Abstract
This article describes three rare syndromes in which the presence of α-thalassemia provided an important clue to the molecular basis of the underlying condition. It exemplifies how rare diseases allied with careful clinical observation can lead to important biological principles. Two of the syndromes, ATR-16 and ATR-X, are characterized by α-thalassemia in association with multiple developmental abnormalities including mental retardation. The third condition, ATMDS, is an acquired disorder in which α-thalassemia arises in the context of myelodysplasia. Intriguingly, mutations in the chromatin remodeling factor, ATRX, are common to both ATR-X syndrome and ATMDS.
In three rare syndromes, α-thalassemia occurs with mental retardation and developmental abnormalities (ATR-16 and ATR-X) or with myelodysplasia (ATMDS). ATR-X and ATMDS involve mutations in the chromatin remodeling factor ATRX.
The rare association of α-thalassemia and mental retardation (MR) was recognized over 25 years ago by Weatherall and colleagues (Weatherall et al. 1981). It was known that α-thalassemia arises when there is a defect in the synthesis of the α globin chains of adult hemoglobin (HbA, α2β2). When these investigators encountered three mentally retarded children with α-thalassemia and a variety of developmental abnormalities, their interest was stimulated by the unusual nature of the α-thalassemia. The children were of northern European origin, where α-thalassemia is uncommon, and although one would have expected to find clear signs of this inherited anemia in their parents, it appeared to have arisen de novo in the affected offspring. It was thought that the combination of α-thalassemia with MR (ATR), and the associated developmental abnormalities represented a new syndrome and that a common genetic defect might be responsible for the diverse clinical manifestations. What emerged was the identification of two quite distinct syndromes in which α-Thalassemia is associated with mental retardation (Wilkie et al. 1990a, b). In the first condition (ATR-16, OMIM: 141750) there are large (1–2 Mb) chromosomal rearrangements that delete many genes, including the α globin genes from the tip of the short arm of chromosome 16 and this is an example of a contiguous gene syndrome (Schmickel 1986). In the second syndrome (ATR-X, OMIM 301040), a complex phenotype, including α-Thalassemia, results from mutations in an X-encoded factor (now called the ATRX protein), which is a putative regulator of gene expression. Mutations in this gene down-regulate α gene expression and also perturb the expression of other, as yet unidentified, genes.
THE ATR-16 SYNDROME
ATR-16 is rare and sporadic contiguous gene syndrome. Often one is alerted to this condition by observing the unusual association of α-Thalassemia and mental retardation in individuals originating from outside of the areas where thalassemia commonly occurs. However, the widespread use of array-based screening for genomic deletions is identifying cases with little regard for the phenotype. In addition to α-Thalassemia, these patients have variable degrees of facial dysmorphism, most commonly featuring a high forehead, mild hypertelorism, and a broad or prominent nasal bridge. A wide spectrum of associated developmental abnormalities has also been described of which clubfoot is the most frequent. In all such cases, initial molecular genetic analyses have shown that affected individuals fail to inherit the entire ζ-α globin cluster from one or other of the parents.
Defining the Genetic Abnormalities in Patients with ATR-16 Syndrome
In some cases, conventional cytogenetic analysis shows the underlying genetic abnormality. Because the α globin complex lies very close to the 16p telomere any chromosomal abnormality affecting this region may give rise to α-Thalassemia. In some patients with ATR-16 syndrome, gross chromosomal abnormalities such as ring chromosomes and translocations are visible on the karyotype. However, in the case of deletions and subtelomeric translocations, initial high-resolution cytogenetic analysis appears entirely normal. Although such abnormalities may arise as de novo genetic events, often one parent carries a pre-existing balanced translocation, which the child inherits in an unbalanced fashion resulting in monosomy for 16p and loss of the α cluster.
Fluorescence in situ hybridization (FISH) studies have also been used to analyze ATR-16 families. In this type of study, large segments (∼40 kb) of chromosome 16 in cosmid vectors are used as probes to show the presence or absence of the corresponding sequences in the 16p telomeric region using fluorescence microscopy (Buckle and Kearney 1994). By analyzing the chromosomes of both parents and the affected child it has been possible to define the extent of 16p monosomy and the mechanism by which it has arisen. The additional use of panels of subtelomeric probes from each human chromosome can identify trisomic segments in unbalanced translocations.
As a first line investigation, FISH is currently being superceded by technologies enabling high-resolution analysis of the entire human genome. In the first of these types of analyses, the genome is interrogated using microarrays consisting of oligonucleotides spanning the genome. Using a competitive genome hybridization (CGH) approach, comparing DNA from one genome with another, it has been possible to detect large regions of monosomy in patients with ATR-16 syndrome (Price et al. 2005). Such oligo-based microarrays or bead technologies (originally made to identify single nucleotide polymorphisms) with a modified analysis have a resolution in the range 5–50 kb (Redon et al. 2006). One limitation of the technology is that it does not provide information of the structural origin of any detected chromosomal imbalance and FISH is commonly used as a follow-up investigation to supplement this. In addition to these approaches, Harteveld et al. (2007) have developed a rapid and simple high-resolution approach to identify and characterize deletions of the terminal 2 Mb of 16p13.3 using the multiple ligation-dependent probe amplification (MLPA) technique. An established panel of specific synthetic oligonucleotides can now be used to detect deletions in 16p13.3 (Harteveld et al. 2005, 2007). Clearly the resolution of this technique is only limited by the spacing of the appropriate oligonucleotides.
To date, few breakpoints have been fully characterized. However, where this has been fully analyzed it appears that the chromosome has been broken, truncated, and “healed” by the direct addition of telomeric repeats (TTAGGG)n (Lamb et al. 1993).
How Do Chromosomal Abnormalities Give Rise to the ATR-16 Syndrome?
Because individuals with ATR-16 syndrome may have quite different degrees of chromosomal imbalance, there is considerable variation in the associated phenotypes. The degree of monosomy in 16p13.3 varies from 0.3–2 Mb but many patients have additional chromosomal aneuploidy and in some cases imbalance of the non-16 material may dominate the clinical picture. For example, in one (DA) loss of material from chromosome 15 while forming the abnormal derivative, t(15:16) chromosome produced the striking phenotype associated with the Prader-Willi syndrome.
