
| Mmp1609 | ftr | 0.10 (1.1) | 0.18 | 0.42 (1.3) | 6.90E-07 |
| Mmp1448 | ehaA | 0.91d (1.9) | 1.36 | 0.24 (1.2) | 1.20E-02 |
| Mmp1696 | vhuD | 1.61 (3.0) | 2.31 | n.a. | n.a |
| Mmp0822 | vhcG | −2.12 (0.23) | 1.26 | 0.12 (1.1) | 2.30E-02 |
| Mmp1567 | mtrE | 1.05 (2.8) | 1.43 | −0.65 (0.64) | 4.60E-01 |
| Mmp1247 | fwdD | 0.93 (1.9) | 1.15 | 0.12 (1.1) | 3.00E-01 |
| Anabolic pathway proteins (energy-consuming) | |||||
| Mmp0984 | cdh | −1.39 (0.38) | 1.94 | 0.43 (1.3) | 4.30E-06 |
| Mmp1504 | porB | −0.86 (0.55) | 1.33 | 0.38 (1.3) | 7.50E-06 |
| Mmp1622 | ehbM | −1.09e (0.47) | 1.46 | 0.66 (1.6) | 5.00E-08 |
| Mmp1512 | alr | 2.74 (6.7) | 2.85 | −0.73 (0.60) | 3.30E-10 |
| Mmp1513 | ald | 3.85 (14.4) | 2.63 | −0.55 (0.68) | 5.50E-09 |
Data from Hendrickson et al. (2007).
Values in parantheses are the R values.
Data available only for mcrD (Mmp1556).
Mean value for ehaACD. Data not available for ehaBEFGHIJKLNO (low Z-scores).
Mean value for ehbBCDNFG. Data not available for ehbOMLKJL (low Z-scores).
Table 3. Normalized relative ratios for significantly changed M. maripaludis proteins.
| Gene number | ID | Description | Log2 (coculture/synthetic coculture)a |
|---|---|---|---|
| MMP1382 | FruA | Selenocystein-containing coenzyme F420-reducing hydrogenase, alpha subunit | 2.93 |
| MMP0802 | FrcA | Coenzyme F420-reducing hydrogenase, alpha subunit | 1.92 |
| MMP0817 | FrcB | Coenzyme F420-reducing hydrogenase, beta subunit | 2.09 |
| MMP1301 | FdhC | Formate dehydrogenase, alpha subunit | 2.03 |
| MMP1513 | ald | Alanine dehydrogenase | 3.46 |
| MMP1302 | Hypothetical protein MMP1302 | 2.18 | |
| MMP1156 | Hypothetical protein MMP1156 | 2.18 | |
| MMP1161 | Hypothetical protein MMP1161 | 2.08 |
ITRAQ proteomics data normalized as described in supplementary section based on D. vulgaris: M. maripaludis ratio of 4: 1. Data average of technical replicates.
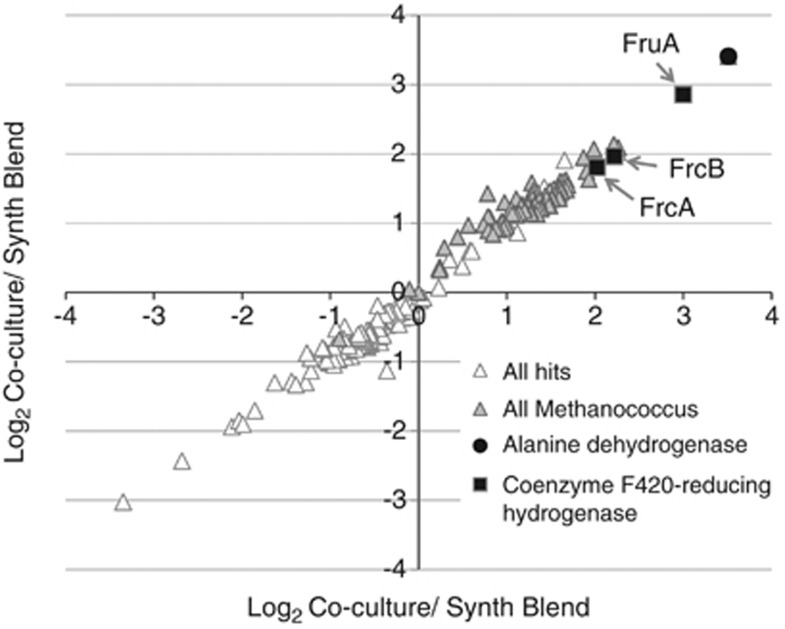
Figure 2.
