Abstract
Family history is the second strongest risk factor for Alzheimer disease (AD) following advanced age. Twin and family studies indicate that genetic factors are estimated to play a role in at least 80% of AD cases. The inheritance of AD exhibits a dichotomous pattern. On one hand, rare mutations in APP, PSEN1, and PSEN2 virtually guarantee early-onset (<60 years) familial AD, which represents ∼5% of AD. On the other hand, common gene polymorphisms, such as the ε4 and ε2 variants of the APOE gene, can influence susceptibility for ∼50% of the common late-onset AD. These four genes account for 30%–50% of the inheritability of AD. Genome-wide association studies have recently led to the identification of 11 additional AD candidate genes. This paper reviews the past, present, and future attempts to elucidate the complex and heterogeneous genetic underpinnings of AD.
After advanced age, family history is the second greatest risk factor for Alzheimer disease (AD). Well-described mutations in APP, PSEN1, PSEN2, and APOE account for only 30%–50% of the heritability of AD.
GENETICS OF EARLY-ONSET FAMILIAL ALZHEIMER DISEASE
Following advanced age, family history is the second greatest risk factor for Alzheimer disease (AD). AD is considered to be a genetically dichotomous disease presenting in two forms: early-onset familial cases usually characterized by Mendelian inheritance (EO-FAD), and late-onset (≥60 years), with no consistent mode of transmission (LOAD; Bertram and Tanzi 2005; Tanzi and Bertram 2005). Familial clustering of AD cases is more obvious for the early-onset form that strikes under the age of 60 years. However, it is estimated that up to 80% of AD involves the inheritance of genetic factors, based on twin and family studies (Gatz et al. 2006). So-called “sporadic” AD is strongly influenced by genetic variants combined with life exposure factors. EO-FAD is most often caused by rare, fully penetrant mutations in three different genes.
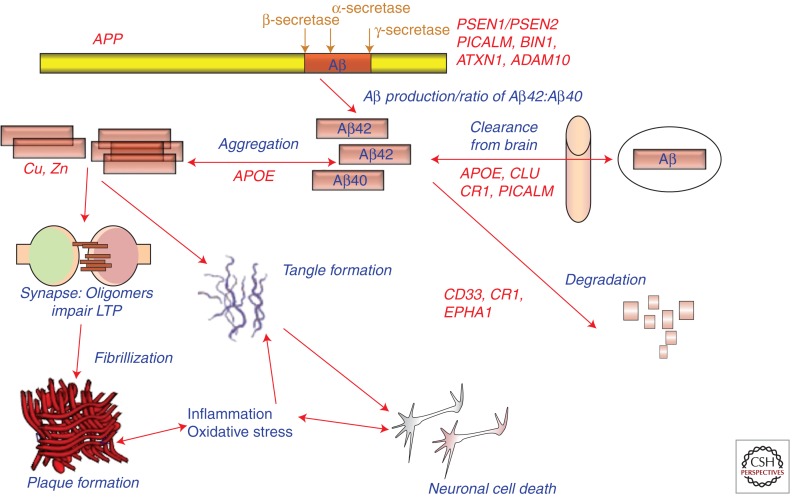
In 1987, we as well as others isolated the amyloid β (A4) protein precursor (APP; Goldgaber et al. 1987; Kang et al. 1987; Tanzi et al. 1987a, 1988) and mapped it to chromosome 21. The cloning of APP was carried out based on the hypothesis that it would be an AD gene. Glenner and Wong (1984) first put forward the prediction that the gene responsible for the production β-amyloid, which makes up senile plaques and cerebral blood vessel deposits of amyloid, would be on chromosome 21 and carry mutations causing AD. In that same seminal study, they were able to analyze cerebral β-amyloid deposits to derive the partial amino acid sequence of the amyloid β protein (Aβ). Glenner also proposed that AD is a cerebral amyloidosis, in which cerebral β-amyloid drives all subsequent pathology (see Fig. 1). This central thesis was later reinterpreted and reported as the “amyloid cascade hypothesis” of AD (Hardy and Higgins 1992; Hardy and Selkoe 2002; Tanzi and Bertram 2005).
Figure 1.
Potential roles of select Alzheimer disease (AD) genes in Aβ-related pathogenesis of AD. (Modified from Bertram and Tanzi 2008; reprinted with permission from the author.)
In the same issue of Science reporting the isolation of APP (Goldgaber et al. 1987; Tanzi et al. 1987a), we simultaneously reported genetic linkage of EO-FAD to genetic markers on chromosome 21 in the vicinity of APP (St George-Hyslop et al. 1987). Interestingly, we would later show that the four large EO-FAD families used in that study were not genetically linked to APP (Tanzi et al. 1987b); they were ultimately demonstrated to carry PSEN1 mutations (Sherrington et al. 1995). The concurrent genetic linkage of these four EO-FAD families to both PSEN1 and markers on the proximal portion of chromosome 21 remains to be resolved.