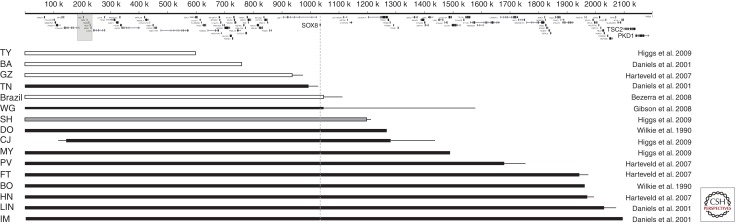
Because many patients with ATR-16 have complex genomic rearrangements, to determine the role played by any gene(s) within 16p13.3 in the developmental abnormalities associated with ATR-16 syndrome, future analysis should focus on patients who have pure monosomy for 16p13.3 (Fig. 1). Surprisingly, these studies have shown that patients with deletions of up to 1000 kb of 16p13.3 (including 55 genes) have α-Thalassemia, but may be otherwise unaffected. Those with deletions of between 1000 kb and 1200 kb may or may not have mild MR. At the other extreme, patients with 16p monosomy for the entire terminal region of 16p (e.g., BO, HN, LIN, and IM) have a α-Thalassemia, mild MR, and dysmorphic features. In a patient whose deletion (associated with unbalanced translocation) extends beyond 2000 kb the clinical picture is dominated by more severe MR, tuberous sclerosis, and polycystic kidney disease (European Polycystic Kidney Disease Consortium 1994).
Figure 1.
Summary of known ATR-16 deletions that are >500 kb and associated with pure monosomy. Initials represent the patient code. Open boxes represent cases without learning difficulties; grey boxes, families in which some cases have learning difficulties whereas other do not; black boxes represent cases with learning difficulties. The length of the box shows the known extent of the deletion and the fine line indicates the region of uncertainty of the breakpoints. The positions of the genes in this region are shown at the top; the α-globin cluster is marked with a pale gray box; SOX8, TSC2, and PKD1 are shown in a larger font.
How might monosomy for 16p13.3 cause such developmental abnormalities? One possibility is that deletion of a large number of genes from one copy of chromosome 16 may unmask mutations in its homolog; the more genes that are deleted the greater the probability of this occurring. However, this is unlikely to be the explanation for most ATR-16 cases because whole genome sequencing indicates that normal individuals carry a relatively small number of harmful mutations of this type. A further possibility is that some genes in 16p are imprinted (Hall 1990) so that deletions could remove the only active copy of the gene. At present, there is no evidence for imprinting of the 16p region (reviewed in Schneider et al. 1996), and in the relatively few ATR-16 cases analyzed there appears to be no major clinical differences between patients with deletions of the maternally or paternally derived chromosomes. It therefore seems more likely that there are some genes in the 16p region that encode proteins whose effect is critically determined by the amount produced, so-called dosage-sensitive genes (see Fisher and Scambler 1994). Examples of such genes include those encoding proteins that form heterodimers, those required at a critical level for a rate-determining step of a regulatory pathway, and tumour suppressor genes (e.g., TSC2). If the deletion includes one or more dosage-sensitive genes this could account for the clinical effects seen in ATR-16 patients.
The region lying between 900–1700 kb from the 16p telomere, deleted in all patients with the characteristic features of ATR-16 syndrome, contains 14 genes and gene families of known function (Daniels et al. 2001) that have been implicated in a wide range of disorders with few or no features in common with ATR-16. One of these (SOX8) was considered a strong candidate because it is involved in the regulation of embryonic development and is strongly expressed in the brain (Pfeifer et al. 2000). However, although deleted in case WG (Fig. 1) (Gibson et al. 2008), the deletion in TN does not extend to SOX8 (Daniels et al. 2001). Furthermore, a recently described Brazilian patient with a deletion that removes both the α globin locus and SOX8 was not associated with MR or any dysmorphism (Bezerra et al. 2008). This suggests that loss of SOX8 is neither necessary nor sufficient to cause MR and dysmorphism. It is conceivable that the genetic background plays a role and this might explain why the deletion in family SH is associated with MR in one individual and a normal phenotype in another. Clearly, further examples of ATR-16 because of monosomy for 16p13.3 must be characterized to identify the gene(s) responsible for the MR and other developmental abnormalities associated with this condition.
THE ATR-X SYNDROME
As additional patients with α-Thalassemia and mental retardation were identified throughout the 1980s, it became clear that a second group of affected individuals existed in whom no structural abnormalities in the α globin cluster or 16p could be found. In contrast to ATR-16 syndrome, this group of patients was all male, presented with a much more uniform phenotype, and had a remarkably similar facial appearance (Wilkie et al. 1990b). That this group had a distinct and recognizable dysmorphism was underscored when additional cases were identified on the basis of their facial features alone (Gibbons et al. 1991; Wilkie et al. 1991). Ultimately, it was shown that this unusual syndrome of α-Thalassemia with severe mental retardation results from an X-linked abnormality (see later) and the condition is now referred to as the ATR-X syndrome (OMIM: 301040).
The Clinical Findings of the ATR-X Syndrome
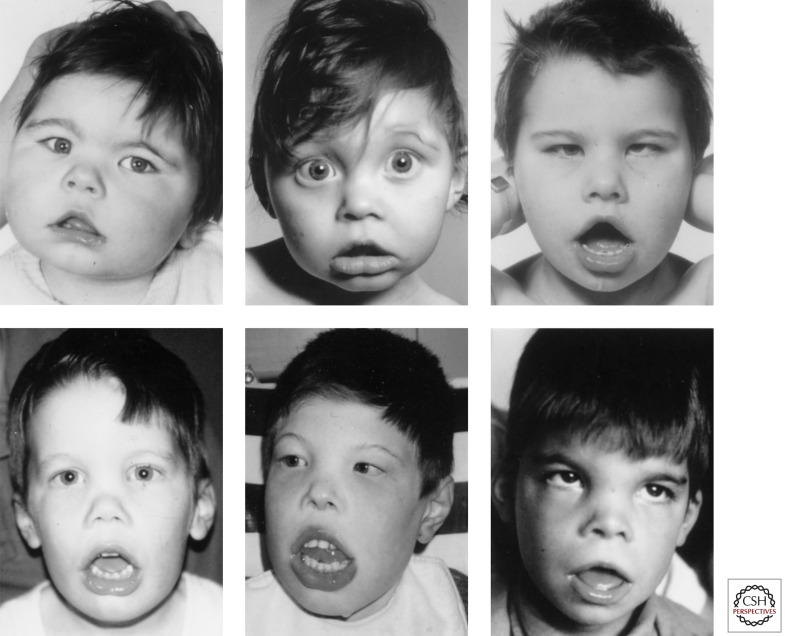
Cases of ATR-X syndrome from over 180 families have now been characterized, and in contrast to ATR-16 a definite phenotype is emerging. The cardinal features of this condition are severe mental retardation and developmental abnormalities associated with a characteristic facial appearance (Fig. 2) and α-Thalassemia. The frequency and nature of the most commonly encountered clinical features are summarized in Tables 1 and 2.
