Video abstract
A video abstract by the authors of this paper is available. video-abstract10324.mov
Keywords: Ac/Ds transposon, exonization, alternative splicing, nonsense-mediated decay pathway
Abstract
Insertion of transposable elements (TEs) into introns can lead to their activation as alternatively spliced cassette exons, an event called exonization which can enrich the complexity of transcriptomes and proteomes. Previously, we performed the first experimental assessment of TE exonization by inserting a Ds element into each intron of the rice epsps gene. Exonization of Ds in plants was biased toward providing splice donor sites from the beginning of the inserted Ds sequence. Additionally, Ds inserted in the reverse direction resulted in a continuous splice donor consensus region by offering 4 donor sites in the same intron. The current study involved genome-wide computational analysis of Ds exonization events in the dicot Arabidopsis thaliana and the monocot Oryza sativa (rice). Up to 71% of the exonized transcripts were putative targets for the nonsense-mediated decay (NMD) pathway. The insertion patterns of Ds and the polymorphic splice donor sites increased the transcripts and subsequent protein isoforms. Protein isoforms contain protein sequence due to unspliced intron-TE region and/or a shift of the reading frame. The number of interior protein isoforms would be twice that of C-terminal isoforms, on average. TE exonization provides a promising way for functional expansion of the plant proteome.
Introduction
Insertion of transposed elements (TEs) within eukaryotic genes is thought to be an important contributor to evolution and speciation.1 A well-known effect of TEs is disruption of the function of the inserted gene, mostly in exons. However, TEs inserted into intronic sequences may not disrupt the target gene; rather, by alternative splicing (AS) and exonization, they alter the regular splicing pattern of a pre-mRNA and result in the translation of new protein isoforms.2 With AS, the inserted TE interferes with the normal splicing of a gene’s transcribed region. With exonization, the inserted TE offers cryptic splice sites incorporated (exonized) as an alternative exon. While the prevailing original splice variant maintains functionality, the additional sequence, free from selection pressure, evolves a new function or eventually vanishes. If the new splice variant is advantageous, selection might operate to optimize the new splice sites and consequently increase the proportion of the alternative splice variant.3
Even without TE, AS is a widespread phenomenon in higher eukaryotes. Yet, Severing et al4 performed a detailed comparison of AS events in alternative spliced orthologs from the dicot Arabidopsis thaliana and the monocot Oryza sativa (rice) and revealed that AS has a limited role in functional expansion of the plant proteome. Unlike with AS, with exonization the resulting transcripts of the inserted gene contain portions of the TE transcripts and may, in that way, alter the reading frames to enrich the complexity of proteomes. Studies of exonization have mostly involved mammalian TEs in silico.5–8 Many results also provided mechanistic insights into the process of exonization, especially 5′ and 3′ splice sites (ie, splice donor/acceptor) formation in Alu exons.9–12 We assessed the ability of a TE to provide splice/acceptor sites by inserting a mini Ds transposon into each intron of the epsps gene.
Ds is a non-autonomous (transposase defective) transposon which is composed of 11 bp terminal-inverted repeats and about 250 bp of both ends (terminal regions) of its full form transposon, Activator (Ac). Previously, we found the first exonization event of Ds in transgenic tobacco containing an inducible transposon system to terminate the marker of transgenic plants. In this system, the marker epsps gene was accompanied by the Ac-based inducible transposon, KCEH. However, the 5′ end of the Ac transposon was located in intron 1 of the modified epsps marker gene.13 We observed abundant exonized transcripts, with the 5′ Ac end of KCEH providing a splice donor site instead of the original site. Since a truncated (one end of) TE located in a plant gene’s intron occurs rarely, we assessed the exonization potential of an intact TE, specifically a mini Ds transposon inserted in the forward or reverse direction in each intron of the epsps gene. Exonization of Ds in epsps was biased in favor of providing splice donor sites from the beginning of the inserted Ds sequence.14 Furthermore, Ds existing in an intron in a reverse pattern could offer 4 donor sites, which can result in the new transcript isoforms having different reading frames according to the different splice junction sites (Fig. 1). However, exonized transcripts may contain a premature termination codon (PTC), which can trigger the decay of the transcript through the nonsense-mediated mRNA decay (NMD) pathway.15 Although our RT-PCR analysis indicated that many PTC-containing transcripts remained abundant,14 the fact that NMD limits AS in expanding the proteome encouraged us to study the role of TE exonization.