These studies prompted other groups, including that of John Hardy, to test and confirm the genetic linkage of other independent EO-FAD families to our markers on chromosome 21. In 1991, the first EO-FAD mutation was found in APP by resequencing these independent EO-FAD families that were tightly linked to chromosome 21 (Goate et al. 1991). However, prior to the Goate et al. (1991) paper, Frangione and colleagues had already reported the first pathogenic mutation in APP (Levy et al. 1990). Their sequencing of exons 16 and 17 of APP, encoding the Aβ portion, uncovered a pathogenic mutation in APP responsible for Dutch hereditary cerebral hemorrhage with amyloidosis. Goate et al. (1991), then resequenced the same two APP exons to reveal the first EO-FAD mutation (London mutation; V717I) in APP.
By 1992, it was clear that mutations in APP accounted for only a tiny proportion of EO-FAD. In fact, none of the 50 EO-FAD families that we resequenced in our laboratory between 1987 and 1988 revealed APP mutations. In 1995, several EO-FAD mutations were found in presenilin 1 (PSEN1; Sherrington et al. 1995) on chromosome 14. This was reported in a study led by Peter St. George-Hyslop and a large host of collaborators, including our laboratory. Shortly thereafter, we first reported a homolog of PSEN1 called presenilin (PSEN2; Levy-Lahad et al. 1995) on chromosome 1 and showed that it harbors the N141I mutation in Volga-German EO-FAD families. This was confirmed by Rogaev et al. (1995) shortly afterward.
To date, 24 mutations (plus duplications) have been reported for APP, 185 for PSEN1, and 13 for PSEN2 (Table 1; Alzheimer Disease and Frontotemporal Dementia Mutation Database; http://www.molgen.ua.ac.be/ADMutations). All but one of these mutations (PSEN2-N141I) is inherited with EO-FAD in a fully penetrant, autosomal-dominant manner. Another mutation in APP is transmitted in a recessive manner.
Table 1.
Early-onset familial Alzheimer disease genes and their pathogenic effects
| Gene | Protein | Chromosome | Mutations | Molecular phenotype |
|---|---|---|---|---|
| APP | Amyloid β (A4) protein precursor | 21q21 | 24 (duplication) | Increased Aβ42/Aβ40 ratio |
| Increased Aβ production | ||||
| Increased Aβ aggregation | ||||
| PSEN1 | Presenilin 1 | 14q24 | 185 | Increased Aβ42/Aβ40 ratio |
| PSEN2 | Presenilin 2 | 1q31 | 14 | Increased Aβ42/Aβ40 ratio |
All but a handful of the more than 200 EO-FAD mutations in APP, PSEN1 and PSEN2 lead to a common molecular phenotype: an increase in the ratio of Aβ42:Aβ40 (Scheuner et al. 1996; Tanzi and Bertram 2005). One mutation (Swedish) in APP increases all species of Aβ and several others, resulting in amino acid substitution within the Aβ domain and leading to increased aggregation of Aβ. However, most EO-FAD mutations increase the ratio of Aβ42:Aβ40 (Table 2). The relative increase in Aβ42 promotes the aggregation of the peptide into oligomers and ultimately amyloid fibrils (Jarrett et al. 1993).
Table 2.
Results of select genome-wide association studies of late-onset Alzheimer disease
| Genome-wide association studies | Study design | Population | Major genes identified |
|---|---|---|---|
| Reiman et al. 2007 | Case–control | US | APOE, GAB2 |
| Bertram 2008 | Family-based | US | APOE, ATXN1, CD33, GWA_14q31 |
| Lambert et al. 2009 | Case–control | US, Europe | APOE, CLU, CR1 |
| Harold et al. 2009 | Case–control | US, Europe | APOE, CLU, PICALM |
| Seshadri et al. 2010 | Case–control | US, Europe | APOE, BIN1 |
| Naj et al. 2011 | Case–control | US, Europe | MS4A6A/MS4A4E, EPHA1, CD33, CD2AP |
| Hollingworth et al. 2011 | Case–control | US, Europe | ABCA7, MS4A6A/MS4A4E, EPHA1, CD33, CD2AP |
Three years after their discovery in 1995, the presenilins were shown to be necessary for the generation of Aβ from APP (De Strooper et al. 1998). One year later, Wolfe et al. (1999) showed that the presenilins act as aspartyl proteases that carry out γ-secretase cleavage of APP to produce Aβ. γ-Secretase can generate a variety of Aβ species ranging in size from 37 to 46 amino acids. Whether EO-FAD mutations appear in APP near the γ-secretase cleavage site or in the presenilins, the vast majority of them serve to increase the ratio of Aβ42:Aβ40. γ-Secretase modulators are aimed at reversing this ratio and represent very promising compounds for treating and preventing AD based on what has been garnered from studies addressing the molecular mechanism of EOFAD mutations (Table 1) in APP, PSEN1 and PSEN1 (Kounnas et al. 2010).