Figure 2.
The facial appearance of patients with ATR-X syndrome. (From Gibbons et al. 1995a; adapted, with permission, from the investigator.)
Table 1.
Summary of the major clinical manifestations of the ATR-X syndrome
| Clinical feature | % |
|---|---|
| Profound mental retardation | 96 |
| Characteristic face | 94 |
| Skeletal abnormalities | 90 |
| HbH inclusions | 88 |
| Neonatal hypotonia | 84 |
| Genital abnormalities | 79 |
| Microcephaly | 77 |
| Gut dysmotility | 76 |
| Short stature | 65 |
| Seizures | 34 |
| Cardiac defects | 21 |
| Renal/urinary abnormalities | 15 |
Table 2.
Clinical manifestations of the ATR-X syndrome
| Genital abnormalities | Small/soft testes, cryptorchidism, gonadal dysgenesis, inguinal hernia, micropenis, hypospadias, deficient prepuce, shawl scrotum, hypoplastic scrotum, ambiguous genitalia, female external genitalia (male pseudo-hermaphroditism) |
| Skeletal abnormalities | Delayed bone age, tapering fingers, drumstick distal phalanges, brachydactyly, clinodactyly, bifid thumb, fixed flexion deformities of joints, overriding toes, vaus or valgus deformities of feet, scoliosis, kyphosis, hemivertebra, spina bifida, coxa valga, chest wall deformity |
| Renal/urinary abnormalities | Renal agenesis, hydonephrosis, small kidneys, vesico-ureteric reflux, pelvo-ureteric junction obstruction, exstrophy of bladder, urethral diverticulum, urethral stricture |
| Cardiac defects | Atrial septal defect, ventricular septal defect, patent ductus arteriosus, tetralogy of Fallot, transposition of the great arteries, dextracardia with situs solitus, aortic stenosis, pulmonary stenosis |
| Gut dysmotility | Discoordinated swallowing, eructation, gastro-esophageal reflux, vomiting, gastric pseudo-volvulus, hiatus hernia, haematemesis, recurrent ileus/small bowel obstruction, volvulus, intermittent diarrhoea, severe constipation, ultra short segment Hirschsprung disease |
| Miscellaneous | Apneoic episodes, cold/blue extremities, blepharitis, conjunctivitis, entropion, cleft palate, pneumonia, umbilical hernia, encephalitis, iris coloboma, optic atrophy, blindness, sensorineural deafness, prolonged periods of screaming/laughing, self-injury, asplenia |
The Hematologic Findings of the ATR-X Syndrome
The presence of α-Thalassemia (in the form of thalassemia trait or mild HbH disease) with HbH inclusions was one of the original diagnostic criteria for ATR-X syndrome. However, with the identification of further cases, it became clear that the hematologic findings (e.g., levels of Hb, MCV, and MCH) were different from those seen in the common types of α-Thalassemia. Now that the disease can be identified via the clinical phenotype and/or the ATRX genotype, it is clear that there is considerable variation in the hematologic manifestations associated with mutations of the ATRX gene. In fact, a number of families have been identified in which some or all of the affected members with mutations of ATRX, and the characteristic phenotype described previously, have no signs of α-Thalassemia (Villard et al. 1996a,b). Nevertheless, when the family history and phenotype are strongly suspect, a careful search for HbH inclusions should be made in all affected individuals and repeated if necessary as they may be very infrequent.
The hematology is often surprisingly normal considering the presence of α-Thalassemia. In many cases, neither the level of hemoglobin nor the mean cell hemoglobin is as severely affected as in the classical forms of α-Thalassemia associated with cis-acting mutations in the α-globin complex, and this probably reflects the different pathophysiology of the conditions. In most cases of ATR-X, there is insufficient HbH to be detected by electrophoresis and the number of HbH inclusions is quite variable between different patients, although relatively constant over time in any affected individual.
The ATR-X Syndrome Is an X-Linked Condition
Evidence that the defect in globin synthesis lay in trans to the globin cluster was provided by a study of a family with four affected sibs in whom the condition segregated independent of the α globin cluster (Donnai et al. 1991).
Because affected individuals were always related via the female line, this suggested that the syndrome mapped to the X chromosome and hence the condition was named the ATR-X syndrome. Subsequently linkage analysis in 16 families mapped the disease interval to Xq13.1–q21.1 confirming that the associated α-Thalassemia results from a trans-acting mutation (Gibbons et al. 1992).
Early genetic studies showed that the ATR-X syndrome behaves as an X-linked recessive disorder; boys are affected almost exclusively. Furthermore, almost all female carriers have a normal appearance and intellect, although approximately one in four carriers has subtle signs of α-Thalassemia with very rare cells containing HbH inclusions (Gibbons et al. 1992). The majority of carriers have a highly skewed pattern of X inactivation in leukocytes (derived from mesoderm), hair roots (ectoderm), and buccal cells (endoderm). In each case, the disease-bearing X chromosome is preferentially inactivated. There is evidence from a recently reported mouse model of ATR-X syndrome (Garrick et al. 2006) that skewed X inactivation results from selection, at key steps during development, against cells that are deficient for ATRX (Muers et al. 2007).
Together, these findings showed that ATR-X is an X-linked disease and when the gene is mutated, among many other effects, this leads to down-regulation of expression of the α globin genes on chromosome 16.