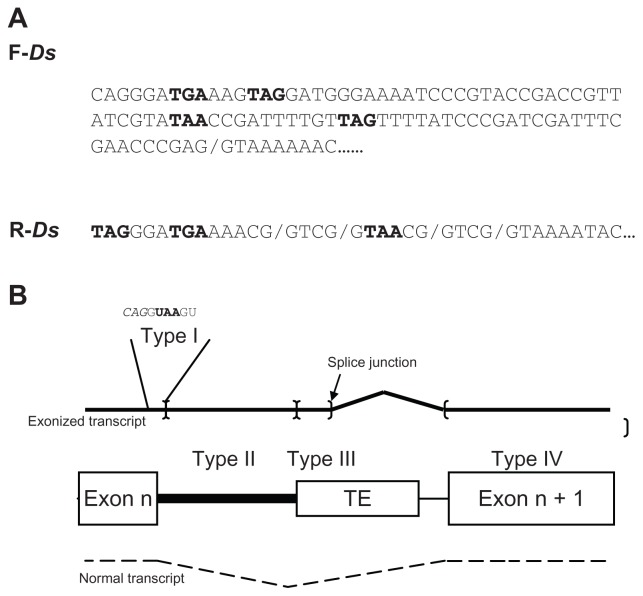
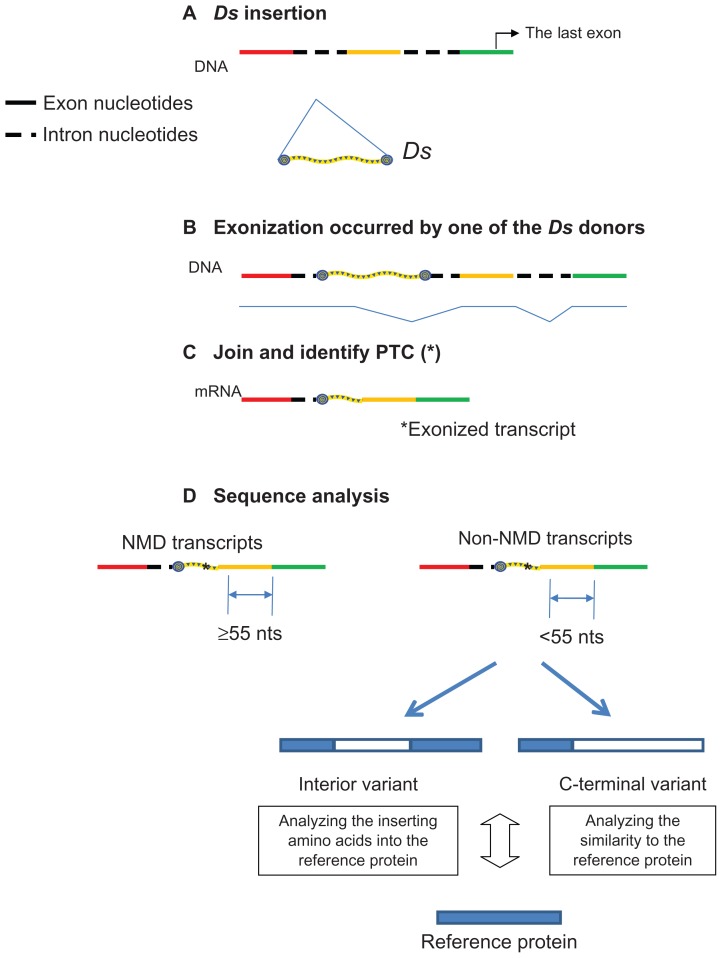
Figure 1.
(A) Ds termini sequences (forward and reverse) providing splice donor junction (slash) and premature termination codons (PTCs) in exonized transcripts (bold). (B) Classification of exonized transcripts (black line) according to location of the PTC.
Notes: Normal transcript is shown as a dashed line. The corresponding DNAs of the TE-inserted target are shown in the center. Black box indicates the unspliced intron. Exonization occurs by using the splice donor (arrow) of TE to join the upcoming exon. The existence and location of an in-frame PTC determines the type of the exonized transcripts, classified into 5 types. As an example, for type I, a PTC (UAA in bold) locates in the skipped exon/intron consensus: italics indicate exonic sequences. Brackets indicate the boundaries of PTC location for classification. Type V transcripts have no in-frame PTC until the end of the gene.
In this study, we performed a detailed analysis of exonized transcript orthologs from the dicot Arabidopsis thaliana and the monocot Oryza sativa (rice) according to the behavior of Ds exonization. These two organisms are widely used model plant systems for functional genomic studies because of their relative small genome sizes, availability of whole genome sequencing, and well-characterized exon/intron annotations. We assumed that Ds inserts after each nucleotide in the genome, with equal chance.16 The resulting exonized transcripts in each genome were classified into 5 types by location of the termination codon: (1) the skipped exon/intron consensus; (2) the intron where Ds exists; (3) the Ds sequence; (4) the original transcripts; and (5) the no in-frame termination codon (Fig. 1). We calculated how many exonized transcripts can bypass the NMD pathway to produce protein isoforms and classified the protein isoforms as C-terminal or interior variants in order to reveal the possible complexity of the proteome caused by TE exonization.