ADDITIONAL EO-FAD CANDIDATE GENES
Beyond APP, PSEN1, and PSEN2 attempts to identify additional EO-FAD genes have led to potentially pathogenic mutations in three other genes. A missense mutation (R406W) in the tau gene (MAPT) on chromosome 17q was reported to be tightly linked to AD in a Belgian family (Rademakers et al. 2003; Ostojic et al. 2004). However, before, this mutation can be validated to cause AD, autopsy confirmation will be necessary to rule out a diagnosis of frontotemporal lobe dementia (FTDP-17). EO-FAD in a family from the Netherlands was also linked to chromosome 7q36 (Rademakers et al. 2005) near the PAX transcription activation domain interacting protein gene (PAXIP1). Finally, the gene for PEN2 on chromosome 19, encoding the γ-secretase component, pen-2, was reported to harbor a missense mutation, D90N, in an AD family (Frigerio et al. 2005). To date, no functional consequences have been demonstrated for this mutation. Clearly, all three of these additional EO-FAD candidate genes will require further confirmation.
GENETICS OF LATE-ONSET AD: APOE
Late-onset Alzheimer is the most common form of the disease and is defined by onset age >65 years. Whereas EO-FAD is characterized by classic Mendelian inheritance usually in an autosomal-dominant manner, for LOAD, we observe a genetically complex pattern of inheritance in which genetic risk factors work together with environmental factors and life exposure events to determine lifetime risk for AD. Consequently, it is much more difficult to reliably identify novel LOAD loci, especially because efforts at replication are often plagued by a mixture of replications and refutations.
Thus far, the only gene variant considered to be an established LOAD risk factor is the ε4 allele of the apolipoprotein E gene (APOE; Strittmatter et al. 1993) on chromosome 19q13. The road to the identification of APOE as an AD gene began with genetic linkage of LOAD to chromosome 19 in the vicinity of APOE followed by the finding that apoE binds Aβ in cerebrospinal fluid leading. Thus, APOE represented a viable AD candidate gene based on both functional data and genomic position. The three major alleles of APOE correspond to combinations of amino acids at residues 112 and 158 (ε2: Cys112/Cys158; ε3: Cys112/Arg158; ε4: Arg112/Arg158). The ε4-allele of APOE increases AD risk by approximately fourfold when inherited in one copy and by greater than 10-fold for two doses of the allele. In contrast, the ε2 allele of APOE exerts “protective” effects (Corder et al. 1994).
Whereas the EO-FAD mutations are sufficient, but not necessary, to cause AD, the APOE ε4 allele is neither necessary nor sufficient to cause AD. Instead, it operates as a genetic risk factor for AD that decreases the age of onset in a dose-dependent manner. Functionally, APOE normally plays a role in lipid metabolism and transport. However, in AD, it is believed to play a role in the clearance of Aβ from brain.
Roses et al. (2010) has proposed that other genes in the genomic neighborhood of APOE may actually confer risk for AD, for example, a polymorphic poly-T variant in the TOMM40 gene encoding the translocase of outer mitochondrial membrane 40 homolog. This gene is in complete linkage disequilibrium with APOE as it maps only ∼2000 base pairs away. “Long” poly-T repeats have been reported to be associated with earlier onset regardless of APOE genotype. Whereas it is theoretically possible that both APOE and TOMM40 contribute to AD risk and just happen to reside next to each other in the genome, “Occam’s razor” and simple reason would dictate that only one of these genes is responsible for AD risk in this region of chromosome 19. Because functional studies have shown that APOE genotype influences cerebral Aβ clearance, APOE would appear to be the more likely candidate versus TOMM40.
It should also be noted that several other genes in linkage disequilibrium with APOE, in addition to TOMM40, also exhibit significant association with AD including BCAM, PVRL2, and APOC1. However, in our own family-based association studies of AD, the strength of association of APOE with AD is 15 orders of magnitude higher than that observed for any of these other genes in this region, including TOMM40 (unpublished data). This adds further support to the notion that APOE is the sole AD genetic risk factor in this region of linkage disequilibrium on chromosome 19.
GENETICS OF LATE-ONSET AD: ALZGENE
Since the original report of APOE as a genetic risk factor for LOAD, hundreds of genes have been tested for association with AD and reported in the literature. With the rapidly growing number of AD genetic studies published every month, it has become increasingly difficult to follow and interpret the published data. To address this problem, Lars Bertram, colleagues, and I started the online database, AlzGene.org, which provides a routinely updated online database and meta-analyses for the growing list of AD candidate genes (“AlzGene”; URL: www.alzgene.org; Bertram 2007; Bertram and Tanzi 2008).