Quantitative proteomic data. Analysis of coculture protein samples using iTRAQ labeling and shotgun liquid chromatography–mass spectrometry methods identified 82 proteins from M. maripaludis (out of 207 total coculture proteins) by at least two unique, high-confidence peptides in replicate runs. Among these 82 M. maripaludis proteins (Supplementary Table S1), 8 exhibited significant abundance increases (Table 3). Plot shows log2 ratios of the iTRAQ ratios of proteins from coculture versus the synthetic blend. The synthetic blend contains a 50:50 mixture of D. vulgaris to M. maripaludis, whereas the coculture is a 80:20 mixture of the same. Normalized data was used. For complete data see Supplementary Table S1.
Carbon assimilation and methanogenesis
A number of genes associated with carbon assimilation and biosynthesis displayed differences in transcript abundance during syntrophic growth. Most notably, lower transcript abundance was observed for many genes coding for subunits of Ehb (ehbC, ehbD, ehbA, ehbN and ehbF, Table 2), the energy-conserving hydrogenase proposed to be involved in CO2 assimilation (Porat et al., 2006), and genes coding for acetyl-CoA decarbonylase/synthase (Mmp0979-85, Supplementary Table S2) and Vor (Mmp1271-2, Supplementary Table S2), both associated with autotrophic growth. Fewer transcripts were also observed for the adenosine diphosphate-forming acetyl-CoA synthetase (acd, Mmp0253, Supplementary Table S2), one of the two enzymes responsible for acetyl-CoA production via acetate.
Transcripts for genes encoding a formate dehydrogenase (fdh)(Mmp1297-8, Table 2), previously shown to be important in both monoculture (Wood et al., 2003) and coculture (Stolyar et al., 2007), were increased. Increased transcript abundance was accompanied by a significant increase in the protein abundance of the FdhC subunit (Figure 1, Table 3). Additionally, genes for an associated transporter and carbonic anhydrase (Mmp1299-301, Supplementary Table S2) displayed transcript increases. Despite the observed increases in both transcript and protein levels, formate was not detected in the culture medium (limit of detection, 0.1 mℳ).
Differential expression was observed for several of the genes coding for proteins involved either directly or indirectly in the seven steps in hydrogenotrophic methanogensis. Transcripts of the mch gene (Mmp1191) and most of the mtr operon (Mmp1560-7, Table 2) increased slightly, as did transcripts for a single gene in the methyl coenzyme M reductase operon (mcrC, Mmp1556, Table 2). The small increase in the transcript levels for the mtr and mcr operons were not reflected in the proteomics data (Supplementary Table S3).
Hydrogenases
The M. maripaludis genome encodes seven potential hydrogenases involved in the various pathways whose expression depends upon growth and nutrient conditions. Two of the hydrogenases (the Eha and Ehb complexes) are membrane-bound and function during ferredoxin reduction and energy conversion. In contrast to decreased transcripts observed for several Ehb subunits, a small number of the genes from the energy-conserving hydrogenase complex Eha showed increased transcript levels (Mmp1447; ehaA, Mmp1448; ehaC, Mmp1450; and ehaD, Mmp1451, Table 2), although no statistically significant differences were observed for the remaining Eha subunits.
Of the two isofunctional cytoplasmic F420-reducing hydrogenases (Frc and Fru), transcripts for the selenocysteine-containing F420-reducing hydrogenase (Fru, Mmp1382-5, Table 2) increased during syntrophic growth, a phenomenon also observed under hydrogen limitation when selenium is present in the culture medium (Noll et al., 1999; Hendrickson et al., 2007; Baidoo et al., 2008). A corresponding increase in the FruA (Table 3, Figure 2) protein was also observed. Transcripts for the cysteine-containing F420-reducing hydrogenase (Frc, Mmp0817-20) did not show a statistically significant change, except for a reduced transcript level for a gene coding for FrcG (Table 2). However, increases in protein abundance were measured for two of the Frc subunits (FrcA and FrcB, Table 3, Figure 2).
A second set of isofunctional enzymes, the F420 non reducing hydrogenases have been proposed to transfer electrons from H2 to the heterodisulfide complex formed in the last steps of methanogenesis (Afting et al., 2000). Transcript levels for genes coding for the cysteine-containing coenzyme F420-nonreducing hydrogenase were decreased (Vhc, Mmp0821-3). The heterodisulfide reductase subunit HdrAV, predicted to be within the same operon, also showed a decrease at the transcript level (Mmp0825, Table 2). In contrast, genes coding for the Vhu and the HdrAU subunit of Hdr showed an increase in transcript levels (Table 2). No significant changes were measured for VhcA,VhcD (Mmp0823, Mmp0821) and Vhu (Mmp1694) at the protein level (Supplementary Table S3).
