Abstract
The mammalian gut harbors complex and variable microbial communities, across both host phylogenetic space and conspecific individuals. A synergy of host genetic and environmental factors shape these communities and account for their variability, but their individual contributions and the selective pressures involved are still not well understood. We employed barcoded pyrosequencing of V1-2 and V4 regions of bacterial small subunit ribosomal RNA genes to characterize the effects of host genetics and environment on cecum assemblages in 10 genetically distinct, inbred mouse strains. Eight of these strains are the foundation of the Collaborative Cross (CC), a panel of mice derived from a genetically diverse set of inbred founder strains, designed specifically for complex trait analysis. Diversity of gut microbiota was characterized by complementing phylogenetic and distance-based, sequence-clustering approaches. Significant correlations were found between the mouse strains and their gut microbiota, reflected by distinct bacterial communities. Cohabitation and litter had a reduced, although detectable effect, and the microbiota response to these factors varied by strain. We identified bacterial phylotypes that appear to be discriminative and strain-specific to each mouse line used. Cohabitation of different strains of mice revealed an interaction of host genetic and environmental factors in shaping gut bacterial consortia, in which bacterial communities became more similar but retained strain specificity. This study provides a baseline analysis of intestinal bacterial communities in the eight CC progenitor strains and will be linked to integrated host genotype, phenotype and microbiota research on the resulting CC panel.
Keywords: Collaborative Cross, intestinal microbial diversity, microbiome, microbiota, pyrosequencing, SSU rRNA gene
Introduction
Throughout their evolutionary history, animals have been in continuous, direct contact with the microbial diversity that thrives in all environments on earth. Specific microbial eco-physiological traits have led to a wide range of associations between metazoan taxa and members of the bacterial and archaeal domains. In some cases, extensive genetic coevolution between the animal host and microbes has resulted in obligate, highly specific, nutritional symbioses involving one or a few vertically transmitted microbial species, such as the endosymbionts of some hydrothermal vent invertebrates and those of plant sap-feeding insects (Moran, 2007; Dubilier et al., 2008). Even for more complex animal gut microbial communities, acquired and maintained dynamically after hatching or birth, there are likely host-microbe specificity determinants, as revealed by natural colonization and experimental microbiota transplantation across host species (Rawls et al., 2004; Rawls et al., 2006; Palmer et al., 2007; Morowitz et al., 2011). Distinct community structure and composition characterizes different vertebrate and invertebrate species in their natural environments, global microbiota and interspecies relatedness, reflecting host phylogeny and incorporating elements of developmental and nutritional specialization (Ley et al., 2008a, 2008b; Ochman et al., 2010; Yidirim et al., 2010). Such complex interactions between deterministic (genetic and developmental), environmental and stochastic factors in the assembly and dynamics of vertebrate gut microbiota are being studied intensely, from fundamental ecological perspectives to its impact on host health and disease (Dethlefsen et al., 2006; Ley et al., 2006; Dethlefsen et al., 2007; Palmer et al., 2007; Ley et al., 2008a; Turnbaugh et al., 2009; Reid et al., 2011; Spor et al., 2011).
Significant advances in understanding the individual roles of host and environmental factors on the composition of vertebrate gut microbiota have resulted from studies on genetically inbred mouse lines (reviewed in Spor et al. (2011)). Such studies have used both conventionally reared and germ-free animals inoculated selectively with different bacterial isolates or natural microbiota samples. Strong evidence exists that the global host genotype influences specific microbiota composition (beta diversity) (Benson et al., 2010; Kovacs et al., 2011), and mutations and inactivation of specific genes have been associated with discrete community changes, in some cases linked to metabolic diseases (for example, obesity, diabetes and metabolic syndrome) (reviewed in Spor et al. (2011)). At the same time, however, studies using embryo transplantation, litter cross-fostering and other variation in mouse rearing and housing have shown that experimental manipulations, environmental and stochastic factors (for example, founder effects) can exert dominating contributions in microbiota taxonomic composition (Friswell et al., 2010). Metagenomic sequencing studies have revealed functionally equivalent gut communities, with similar gene composition, that have quite diverse taxonomic structure (Turnbaugh et al., 2009). Such results suggest that physiological interactions, both with the host and between the microbes, may have a dominant role over phylogenetic composition (alpha diversity) of the community. Therefore, linking host genetic background with discrete units of the microbiome (microbial taxa or genes) relies upon a combination of diversity and functional genomic/physiological measurements. With thousands of segregating genes and millions of segregating polymorphisms in mouse populations, comprehensive mapping of potential deterministic associations between host genotype and the hundreds of bacterial taxonomic or functional units, as well as distinguishing environmental and stochastic effects, requires an extensive population genetics and statistical framework. A recent study using quantitative trait locus analysis of advanced intercross lines identified a subset of microbial lineages that cosegregate with host genetic loci (Benson et al., 2010).
The Collaborative Cross (CC), a large panel of recombinant, inbred mouse strains designed by the Complex Trait Consortium, offers a standardized and reproducible foundation for complex trait analysis, including microbiota heritability factors (Churchill et al., 2004). The CC will encompass a large number of inbred strains resulting from systematic crossing of eight genetically diverse founder strains that capture ∼90% of the known mouse genetic variability (Roberts et al., 2007). The CC was initiated at several research institutions including Oak Ridge National Laboratory (ORNL) (Chesler et al., 2008; Philip et al., 2011) and recently has been employed in quantitative trait analysis of a wide range of phenotypes (Aylor et al., 2011; Philip et al., 2011).
Here we present an analysis of gut microbial community structure associated with the eight founder strains of the CC and two additional strains to assess environmental and founder population effects using pyrosequencing of two separate regions of the small subunit ribosomal RNA (SSU rRNA) gene. This study lays the foundation for determining the community structure variability in mouse lines resulting from controlled crossing of the founder populations at different levels of inbreeding and correlating with quantitative host physiological and genetic markers.
Materials and methods
Mice
Mice were bred and housed at the William L and Liane B Russell vivarium at ORNL and at the University of Tennessee (UTK), Knoxville, TN, USA. Mice at ORNL profiled in this study were bred at the facility and weaned at 3–4 weeks after birth and distributed in separate cages either individually or with same-gender siblings or non-siblings based on experimental design (Supplementary Figure S1) until adult (8–10 weeks of age). The eight parental mouse lines of the CC were used: A/J, C57BL/6J, 129S1/SvImJ, NOD/LtJ, NZO/HILtJ, CAST/EiJ, PWK/PhJ and WSB/EiJ (abbreviated AJ, BL6J, 129S1, NOD, NZO, CAST, PWK and WSB, respectively). Strains were originally obtained from The Jackson Laboratory and maintained over no longer than 10 generations. Because of difficulties in breeding, mice from the NZO line were the only age exception, with some >1 year. C3H/Ri and DBA/2JR mice (abbreviated C3HRI and DBAJR, respectively) were also profiled. Replicates of 7–10 mice were used per strain. Cecum content samples were collected as described in the Supplementary Methods.