Meta-analysis results on AlzGene reveal more than three dozen loci that show nominally significant association with risk for AD and 10 with nominal p-values < 10−5. The 10 genes with the strongest signals for association with AD include APOE and nine others, all of which came from genome-wide association studies (GWASs). The GWAS hits are discussed in a separate section below.
Many genes with weak but significant meta-analysis signals have in common that they did not derive from GWASs. These include, for example, SORL1 (sortilin-related receptor), which is a sorting receptor that regulates the intracellular trafficking of a variety of proteins, including APP (Andersen et al. 2005; Rogaeva et al. 2007). Another is ACE (angiotensin converting enzyme 1; Kehoe et al. 1999), a zinc metalloprotease involved in controlling blood pressure, which may increase the risk for AD. ACE is also able to degrade Aβ in vitro (Hu et al. 2001; Hemming et al. 2005). A third example is IL8 (interleukin 8), a cytokine mediating inflammatory responses (Mines et al. 2007). A final example is LDLR (low-density lipoprotein receptor), which serves as a receptor for APOE and removes low-density lipoproteins (LDL and VLDL) from circulation (Kim et al. 2009).
GENETICS OF LATE-ONSET AD: GENOME-WIDE ASSOCIATION STUDIES
Over the past several years, the most common strategy for finding novel AD gene candidates has been the genome-wide association study. In a GWAS, as many as one million genetic markers (single nucleotide polymorphisms, SNPs) are tested for genetic association with disease risk and/or phenotypic endophenotypes such as age-of-onset, biomarkers, imaging results, and neuropathological endpoints. More than a dozen GWASs have been reported for AD (reviewed in Bertram et al. 2009, 2010; Table 2). The first genome-wide significant finding was reported for GAB2 (GRB2-associated binding protein 2) by (Reiman et al. 2007); however, it required post hoc stratification by APOE to reach significance. This association was followed up with a series of replications and refutations. GAB2 has been proposed to influence tau phosphorylation (Reiman et al. 2007) and to modulate Aβ production by binding growth factor receptor-bound protein 2 (GRB2), which binds APP and the presenilins (Nizzari et al. 2007).
In 2008, we reported the first genes to directly exhibit genome-wide significant association with AD without the need for stratification on APOE. We employed a family-based GWAS approach (Bertram et al. 2008) and discovered three novel AD genes candidates including ATXN1 (ataxin 1), CD33 (siglec 3), and an uncharacterized locus on chromosome 14 (GWA_14q31.2). ATXN1 can harbor an expanded polyglutamine repeat that causes spinocerebellar ataxia type 1. With regard to AD, we later showed that ATXN1 affects Aβ levels by modulating β-secretase levels and cleavage of APP (Zhang et al. 2010). This finding was confirmed in vivo in ATXN1 knockout mice, which also suffer cognitive impairment after 6 mo of age (unpublished data). CD33 is a member of the family of sialic acid–binding, immunoglobulin-like lectins. These molecules promote cell–cell interactions that regulate the innate immune system (Crocker et al. 2007), including inflammation. Interestingly, the innate immune system has recently gained increased attention in AD, with our discovery that Aβ may act as an antimicrobial peptide in the brain’s innate immune system (Soscia et al. 2010). CD33 also resides in the top ten meta-analysis hits on AlzGene.org.
In 2009, two large case–control GWASs (Harold et al. 2009; Lambert et al. 2009) reported three novel AD genes: CLU (clusterin; apolipoprotein J), CR1 (complement component (3b/4b) receptor 1), and PICALM (phosphatidylinositol binding clathrin assembly protein). These three loci have been widely replicated in independent follow-up studies and are listed in the top 10 of AlzGene.org meta-analysis hits. Like APOE, two of these genes (CLU on chromosome 8 and CR1 on chromosome 1) map in previously implicated LOAD genetic linkage peaks (Butler et al. 2009). Despite the strong statistical support for these three candidates, the effects of these genes on risk for AD are exceedingly small with allelic odds ratios (ORs) of ∼1.15 for all three genes, that is, increasing or decreasing risk by ∼1.15×. In contrast, the OR for APOE ε4 is ∼4 or 15 for one or two alleles, respectively.
At the functional level, clusterin (CLU) has been reported to be involved in transport of Aβ from plasma to brain and Aβ fibrillization (DeMattos et al. 2004; Nuutinen et al. 2009). PICALM is involved in clathrin-mediated endocytosis (Tebar et al. 1999), which is necessary for APP to be processed by γ-secretase into Aβ (Koo and Squazzo 1994). CR1 is the receptor for complement C3b, a key inflammatory molecule that is activated as part of the brain’s innate immune system in AD (Khera et al. 2009), and may be able to protect against Aβ-induced neurotoxicity (Rogers et al. 2006).