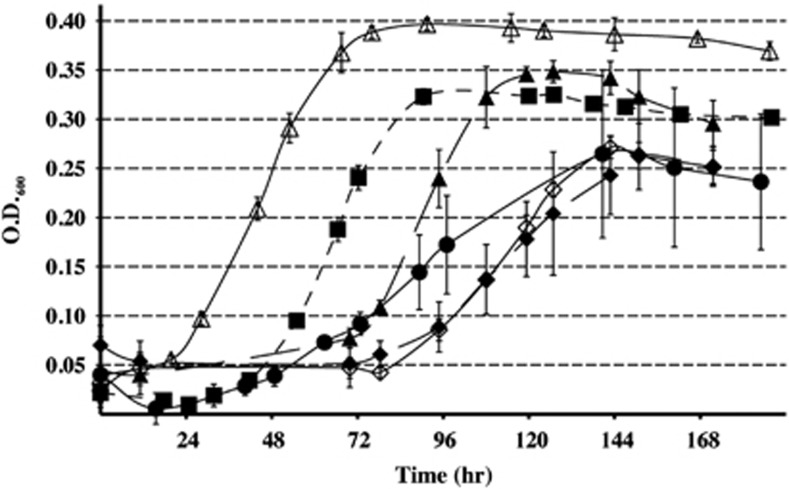
Given the significant increases of both transcript and protein abundance, the roles of the Fru/c hydrogenases were examined using the corresponding gene deletion mutants in syntrophic growth with D. vulgaris. The ΔfruA and ΔfrcA mutants each exhibited diminished growth rates and maximum cell densities during syntrophic growth on lactate (Figure 3). A deletion in both genes (a ΔfruΔfrcA mutant) produced highly variable maximum cell densities, but syntrophic growth still occurred, albeit at generally slower rates than in wild-type cocultures. All the three mutants demonstrated an increase in lag time prior to initiating exponential growth. In contrast, when pyruvate was the substrate for syntrophic growth, there were no differences in growth rate or maximum cell densities between the wild-type and the ΔfruA or ΔfrcA mutants, and there was only a slight decrease in maximum cell density for the ΔfrcAΔfruA double mutant (Supplementary Figure S2).
Figure 3.
Growth curves for wild-type and mutant M. maripaludis cultures on lactate. The error bars indicate s.d. of triplicate cultures. Wild type (filled squares); Δmtd (open triangles); Δhmd (closed triangles); ΔfruA (open diamonds); ΔfrcA (closed diamonds); and ΔfrcAΔfruA (closed circles).
Although there were no statistically significant changes in transcript or protein, abundance for either the hydrogen-dependent (Hmd, Mmp0127) or the F420-dependent (Mtd, Mmp0372) methylene-H4MPT dehydrogenases, both the Δmtd and Δhmd mutant strains, affected syntrophic growth. Unexpectedly, the Δmtd strain growing in syntrophic association on lactate had a reduced lag time and achieved a greater maximum cell density than the wild type (Figure 3). Thus, restricting this step in methanogenesis to a hydrogen-dependent enzyme (Hmd) improved overall growth performance of the coculture, possibly by enhancing Desulfovibrio growth by maintaining a lower concentration of H2. Although the Δhmd strain showed a longer lag period, there was no difference in maximum cell density. However, deletion of hmd (Mmp0127) may be compensated for by its paralog hmdII (Mmp1716). These genes responded differentially to syntrophic growth, with a two-fold increase in Mmp1716 transcripts and a reduction in Mmp0127 transcripts (Table 2). As with the ΔfruA or ΔfrcA mutants, no observable differences in growth rate or maximum cell density were noted for either the Δmtd or the Δhmd mutant strains compared with the wild type when grown in coculture using pyruvate as carbon source (Supplementary Figure S2).