For the interstrain cohabitation study, 3-week-old BL6J and C3HRI mice were purchased from The Jackson Laboratory and were housed in a separate facility (UTK) until they reached 10 weeks of age, at which time they were all euthanized. Thoren cages with microisolator tops and individual water bottles were used for this experiment. Separate cages contained five individuals of only BL6J (cage 1) or C3HRI (cage 4). Cage 2 contained three BL6J and two C3HRI mice. Cage 3 contained two BL6J and three C3HRI mice (Supplementary Figure S1). All the mice were fed Harlan Laboratories (Indianapolis, IN, USA) Teklad Rodent Diet 8604, which is similar to Purina Rodent Chow 5053 (high-protein, low-carbohydrate content).
SSU rRNA gene amplification and pyrosequencing
DNA was extracted from cecum contents using a protocol modified from that of Ley et al. (2008a) (Supplementary Methods). Amplicon libraries of both V1-2 and V4 regions of 16S SSU rRNA genes were obtained using barcoded primers and sequenced using a 454-FLX instrument (Roche, Indianapolis, IN, USA), using 40 samples per plate. Resulting sequences were filtered for length, quality and chimera removal using the software package mothur (Schloss et al., 2009). High-quality sequences were subjected to operational taxonomic unit (OTU)-based clustering (Huse et al., 2010) and phylogeny-based analysis using Fast UniFrac (Hamady et al., 2010) to evaluate the effects of host genetics on bacterial community composition. Details of sequencing and data processing steps are provided in the Supplementary Methods.
Statistical analyses
Matrices of OTU-by-sample were imported into PRIMER-E v6 (Clarke and Gorley, 2006) for downstream statistical analyses. Raw sequence counts of each OTU within each sample were converted into percentages, square-root transformed and a Bray–Curtis resemblance matrix was calculated. This matrix was used for nonmetric multidimensional scaling plots, hierarchical clustering, analysis of similarity and similarity percentage (SIMPER). Permuted (n=9999) multivariate analyses of variance were performed on Bray–Curtis matrices using the PERMANOVA+ add-on package for PRIMER-E. Permuted calculations of P were used when unique permutation values were >100 and Monte Carlo calculations of P were used when unique permutations were <100 (Clarke and Gorley, 2006). Retrospective power analyses were performed for each within strain comparison of the sexes. Briefly, critical values (α=0.05) along a t distribution were determined for one population of mice in the comparison and used to determine the overlapping section of the second population of mice (Sokal and Rohlf, 1981). This P-value equals β, and power was calculated from β (power=1−β).
We used a two-step approach to identify OTUs that were most influential in differentiating mouse strains in each of the V1-2 and V4 amplicon libraries. First, SIMPER was used to calculate the relative contribution of each OTU to the overall dissimilarity in each pair of mice. Because of the large number of pairwise comparisons, it is difficult to elucidate clear trends. However, we used SIMPER as a data reduction technique to discard OTUs that did not contribute at least 0.5% to the dissimilarity of any pair of mice. OTUs in V1-2 and V4 data found to contribute at least 0.05% to any pairwise difference in SIMPER comparisons were further screened for differential abundances across strains using a discriminant function analysis (DFA) in Matlab (v7.10) using a freely available statistics toolbox (Strauss, 2010). DFA is a multivariate technique used to identify variables (OTUs) that distinguish a priori groups (mouse strain). Thus, DFA was used to further reduce V1-2 and V4 OTU matrices to a suite of OTUs that could be used to predict mouse strain membership. Hierarchical clustering of strains based upon these predictive OTUs was performed on Euclidean distances in Matlab.
Sequence deposition
Nucleotide sequences generated in this study have been deposited in the NCBI Sequence Read Archive (Accession no. SRPO12588.1).
Results
Mice representing 10 inbred mouse lines, including the 8 progenitors of the CC project, were used to determine differences in gut microbial diversity linked to distinct host genetic background. Embedded in this, maternal, sex and cage-sharing effects were also explored. The mouse lines were maintained separately but under the same conditions at the ORNL facility. Second, to compare effects of environmental exposure and interstrain contact, we analyzed the gut microbial diversity in two of the strains raised at a different location and exposed to one another (Supplementary Figure S1).
For the primary study, the cecum microbiota of 94 mice were profiled by SSU rRNA gene pyrosequencing (Supplementary Table S1). Two regions of SSU rRNA gene (V1-2 and V4) were analyzed to complement differences in taxonomic representation due to primer bias (Griffen et al., 2012), as well as to compare and contrast inferred relationships between the microbiome and the host genetic background. After sequence processing, V1-2 amplicon libraries contained 293 928 reads (mean of 4982 reads/mouse) and V4 libraries contained 605 397 reads (mean of 6640 reads/mouse).
Taxonomic analysis of all the sequences using the RDP Naïve Bayesian rRNA Classifier (Cole et al., 2009) revealed similar bacterial diversity to previously observed communities in mouse ceca (Ley et al., 2005), with a dominance of Firmicutes (53–89%) (Supplementary Figure S2). A large difference was observed for detection of Bacteroidetes, with the V4 data set containing many fewer sequences mapped to that phylum relative to the V1-2 data set (∼2% vs 30% median, respectively). Conversely, phyla generally present at low abundance in the mouse cecum (<2%), such as Proteobacteria, Verrucomicrobia, TM7, Deferribacteria and Tenericutes, were detected more efficiently by the V4 than by the V1-2 primer set. Additional taxa were detected at much lower abundance. For example, the Cyanobacteria-like group (Ley et al., 2005) was present in only mice 129S1-352 (39 sequences; 0.45%) and 129S1-353 (25 sequences; 0.58%) of the V4-amplified microbiota. This same Cyanobacteria-like group was only detected as a single sequence in V1-2 amplicon libraries (mouse 129S1-352). Differences in taxonomic coverage and efficiency of detection are known to occur between primer sets (Hong et al., 2009; Engelbrektson et al., 2010). In many cases, these discrepancies are not predictable based on sequence complementarity analysis (such as V4 detection of Bacteroidetes), highlighting the advantage in targeting more than one SSU rRNA gene region for analyses of taxonomic diversity (Griffen et al., 2012). Analysis of gut microbiota based on taxonomic classification is limited by the high diversity of taxa below the genus level, many with uncultured relatives, which reduces resolution of sequence assignment. Therefore, in this study, we primarily used a taxonomy-independent analysis approach by classifying the sequences into OTUs based on sequence similarity (genetic distance).
Amplicon libraries of V1-2 hypervariable regions of bacterial SSU rRNA gene produced 3821 OTUs across all samples at 0.03 genetic distance, whereas libraries of the V4 region produced 1142 OTUs across all samples at the same genetic distance. Variation observed in the two hypervariable regions and current analytical methods for such microbial community data led us to adopt a consensus approach for data analysis. Both OTU-based clustering and phylogenetic (Fast UniFrac) analyses were pursued for all data to ensure that overarching trends were not dependent on analytical method.