In 2010, the results of another large case–control GWAS were reported, suggesting the existence of additional AD genetic risk factors (Seshadri et al. 2010). Among them was the gene BIN1 (bridging integrator 1), which had previously been reported to be associated with AD with subgenome-wide significance (Lambert et al. 2009). BIN1 is also a top 10 meta-analysis hit on AlzGene and has a tiny effect on AD risk with an allelic OR of ∼1.15. BIN1 (amphiphysin II) is highly expressed in the central nervous system and plays a role in receptor-mediated endocytosis (Pant et al. 2009), which could theoretically affect APP processing and Aβ production or Aβ clearance from brain.
In 2011, two more large case–control GWASs were reported, leading to four more Alzheimer gene candidates: CD2AP, MS4A6A/MS4A4E, EPHA1, and ABCA7 (Hollingworth et al. 2011; Naj et al. 2011). These studies also identified an additional SNP in CD33 that influenced risk for AD. Whereas we had previously reported a CD33 SNP that increased risk for AD with genome-wide significant association (Bertram et al. 2008) using family-based GWASs, these two case–control GWASs reported an SNP roughly 1400 base pairs away that conferred protection against AD. Collectively, our 2008 study along with this 2011 study shows that CD33 is now the only gene besides APOE that exhibits genome-wide significance for association with AD in both case–control and family-based GWASs. Additionally, both genes harbor SNPs that can confer either risk or protection for AD.
When one considers the three EO-FAD genes (APP, PSEN1, and PSEN2), the LOAD gene (APOE), and the 11 genes (CD33, GWA_14q31.2, ATXN1, CLU, PICALM, CR1, BIN1, ABCA7, MS4A6E/MS4A4E, CD2AP, and EPHA1) exhibiting genome-wide significance for association with LOAD, we now have 15 established AD genes (Table 2). However, we only know the pathogenic gene variants/mutations for the original four. With regard to function, roles for these genes in AD pathogenesis can be divided into three basic categories: production, degradation, and clearance of Aβ, lipid metabolism, innate immunity, and cellular signaling (Table 3 and Fig. 1).
Table 3.
Predicted pathogenic mechanisms of late-onset Alzheimer disease genes from GWASs
| Gene | Protein | Location | Risk change (%) | Proposed molecular phenotype |
|---|---|---|---|---|
| APOE | Apolipoprotein E | 19q13 | ∼400%–1500% | Clearance of Aβ; lipid metabolism |
| CD33 | CD33 (Siglec 3) | 19q13.3 | ∼10% | Innate immunity; degradation of Aβ |
| CLU | Clusterin | 8p21.1 | ∼10% | Clearance of Aβ; innate immunity |
| CR1 | Complement component (3b/4b) receptor 1 | 1q32 | ∼15% | Clearance of Aβ; innate immunity |
| PICALM | Phosphatidylinositol binding clathrin assembly molecule | 11q14 | ∼15% | Production and clearance of Aβ; cellular signaling |
| BIN1 | Bridging integrator 1 | 2q14 | ∼15% | Production and clearance of Aβ; cellular signaling |
| ABCA7 | ATP-binding cassette subfamily A member 7 | 19p13.3 | ∼20% | Lipid metabolism; cellular signaling |
| CD2AP | CD2-associated protein | 6p12.3 | ∼10% | Cellular signaling |
| EPHA1 | EPH receptor A1 | 7q34 | ∼10% | Cellular signaling; innate immunity |
| MS4A6A/MS4A4E | Membrane-spanning 4- domains, subfamily A, members 6A and 4E | 11q12.1 | ∼10% | Cellular signaling |
| ATXN1 | Ataxin 1 | 6p22.3 | NA | Production of Aβ |
GENETICS OF LATE-ONSET AD: MISSING HERITABILITY AND ADAM10
The GWAS approach involves genotyping common ancestral polymorphisms that usually occur in >5% of the general population. The risk effects exerted by the GWAS-derived genes discussed above are tiny, that is they confer only an ∼0.10- to 0.15-fold increase or decrease in AD risk in carriers versus noncarriers of the associated alleles, as compared with a four- to 15-fold increase in AD risk owing to the inheritance of APOE ε4. A substantial proportion of the genetic variance of LOAD remains unexplained by the currently known susceptibility genes.
The “missing heritability” of complex and heterogeneous disease has been coined the “dark matter” of GWASs because we know it exists but cannot yet observe it (Manolio et al. 2009). The “dark matter” may be explained by common variants that are not included on existing microarrays, copy number variants including genomic insertion/deletions and rearrangements, and rare sequence variants. With regard to rare variants conferring large effects on risk for LOAD, we recently reported two rare LOAD mutations in the ADAM10 gene that caused AD at an average age of 70 years in seven of 1000 LOAD families tested (Kim et al. 2009).