Alanine utilization
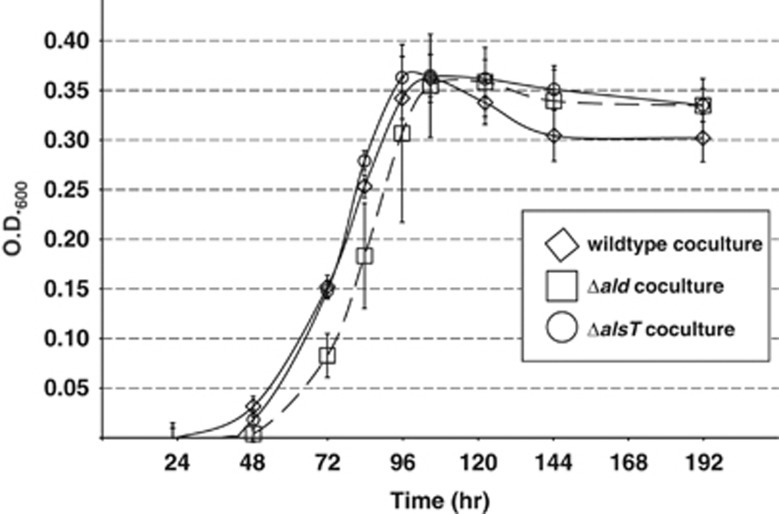
Transcript levels for alanine dehydrogenase (ald) and alanine racemase (alr) genes in the methanogen (both involved in alanine utilization) were greatly elevated during syntrophic growth, with corresponding increases in protein abundances in the case of ald (Tables 2 and 3, Figure 2). The associated alanine transport gene, annotated as a sodium:alanine symporter (alsT, Mmp1511), exhibited a significant downregulation at the transcript level that may have resulted from repression by the nrpR nitrogen-regulation gene (Xia et al., 2009). AlsT was not identified in the proteomics data set. Previous characterization of the agcS (alsT) and ald deletion mutants indicated that these genes are essential when alanine is the sole source of nitrogen (Moore and Leigh 2005). However, in an ammonia-containing lactate medium, no significant differences in growth rate or cell density were observed for either deletion mutant in coculture as compared with wild type (Figure 4). Nonetheless, because the alanine dehydrogenase and racemase transcripts were among those most highly elevated in coculture, we further examined intra- and extracellular concentrations of alanine and its conversion product, pyruvate. Concentrations were determined for both cocultures and monocultures of the ΔalsT mutant and wild-type M. maripaludis. Values for cocultures reflect aggregate contributions from both D. vulgaris and M. maripaludis. Notably for both co- and monocultures, the cultures containing the mutant strain had significantly higher intra- and extracellular concentrations of both alanine and pyruvate (Table 4). Additionally, higher internal concentrations of both metabolites were observed in wild-type monocultures of M. maripaludis when compared with equivalent amounts of coculture cells.
Figure 4.
Growth curves for wild-type and alanine-related mutant M. maripaludis cultures on lactate. The error bars indicate s.d. of triplicate cultures.
Table 4. Measured intra- and extracellular alanine and pyruvate concentrations. Alanine and pyruvate concentrations (pmol/mg of dry cells) observed for cocultures (CC) and monocultures of M. maripaludis (MmS) and D. vulgaris (DvH). Wild type (WT) or alanine transporter deficient (ΔalsT) strains were used.
|
Average |
s.d. |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CCWT | CCΔalsT | MmSWT | MmSΔalsT | DvHWT | CCWT | CCΔalsT | MmSWT | MmSΔalsT | DvHWT | |
| Internal alanine | 0.13 | 1.60 | 0.58 | 2.46 | 7.16 | 0.04 | 0.08 | n.a. | 0.74 | 0.72 |
| External alanine | 82.27 | 442.61 | 382.8 | 772.14 | 1395.98 | 58.52 | 43.67 | n.a. | 135.32 | 109.36 |
| Internal pyruvate | 0.18 | 0.85 | 0.22 | 0.34 | 2.53 | 0.03 | 0.02 | n.a. | 0.09 | 0.8 |
| External pyruvate | 4.74 | 25.00 | 10.43 | 20.32 | 34.49 | 0.36 | 5.38 | n.a. | 0.93 | 3.42 |
Abbreviation: n.a., not applicable.
Discussion
Syntrophic growth in the Desulfovibrio/Methanococcus model community requires that the two participating organisms share the free energy available from the fermentation of lactate. Assuming that molecular hydrogen is the primary mediator of reduced metabolite exchange, the fraction of energy available to each organism is determined primarily by H2 concentration. A low H2 concentration favors the Desulfovibrio, but limits the methanogen. Conversely, a high H2 concentration favors the methanogen, but limits the Desulfovibrio. Although during steady-state growth the H2 concentration must be maintained at a concentration that satisfies the energetic needs of both organisms, the energy need not be divided equally (Worm et al., 2011). Using the average concentrations measured during the steady-state growth experiments (4 mℳ lactate, 26 mℳ acetate, 2.5 Pa H2, 5,100 Pa CO2 and 61 Pa CH4; temperature 310 K), the total free energy available from lactate fermentation to methane, acetate and CO2 (−82.8 kJ mol−1) is not equally shared between Desulfovibrio (∼ 60 KJ mol−1) and Methanococcus (∼20 kJ mol−1). Thus, the methanogen appears to be close to the thermodynamic threshold for growth defined by the minimum increment of energy (−15 to −20 kJ mole−1) required for adenosine triphosphate synthesis (Schink and Stams, 2006). In the current study, we examined the adaptive response of the methanogen to the energetic constraints of extreme hydrogen limitation imposed by syntrophic growth.