Results
More than half of rice Ds exonized transcripts undergo the NMD pathway or yield truncated protein isoforms without a TE genetic message
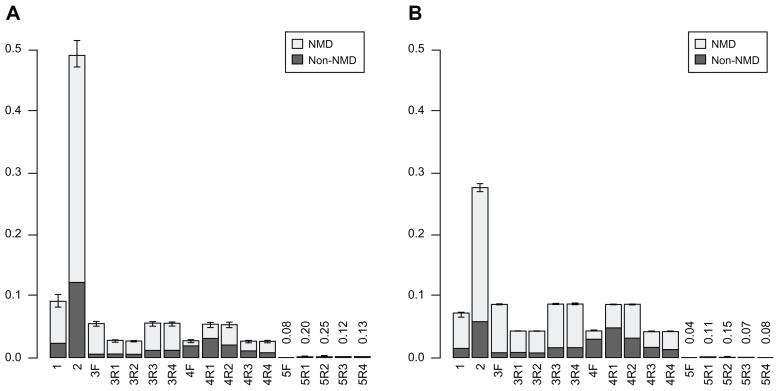
Our previous results revealed that Ds is biased toward providing splice donor sites for exonization; the original exon donor site is skipped and transcription proceeds to the donor of Ds for joining the upcoming exons. With the determined donor sites of Ds (Fig. 1A), a genome-wide analysis of TE exonized transcripts in each intron of rice and Arabidopsis genes yielded 58,016,056 and 37,285,244 exonized transcripts, respectively (Supplementary Tables 1 and 2). The resulting exonized transcripts in each genome were classified into 5 types by location of PTC (Fig. 1B). A PTC in the splice donor consensus is represented as CAG/GTAAGT in plants.17,18 Therefore, about one third of the exonized transcripts created by skipping this donor consensus carry a TAA termination codon in frame. For example, the rice epsps gene has 7 introns, with 5 of the splice donor consensuses containing a PTC while 2 (in introns 4 and 5) are in frame. Because Ds is biased to provide a splice donor site, most exonization events for Ds in introns 4 and 5 will undergo NMD. Genome-wide computational analysis of rice revealed that when the TE upstream splice donor sites are skipped, 9.2% of the exonized transcripts contain a PTC in frame, including 6.8% that are putative targets of NMD (Supplementary Table 1 and Fig. 2A). A similar result was obtained for Arabidopsis (Supplementary Table 2 and Fig. 2B). This consequence was termed as type I exonization of Ds. In such case, the PTC in the skipped consensus is the first limitation in expanding the proteome. The resulting transcripts of Type I exonization may undergo NMD or yield truncated protein isoforms without any genetic message of TE. In type II exonization, the PTCs in the TE inserting introns, but outside the original donor consensus, are in frame and allow the resulting exonized transcripts to become targets of NMD. In rice, 49.1% of exonization events are type II, including 36.9% that will undergo NMD. With type I and II events combined, 58.3% of Ds exonized transcripts will undergo the NMD pathway or yield truncated protein isoforms without a genetic message of the TE. In Arabidopsis, type I and II events present 7.2% (1.5% non-NMD) and 27.6% (5.8% non-NMD), respectively, of all events. In Figure 2, the portion of type I and II in sum in rice differs in Arabidopsis (see Discussion section). In Arabidopsis, a larger portion of events are of subsequent types, which show a similar pattern to those events of rice. We present only the results for subsequent types from Arabidopsis in figures.
Figure 2.
Proportions of 5 types (named 1 to 5) of exonized transcripts for Ds insertion on different chromosomes leading to the nonsense-mediated dacay (NMD) or non-NMD pathway in rice (A) and Arabidopsis (B).
Notes: Data are mean (whiskers are range). R = reverse Ds insertion; F = forward Ds insertion. Numbers above 5F and Rs indicate the percentage of corresponding transcripts.
Ds offers the termination codons of the exonized transcripts
Our results show that the introns’ PTCs are major limits to enhancing proteomes by TE exonization. With a TE acting like Ds, which provides only splice donor sites for exonization, for rice, 43.7% (Arabidopsis, 34.8%) of the exonized transcripts will undergo NMD and 14.6% (7.3%, Arabidopsis) will yield truncated protein isoforms without any TE genetic message. The TE itself may contain PTCs upstream of the donor sites, which leads to the exonized transcripts undergoing the NMD pathway—for example, the reverse Ds element begins with a termination codon (TAG). Therefore, we considered type III exonization events whereby the inserted TE offers the termination codon of the exonized transcripts. According to our previous observation, the forward-inserted Ds provides the splice donor junction at position 91 bp. Four termination codons with 2 reading frames were located upstream of this site (Fig. 1A). In rice, about 5.5% of the total exonized transcripts are type III exonization events of forward-inserted Ds, including 5.0% that are targets of NMD. For reverse-inserted Ds, 2 different patterns of type III exonized transcripts may occur. As shown in Figure 1A, Ds begins with 2 discontinuous but in-framed PTCs. When the exonization in rice occurred at the first 2 donor junction sites (ie, 3R1 and 3R2), the frequency of type III exonized transcripts showed 2.7% (Fig. 2). Because an additional PTC locates after the second donor junction site, the exonization that occurred at the latter 2 donor junction sites (3R3 and 3R4) showed 5.6%. Among all type III transcripts, about 81% are targets of the NMD pathway. Even though the other 19% are non-NMD targets, they would yield protein isoforms with few TE genetic messages. Forward Ds contributed 20 amino acids to the new protein isoforms and reverse Ds a maximum of only 7.
With type IV Ds exonization, the hidden stop codons become in frame
The results above indicate that, in rice, about 80% of the Ds exonized transcripts contain a stop codon upstream of the Ds donor junction sites and may undergo NMD. Even if that is not the case, the translated protein isoforms are either truncated or C-terminal variants. Thus, the potential protein isoforms caused by Ds exonization would depend on the location of termination codons downstream of the Ds donor junction. Therefore, we classified 2 additional types of Ds exonization events. Type IV involves events in which PTCs in the original transcripts become in-frame in the exonized transcripts. The frequency of this type, when Ds is forward-inserted in a gene intron, is 2.7%, including only 0.8% that may undergo the NMD pathway. Ds reverse-inserted in a gene introns would yield 4 different patterns of type IV exonized transcripts of a single Ds insertion site, termed 4R1 (for a frequency of 5.4%), 4R2 (5.3%), 4R3 (2.7%) and 4R4 (2.7%), by order of the Ds donor junctions located on the consensus (Fig. 1A). Among all type IV transcripts, only 1.9% (4F), 3.1% (4R1), 2.1% (4R2), 1.2% (4R3) and 0.9% (4R4) were non-NMD targeted and would yield protein isoforms. The translated products of 4R3 and 4R2 differ by only 2 additional amino acids because the splice junction of 4R3 locates downstream of 4R2 by 6 bp.
Type V Ds exonized transcripts may yield functional protein isoforms
Type V exonization events represent those in which resulting transcripts harbor no in-frame termination codon until the end of the target gene. In rice, these occur with low frequencies, of 0.08%, 0.20%, 0.25%, 0.12% and 0.13% for 5F, 5R1, 5R2, 5R3 and 5R4, respectively (Fig. 2). Exonized transcripts of Type V potentially enhanced the functional protein isoforms of the Ds-inserted genes.