Strain-wise comparisons
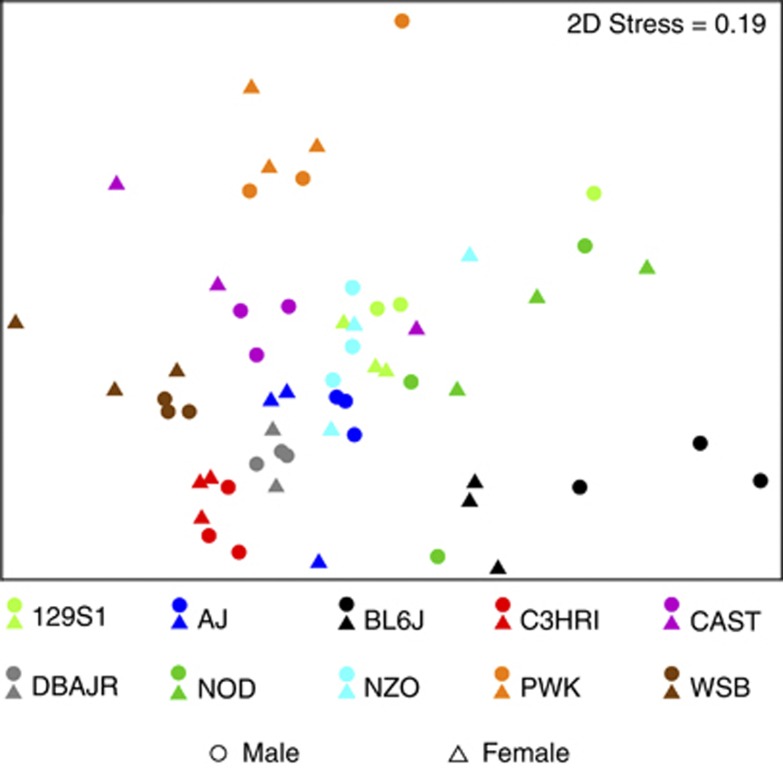
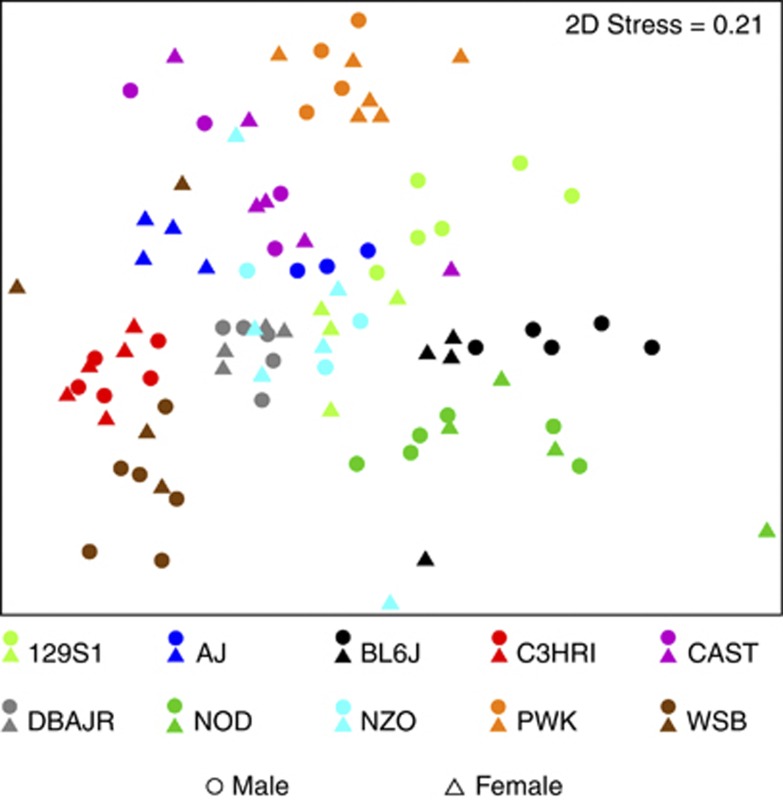
Conceptually, analysis of mouse cecum communities by clustering sequences into operational units (OTUs) differs from analysis based on phylogenetic sequence information, but both methods produced similar results for both SSU rRNA gene hypervariable regions, and consistent differences between strains were found. Nonmetric multidimensional scaling (NMDS) of Bray–Curtis similarity matrices for OTU-based clustering (Figures 1 and 2) provided similar visual separation by strain as principal coordinates analysis of UniFrac distance matrices (Supplementary Figures S3 and S4). Subsampling to achieve equal sequencing depth for each sample resulted in slightly lower explained variation for either hypervariable region, but mice appeared to separate more clearly by strain with equal sequencing depth (Supplementary Figures S3 and S4). Although explained variation was enhanced in UniFrac analysis of V4 sequences, groupwise separation of strains was reduced. It is possible that this is a result of the larger number of samples sequenced and number of sequences per sample in the V4 data set. However, it is evident from both analyses that BL6J, C3HRI, DBAJR, PWK and WSB strains harbored distinct microbial assemblages, whereas individual variation appeared higher within 129S1, AJ, CAST, NOD and NZO strains.
Figure 1.
Nonmetric multidimensional scaling (NMDS) representation of OTU-based clustering (0.03 genetic distance) of data from the V1-2 hypervariable region of SSU rRNA gene. Counts of each OTU within each mouse (n=59) were standardized to percentage, square-root transformed and a Bray–Curtis similarity matrix was calculated.
Figure 2.
NMDS representation of OTU-based clustering (0.03 genetic distance) of data from the V4 hypervariable region of SSU rRNA gene. Counts of each OTU within each mouse (n=94) were standardized to percentage, square-root transformed and a Bray–Curtis similarity matrix was calculated.
Hierarchical clustering was also used to visualize relationships of individual mice for OTU-based clustering. Branching of V1-2 based on OTUs largely adhered to strain identification of individuals, with most strains condensing to discreet nodes of the dendrogram. Strains BL6J and PWK appeared to be most distinct from other strains with OTUs, but most strains separated into distinct clades. Some overlap was seen in one individual of both 129S1 and NZO with NOD. NOD mice appeared to be the least cohesive strain (Supplementary Figure S5). Similarly, the V4 region OTUs (Supplementary Figure S6) showed clear separation of strains in good agreement with nonmetric multidimensional scaling plots, with BL6J appearing most distinct. Again, even though several strains had one individual outlier, they were quite different from one another. Strains C3HRI and DBAJR had the lowest intrastrain variation (Supplementary Figure S6).
Strain-wise separation of UniFrac clusters for V1-2 data (Supplementary Figure S7) was comparable to OTU-based data. BL6J and NOD mice were broken into two clusters, and an individual from 129S1 and CAST failed to congregate with their strains. BL6J appeared to separate by sex. WSB individuals were more cohesive in this UniFrac analysis than was observed for OTUs (Figure 4). UniFrac clustering for V4 for individual mice (Supplementary Figure S8) also showed similar results to OTU data, but OTU-based clusters (Supplementary Figure S6) were separated better by strain. However, individuals of 129S1, CAST and NZO fragmented into separate clades. Moreover, 5 out of 10 strains had at least 1 individual that did not congregate with their respective strains, but this could not be linked to either maternal or caging factors and likely reflects stochastic community assembly.