All the transcription data were related to Methanococcus growing under hydrogen limitation in chemostats in metal-replete medium at the same generation time as the coculture, as previously reported by Walker et al. (2009). In that study, we showed that an alternative electron-transfer system was required for lactate oxidation by the Desulfovibrio when growing in syntrophic association with Methanococcus, but not for respiratory growth on sulfate (Walker et al., 2009). Thus, it appeared that specific enzyme systems are dedicated to Desulfovibrio growth under conditions of syntrophy. In the current study, we observe changes in Methanococcus gene expression that also appear of greater relevance to syntrophic growth. Relative to growth under hydrogen limitation in monoculture, the syntrophically grown methanogen showed increased transcription of genes in the central pathway for methanogenesis (mtrH, mcrA and fwdD) and of paralogs coding primarily for different steps in hydrogen uptake (ehaA, hmdII, fruB, vhuD and hdrAU). Notably, transcription of paralogs previously reported to be upregulated with hydrogen limitation in monoculture (Hendrickson et al., 2007) was significantly reduced in coculture (hmd, hdrAV, vhcG and frcG). Thus, different paralogs presumably provide physiological advantages at different H2 concentrations. An increase in hmdII levels in coculture, and a decrease in the levels of its paralog hmd, is particularly noteworthy as HmdII was reported to not function as a hydrogenase/dehydrogenase based on in vitro characterization of the Methanothermobacter marburgensis paralogs (Afting et al., 2000). Because the Hmd hydrogenase has >20-fold higher Km for H2 (0.2 mℳ) than the F420-reducing (NiFe)-hydrogenase (0.01 mℳ), and because the paralog competitively binds a Fe-binding guanylylpyridinol cofactor required by Hmd, it was suggested that the paralog functions primarily to store the cofactor during hydrogen limitation (Goldman et al., 2009; Thauer et al., 2010). However, our observations of downregulation of hmd and coincident upregulation of hmdII during hydrogen-limited syntrophic growth now point to a more direct function in hydrogen metabolism. Direct involvement of HmdII in the reduction of methenyl-H4-MPT is also supported by the unexpected improvement in syntrophic growth of the mtd deletion mutant relative to the wild type (Figure 3), suggesting that HmdII variant is functionally relevant in the reduction of methenyl-H4-MPT at extremely low H2 concentrations (1 Pa).
The upregulation of fruB and the corresponding increase in Fru protein abundance points to the challenge of syntrophically grown M. maripaludis in maintaining reduced coenzyme F420 at low H2 concentrations (Nolling et al., 1995; Morgan et al., 1997; Hendrickson et al., 2007). This is also consistent with the poor growth in coculture of the ΔfrcA and ΔfruA mutant strains. An impaired capacity of these mutants to link hydrogen oxidation with the reduction of F420 is the likely basis for reduced growth rate and cell densities observed in coculture, but not in monoculture (Hendrickson et al., 2008). Importantly, the growth of the ΔfrcAΔfruA mutant in coculture indicates that the previously suggested reverse Hmd-Mtd pathway for coenzyme F420-reduction (Hendrickson et al., 2008) may be sufficient to support syntrophy, although the other unknown pathways cannot be ruled out.
The increased transcription of genes coding for Vhu and HdrAU, paralogs of the heterodisulfide reductase complex, and decrease in Vhc and HdrAV is consistent with formation of a complex between Fdh, Fwd, Vhu and HdrAu in metal-replete medium (Berghofer and Klein 1995; Costa et al., 2010). Within this complex, a flavin-mediated electron bifurcation at HdrA results in reduction of both the CoM-S-S-CoB heterodisulfide and ferredoxin required by Fwd for the first step in methanogenesis. In the presence of formate, or during hydrogen limitation, expression of fdh increases and the protein is incorporated into the protein complex to facilitate electron transfer from formate to Hdr (Costa et al., 2010). The coordinate increase in transcription of the genes coding for all members of this complex (fwd, hdrAU, vhu and fdh) also suggests a requirement for increased expression of the complex under conditions of hydrogen limitation imposed by syntrophic growth.