Identification of the ATR-X Disease Gene
The isolation of cDNA fragments mapping to Xq13.1–q21.1 provided an opportunity to study candidate genes for ATR-X (Gecz et al. 1994). A number of these cDNA fragments were hybridized to DNA from a group of affected individuals. An absent hybridization signal was noted in one patient when an 84 bp cDNA fragment (E4) was used. E4 was shown to be part of a gene known as XH2/XNP (Gibbons et al. 1995b). Subsequently, a 2-kb genomic deletion was shown in this individual with ATR-X syndrome. Subsequently, analysis of a segment of cDNA corresponding to XH2/XNP identified disease-causing mutations in several individuals with the clinical and hematologic features of ATR-X syndrome. The X-linked gene was thus renamed as the ATRX gene.
Characterization of the ATRX Gene and Its Protein Product
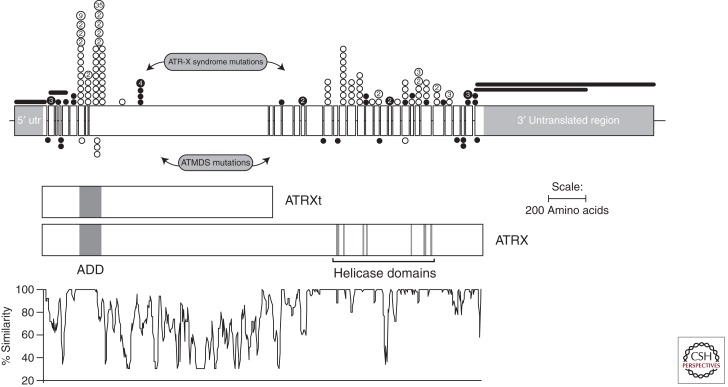
We now know that the ATRX gene spans about 300 kb of genomic DNA and contains 36 exons (Picketts et al. 1996) although exon 7 may be nonfunctional. It encodes at least two alternatively spliced, 10.5-kb mRNA transcripts that differ at their 5′ ends and are predicted to give rise to slightly different proteins of 265 and 280 kDa (Fig. 3). A further transcript of ∼7 kb represents an isoform, which retains intron 11 and truncates at this point. This gives rise to a truncated protein isoform (ATRXt) which is conserved between mouse and man (Garrick et al. 2004).
Figure 3.
Schematic diagram of the ATRX gene: boxes represent the 35 exons (excluding the alternatively spliced exon 7); thin horizontal lines represent introns (not to scale). The 3′ and 5′ untranslated regions (utr) are shown flanking the open reading frame. The two protein products ATRX and ATRXt are shown as rectangles. The principal domains, the zinc finger motif (ADD), and the highly conserved helicase motif are indicated. In the lower part of the figure is a graphical representation of the amino acid similarity between human and mouse ATRX proteins. In the upper part of the figure is illustrated the spectrum of ATRX mutations described in boys with ATR-X syndrome (above the gene) and in ATMDS (below the gene). The positions of the mutations are shown by circles: filled circles represent mutations (nonsense or leading to a frameshift) that would cause protein truncation; open circles represent missense mutations and small deletions that maintain the reading frame; deletions are indicated by horizontal lines. Recurrent mutations are illustrated by larger circles, and the number of independent families is indicated. (From Gibbons et al. 2008; adapted, with permission, from the author.)
Within the amino-terminal region lies a complex cysteine-rich segment (called the ADD domain; Fig. 3). This comprises a PHD-like zinc finger and an additional C2C2 motif just upstream, which is structurally similar to the GATA1 zinc fingers (Gibbons et al. 1997; Argentaro et al. 2007) and is highly related to the zinc finger domains of DNA methyltransferases (Argentaro et al. 2007). The domain mediates binding to the amino-terminal tail of histone H3 trimethylated at lysine 9 (Dhayalan et al. 2011; Eustermann et al. 2011; Iwase et al. 2011). The functional importance of the ADD segment in ATRX is clear. It is highly conserved throughout evolution and it contains over 50% of all mutations found in patients with ATR-X syndrome (Fig. 3 and see below).
The central and carboxy-terminal regions of ATRX show the greatest conservation between murine and human sequences (94%) (Fig. 3) (Picketts et al. 1998). The central portion of the molecule contains motifs that identify ATRX as a member of the SNF2 group of proteins. A central property of this group of proteins is to remodel chromatin during processes such as DNA replication, transcription, mitosis, and DNA repair.
Mutations of the ATRX Gene and Their Associated Phenotype
In addition to ATR-X syndrome, mutations in the ATRX gene have now been found in many other forms of syndromal X-linked MR (reviewed in Gibbons et al. 2008) and it is also the disease gene associated with the occurrence of α-Thalassemia in myelodysplasia (see later section).
To date, 113 different constitutional mutations have been documented in 182 independent families with ATR-X syndrome (reviewed in Gibbons et al. 2008). Missense mutations are clustered in two regions: the ADD domain, and the helicase domain. Analysis of the mutations and their resulting phenotypes allows important conclusions to be drawn. It appears that none of the mutations lead to a true null but rather are hypomorphs.
ATR-X syndrome is frequently caused by missense mutations (Fig. 3) and the levels of ATRX protein were shown to be substantially reduced in patients with such mutations involving the ADD domain (McDowell et al. 1999; Cardoso et al. 2000). The structure of the ADD domain casts light on how mutations might affect protein folding and stability (Argentaro et al. 2007). Most mutations affect zinc-binding cysteine residues or residues in the tightly packed hydrophobic core thus reducing its stability. Of greater interest, there is a small cluster of surface mutations (e.g., R246, G249D), which are associated with higher levels of stable protein. These mutations may interfere with protein function, possibly by disrupting an important protein/protein interaction.
Most missense mutations affecting the helicase domain lead to protein instability although one interesting mutation, L1746S, appears to decouple the ATPase activity from productive work (Mitson et al. 2011).