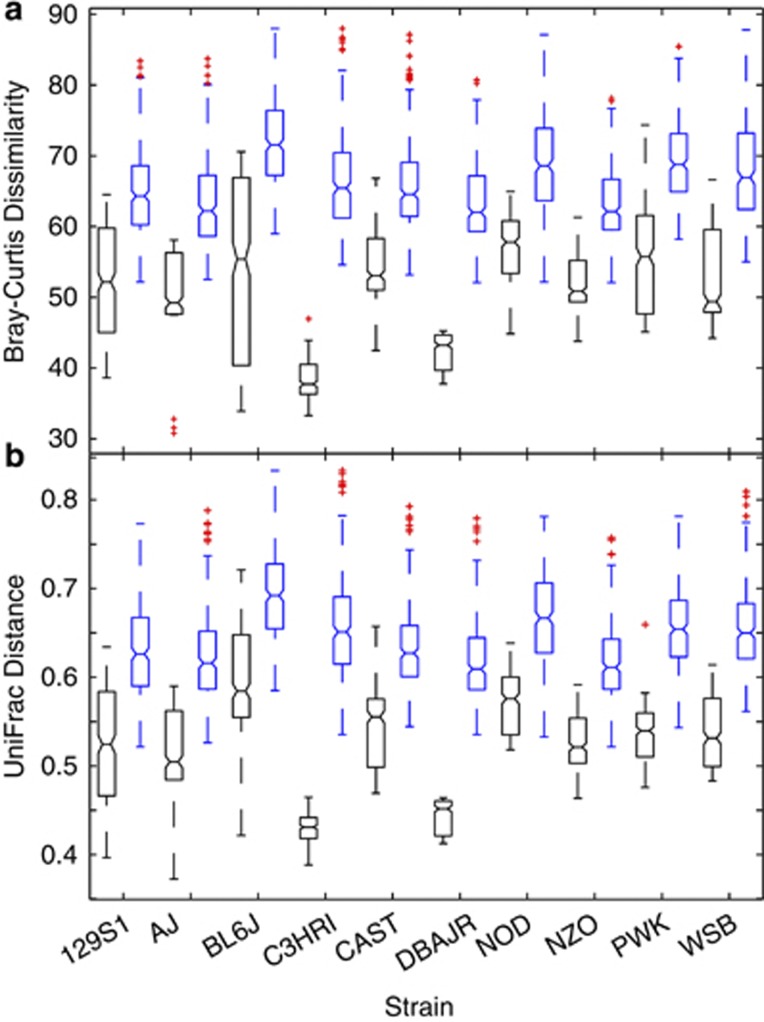
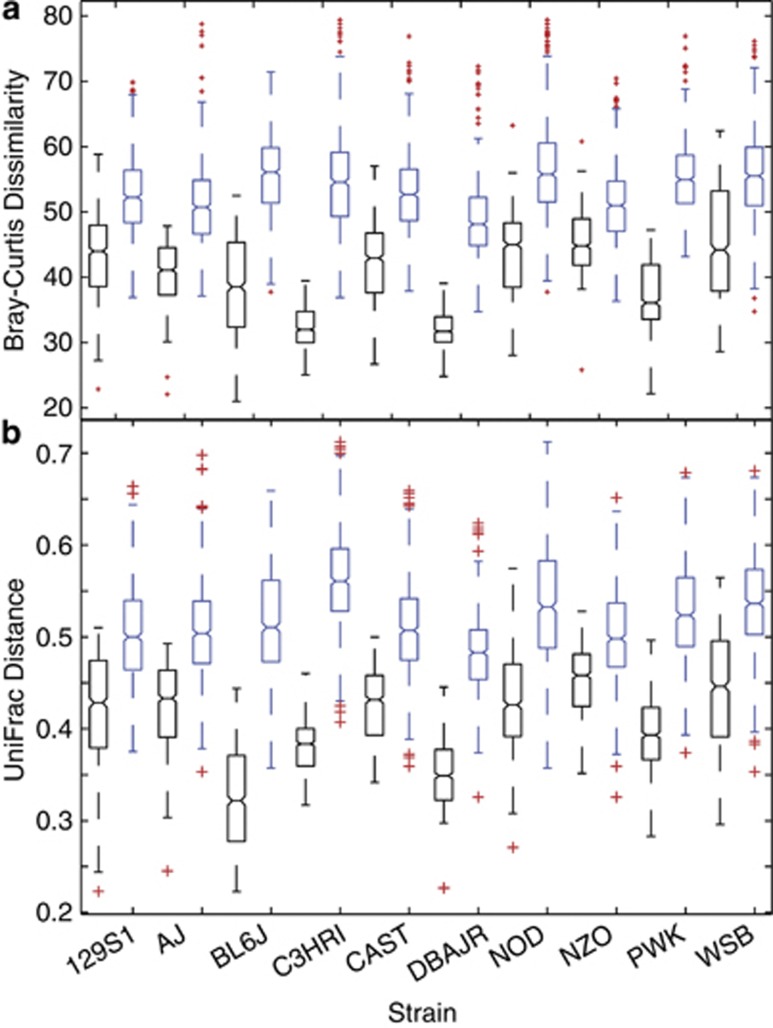
Due to the difficulty in visualizing three-dimensional arrangements, box-and-whisker plots of intra- and interstrain dissimilarities were constructed from both V1-2 (Figure 3) and V4 (Figure 4) distance matrices for Bray–Curtis dissimilarities and UniFrac distances (sub-sampled only), and these analyses indicated that mice within all strains were more similar to one another than to mice from all other strains. Sequences from the V1-2 region displayed greater variation between mice than V4, but both analytical methods produced similar intrastrain and interstrain relationships. Data from both SSU regions were supportive of one another. In these simplified representations, V1-2 and V4 libraries both showed C3HRI and DBAJR strains to be the most distinct.
Figure 3.
Box-and-whisker plots of intrastrain (black) and interstrain (blue) distributional comparisons within V1-2 data. Distributions were formed by parsing strain-wise data from larger (a) Bray–Curtis dissimilarity and (b) UniFrac distance matrices of mouse-by-mouse comparisons. Outliers are denoted by red plus characters (+).
Figure 4.
Box-and-whisker plots of intrastrain (black) and interstrain (blue) distributional comparisons within V4 data. Distributions were formed by parsing strain-wise data from larger (a) Bray–Curtis dissimilarity and (b) UniFrac distance matrices of mouse-by-mouse comparisons. Outliers are denoted by red plus characters (+).