The prominent increase in alanine metabolism genes highlights a potentially unexplored mechanism of syntrophic coupling. M. maripaludis is capable of utilizing alanine as a nitrogen source, with the dehydrogenase and transporter being essential when alanine is the sole nitrogen source, but not in the presence of ammonia (Moore and Leigh, 2005). Production of alanine by D. vulgaris via reduction of pyruvate (presumably by an alanine dehydrogenase) could complement H2 as a mediator of interspecies electron transfer. This form of reduced product exchange would also benefit the methanogen by providing fixed carbon and nitrogen. A variety of Methanococcus strains have capacity to assimilate alanine (Whitman et al., 1987). However, no appreciable differences in growth rates or yields were observed relative to the wild type for cocultures established with either the M. maripaludis Δald or ΔalsT mutants. This might mean that the cost of alanine secretion to Desulfovibrio is compensated for by an energy advantage provided by the methanococcus, such that changes in growth rate or yield are not discernable by the aggregate measure of optical density. Desulfovibrio would forfeit one adenosine triphosphate for each pyruvate not further oxidized to acetate, whereas Methanococcus would benefit from a reduced energy investment in the autotrophic synthesis of pyruvate. However, another feature of alanine export via a Na+/alanine symporter would be the coincident export of Na+, contributing to a sodium motive force that might be used to drive energetically unfavorable reactions (Figure 1). Although it is also possible that D. vulgaris uses alanine primarily as a compatible osmolyte to alleviate salt stress imposed by the brackish CCM (He et al., 2010) and Methanococcus benefits from alanine leakage, the data are most consistent with a flux of alanine between the two species. First, transcripts of genes for autotrophic growth (por, ehb and cdh) are all reduced in coculture relative to H2-limited monoculture. Second, when compared with monocultures and ΔalsT mutant strains, the lowest concentration of internal alanine was observed for the wild-type coculture sample. Finally, the introduction of the ΔalsT mutant to the coculture, preventing uptake of alanine by M. maripaludis, resulted in a greater concentration of both external and internal alanine.
Our study also adds to a number of previous theoretical and experimental studies of the metabolic basis of syntrophic association. Those studies primarily examine alternative mechanisms for generating hydrogen or formate from the high-potential oxidation reactions mediated by the bacterial syntroph. It is now apparent that syntrophs use a variety of mechanisms to couple exergonic reactions with energetically unfavorable oxidation of substrates such as propionate, butyrate or lactate (McInerney et al., 2009; Stams and Plugge 2009; Walker et al., 2009; Müller et al., 2010). Fewer studies have considered both the metabolism and gene expression of syntrophically associated methanogens (Luo et al., 2002, Enoki et al., 2011; Worm et al., 2011). However, the use of phylogenetically divergent methanogens in these studies complicates direct comparison with our results. Worm and colleagues focused on changes in expression of alternative fdh and hydrogenases (hyd) in Methanospirillum hungatei growing in syntrophic association with Syntrophobacter fumaroxidans, showing that fdh and hyd genes were transcribed in both organisms in either monoculture or coculture. Notably, although expression levels did vary with culture conditions, the fdh and hyd genes in M. hungatei were both transcribed in monoculture on either formate or hydrogen (Worm et al., 2011). Thus, the increased transcription of fdh genes we observed in M. maripaludis during syntrophic growth may not be in direct response to production of formate by D. vulgaris. In other studies, proteome and transcript analysis of Methanothermobacter thermautotrophicus in coculture with a fatty acid oxidizing syntroph showed primarily differential expression of two variants of the methyl coenzyme M reductase (Mcr), one variant (MCRI) was preferentially expressed in coculture and the other (MCRII) in monoculture. However, M. maripaludis contains only a single variant of Mcr.
These reports and our study point to a tremendous variety of strategies for the metabolic coupling of different syntrophic partners. The increased transcription of paralogs feeding electrons derived from hydrogen into the methanogenesis pathway and indications for novel exchange of alanine between species observed in our study seem to be of significance to the adaptive response and habitat preference (niche) of Methanococcus species. Thus, there is now added impetus for studies designed to further evaluate the biochemical properties and physiological significance of the adaptive changes observed in this syntropic pair, and more generally how different syntrophic pairings influence anaerobic food-web structure and function.