Analysis of protein isoforms created by Ds exonized transcripts
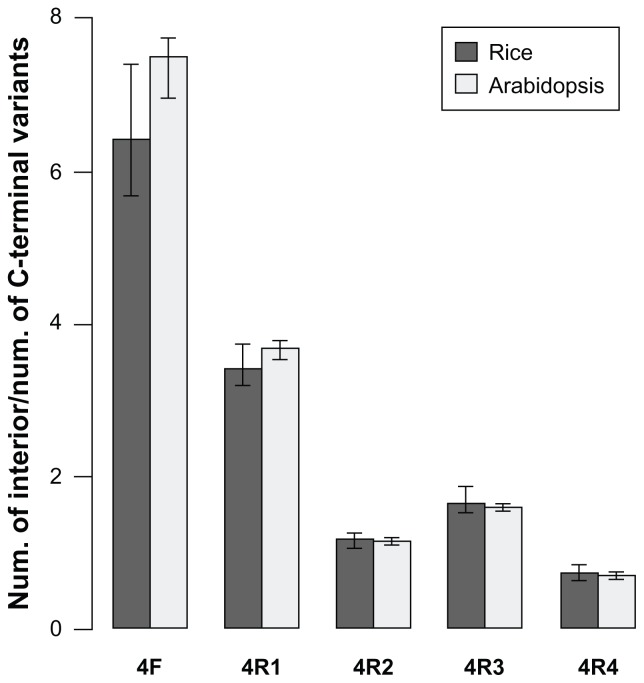
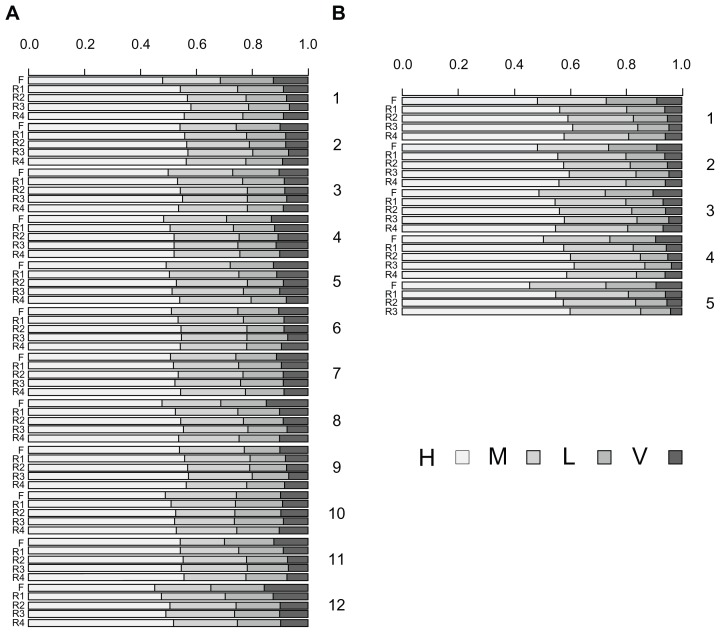
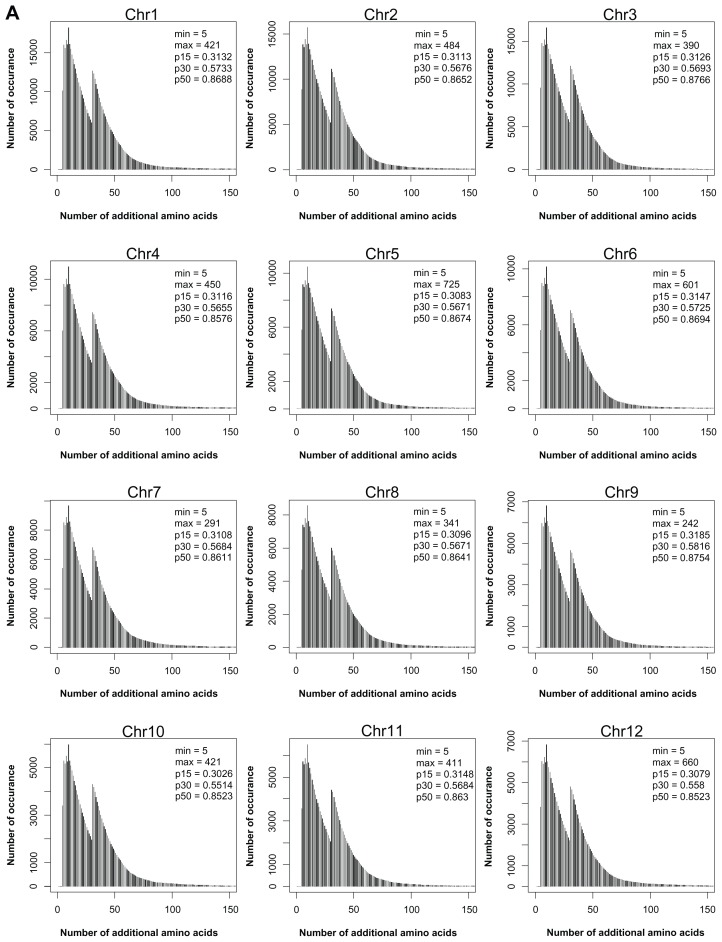
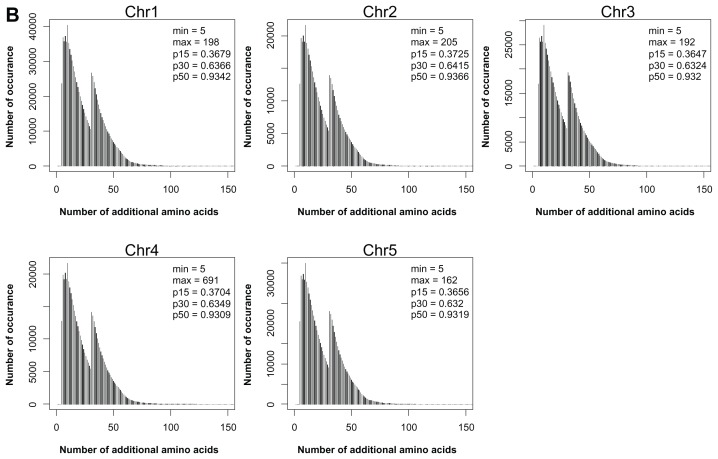
Although many exonization events can produce different protein isoforms, the extent of this production needs further analysis. We analyzed the translated products yielded by type V transcripts and non-NMD transcripts of type IV. The translated protein isoforms were characterized as C-terminal or interior variants. For C-terminal isoforms, peptides from the new reading frame replace the C terminus of the reference protein. All type V transcripts yielded this kind of isoforms. For interior isoforms, the exonized transcripts have the same termination codon as the reference transcript. In these transcripts, the upcoming transcripts of the exonized junction have the same reading frame as the reference gene, and therefore, the translation products of the TE and intron transcripts act as a “peptide insertion” in the reference gene products. Figure 3 shows the ratio of interior to C-terminal isoforms of type IV non-NMD proteins in rice and Arabidopsis. Interestingly, the number of 4F interior variants are about 6-fold that of C-terminal variants. To analyze the similarity to the reference proteins, C-terminal variants were further graded by the proportion of amino acids that were identical to the reference protein. The proportions are classified as <25% (very low), 25% to 50% (low), 50% to 75% (medium) and >75% (high). Figure 4 shows that in most chromosomes, the proportion of H and M variants was more than 40% and 20%, respectively, in Ds inserted at forward or reverse direction. For the interior variants, Figure 5 shows the length of inserted peptides for all interior isoforms from each chromosome. The number of amino acids of this type of insertion can vary from 5 to 725. The extremely long insertion of peptide resulted from Ds reverse-inserted in intron 2, position 2147, of rice gene Os05s0162500, which originally created a translation product of 126 amino acids. Interestingly, more than 30% of interior isoforms differ from the reference proteins by ≤15 amino acids and among these, 70,359 isoforms (about 2%) differ from the reference proteins by an additional 5 amino acids. Therefore, many interior isoforms contain short insertions. Specifically, a 5 amino acid insertion may occur in different introns of a single gene and result in fine modification of different domains of the reference protein. As an example, Ds reverse-inserted in each intron of position 1 of