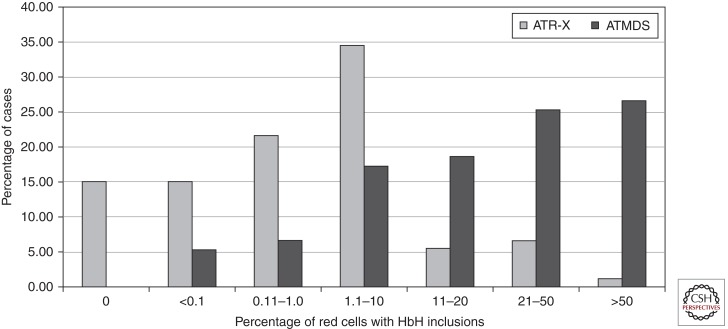
Variability in the Severity of α-Thalassemia
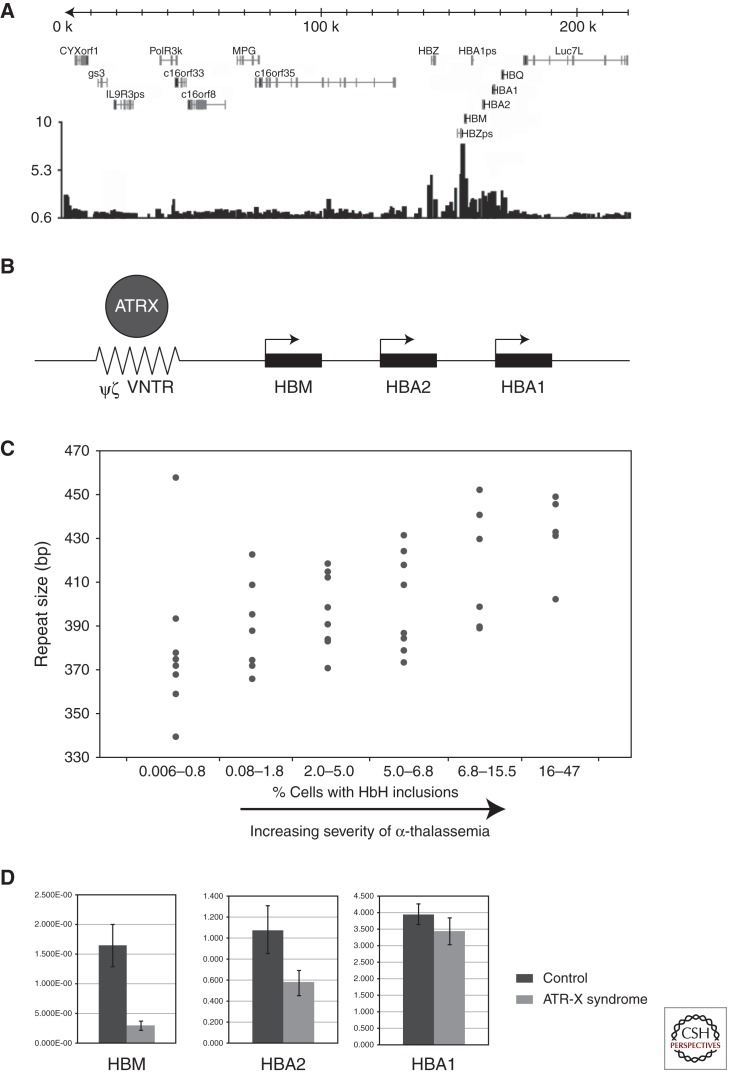
There is considerable variability in the degree to which α-globin synthesis is affected in ATR-X syndrome, as judged by the frequency of cells with HbH inclusions. Up to 15% of patients do not have HbH inclusions (Fig. 4) (Villard et al. 1996a,b,c). It is interesting that patients with identical mutations may have very different, albeit stable, degrees of α-Thalassemia, suggesting that the effect of the ATRX protein on α-globin expression may be modified by other genetic factors (Fig. 4). This is most clearly illustrated by comparing the hematology of cases with identical mutations. Comparison of the 41 cases from 34 pedigrees with the common 736C > T; R246C mutation shows a wide range in the frequency of cells with HbH inclusions (0%–14%). Recent studies into the basis of this variability have led to an important insight into the function of ATRX. Chromatin immunoprecipitation (ChIP) has indicated the ATRX binds to a G-rich tandem repeat (ψζ VNTR) adjacent to the α-globin cluster (Fig. 5A,B) (Law et al. 2010). The length of the repeat is variable and in individuals with ATR-X syndrome, the severity of the thalassemia (as measured by the frequency of cells with HbH inclusions) was related to the size of the repeat: the longer the repeat, the more down-regulated were the genes of the α-globin cluster (Fig. 5C). Furthermore, the closer the gene to the ψζ VNTR, the more down-regulated was the expression so that HBM was the most affected gene in the cluster (Fig. 5D). It appeared that normally ATRX in some way was preventing the G-rich tandem repeat from perturbing local gene expression.
Figure 4.
Percentage of cells with HbH inclusions in individuals with ATR-X syndrome or ATMDS.
Figure 5.
(A) Microarray analysis of ATRX ChIP DNA enrichment from human erythroblasts across the 220-kb terminal region of chromosome 16p containing the α-globin genes (HBM, HBA2, HBA1, HBQ). (B) A schematic showing the relationship of the ψζ VNTR (peak of ATRX binding) to the adjacent α-globin genes. (C) ψζ VNTR length was measured in ATR-X patients with α-Thalassemia and the average length of the two alleles was plotted against the degree of α-Thalassemia as measured by percent red cells showing HbH inclusions (From Law et al. 2010; adapted, with permission, from the author). (D) Comparison of α-globin gene expression between a cohort of normal controls (n = 15) and individuals with ATR-X syndrome (n = 25) showing that the degree of down-regulation is related to the distance from the ψζ VNTR.
What Is the Normal Functional Role of ATRX?
In the adult, ATRX mRNA is widely expressed early in development, and continues to be widely expressed throughout development with particularly high expression in the brain, heart, and skeletal muscle (Stayton et al. 1994). Both isoforms, ATRX (280 kDa) and ATRXt (200 kDa) are readily detected on Western blots (McDowell et al. 1999; Garrick et al. 2004). Immunocytochemical analysis and indirect immunolocalization show that both isoforms are nuclear proteins that predominantly associate with heterochromatin (in interphase and metaphase) (McDowell et al. 1999; Garrick et al. 2004). A significant proportion of ATRX (but not ATRXt) is also found in nuclear speckles (called promyelocytic leukemia [PML] bodies) in human cells (Xue et al. 2003). One additional, striking finding in human metaphase preparations is that ATRX antibodies localize to the short arms of acrocentric chromosomes associated with the rDNA arrays (Gibbons et al. 2000). Genome-wide ChIP indicates that ATRX is most commonly associated with tandem repeats (G-rich rDNA, telomeres and interstitial tandem repeats, as well as A-T rich pericenric heterochromatin) and to a lesser extent CpG islands. Some G-rich repetitive sequences can take up non-B form DNA structures such as G-quadruplex structures in vitro and many ATRX targets are predicted to do so (Law et al. 2010). Furthermore, in a competition assay, recombinant ATRX protein binds to a G4 structure in preference to the unfolded DNA form. It is possible that ATRX plays a role to recognize and resolve non-B forms of DNA.