Acknowledgments
We thank Professor Michael J McInerney (University of Okhlahoma) for valuable discussion and review of an earlier draft of this paper, Dr Christopher Petzold (LBNL) for help with iTRAQ data analysis on Protein Pilot and Jason Baumohl (LBNL) for help with submitting data to the GEO database. This work is part of ENIGMA, a Scientific Focus Area Program supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomics: GTL Foundational Science through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Afting C, Kremmer E, Brucker C, Hochheimer A, Thauer RK. Regulation of the synthesis of H2-forming methylenetetrahydromethanopterin dehydrogenase (Hmd) and of HmdII and HmdIII in Methanothermobacter marburgensis. Arch Microbiol. 2000;174:225–232. doi: 10.1007/s002030000197. [DOI] [PubMed] [Google Scholar]
- Baidoo EE, Benke PI, Neususs C, Pelzing M, Kruppa G, Leary JA, et al. Capillary electrophoresis-fourier transform ion cyclotron resonance mass spectrometry for the identification of cationic metabolites via a pH-mediated stacking-transient isotachophoretic method. Anal Chem. 2008;80:3112–3122. doi: 10.1021/ac800007q. [DOI] [PubMed] [Google Scholar]
- Berghofer Y, Klein A. Insertional Mutations in the Hydrogenase Vhc and Frc Operons Encoding Selenium-Free Hydrogenases in Methanococcus voltae. Appl Environ Microbiol. 1995;61:1770–1775. doi: 10.1128/aem.61.5.1770-1775.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa KC, Wong PM, Wang T, Lie TJ, Dodsworth JA, Swanson I, et al. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc Natl Acad Sci USA. 2010;107:11050–11055. doi: 10.1073/pnas.1003653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoki M, Shinzato N, Sato H, Nakamura K, Kamagata Y. Comparative proteomic analysis of Methanothermobacter themautotrophicus DeltaH in pure culture and in co-culture with a butyrate-oxidizing bacterium. PLoS One. 2011;6:e24309. doi: 10.1371/journal.pone.0024309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman AD, Leigh JA, Samudrala R. Comprehensive computational analysis of Hmd enzymes and paralogs in methanogenic Archaea. BMC Evol Biol. 2009;9:199. doi: 10.1186/1471-2148-9-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydock AK, Porat I, Whitman WB, Leigh JA. Continuous culture of Methanococcus maripaludis under defined nutrient conditions. Fems Microbiol Lett. 2004;238:85–91. doi: 10.1016/j.femsle.2004.07.021. [DOI] [PubMed] [Google Scholar]
- He Z, Zhou A, Baidoo E, He Q, Joachimiak MP, Benke P, et al. Global transcriptional, physiological, and metabolite analyses of the responses of Desulfovibrio vulgaris Hildenborough to salt adaptation. Appl Environ Microbiol. 2010;76:1574–1586. doi: 10.1128/AEM.02141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Seshadri R, Haveman SA, Hemme CL, Paulsen IT, Kolonay JF, et al. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat Biotechnol. 2004;22:554–559. doi: 10.1038/nbt959. [DOI] [PubMed] [Google Scholar]
- Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, et al. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J Bacteriol. 2004;186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson EL, Haydock AK, Moore BC, Whitman WB, Leigh JA. Functionally distinct genes regulated by hydrogen limitation and growth rate in methanogenic Archaea. Proc Natl Acad Sci USA. 2007;104:8930–8934. doi: 10.1073/pnas.0701157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson EL, Liu Y, Rosas-Sandoval G, Porat I, Soll D, Whitman WB, et al. Global responses of Methanococcus maripaludis to specific nutrient limitations and growth rate. J Bacteriol. 2008;190:2198–2205. doi: 10.1128/JB.01805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillesland KL, Stahl DA. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci USA. 2010;107:2124–2129. doi: 10.1073/pnas.0908456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WJ, Whitman WB, Fields RD, Wolfe RS. Growth and plating efficiency of methanococci on agar media. Appl Environ Microbiol. 1983;46:220–226. doi: 10.1128/aem.46.1.220-226.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller KL, Bender KS, Wall JD. Development of a markerless genetic exchange system for Desulfovibrio vulgaris Hildenborough and its use in generating a strain with increased transformation efficiency. Appl Environ Microbiol. 2009;75:7682–7691. doi: 10.1128/AEM.01839-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HW, Zhang H, Suzuki T, Hattori S, Kamagata Y. Differential expression of methanogenesis genes of Methanothermobacter thermoautotrophicus (formerly Methanobacterium thermoautotrophicum) in pure culture and in cocultures with fatty acid-oxidizing syntrophs. Appl Environ Microbiol. 2002;68:1173–1179. doi: 10.1128/AEM.68.3.1173-1179.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major TA, Liu Y, Whitman WB. Characterization of energy-conserving hydrogenase B in Methanococcus maripaludis. J Bacteriol. 