the rice gene Os11g0446500 can create 22 interior protein isoforms that have 5 additional amino acids along the corresponding introns of this gene, which is characterized as a P-type ATPase. Blast analysis revealed that all isoforms retain the conserved domains, E1-E2 ATPase, HAD_ like and ATPase-Plipid (data not shown). Comparison of each isoform to the reference protein revealed that all interior variants showed a slight modification in the secondary structure, either a helix or sheet strand (Supplementary Fig. 1: http://homepage.ntu.edu.tw/~lyliu/Exon2011/).
Figure 3.
Ratio of interior to C-terminal protein isoforms for type IV transcripts in rice (dark gray) and in Arabidopsis (light gray) with Ds insertion.
Notes: Data are ratios; whiskers indicate the minimal and the maximal ratios in each case.
Figure 4.
Distribution of C-terminal variants among Type V and Type IV non-NMD transcripts in rice (A) and Arabidopsis (B).
Note: <25% (very low), 25%–50% (low), 50%–75% (medium) and >75% (high).
Figure 5.
Number of additional amino acids in the interior protein isoforms in rice (A) and Arabidopsis (B) genomes.
Notes: ‘Min’ and ‘max’ are the minimum and maximum number of additional amino acids, respectively; p15, p30, and p50 are the cumulative proportions of interior variants with additional number of amino acids ≤15, ≤30, and ≤50, respectively.
Finally, we analyzed the protein isoforms created with a reverse Ds single insertion, which can offer 4 splice junctions and may therefore create 4 different isoforms at once. Because the exonized transcripts using R2 and R3 splice sites may result in the proteins differing by 2 amino acids, their products may act similarly and were also considered the same in subsequent analyses. As a result, exonized transcript isoforms caused by a reverse Ds insertion could yield a maximum of 3 different isoforms. From a total number of 3,534,907 insertion sites, 16.9% and 25.2% of reverse Ds insertions may yield 3 and 2 additional isoforms respectively (Supplementary Table 3). These results revealed the potential of TE exonization to enrich the transcriptome and subsequent proteome in plants.
Discussion
The contribution of TE exonization to evolution has been previously indicated.1 In mammalian cells, most exonization events were caused by retrotransposons suh as the Alu element in human and the B1 element, homologous to the left arm of the Alu element, in mouse.6 In plants, the exonization may be alternatively triggered by DNA transposons (eg, Ds element).14 More interestingly, exonization of Ds may yield 3 different reading frames of upcoming transcripts.