ADAM10 encodes the major α-secretase in the brain. This enzyme cleaves within the Aβ domain of APP to preclude the formation of β-amyloid. The two novel ADAM10 LOAD mutations, Q170H and R181G, are located in the prodomain region, and dramatically impair the ability of ADAM10 to carry out α-secretase cleavage of APP both in vitro (Kim et al. 2008) and in vivo (unpublished observation). Thus, the two LOAD-associated mutations in ADAM10 would appear to be strong candidates for the first rare, highly penetrant pathogenic mutations to be genetically associated with late-onset AD (Kim et al. 2009). This study then becomes the first (and to date, only) to challenge the rare mutation–rare disease (EO-FAD)/common variant–common disease (LOAD) paradigm that postulates that LOAD is only associated with common variants, (e.g., APOE ε4).
The two LOAD mutations in ADAM10 also argue that, in addition to GWASs, resequencing, for example, using novel high-throughput (“next-generation”) technologies and whole-genome or exome sequencing, will be necessary to search for additional rare variants causing LOAD. Along these lines, large-scale resequencing has been employed to create a comprehensive array of all rare mutations in the human genome, known as the “1000 Genomes project” (http://www.1000genomes.org/). This database will aid immensely in determining whether rare mutations found in disease cases are novel and pathogenic. Ultimately, the validation of bona fide pathogenic mutations will require comprehensive functional studies in in vitro systems, in vivo animal models, and clinical samples.
SUMMARY
Our understanding of the etiology and pathogenesis of AD can be arguably be divided into two eras: before and after the identification of the AD genes. The past three decades of genetic research in AD have revolutionized our understanding of the causes of AD and accelerated the discovery and development of novel therapeutics aimed at the treatment and prevention of AD (Tanzi and Bertram 2005). The modern era of AD research guided by genetics began in the 1980s and 1990s with genetic linkage studies and positional cloning efforts that led to the identification of the three EO-FAD genes: APP, PSEN1, and PSEN2. The late-onset AD gene APOE was discovered using a similar strategy, but was ultimately validated by genetic association studies. Genetic association studies remain the most common method for identifying novel LOAD genes and are most often employed in the form of large-scale GWASs that screen the entire human genome for novel disease loci. Recently, our family-based and others’ case–control GWASs (reviewed in Bertram et al. 2009, 2010) led to the elucidation of novel susceptibility variants, all of which confer only small effects on risk. Moreover, up to 50% the heritability of AD remains unexplained by the known genes and AD candidate loci that have been derived from GWASs.
Our initial understanding of the etiology of AD began with the identification of rare causal mutations in the three EO-FAD genes. More recently, we discovered two rare pathogenic mutations in ADAM10 that impair ADAM10 enzyme activity and lead to AD by 70 years old with high penetrance (Kim et al. 2009). Based on the roughly 200 EO-FAD gene mutations and two ADAM10 LOAD mutations, it will be important to search for additional rare sequence variants in other genes that predispose to AD. Ultimately, the insights into the etiology and pathogenesis of AD garnered from genetic studies of AD will continue to enhance our understanding of the pathological mechanisms leading to AD. Moreover, these findings will aid in the development of novel therapeutic strategies for preventing, stopping and even reversing AD.
Footnotes
Editors: Dennis J. Selkoe, Eckhard Mandelkow, and David M. Holtzman
Additional Perspectives on The Biology of Alzheimer Disease available at www.perspectivesinmedicine.org
REFERENCES
- Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, von Arnim CAF, Breiderhoff T, Jansen P, Wu X, et al. 2005. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci 102: 13461–13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE 2005. The genetic epidemiology of neurodegenerative disease. J Clin Invest 115: 1449–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE 2008. Thirty years of Alzheimer’s disease genetics: Systematic meta-analyses herald a new era. Nat Rev Neurosci 9: 768–778 [DOI] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE 2009. Genome-wide association studies in Alzheimer’s disease. Hum Mol Genet 18: R137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE 2007. Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat Genet 39: 17–23 [DOI] [PubMed] [Google Scholar]
- Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BMM, Hooli B, Divito J, Ionita I, et al. 2008. Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am J Hum Genet 83: 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lill CM, Tanzi RE 2010. The genetics of Alzheimer disease: Back to the future. Neuron 68: 270–281 [DOI] [PubMed] [Google Scholar]