Significance measures calculated around separation of mouse cecum communities (multivariate analyses of variance, MANOVA, and analysis of similarity, ANOSIM) reinforced that mouse strains harbored distinctly different assemblages. The effect of strain (Fpseudo=5.48; Ppermuted=0.0001) on the V1-2 data was significant (Table 1), accounting for 38.9% of all variation. Similarly, strain effects (Fpseudo=8.55; Ppermuted=0.0001) were significant for V4 data (Table 2), accounting for 41.1% of all variation. Pairwise t-tests for individual strains indicated that each strain differed significantly from other strains for both V1-2 and V4 regions (Supplementary Tables S2 and S3). These analyses were supported by analysis of similarity, in which the V1-2 region (global R=0.818; P=0.001) separated strains with higher resolution than did the V4 region (global R=0.795; P=0.001). V1-2 comparisons (Supplementary Table S4) showed C3HRI and PWK to be strongly separated from most other strains, and BL6J, DBAJR and NOD also showed little overlap with other strains. V4 data (Supplementary Table S5) supported clear distinction of C3HRI and PWK microbiota from other strains. However, bacteria detected using this region of SSU rRNA gene did not strongly separate NZO from most other mouse strains.
Table 1. Permuted multivariate analysis of variance (MANOVA) tests of significance of mouse strain and sex for the V1-2 region.
| Source | Dfa | SSb | MSc | Pseudo-F | Unique permutations | Pd |
|---|---|---|---|---|---|---|
| Strain | 9 | 59701 | 6633.5 | 5.4829 | 9747 | 0.0001 |
| Sex | 1 | 1858.9 | 1858.9 | 1.5364 | 9862 | 0.0152 |
| Strain × Sex | 9 | 16589 | 1843.3 | 1.5235 | 9605 | 0.0001 |
| Residual | 39 | 4.72E+04 | 1209.8 | |||
| Total | 58 | 1.26E+05 |
Degrees of freedom.
Sum of squares.
Mean squared.
Bolded P-values indicate statistical significance.
Table 2. Permuted multivariate analysis of variance (MANOVA) tests of significance of mouse strain and sex for the V4 region.
| Source | Dfa | SSb | MSc | Pseudo-F | Unique permutations | Pd |
|---|---|---|---|---|---|---|
| Strain | 9 | 58297 | 6477.4 | 8.5513 | 9766 | 0.0001 |
| Sex | 1 | 1225.7 | 1225.7 | 1.6181 | 9873 | 0.01 |
| Strain × Sex | 9 | 12357 | 1373 | 1.8126 | 9665 | 0.0001 |
| Residual | 74 | 5.61E+04 | 757.48 | |||
| Total | 93 | 1.30E+05 |
Degrees of freedom.
Sum of squares.
Mean squared.
Bolded P-values indicate statistical significance.
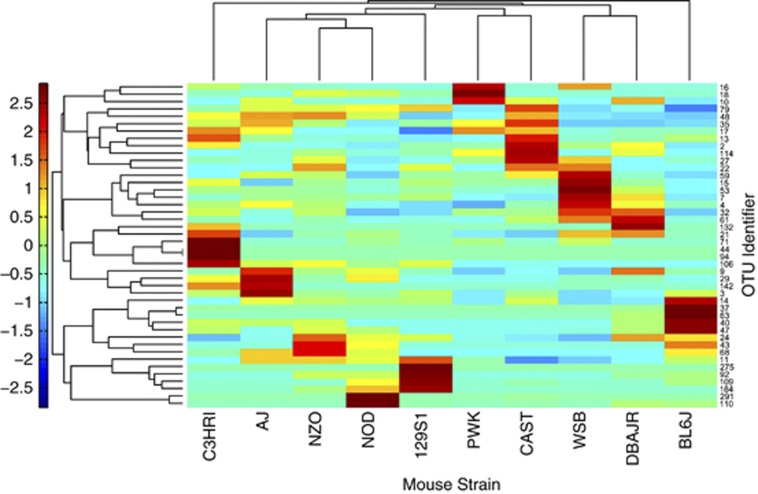
DFA indicated that relatively few OTUs could be used to reliably predict strain membership. Within all OTUs detected in V1-2 libraries, SIMPER analysis found 80 OTUs that explained ⩾0.5% of the difference between any two pairwise comparisons of strains. DFA reduced these OTUs to 44 that differed significantly across mouse strain (Supplementary Table S6). Discriminating OTUs were dominated by uncultured phylotypes among the firmicutes (59%) and Bacteroidetes (36%), but one Proteobacteria and one Deferribacteres were also differential across strains. Clustering of only those discriminating OTUs (Figure 5) indicated that subsets of at least two OTUs could be positively associated with each strain. OTUs showing consistently high abundances within a strain usually showed a phylogenetic association, as well. Strains AJ and BL6J contained differential OTUs found in the Bacteroidetes. In particular, BL6J contained the highest abundances of five OTUs most closely related to the genus Barnesiella. Conversely, differentially abundant OTUs of strains 129S1, CAST, NOD, NZO and WSB were from the Firmicutes. CAST and NOD mice were enriched for members of the Clostridiales. Differential OTUs of C3HRI, DBAJR and PWK lacked phylogenetic relationships.
Figure 5.
Heatmap of V1-2 OTUs found to vary across strains by discriminant function analysis (DFA). Means of each OTU (n=44) were calculated for each strain (n=10). Hierarchial clustering was determined for both dimensions of the heatmap using Euclidean distances. Taxonomic assignments of OTUs can be found in Supplementary Table S6.
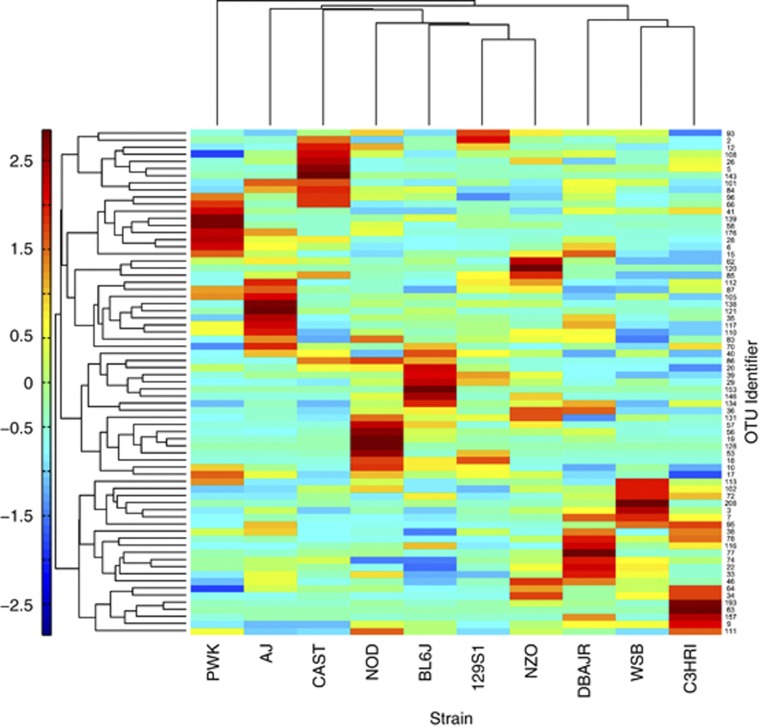
The same approach was applied to detect differentially abundant OTUs for V4 data (Supplementary Table S7), and again a relatively few OTUs could be used to reliably predict strain membership. This region of the 16S rRNA gene produce 71 differential OTUs dominated by the phylum Firmicutes (93%), many of which matched most closely to uncharacterized members of family Lachnospiraceae. A single representative cluster of the phyla Bacteroidetes, Deferribacteres, Proteobacteria, Tenericutes and TM7 also displayed unequal distributions across mouse strains. Again, hierarchical clustering of just these OTUs (Figure 6) indicated that subsets of at least two OTUs could be positively associated with a mouse strain. With a narrower phylogenetic scope than discriminating OTUs in V1-2 regions, a pattern of taxonomic associations by strain is not clear. It is possible that these OTUs are interchangeable with closely related bacteria across the strains survey in this study.