In this study, we created genome-wide exonized transcript orthologs with Ds insertion from the dicot A. thaliana and the monocot O. sativa (rice) and studied their impact on proteome complexity in plants. In general, exonization is defined as an event in which TEs can create a new exon when inserted into the introns of the target gene.19 Nonetheless, an HSmar1 transposon could contribute to a functional anthropoid protein when inserted downstream of the target gene, and the target’s stop codon was subsequently removed during evolution.20 Nevertheless, in our study the exonization events were presumed to occur with intronic Ds. The process yielded 58,016,056 and 37,285,244 exonized transcripts for rice and Arabidopsis respectively (Supplementary Tables 1 and 2). Indeed, the abundance of the exonized transcripts depended on the TE exonization efficiency, which is affected by many factors that induce or inhibit splicing process. These factors include the sequences of the inserted TE and the flanking intron, the locations/sequences of other introns within the gene, and spatial, temporal and environmental conditions of the organism.21–22 Still, genome-wide analysis of all possible exonized transcripts allows for assessing the extent to which it leads to proteome expansion.
To yield the translation products, the first step is to exclude exonized transcripts that contain a PTC, which can trigger the decay of the transcript through the NMD pathway. According to the location of termination codons, we classified the exonized transcripts into 5 types, which with the exception of type II showed similar distribution in rice and Arabidopsis. In Figure 2, the sum portion of type I and II in rice differs to that of Arabidopsis because of the shorter introns in the latter. Thus, we discuss only the results obtained in rice in this section. About 80.5% of all exonized transcripts are types I, II, and III, which would yield no translation product or a truncated protein isoform with little TE genetic message. Thus, further analysis of the potential protein isoforms caused by Ds exonization would be based on the remaining exonized transcripts, types IV (18.8%) and V (0.8%). The former type has a hidden termination codon of the original transcript, which becomes in-frame in exonized transcripts. Half of the type IV (which are non-NMD) and type V transcripts should be the main sources of new protein isoforms of the reference genes. Different NMD levels had been observed in different types of exonization. The proportions of transcripts that undergo NMD for a gene depend on the number of introns, the intron lengths, and the length of the transposon. We further characterized the translation products of these transcripts. Isoforms generated by exonization often contain an additional protein sequence due to an unspliced intron-TE region and/or a shift of the reading frame. Thus, we characterized the translated protein isoforms as C-terminal or interior variants. From a total of 2,125,865 C-terminal variants, about 40% and 20% show high and medium similarity to their reference protein, respectively (Fig. 4). Therefore, by retaining the major portion of the reference protein, these variants may provide modified peptides at C-terminal yet functional isoforms for selective advantage. The other 40% of the C-terminal variants, designated to have low (L) or very low (V) similarity with the reference protein, mostly result from Ds insertions in the first few introns. These variants may retain the less functional domains of their reference proteins. However, the second intronic Alu-exonized C-terminal variants code for functional isoforms, which were determined as new members of the reference protein.23–25 Therefore, these L and V variants may still act as functional proteins.
Interior variants showed 2 interesting features. In Figure 3, the ratio of the number of interior to C-terminal variants showed significantly different for each sub-type; specifically for 4F transcripts, the number of interior translation products was about 6-fold to that of C-terminal variants. Therefore, the inserted peptide for most 4F translation protein isoforms was composed of at least 30 amino acids encoded by Ds transposon. As well, the abundance of products contributed to the second peak of additional amino acids in Figure 5, which indicates that the additional amino acids in all interior variants showed a bimodal distribution pattern. Figure 5 shows the composition of inserted peptides culminating in 2 groups, of 31 and about 10 amino acids. The former consists mostly of 4F transcripts and the latter 4R transcripts. Because the TE exonization yielded interior variants acting as “peptide insertions” to the reference protein, the peptide with more than 31 amino acids may affect the reference protein domains or even a new functional domain. Indeed, previous reports indicated that a reference protein, replaced by a TE-coding peptide of 28 amino acids at the C terminus, generated a new domain and contributed to the proapoptotic capability.25 As compared with “medium-sized” inserting peptides, more than 30% of the interior variants, or a total of 1,153,978, contained an inserted peptide of equal to or less than 15 amino acids. Specifically, 5 amino acids inserts presented as 1.9%, which may finely modify the reference gene. Unlike the function of an AS protein, which is generally expected to modulate the addition or deletion of complete functional domains, these small amino acid sequence changes on the protein function need additional structural analysis.4
A single reverse-insertion of Ds may yield 4 exonized transcript isoforms and subsequently 4 protein isoforms. Our previous study indicates the 4 splice donors were used for exonization with different efficiencies: 1st (10.7%), 2nd (6.7%), 3rd (44.0%) and 4th (38.7%). This implies the chances of yielding exonized transcripts by using the latter two donors are about 5-fold to those using the former two. For simplicity, in this study, the results were presented under the assumption that the donors are used with the same efficiency. Whether or not this feature coincides, the proportion of each translated protein isoform needs further studies. Specifically, if the new splice variant is advantageous, selection might operate to optimize the new splice sites and consequently increase the proportion of the alternative splice variant.3 In order to clarify the significant proteomic diversity resulting from the 4 distinctive transcripts obtained from a single insertion, we characterized the products of R2 and R3 as the same isoform. According to this definition, exonization of a single reverse-insertion of Ds may yield a maximum of 3 unique isoforms for selective advantage, in addition to the reference protein. From a total 3,534,907 insertion sites, 16.9% and 25.2% of reverse Ds insertions can yield 3 and 2 additional isoforms, respectively (Supplementary Table 3). Therefore, a single TE insertion may yield multiple protein isoforms for the target gene. These results allow us to perform more evolutional studies of Ds at molecular level.
In conclusion, we show that Ds exonization can yield abundant new protein isoforms, which further reveals the role of TEs in expanding plant proteomes.