- Butler AW, Ng MYM, Hamshere ML, Forabosco P, Wroe R, Al-Chalabi A, Lewis CM, Powell JF 2009. Meta-analysis of linkage studies for Alzheimer’s disease—A web resource. Neurobiol Aging 30: 1037–1047 [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC Jr, Rimmler JB, Locke PA, Conneally PM, Schmader KE, et al. 1994. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet 7, 180–184 [DOI] [PubMed] [Google Scholar]
- Crocker PR, Paulson JC, Varki A 2007. Siglecs and their roles in the immune system. Nat Rev Immunol 7: 255–266 [DOI] [PubMed] [Google Scholar]
- DeMattos RB, Cirrito JR, Parsadanian M, May PC, O’Dell MA, Taylor JW, Harmony JA, Aronow BJ, Bales KR, Paul SM, et al. 2004. ApoE and clusterin cooperatively suppress Aβ levels and deposition: Evidence that ApoE regulates extracellular Aβ metabolism in vivo. Neuron 41, 193–202 [DOI] [PubMed] [Google Scholar]
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F 1998. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature 391, 387–390 [DOI] [PubMed] [Google Scholar]
- Frigerio C, Piscopo P, Calabrese E, Crestini A, Malvezzi Campeggi L, Civita di Fava R, Fogliarino S, Albani D, Marcon G, Cherchi R, et al. 2005. PEN-2 gene mutation in a familial Alzheimer’s disease case. J Neurol 252: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL 2006. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 63, 168–174 [DOI] [PubMed] [Google Scholar]
- Glenner GG, Wong CW 1984. Alzheimer’s disease and Down’s syndrome: Sharing of a unique cerebrovascular amyloid fibril protein. Biochem Biophys Res Commun 122, 1131–1135 [DOI] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, et al. 1991. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 349: 704–706 [DOI] [PubMed] [Google Scholar]
- Goldgaber D, Lerman MI, McBride OW, Saffiotti U, Gajdusek DC 1987. Characterization and chromosomal localization of a cDNA encoding brain amyloid of Alzheimer’s disease. Science 235, 877–880 [DOI] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA 1992. Alzheimer’s disease: The amyloid cascade hypothesis. Science 256, 184–185 [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ 2002. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. 2009. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41: 1088–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming ML, Selkoe DJ 2005. Amyloid β-protein is degraded by cellular angiotensin-converting enzyme (ACE) and elevated by an ACE inhibitor. J Biol Chem 280: 37644–37650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, et al. 2011. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43: 429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Igarashi A, Kamata M, Nakagawa H 2001. Angiotensin-converting enzyme degrades Alzheimer amyloid β-peptide (Aβ); retards Aβ aggregation, deposition, fibril formation; and inhibits cytotoxicity. J Biol Chem 276: 47863–47868 [DOI] [PubMed] [Google Scholar]
- Jarrett JT, Berger EP, Lansbury PT Jr 1993. The carboxy terminus of the β amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer’s disease. Biochemistry 32, 4693–4697 [DOI] [PubMed] [Google Scholar]
- Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B 1987. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature 325, 733–736 [DOI] [PubMed] [Google Scholar]
- Kehoe PG, Russ C, McIlory S, Williams H, Holmans P, Holmes C, Liolitsa D, Vahidassr D, Powell J, McGleenon B, et al. 1999. Variation in DCP1, encoding ACE, is associated with susceptibility to Alzheimer disease. Nat Genet 21: 71–72 [DOI] [PubMed] [Google Scholar]
- Khera R, Das N 2009. Complement receptor 1: Disease associations and therapeutic implications. Mol Immunol 46: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Castellano JM, Jiang H, Basak JM, Parsadanian M, Pham V, Mason SM, Paul SM, Holtzman DM 2009. Overexpression of low-density lipoprotein receptor in the brain markedly inhibits amyloid deposition and increases extracellular Aβ clearance. Neuron 64: 632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo EH, Squazzo SL 1994. Evidence that production and release of amyloid β-protein involves the endocytic pathway. J Biol Chem 269: 17386–17389 [PubMed] [Google Scholar]
- Kounnas MZ, Danks AM, Cheng S, Tyree C, Ackerman E, Zhang X, Ahn K, Nguyen P, Comer P, Mao L, et al. 2010. Modulation of γ-secretase reduces β-amyloid deposition in a transgenic mouse model of Alzheimer’s disease. Neuron 67: 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, et al. 2009. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41: 1094–1099 [DOI] [PubMed] [Google Scholar]
- Levy E, Carman MD, Fernandez-Madrid IJ, Power MD, Lieberburg I, van Duinen SG, Bots GT, Luyendijk W, Frangione B 1990. Mutation of the Alzheimer’s disease amyloid gene in hereditary cerebral hemorrhage, Dutch type. Science 248, 1124–1126 [DOI] [PubMed] [Google Scholar]
- Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K 1995. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science 269: 973–977 [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mines M, Ding Y, Fan G 2007. The many roles of chemokine receptors in neurodegenerative disorders: Emerging new therapeutical strategies. Curr Med Chem 14: 2456–2470 [DOI] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, et al. 2011. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43: 436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizzari M, Venezia V, Repetto E, Caorsi V, Magrassi R, Gagliani MC, Carlo P, Florio T, Schettini G, Tacchetti C, et al. 2007. Amyloid precursor protein and Presenilin1 interact with the adaptor GRB2 and modulate ERK 1,2 signaling. J Biol Chem 282: 13833–13844 [DOI] [PubMed] [Google Scholar]