Figure 6.
Heatmap of V4 OTUs found to vary across strains by DFA. Means of each OTU (n=71) were calculated for each strain (n=10). Hierarchial clustering was determined for both dimensions of the heatmap using Euclidean distances. Taxonomic assignments of OTUs can be found in Supplementary Table S7.
Maternal effects
We used V4 data to investigate the effects of maternal lineage on gut microbial communities because it contains a larger sample size and can, therefore, be considered more comprehensive. To illustrate the effects of maternal lineage, intrastrain dissimilarities (Bray–Curtis) were separated into two groups: (1) pairwise distances of siblings and (2) pairwise distances of all non-siblings within a strain. Distributions of non-siblings were plotted (Supplementary Figure S9), and distances of siblings were superimposed onto these distributions. Gut communities from siblings were spread without a clear pattern along strain-wise distributions of non-siblings. This suggests that inclusion of siblings with non-siblings had little effect on strain-wise bacterial communities. In support of this conclusion, siblings did not appear to cluster (Supplementary Figure S6) more closely than any other individuals.
Sex-based comparisons
Distributions of males and females within most strains overlapped broadly and did not indicate strongly differential microbial communities in males and females. The effect of sex was significant for both V1-2 (Fpseudo=1.54; Ppermuted=0.015) and V4 (Fpseudo=1.62; Ppermuted=0.01), but explained only 0.9% and 0.7% of variation in the data, respectively (Tables 1 and 2). Strain-by-sex interactions were also significant for each data set, indicating that males and females of some mouse strains contained divergent cecal communities. Retrospective power analysis of each comparison (Supplementary Tables S8 and S9) indicated that most t-tests were robust, but some had low sex resolution. Differential cecal communities within sex were detected for the BL6J strain in V1-2 data (Supplementary Table S8) and for 129S1, AJ, BL6J, C3HRI and PWK in V4 data (Supplementary Table S9). All individuals of BL6J were cocaged with at least one other mouse of the same sex (Supplementary Table S1), indicating that separation of sexes was potentially an artifact. Similarly, one pair of males and females was cocaged within strains 129S1, AJ and C3HRI, and PWK contained a pair of cocaged females. However, strains CAST, NOD, NZO and WSB also had cocaged pairs of the same sex, but did not show significant differences in microbial communities. Therefore, sex-based differences could vary with strain, but more replication is needed for some strains to answer this definitively.
Cagemate comparisons
Some mice of the same sex and strain were caged together (Supplementary Table S1) and compared with mice housed separately to test the effects of cage environment on the gut microbial community. To analyze this effect, intrastrain dissimilarities (Bray–Curtis) were separated into two groups: (1) pairwise distances of cagemate mice and (2) pairwise distances from all mice kept separately. Distributions were plotted only for mice that were not cocaged, and distances of cagemates were superimposed onto these distributions (Supplementary Figure S10). Cagemates tended to be more similar to one another than the majority of the isolated mice. This was most evident within strains 129S1, AJ and NZO. However, dissimilarity measures of most cagemates fell within the ranges of those observed for isolated mice. Therefore, overall variation within strains is of greater magnitude than cocaging effects.
Interstrain cohabitation
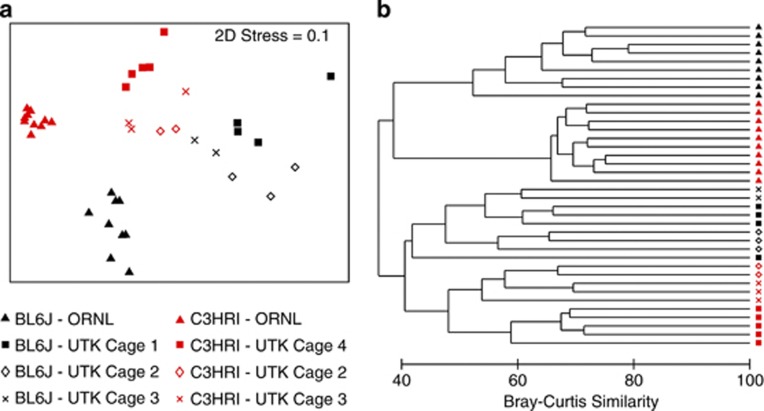
We also tested the effects of cohabitation of adult BL6J and C3HRI mice in varied ratios, and again strain effects appeared to dominate caging effects. Four cages were used for this experiment. Separate cages contained five individuals of only BL6J (cage 1) or C3HRI (cage 4). Cage 2 contained three BL6J and two C3HRI mice. Cage 3 contained two BL6J and three C3HRI mice. Mice were purchased specifically for this purpose and housed in a separate facility (UTK) for 8 weeks prior to euthanization. Gut communities of mice housed at ORNL differed from those at UTK (Figure 7), similar to previous reports (Friswell et al., 2010). OTU-based clustering of V4 amplicon libraries found 483 OTUs at a genetic distance of 0.03. An individual mouse in BL6J was not closely positioned with any other mice in the experiment. Therefore, it (and all of its unique OTUs) was removed from further analyses. An NMDS plot and hierarchial clustering (Figure 7) of these data indicated clear separation of mice by strain and cage. Hierarchical clustering also showed clear delineation of mice primarily by strain and secondarily by cage. Interestingly, cohabitation influenced gut microbial communities, but host genetics appeared to outweigh this environmental influence. However, mice were cohoused post weaning, possibly rendering their microbiota more resistant to change. Therefore, host genetic effects and maternal inoculation could not be simultaneously addressed. Further studies employing larger populations of mice, temporal sampling and strain cross-fostering would better determine the resilience of established gut communities and the effects of initial colonization.
Figure 7.
Effects of interstrain cohabitation and housing facility on cecum bacterial communities. BL6J and C3HRI mice were housed separately (cages 1 and 4) or in cohabitation (cages 2 and 3). Only the V4 hypervariable region was sequenced and OTUs were calculated (0.03 genetic distance) for all mice. Counts of each OTU within each mouse (n=19) were standardized to percentage, square-root transformed and a Bray–Curtis similarity matrix was calculated and used to produce an (a) NMDS and (b) hierarchical clustering of the gut communities.