Materials and Methods
Data sources and exonized transcript construction
Arabidopsis and rice chromosome Genbank data and whole-genome sequences were downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/genomes/PLANTS/PlantList.html), and the amino acid coding region (CDS) for each gene was extracted. For rice, every gene has only one CDS record. However, for Arabidopsis, some genes have multiple CDS records, so we used only the first CDS record to avoid redundancy. Exonization was defined as an event in which a transcript variant was created with insertion of a TE in the intronic sequence of a gene. Therefore, we considered only genes that were completely sequenced and had at least 1 intron.
The construction of the exonized transcripts involved use of R.26 For each gene, we simulate Ds insertion events for every intron. A flow chart for the analytic steps was shown in Figure 6. Let a target gene, G, have I introns (and, of course, I + 1 exons), with the ith intron of length ni. First, the sequence of the 512 bp Ds was inserted in a forward or reverse direction after the jth nt (j = si, ..., ni) of the ith intron of G. Usually, si = 0, meaning “before the first nucleotide.” When the ith exon of G has length mi < 9, let si be 9 − mi to avoid exonic insertions. Furthermore, if mi > ni, the ith intron of G was skipped for the analyses. The insertion we describe here refers to the insertion of the letters of Ds after the jth nucleotide. This insertion was equivalent to a biological event of Ds inserted at 8 bp before the assigned position. Biologically, the insertion of Ds causes the duplication of 8 bp of G right after the insertion position, and the sequence of the Ds starts at the 9th nt after the insertion position.
Figure 6.
Flow chart of the steps for analyzing the exonized transcripts.
Second, we obtained all exonized sequences by recognizing appropriate splice donor/acceptor sites. From our previous observations, the insertion of Ds would affect the recognition of the donor sites. More specifically, the original donor site located at the nearest upstream would be skipped and replaced by the donor site provided by Ds. The Ds preferred to provide early splice donor sites in an exonized event. With the Ds inserted in a forward direction in the target, the splice donor junction occurs at position 91 bp; with the Ds inserted in a reverse direction, the splice donor junction may position at 14, 18, 24 and 28 bp (Fig. 1). Thus, one TE may result in 1 and 4 exonized transcript variants for forward and reverse insertions, respectively.
Finally, the exonized transcripts were constructed by joining the sequences of the 1st to the ith exons, the first j nt of the ith intron, the Ds sequences until the junction site, and the sequences of the (i + 1)th to the (I + 1)th exons.
Analysis of exonized transcript variants and prediction of isoforms
All exonized transcripts were assigned for open reading frame (ORF) analysis, starting at the original start codon and terminating at the first in-frame stop codon. The transcripts were designated type I, II, III, or IV. This designation was based on whether the in-frame stop codon occurred at the conserved region in the original splice junction, the intron inserted by Ds, the Ds, or any exon after Ds insertion, respectively. If no in-frame stop codon was found during ORF analysis, the corresponding transcript was designated type V and the incomplete transcript without a stop codon was output directly. All transcripts containing a termination codon more than 55 nt upstream of the last exon/exon junction were considered putative targets for the NMD pathway15,27 and were excluded from isoform prediction.
The proteins for transcripts not targeted to the NMD pathway were further classified into 2 subtypes: an interior isoform if the termination codon was the same as the reference transcript (the transcript without Ds insertion); otherwise, a C-terminal isoform. For an interior variant, the number of additional peptides inserted in the middle was recorded. For a C-terminal variant, its similarity to the corresponding reference protein was defined as the number of peptides in the isoform being identical to the reference protein divided by the total number of peptides in the reference protein. Analysis of secondary structure prediction of each isoform was performed with PSIPRED server (http://bioinf.cs.ucl.ac.uk/psipred/).
Supplementary Data
Additional files 1–2: supplementary tables 1 and 2
Description of data: Analysis of Ds exonized transcripts in each intron of rice (Supplementary Table 1) and Arabidopsis (Supplementary Table 2) genes yielded 58,016,056 and 37,285,244 exonized transcripts, respectively.
Additional file 3: supplementary table 3
Description of data: The number of positions where at least one transcript can be yielded after the Ds is inversely inserted. Some of the positions can be related to more than one transcript products due to different splice donor junctions, respectively. available. video-abstract10324.mov
Footnotes
Competing Interests
Author(s) disclose no potential conflicts of interest.
Author Contributions
YCC conceived and designed the experiments. LDL analyzed the data. YCC wrote the first draft of the manuscript. Both authors contributed to the writing of the manuscript and jointly developed the structure and arguments for the paper. The final manuscript was reviewed and approved by both authors.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Funding
This project was supported by the National Science Council (Grant No. NSC98-2313-B-002-041-MY3) of Taiwan.