- Nuutinen T, Suuronen T, Kauppinen A, Salminen A 2009. Clusterin: A forgotten player in Alzheimer’s disease. Brain Res Rev 61: 89–104 [DOI] [PubMed] [Google Scholar]
- Ostojic J, Elfgren C, Passant U, Nilsson K, Gustafson L, Lannfelt L, Froelich Fabre S 2004. The tau R406W mutation causes progressive presenile dementia with bitemporal atrophy. Dement Geriatr Cogn Disord 17: 298–301 [DOI] [PubMed] [Google Scholar]
- Pant S, Sharma M, Patel K, Caplan S, Carr CM, Grant BD 2009. AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol 11: 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Dermaut B, Peeters K, Cruts M, Heutink P, Goate A, Van Broeckhoven C 2003. Tau (MAPT) mutation Arg406Trp presenting clinically with Alzheimer disease does not share a common founder in Western Europe. Hum Mutat 22: 409–411 [DOI] [PubMed] [Google Scholar]
- Rademakers R, Cruts M, Sleegers K, Dermaut B, Theuns J, Aulchenko Y, Weckx S, De Pooter T, Van den Broeck M, Corsmit E, et al. 2005. Linkage and association studies identify a novel locus for Alzheimer disease at 7q36 in a Dutch population-based sample. Am J Hum Genet 77: 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, et al. 2007. Papassotiropoulos A, Stephan DA. GAB2 alleles modify Alzheimer’s risk in APOE ε4 carriers. Neuron 54: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T 1995. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature 376: 775–778 [DOI] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, et al. 2007. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet 39: 168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Li R, Mastroeni D, Grover A, Leonard B, Ahern G, Cao P, Kolody H, Vedders L, Kolb WP, et al. 2006. Peripheral clearance of amyloid β peptide by complement C3-dependent adherence to erythrocytes. Neurobiol Aging 27: 1733–1739 [DOI] [PubMed] [Google Scholar]
- Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS, Huentelman MJ, Welsh-Bohmer KA, Reiman EM 2010. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease. Pharmacogenom J 10, 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W, et al. 1996. Secreted amyloid β-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med 2: 864–870 [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, et al. 2010. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303: 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. 1995. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature 375: 754–760 [DOI] [PubMed] [Google Scholar]
- Soscia SJ, Kirby JE, Washicosky KJ, Tucker SM, Ingelsson M, Hyman B, Burton MA, Goldstein LE, Duong S, Tanzi RE, et al. 2010. The Alzheimer’s disease-associated amyloid β-protein is an antimicrobial peptide. PLoS ONE 5: e9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. George-Hyslop PH, Tanzi RE, Polinsky RJ, Haines JL, Nee L, Watkins PC, Myers R, Feldman R, Pollen D, Drachman D, et al. 1987. The genetic defect causing familial Alzheimer’s disease maps on Chromosome 21. Science 235: 885–890 [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD 1993. Apolipoprotein E: High-avidity binding to β-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci 90: 1977–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi RE, Bertram L 2005. Twenty years of the Alzheimer’s disease amyloid hypothesis: A genetic perspective. Cell 120: 545–555 [DOI] [PubMed] [Google Scholar]
- Tanzi RE, Gusella JF, Watkins PC, Bruns GA, St George-Hyslop P, Van Keuren ML, Patterson D, Pagan S, Kurnit DM, Neve RL 1987a. Amyloid β protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science 235: 880–884 [DOI] [PubMed] [Google Scholar]
- Tanzi RE, St. George-Hyslop PH, Haines JL, Polinsky RJ, Nee L, Foncin J-F, Neve RL, McClatchey AI, Conneally PM, Gusella JF 1987b. The genetic defect in familial Alzheimer’s disease is not tightly linked to the amyloid β protein gene. Nature 329: 156–157 [DOI] [PubMed] [Google Scholar]
- Tanzi RE, McClatchey AI, Lamperti ED, V-Komaroff L, Gusella JF, Neve R 1988. Protease inhibitor domain encoded by an amyloid protein precursor mRNA associated with Alzheimer’s disease. Nature 331: 528–530 [DOI] [PubMed] [Google Scholar]
- Tebar F, Bohlander SK, Sorkin A 1999. Clathrin assembly lymphoid myeloid leukemia (CALM) protein: Localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol Biol Cell 10: 2687–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ 1999. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature 398, 513–517 [DOI] [PubMed] [Google Scholar]
- Zhang C, Browne A, Child D, Divito JR, Stevenson JA, Tanzi RE 2010. Loss of function of ATXN1 increases amyloid β-protein levels by potentiating β-secretase processing of β-amyloid precursor protein. J Biol Chem 285: 8515–8526 [DOI] [PMC free article] [PubMed] [Google Scholar]