ATRX plays an important role in the incorporation of the histone variant H3.3 into telomeric and pericentromeric DNA (Goldberg et al. 2010; Wong et al. 2010). It does so with its partner protein Daxx, which directly binds H3.3 and acts as a histone chaperone (Drane et al. 2010; Goldberg et al. 2010); ATRX both directs where this occurs and facilitates the incorporation of H3.3 into chromatin. Tandem repetitive DNA is predicted to be an inherently unfriendly substrate for nucleosomes leading to rapid turnover (Kaplan et al. 2009). Unlike the canonical histones, H3.3 can be incorporated into chromatin in a replication-independent manner so is available for re-establishing the nucleosomal organization of tandem repetitive sequences (Ahmad and Henikoff 2002). An attractive, but as yet unproven, model is that ATRX acts at tandem repeats to prevent them taking up unusual structures by restablishing nucleosomal organization through the insertion of histone H3.3.
α-THALASSEMIA-MYELODYSPLASIA (ATMDS)
Occasionally, patients who previously showed normal erythropoiesis will develop an acquired form of α-Thalassemia, which most commonly arises within the context of hematologic malignancy (Steensma et al. 2005). The first cases of this “acquired hemoglobin H (HbH) disease” were described in 1960 (White et al. 1960). Over the past half century, it has become clear that all cases of acquired α-Thalassemia arise in the context of an underlying clonal disorder of hematopoiesis, most commonly a form of the myelodysplastic syndromes (MDS). Therefore, this condition is now commonly referred to as α-Thalassemia-myelodysplasia (ATMDS, OMIM: 300448). In its most characteristic manifestation there is a severe form of HbH disease, characterized by striking hypochromic microcytic anemia, numerous HbH inclusions detectable by supravital staining of peripheral blood (Fig. 4), and measurable amounts of HbH in the hemolysate (1%–70%).
Other than the disease-defining red cell changes, there are no clinical features that clearly distinguish ATMDS from MDS more generally, although there may be a greater male predominance than is observed in chronic myeloid disorders overall (the male:female ratio in ATMDS is >5:1 whereas in MDS in general it is about 1.5:1) (Haas et al. 2009). The reason for this dramatic sex imbalance in ATMDS is unclear. Patients with ATMDS are diagnosed at similar ages to patients with chronic myeloid disorders who lack thalassemia (median age, 68 years), have similar marrow findings and karyotypic results, have a median survival typical for MDS overall (2–3 years), and die of the same complications—primarily death from infection and, in about 25% of cases, of complications of progression to acute myeloid leukemia (AML) (Steensma et al. 2005). When ATMDS progresses to AML, and the ability of hematopoietic cells to differentiate is further impaired, some patients still continue to have detectable HbH, whereas in other cases the HbH inclusions disappear entirely.
The reticulocyte α/β-globin chain synthesis ratio (and α- to β-globin mRNA ratio) is severely reduced and in the most severely affected individuals, α-chain synthesis is almost completely abolished.
Patients with ATMDS are always anemic at presentation (mean hemoglobin 8.5 g/dL), with a reduced RBC count (mean 4.3 × 1012/L) and markedly hypochromic with microcytic red cell indices (average MCH 22 pg and MCV 75 fL) (Steensma et al. 2005). These abnormalities are even more striking when compared with patients with MDS without thalassemia, who at presentation tend to have slightly higher than normal MCH and MCV values (average MCH 31 pg and MCV 97 fL).
ATRX Mutations in ATMDS
It is now clear that most patients with ATMDS have an acquired, somatic point mutation or an mRNA intron-exon splicing abnormality involving ATRX (Fig. 3). In one exception, the telomeric end of chromosome 16p, including the α-globin cluster, was deleted in a clonally restricted fashion as part of a complex MDS-associated karyotype with numerous large chromosomal rearrangements (Steensma et al. 2004b).
In general, patients with ATMDS have a more severe form of α-Thalassemia than boys with inherited ATR-X syndrome. More than 30% of described ATMDS patients have had >50% of circulating erythrocytes containing HbH inclusions, whereas 90% of boys with ATR-X syndrome have less than 10% HbH-containing cells (Fig. 4) (Gibbons and Higgs 2000; Higgs 2004). This is not explained by the underlying ATRX mutation; even when an identical or similar ATRX mutation is detected in the germline in ATR-X syndrome and as a somatic change in the hematopoietic cells in ATMDS, the thalassemic phenotype is consistently more extensive in ATMDS than in ATR-X. For instance, a boy with germline c.576G > S, p.L192F ATRX mutation had 0.1% HbH, whereas a man with ATMDS who had the identical mutation as an acquired phenomenon had 50% HbH. The reason for this striking difference is unclear, and is suspected to be dependent on the cellular context, perhaps pointing to an interaction between ATRX mutations and a disturbed genetic or epigenetic background in MDS cells that is due to other mutations not present in boys with inherited ATR-X syndrome.
Role of ATRX Mutation in ATMDS and Other Neoplasms
It is debatable whether the ATRX mutations are driver or passenger changes. The loss of detectable HbH and loss of the ATRX mutation with time in an individual with ATMDS suggested the latter (Steensma et al. 2004a). The recent identification in a variety of cancers of frequent mutations in ATRX, its partner protein Daxx, or the histone variant H3.3 that ATRX/Daxx deposit into chromatin has reignited the issue (Heaphy et al. 2011; Jiao et al. 2011; Schwartzentruber et al. 2012). What links the affected cancers is that they appear to show a telomerase independent mechanism for maintaining their telomeres—alternative lengthening of telomeres—involving an aberrant from of homologous recombination. It is of course of considerable interest to determine whether ATMDS shares this characteristic and how the absence of ATRX is associated with this phenomenon.
CONCLUDING REMARKS
Both ATR-X syndrome and ATMDS have revealed the pivotal role of chromatin remodeling in the expression of the α-globin genes. This emphasizes the value of studying orphan diseases to reveal previously unknown biology. Furthermore, this pathway may provide a therapeutic target for manipulating α-globin expression with a view to reducing the toxic effects of the excess α-globin chains seen in β thalassemia.