Discussion
Studies of genetic effects on microbiota are accumulating in the literature. Some of these studies address fine genetic scales, such as monozygotic, human twins (Turnbaugh et al., 2009) and well-characterized host mutations (Vaahtovuo et al., 2005; Khachatryan et al., 2008). Others have addressed the effects of host genetics on the gut microbiome on a larger scale with studies of species of primates (Ley et al., 2008a; Ochman et al., 2010) and various animals in captivity or the wild (Ley et al., 2008a). In this study, we investigated the effects of host genetics on cecum microbiota in 10 commonly used, inbred strains of laboratory mice, 8 of which are progenitor strains of the CC (Consortium, 2004). Therefore, this study serves as a baseline for determining the nature and extent of genetic effects on microbial diversity of these mouse lines for future studies of the CC.
Individual variation within strains was observed for all mouse lines used in this study, but the influence of host genetics on bacterial communities in the cecum is apparent. This observation was supported by independent analyses of two regions of SSU rRNA gene sequence libraries. Individuals within several strains appear to be more cohesive than others (for example, C3HRI, DBAJR and WSB), indicating that a gradient of host genetic factors produces varied levels of strain-level conformity. Unlike microbial communities of wild primates (Ochman et al., 2010), dendrograms of strain-wise relationships based on cecum microbiota failed to recapitulate apparent natural histories of the hosts (Petkov et al., 2004; Kirby et al., 2010). Mice of the same strain purchased from different vendors also harbor different microbial communities (Friswell et al., 2010). Therefore, lack of a reflection of the natural history of the strains in their cecal communities was not surprising.
Other studies have also reported that host genetics shape gut communities in mice. Two such studies (Benson et al., 2010; Buhnik-Rosenblau et al., 2011) found ties between host genetics and Lactobacillus in mice. Another study (Alexander et al., 2006) in which mice from 23 inbred strains were inoculated with and tested for the altered Schaedler's flora using specific quantitative PCR assays noted significant differences for these species. Also, it was noted that different strains of 129 and BALB mice were similar when supplied through a different vendor (Alexander et al., 2006). However, Friswell showed that obtaining the same strain from different vendors produced varied microbial flora (Friswell et al., 2010). Moreover, Friswell et al. (2010) found that C3HRI and BL6J mice harbored distinct microbial communities that were more strongly regulated by host genetics than changes in environment. Interestingly, host genetics could be overcome by implanting embryos of different strains into a surrogate mother, producing microbial communities within offspring that resembled the surrogate mother (Friswell et al., 2010). Similar to Friswell et al. (2010), we found C3HRI to have low intrastrain variation and BL6J mice to have high intrastrain variation. Gut communities in BL10J mice (Loh et al., 2008) have also been shown to vary among individuals.
These studies provided a structural basis for us to more deeply investigate effects of host genetics on gut microbiota. In contrast to employing quantitative PCR (Alexander et al., 2006) or DGGE (Friswell et al., 2010), we used pyrosequencing to compile libraries of two regions of SSU rRNA gene, allowing us to identify bacterial taxa that were specific to mouse strain. Identification of differentially abundant OTUs across mouse strains makes it tempting to speculate as to their potential roles within each strain. However, many of the discriminating OTUs have neither cultured relatives nor genomic data available. Moreover, definitive trends in health and disease cannot be discerned for many taxa closely related to these OTUs. For instance, the genus Oscillibacter has been linked to diet in humans (Walker et al., 2011), but this genus appeared to be differential in several of our strains, which were fed the same diet. Also, among the four Oscillibacter OTUs found discriminatory in V4 data, no clear trend across strains was found for this genus, as a whole. Correlations of quantitative trait locus and host gene expression to bacterial diversity data presented here will likely shed more light on potential physiological roles of these bacteria in the mouse cecum. Future isolation and physiological studies of bacterial taxa that were discriminatory among mouse strains will also improve our understanding of the role of these bacteria. Linkage of host genetics, host health/disease and microbial flora will be the ultimate goal of such microbiome studies, and these mapping studies will enable the detection of the sources of host molecular variation and impacts on the intestinal micro-environment.
Our study was not designed to quantify effects of maternal lineage on gut microbiota across these 10 strains, but we were able to make some comparisons of siblings to unrelated individuals. Siblings from some strains bore stronger resemblance to one another than to unrelated mice. However, siblings of other strains were markedly dissimilar in microbial flora. Recently, DNA fingerprinting techniques revealed no maternal-derived differences in gut bacteria of CC mice (Kovacs et al., 2011). However, Ley et al. (2005) found lineage influences to extend to more than one generation. In fact, others have (Hufeldt et al., 2010) observed a sufficiently strong effect of maternal lineage that they suggested related individuals be used to reduce microbiome variation for disease studies. Perhaps the most convincing demonstration of maternal effects was provided by Friswell et al. (2010). In this study, a BDF1 female was implanted with embryos from BL6J and Agouti strains, resulting in pups with gut bacteria similar to the surrogate mother (Friswell et al., 2010). At this point, factors driving maternal differences observed in some studies and not others remain unclear. In fact, it appeared that maternal influences could be strain-specific, possibly indicating underlying genetic or behavioral disparities (Alexander et al., 2006).
We did not observe major separation of males and females within most mouse strains in this study, but some minor sex-based differences among individuals in some mouse strains were found. Lack of a predictable response of cecum bacteria to gender differences has been substantiated (Spor et al., 2011). Comparatively few studies have addressed sex-associated changes in mouse gut microbiomes. Although more narrow in taxonomic scope than our study, Alexander's quantitative PCR study (Alexander et al., 2006) detected gender-specific differences in only two species of the altered Schaedler's flora (species of Firmicutes and Clostridium). Moreover, expression of a human caspase conferred differential susceptibility to Listeria monocytogenes infection in male and female transgenic mice, apparently due to estrogen interactions (Yeretssian et al., 2009). However, Kovacs et al. (2011) found no gender differences in the CC mice of their study. As is the case with maternal lineage, undetected genetic or epigenetic factors that were not adequately controlled could manifest in sex-based differences between strains. Quantitative trait locus and gene expression analyses of our mice (to be presented elsewhere) have potential for elucidating such mechanisms.
Controlled cohabitation of mice of the same strain offered the ability to weigh the effects of genetics against environmental pressures. A limited number of individuals of the same strain were cocaged to evaluate this effect on cecum bacterial communities. Similar to previous reports (Alexander et al., 2006; Loh et al., 2008; Terán-Ventura et al., 2010), some cagemates were more similar to one another than to isolated mice, but for many no difference was detected. This response varied by mouse and was not strong for most strains. Terán-Ventura et al. (2010) employed cultivation, fluorescence in situ hybridization and terminal restriction fragment length polymorphism to detect minor variations in abundances of Enterobacteriaceae, Bacteroides, Clostridium and Lactobacillus associated with varied levels of caging isolation (Terán-Ventura et al., 2010). Alexander et al. (2006) also noted that cage effects appeared to vary by strain, and it was suggested that behavioral differences in the strains (such as differential coprophagy) could explain the strain-wise differences.