2010;192:4022–4030. doi: 10.1128/JB.01446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney MJ, Sieber JR, Gunsalus RP. Syntrophy in anaerobic global carbon cycles. Curr Opin Biotechnol. 2009;20:623–632. doi: 10.1016/j.copbio.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BC, Leigh JA. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J Bacteriol. 2005;187:972–979. doi: 10.1128/JB.187.3.972-979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RM, Pihl TD, Nolling J, Reeve JN. Hydrogen regulation of growth, growth yields, and methane gene transcription in Methanobacterium thermoautotrophicum deltaH. J Bacteriol. 1997;179:889–898. doi: 10.1128/jb.179.3.889-898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, He ZL, Alm EJ, Arkin AP, Baidoo EE, Borglin SC, et al. Salt stress in Desulfovibrio vulgaris Hildenborough: An integrated genomics approach. J Bacteriol. 2006;188:4068–4078. doi: 10.1128/JB.01921-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller N, Worm P, Schink B, Stams AJM, Plugge CM. Syntrophic butyrate and propionate oxidation processes: from genomes to reaction mechanisms. Environ Microbiol Rep. 2010;2:489–499. doi: 10.1111/j.1758-2229.2010.00147.x. [DOI] [PubMed] [Google Scholar]
- Noll I, Muller S, Klein A. Transcriptional regulation of genes encoding the selenium-free [NiFe]-hydrogenases in the archaeon Methanococcus voltae involves positive and negative control elements. Genetics. 1999;152:1335–1341. doi: 10.1093/genetics/152.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolling J, Pihl TD, Reeve JN. Cloning, Sequencing, and Growth Phase-Dependent Transcription of the Coenzyme F-420-Dependent N-5,N-10-Methylenetetrahydromethanopterin Reductase-Encoding Genes from Methanobacterium thermoautotrophicum Delta-H and Methanopyrus kandleri. J Bacteriol. 1995;177:7238–7244. doi: 10.1128/jb.177.24.7238-7244.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat I, Kim W, Hendrickson EL, Xia QW, Zhang Y, Wang TS, et al. Disruption of the operon encoding Ehb hydrogenase limits anabolic CO2 assimilation in the archaeon Methanococcus maripaludis. J of Bacteriol. 2006;188:1373–1380. doi: 10.1128/JB.188.4.1373-1380.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding AM, Mukhopadhyay A, Joyner DC, Hazen TC, Keasling JD. Study of nitrate stress in Desulfovibrio vulgaris Hildenborough using iTRAQ proteomics. Brief Funct Genomics Proteomics. 2006;5:133–143. doi: 10.1093/bfgp/ell025. [DOI] [PubMed] [Google Scholar]
- Schink B, Stams AJM.2006Syntrophism Among ProkaryotesIn: Dworkin M, Falkow S, Rosenberg E, Schliefer K-H, Stackebrandt E (eds).The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community Springer-Verlag: New York; 309–335. [Google Scholar]
- Stams AJ, Plugge CM. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol. 2009;7:568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- Stolyar S, Van Dien S, Hillesland KL, Pinel N, Lie TJ, Leigh JA, et al. Metabolic modeling of a mutualistic microbial community. Mol Syst Biol. 2007;3:92. doi: 10.1038/msb4100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- Thauer RK, Shima S. Methane as fuel for anaerobic microorganisms. Ann N Y Acad Sci. 2008;1125:158–170. doi: 10.1196/annals.1419.000. [DOI] [PubMed] [Google Scholar]
- Thauer RK, Kaster AK, Goenrich M, Schick M, Hiromoto T, Shima S. Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu Rev Biochem. 2010;79:507–536. doi: 10.1146/annurev.biochem.030508.152103. [DOI] [PubMed] [Google Scholar]
- Walker CB, He Z, Yang ZK, Ringbauer JA, He Q, Zhou J, et al. The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J Bacteriol. 2009;191:5793–5801. doi: 10.1128/JB.00356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Sohn S, Kuk S, Xing R. Role of Amino Acids and Vitamins in Nutrition of Mesophilic Methanococcus spp. Appl Environ Microbiol. 1987;53:2373–2378. doi: 10.1128/aem.53.10.2373-2378.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood GE, Haydock AK, Leigh JA. Function and regulation of the formate dehydrogenase genes of the methanogenic archaeon Methanococcus maripaludis. J Bacteriol. 2003;185:2548–2554. doi: 10.1128/JB.185.8.2548-2554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worm P, Stams AJ, Cheng X, Plugge CM. Growth- and substrate-dependent transcription of formate dehydrogenase and hydrogenase coding genes in Syntrophobacter fumaroxidans and Methanospirillum hungatei. Microbiology. 2011;157:280–289. doi: 10.1099/mic.0.043927-0. [DOI] [PubMed] [Google Scholar]
- Xia Q, Wang T, Hendrickson EL, Lie TJ, Hackett M, Leigh JA. Quantitative proteomics of nutrient limitation in the hydrogenotrophic methanogen Methanococcus maripaludis. BMC Microbiol. 2009;9:149. doi: 10.1186/1471-2180-9-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.