ACKNOWLEDGMENTS
Many thanks to Liz Rose for help with preparing the manuscript.
Footnotes
Editors: David Weatherall, Alan N. Schechter, and David G. Nathan
Additional Perspectives on Hemoglobin and Its Diseases available at www.perspectivesinmedicine.org
REFERENCES
- Ahmad K, Henikoff S 2002. Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci 99: 16477–16484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argentaro A, Yang JC, Chapman L, Kowalczyk MS, Gibbons RJ, Higgs DR, Neuhaus D, Rhodes D 2007. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc Natl Acad Sci 104: 11939–11944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra MA, Araujo AS, Phylipsen M, Balak D, Kimura EM, Oliveira DM, Costa FF, Sonati MF, Harteveld CL 2008. The deletion of SOX8 is not associated with ATR-16 in an HbH family from Brazil. Br J Haematol 142: 324–326 [DOI] [PubMed] [Google Scholar]
- Buckle VJ, Kearney L 1994. New methods in cytogenetics. Curr Opin Genet Dev 4: 374–382 [DOI] [PubMed] [Google Scholar]
- Cardoso C, Lutz Y, Mignon C, Compe E, Depetris D, Mattei MG, Fontes M, Colleaux L 2000. ATR-X mutations cause impaired nuclear location and altered DNA binding properties of the XNP/ATR-X protein. J Med Genet 37: 746–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels RJ, Peden JF, Lloyd C, Horsley SW, Clark K, Tufarelli C, Kearney L, Buckle VJ, Doggett NA, Flint J, et al. 2001. Sequence, structure and pathology of the fully annotated terminal 2 Mb of the short arm of human chromosome 16. Hum Mol Genet 10: 339–352 [DOI] [PubMed] [Google Scholar]
- Dhayalan A, Tamas R, Bock I, Tattermusch A, Dimitrova E, Kudithipudi S, Ragozin S, Jeltsch A 2011. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum Mol Genet 20: 2195–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnai D, Clayton-Smith J, Gibbons RJ, Higgs DR 1991. The non-deletion a thalassaemia/mental retardation syndrome. Further support for X linkage. J Med Genet 28: 742–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A 2010. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev 24: 1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Polycystic Kidney Disease Consortium 1994. The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell 77: 881–894 [DOI] [PubMed] [Google Scholar]
- Eustermann S, Yang JC, Law MJ, Amos R, Chapman LM, Jelinska C, Garrick D, Clynes D, Gibbons RJ, Rhodes D, et al. 2011. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol 18: 777–782 [DOI] [PubMed] [Google Scholar]
- Fisher E, Scambler P 1994. Human haploinsufficiency—one for sorrow, two for joy. Nat Genet 7: 5–7 [DOI] [PubMed] [Google Scholar]
- Garrick D, Samara V, McDowell TL, Smith AJ, Dobbie L, Higgs DR, Gibbons RJ 2004. A conserved truncated isoform of the ATR-X syndrome protein lacking the SWI/SNF-homology domain. Gene 326: 23–34 [DOI] [PubMed] [Google Scholar]
- Garrick D, Sharpe JA, Arkell R, Dobbie L, Smith AJ, Wood WG, Higgs DR, Gibbons RJ 2006. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet 2: e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gecz J, Pollard H, Conzalez G, Villard L, Stayton C, Millasseau P, Khrestchatisky M, Fontes M 1994. Cloning and expression of the murine homologue of a putative human X-linked nuclear protein gene closely linked to PGK1 in Xq13.3. Hum Mol Genet 3: 39–44 [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Higgs DR 2000. The molecular-clinical spectrum of the ATR-X syndrome. Am J Med Genet 97: 204–212 [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Wilkie AOM, Weatherall DJ, Higgs DR 1991. A newly defined X linked mental retardation syndrome associated with a thalassaemia. J Med Genet 28: 729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, Suthers GK, Wilkie AOM, Buckle VJ, Higgs DR 1992. X-linked α thalassemia/mental retardation (ATR-X) syndrome: Localisation to Xq12-21.31 by X-inactivation and linkage analysis. Am J Hum Genet 51: 1136–1149 [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, Brueton L, Buckle VJ, Burn J, Clayton-Smith J, Davison BCC, Gardner RJM, Homfray T, Kearney L, Kingston HM, et al. 1995a. The clinical and hematological features of the X-linked α thalassemia/mental retardation syndrome (ATR-X). Am J Med Genet 55: 288–299 [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Picketts DJ, Villard L, Higgs DR 1995b. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome). Cell 80: 837–845 [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Bachoo S, Picketts DJ, Aftimos S, Asenbauer B, Bergoffen J, Berry SA, Dahl N, Fryer A, Keppler K, et al. 1997. Mutations in a transcriptional regulator (hATRX) establish the functional significance of a PHD-like domain. Nat Genet 17: 146–148 [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, McDowell TL, Raman S, O’Rourke DM, Garrick D, Ayyub H, Higgs DR 2000. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet 24: 368–371 [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Wada T, Fisher C, Malik N, Mitson M, Steensma D, Goudie D, Fryer A, Krantz I, Traeger-Synodinos J 2008. Mutations in the chromatin associated protein ATRX. Human Mutation 29: 796–802 [DOI] [PubMed] [Google Scholar]
- Gibson WT, Harvard C, Qiao Y, Somerville MJ, Lewis MES, Rajcan-Separovic E 2008. Phenotype-genotype characterization of α-thalassemia mental retardation syndrome due to isolated monosomy of 16p13.3. Am J Med Genet 146A: 225–232 [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas PS, Roy NB, Gibbons RJ, Deville MA, Fisher C, Schwabe M, Bisse E, van Dorsselaer A, Higgs DR, Lubbert M 2009. The role of X-inactivation in the gender bias of patients with acquired α-thalassaemia and myelodysplastic syndrome (ATMDS). Br J Haematol 144: 538–545 [DOI] [PubMed] [Google Scholar]
- Hall JG 1990. Genomic imprinting: Review and relevance to human diseases. Am J Hum Genet 46: 857–873 [PMC free article] [PubMed] [Google Scholar]