Results of an interstrain cohabitation experiment were more informative than observations of intrastrain cohabitation. We did note the same ‘consortial drift' (Friswell et al., 2010) between BL6J and C3HRI populations used in studies at ORNL and UTK. When cages containing only one strain were compared with those containing two strains of mice, genetic strain best separated mice. This is supported by an experiment in Alexander's study (Alexander et al., 2006), in which cages with five inbred strains were monitored for members of the altered Schaedler's flora, demonstrating that host genetics are more influential in determining host mouse microbial flora than the environment. Interestingly, environmental effects were weaker than underlying host genetics in shaping cecum bacterial communities.
Assessing the causal role of host genetic variation in gut microflora composition and dynamics will enable an understanding of the mechanisms of colonization, and in well-characterized mouse strains, the correlation to phenotypes of health and disease, and will enable comparisons with similar studies in the human population. Understanding the mechanisms of community selection and robustness of genetic influences on community structure will have many implications for attempts to alter community structures as a therapeutic intervention. Establishing the relationship of microbial communities to the spectrum of variation in physiological phenotypes will further our understanding of pathological and normal metabolic processes. Emerging mouse resources such as the CC are a powerful system with which to assess these phenomena and widespread variation in microbial structure.
Acknowledgments
We thank J Mosher, M Shakya, C Brandt, C Schadt (ORNL), M Hauser and J Becker (UTK) for many helpful discussions during data collection and analysis. We also thank L Miller (ORNL) for molecular biology assistance. We are also grateful to P Schloss (mothur; University of Michigan) and R Knight (UniFrac; University of Colorado) for guidance with their respective analytical tools. We also thank F Bushman (University of Pennsylvania) for critical evaluation of the manuscript and anonymous reviewers for helpful suggestions. This research was funded by the US Department of Energy Office of Science, Biological and Environmental Research programs at Oak Ridge National Laboratory (ORNL), by the Laboratory Directed Research and Development Program of ORNL and in part by a grant to MP from the National Human Genome Research Institute (NIH R01-HG004857). ORNL is managed by UT-Battelle, LLC, for the US Department of Energy under contract DE-AC05-00OR22725.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alexander AD, Orcutt RP, Henry JC, Baker JrJ, Bissahoyo A, Threadgill DW. Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mammalian Genome. 2006;17:1093–1104. doi: 10.1007/s00335-006-0063-1. [DOI] [PubMed] [Google Scholar]
- Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, et al. Genetic analysis of complex traits in the emerging collaborative cross. Genome Res. 2011;21:1213–1222. doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhnik-Rosenblau K, Danin-Poleg Y, Kashi Y. Predominant effect of host genetics on levels of Lactobacillus johnsonii bacteria in the mouse gut. Appl Environ Microbiol. 2011;77:6531–6538. doi: 10.1128/AEM.00324-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, et al. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome. 2008;19:382–389. doi: 10.1007/s00335-008-9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Clarke KR, Gorley RN.2006. PRIMER v6: User Manual/Tutorial. PRIMER-E, Plymouth.
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TCT. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends Ecol Evol. 2006;21:517–523. doi: 10.1016/j.tree.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol. 2008;6:725–740. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, et al. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010;4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- Friswell MK, Gika H, Stratford IJ, Theodoridis G, Telfer B, Wilson ID, et al. Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS One. 2010;5:e8584. doi: 10.1371/journal.pone.0008584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Bunge J, Leslin C, Jeon S, Epstein SS. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 2009;3:1365–1373. doi: 10.1038/ismej.2009.89. [DOI] [PubMed] [Google Scholar]
- Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, Hansen AK. Family relationship of female breeders reduce the systematic inter-individual variation in the gut microbiota of inbred laboratory mice. Lab Anim. 2010;44:283–289. doi: 10.1258/la.2010.010058. [DOI] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khachatryan ZA, Ktsoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, Aminov RI. Predominant role of host genetics in controlling the composition of the gut microbiota. PLoS One. 2008;3:e3064. doi: 10.1371/journal.pone.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A, Kang HM, Wade CM, Cotsapas CJ, Kostem E, Han B, et al. Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics. 2010;185:1081–1095. doi: 10.1534/genetics.110.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs A, Ben-Jacob N, Tayem H, Halperin E, Iraqi FA, Gophna U. Genotype is a stronger determinant than sex of the mouse gut microbiota. Microb Ecol. 2011;61:423–428. doi: 10.1007/s00248-010-9787-2. [DOI] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008a;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008b;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh G, Brodziak F, Blaut M. The Toll-like receptors TLR2 and TLR4 do not affect the intestinal microbiota composition in mice. Environ Microbiol. 2008;10:709–715. doi: 10.1111/j.1462-2920.2007.01493.x. [DOI] [PubMed] [Google Scholar]
- Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci USA. 2007;104 (Suppl 1:8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morowitz MJ, Denef VJ, Costello EK, Thomas BC, Poroyko V, Relman DA, et al. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci USA. 2011;108:1128–1133. doi: 10.1073/pnas.1010992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Sokoloff G, Ackert-Bicknell CL, Striz M, Branstetter L, Beckmann MA, et al. Genetic analysis in the Collaborative Cross breeding population. Genome Res. 2011;21:1223–1238. doi: 10.1101/gr.113886.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Younes JA, Van der Mei HC, Gloor GB, Knight R, Busscher HJ. Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat Rev Microbiol. 2011;9:27–38. doi: 10.1038/nrmicro2473. [DOI] [PubMed] [Google Scholar]
- Roberts A, De Villena FP-M, Wang W, McMillian L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome. 2007;18:473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open Source, Platform-independent, Community-supported Software for Describing and Comparing Microbial Communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. W. H. Freeman and Company: San Francisco; 1981. [Google Scholar]
- Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Strauss RE.2010. Matlab statistical functions [computer software]. Retrieved 1 April 2011, from http://www.faculty.biol.ttu.edu/Strauss/ Matlab/matlab.htm .
- Terán-Ventura E, Roca M, Martin M, Abarca M, Martinez V, Vergara P. Characterization of housing-related spontaneous variations of gut microbiota and expression of toll-like receptors 2 and 4 in Rats. Microb Ecol. 2010;60:691–702. doi: 10.1007/s00248-010-9737-z. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaahtovuo J, Korkeamaki M, Munukka E, Viljanen MK, Toivanen P. Quantification of bacteria in human feces using 16S rRNA-hybridization, DNA-staining and flow cytometry. J Microbiol Methods. 2005;63:276–286. doi: 10.1016/j.mimet.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeretssian G, Doiron K, Shao W, Leavitt BR, Hayden MR, Nicholson DW, et al. Gender differences in expression of the human caspase-12 long variant determines susceptibility to Listeria monocytogenes infection. Proc Natl Acad Sci USA. 2009;106:9016–9020. doi: 10.1073/pnas.0813362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yidirim S, Yeoman CJ, Sipos M, Torralba M, Wilson BA, Goldberg TL, et al. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS One. 2010;5:e13963. doi: 10.1371/journal.pone.0013963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.