References
- 1.Sela N, Kim E, Ast G. The role of transposable elements in the evolution of non-mammalian vertebrates and invertebrates. Genome Biol. 2010;11(6):R59. doi: 10.1186/gb-2010-11-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9(5):397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitz, Brosius Exonization of transposed elements: A challenge and opportunity for evolution. Biochimie. 2011;93(11):1928–34. doi: 10.1016/j.biochi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Severing E, van Dijk A, Stiekema W, van Ham R. Comparative analysis indicates that alternative splicing in plants has a limited role in functional expansion of the proteome. BMC Genomics. 2009;10(1):154. doi: 10.1186/1471-2164-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy A, Sela N, Ast G. TranspoGene and microTranspoGene: transposed elements influence on the transcriptome of seven vertebrates and invertebrates. Nucleic Acids Res. 2008 Jan;36(Database issue):D47–52. doi: 10.1093/nar/gkm949. Epub Nov 5, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mersch B, Sela N, Ast G, Suhai S, Hotz-Wagenblatt A. SERpredict: Detection of tissue- or tumor-specific isoforms generated through exonization of transposable elements. BMC Genet. 2007 Nov 6;8:78. doi: 10.1186/1471-2156-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mola G, Vela E, Fernández-Figueras MT, Isamat M, Munoz-Mármol AM. Exonization of Alu-generated Splice Variants in the Survivin Gene of Human and Non-human Primates. J Mol Biol. 2007;366(4):1055–63. doi: 10.1016/j.jmb.2006.11.089. [DOI] [PubMed] [Google Scholar]
- 8.Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A, Ast G. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu’s unique role in shaping the human transcriptome. Genome Biol. 2007;8(6):R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krull M, Petrusma M, Makalowski W, Brosius J, Schmitz J. Functional persistence of exonized mammalian-wide interspersed repeat elements (MIRs) Genome Res. 2007 Aug;17(8):1139–45. doi: 10.1101/gr.6320607. Epub Jul 10, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lev-Maor G, Sorek R, Levanon E, Paz N, Eisenberg E, Ast G. RNA-editing-mediated exon evolution. Genome Biol. 2007;8(2):R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ram O, Schwartz S, Ast G. Multifactorial Interplay Controls the Splicing Profile of Alu-Derived Exons. Mol Cell Biol. 2008 May;28(10):3513–25. doi: 10.1128/MCB.02279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorek R, Lev-Maor G, Reznik M, et al. Minimal Conditions for Exonization of Intronic Sequences: 5′ Splice Site Formation in Alu Exons. Mol Cell. 2004 Apr 23;14(2):221–31. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 13.Charng Y-C, Li K-T, Tai H-K, Lin N-S, Tu J. An inducible transposon system to terminate the function of a selectable marker in transgenic plants. Mol Breeding. 2008;21(3):359–68. [Google Scholar]
- 14.Huang K-C, Yang H-C, Li K-T, Liu L-Y, Charng Y-C. Ds transposon is biased towards providing splice donor sites for exonization in transgenic tobacco. Plant Mol Biol. 2012 Jul;79(4–5):509–19. doi: 10.1007/s11103-012-9927-9. Epub May 27, 2012. [DOI] [PubMed] [Google Scholar]
- 15.Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 16.Ito T, Motohashi R, Kuromori T, et al. A Resource of 5,814 Dissociation Transposon-tagged and Sequence-indexed Lines of Arabidopsis Transposed from Start Loci on Chromosome 5. Plant Cell Physiol. 2005 Jul;46(7):1149–53. doi: 10.1093/pcp/pci112. Epub Apr 19, 2005. [DOI] [PubMed] [Google Scholar]
- 17.Baek J-M, Han P, Iandolino A, Cook D. Characterization and comparison of intron structure and alternative splicing between Medicago truncatula, Populus trichocarpa, Arabidopsis and rice. Plant Mol Biol. 2008 Jul;67(5):499–510. doi: 10.1007/s11103-008-9334-4. Epub Apr 27, 2008. [DOI] [PubMed] [Google Scholar]
- 18.Schuler MA, Reddy ASN, Golovkin M. Splice Site Requirements and Switches in Plants Nuclear pre-mRNA Processing in Plants. Vol. 326. Springer; Berlin Heidelberg: 2008. pp. 39–59. [DOI] [PubMed] [Google Scholar]
- 19.Sela N, Mersch B, Hotz-Wagenblatt A, Ast G. Characteristics of Transposable Element Exonization within Human and Mouse. PLoS One. 2010;5(6):e10907. doi: 10.1371/journal.pone.0010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci U S A. 2006 May 23;103(21):8101–6. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3(4):285–98. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 22.Zabala G, Vodkin L. Novel exon combinations generated by alternative splicing of gene fragments mobilized by a CACTA transposon in Glycine max. BMC Plant Biol. 2007;7(1):38. doi: 10.1186/1471-2229-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J-W, Chandra D, Tang S-H, Chopra D, Tang DG. Identification and Characterization of Bimgamma, a Novel Proapoptotic BH3-only Splice Variant of Bim. Cancer Res. 2002 May 15;62(10):2976–81. [PubMed] [Google Scholar]
- 24.Wu M, Li L, Sun Z. Transposable element fragments in protein-coding regions and their contributions to human functional proteins. Gene. 2007 Oct 15;401(1–2):165–71. doi: 10.1016/j.gene.2007.07.012. Epub Jul 26 2007. [DOI] [PubMed] [Google Scholar]
- 25.Yi P, Zhang W, Zhai Z, Miao L, Wang Y, Wu M. Bcl-rambo beta, a special splicing variant with an insertion of an Alu-like cassette, promotes etoposide-and Taxol-induced cell death. FEBS Lett. 2003 Jan 16;534(1–3):61–8. doi: 10.1016/s0014-5793(02)03778-x. [DOI] [PubMed] [Google Scholar]
- 26.R: A Language and Environment for Statistical Computing [computer program] Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 27.Hori K, Watanabe Y. Context Analysis of Termination Codons in mRNA that are Recognized by Plant NMD. Plant Cell Physiol. 2007 Jul;48(7):1072–8. doi: 10.1093/pcp/pcm075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional files 1–2: supplementary tables 1 and 2
Description of data: Analysis of Ds exonized transcripts in each intron of rice (Supplementary Table 1) and Arabidopsis (Supplementary Table 2) genes yielded 58,016,056 and 37,285,244 exonized transcripts, respectively.
Additional file 3: supplementary table 3
Description of data: The number of positions where at least one transcript can be yielded after the Ds is inversely inserted. Some of the positions can be related to more than one transcript products due to different splice donor junctions, respectively. available. video-abstract10324.mov