- Harteveld CL, Voskamp A, Phylipsen M, Akkermans N, den Dunnen JT, White SJ, Giordano PC 2005. Nine unknown rearrangements in 16p13.3 and 11p15.4 causing α- and β-thalassaemia characterised by high resolution multiplex ligation-dependent probe amplification. J Med Genet 42: 922–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harteveld CL, Kriek M, Bijlsma EK, Erjavec Z, Balak D, Phylipsen M, Voskamp A, Capua E, White SJ, Giordano PC 2007. Refinement of the genetic cause of ATR-16. Hum Genet 122: 283–292 [DOI] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, et al. 2011. Altered telomeres in tumors with ATRX and DAXX mutations. Science 333: 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs DR 2004. Ham-wasserman lecture. Gene regulation in hematopoiesis: New lessons from thalassemia. Hematology 2004: 1–13 [DOI] [PubMed] [Google Scholar]
- Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD, Picketts DJ, Patel DJ, Li H, et al. 2011. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol 18: 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. 2011. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331: 1199–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Harris PC, Wilkie AOM, Wood WG, Dauwerse JG, Higgs DR 1993. De novo truncation of chromosome 16p and healing with (TTAGGG)n in the α-thalassemia/mental retardation syndrome (ATR-16). Am J Hum Genet 52: 668–676 [PMC free article] [PubMed] [Google Scholar]
- Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, et al. 2010. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 143: 367–378 [DOI] [PubMed] [Google Scholar]
- McDowell TL, Gibbons RJ, Sutherland H, O’Rourke DM, Bickmore WA, Pombo A, Turley H, Gatter K, Picketts DJ, Buckle VJ, et al. 1999. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci 96: 13983–13988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitson M, Kelley LA, Sternberg MJ, Higgs DR, Gibbons RJ 2011. Functional significance of mutations in the Snf2 domain of ATRX. Hum Mol Genet 20: 2603–2610 [DOI] [PubMed] [Google Scholar]
- Muers MR, Sharpe JA, Garrick D, Sloane-Stanley J, Nolan PM, Hacker T, Wood WG, Higgs DR, Gibbons RJ 2007. Defining the cause of skewed X-chromosome inactivation in X-linked mental retardation by use of a mouse model. Am J Hum Genet 80: 1138–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer D, Poulat F, Holinski-Feder E, Kooy F, Scherer G 2000. The SOX8 gene is located within 700 kb of the tip of chromosome 16p and is deleted in a patient with ATR-16 syndrome. Genomics 63: 108–116 [DOI] [PubMed] [Google Scholar]
- Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OWJ, Gibbons RJ 1996. ATRX encodes a novel member of the SNF2 family of proteins: Mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet 5: 1899–1907 [DOI] [PubMed] [Google Scholar]
- Picketts DJ, Tastan AO, Higgs DR, Gibbons RJ 1998. Comparison of the human and murine ATRX gene identifies highly conserved, functionally important domains. Mamm Genome 9: 400–403 [DOI] [PubMed] [Google Scholar]
- Price TS, Regan R, Mott R, Hedman A, Honey B, Daniels RJ, Smith L, Greenfield A, Tiganescu A, Buckle V, et al. 2005. SW-ARRAY: A dynamic programming solution for the identification of copy-number changes in genomic DNA using array comparative genome hybridization data. Nucleic Acids Res 33: 3455–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. 2006. Global variation in copy number in the human genome. Nature 444: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickel RD 1986. Contiguous gene syndromes: A component of recognizable syndromes. J Pediatr 109: 231–241 [DOI] [PubMed] [Google Scholar]
- Schneider AS, Bischoff FZ, McCaskill C, Coady ML, Stopfer JE, Shaffer LG 1996. Comprehensive 4-year follow-up on a case of maternal heterdisomy for chromosome 16. Am J Med Genet 66: 204–208 [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231 [DOI] [PubMed] [Google Scholar]
- Stayton CL, Dabovic B, Gulisano M, Gecz J, Broccoli V, Giovanazzi S, Bossolasco M, Monaco L, Rastan S, Boncinelli E, et al. 1994. Cloning and characterisation of a new human Xq13 gene, encoding a putative helicase. Hum Mol Genet 3: 1957–1964 [DOI] [PubMed] [Google Scholar]
- Steensma DP, Higgs DR, Fisher CA, Gibbons RJ 2004a. Acquired somatic ATRX mutations in myelodysplastic syndrome associated with α thalassemia (ATMDS) convey a more severe hematologic phenotype than germline ATRX mutations. Blood 103: 2019–2026 [DOI] [PubMed] [Google Scholar]
- Steensma DP, Viprakasit V, Hendrick A, Goff DK, Leach J, Gibbons RJ, Higgs DR 2004b. Deletion of the alpha-globin gene cluster as a cause of acquired α-thalassemia in myelodysplastic syndrome. Blood 103: 1518–1520 [DOI] [PubMed] [Google Scholar]
- Steensma DP, Gibbons RJ, Higgs DR 2005. Acquired α thalassemia in association with myelodysplastic syndrome and other hematologic malignancies. Blood 105: 443–452 [DOI] [PubMed] [Google Scholar]
- Villard L, Gecz J, Mattéi JF, Fontés M, Saugier-Veber P, Munnich A, Lyonnet S 1996a. XNP mutation in a large family with Juberg-Marsidi syndrome. Nature Genetics 12: 359–360 [DOI] [PubMed] [Google Scholar]
- Villard L, Lacombe D, Fontés M 1996b. A point mutation in the XNP gene, associated with an ATR-X phenotype without α-thalassemia. Eur J Hum Genet 4: 316–320 [DOI] [PubMed] [Google Scholar]
- Villard L, Toutain A, Lossi A-M, Gecz J, Houdayer C, Moraine C, Fontès M 1996c. Splicing mutation in the ATR-X gene can lead to a dysmorphic mental retardation phenotype without α-thalassemia. Am J Hum Genet 58: 499–505 [PMC free article] [PubMed] [Google Scholar]
- Weatherall DJ, Higgs DR, Bunch C, Old JM, Hunt DM, Pressley L, Clegg JB, Bethlenfalvay NC, Sjolin S, Koler RD, et al. 1981. Hemoglobin H disease and mental retardation. A new syndrome or a remarkable coincidence? N Engl J Med 305: 607–612 [DOI] [PubMed] [Google Scholar]
- White JC, Ellis M, Coleman PN, Beaven GH, Gratzer WB, Shooter EM, Skinner ER 1960. An unstable haemoglobin associated with some cases of leukaemia. Br J Haematol 6: 171–177 [DOI] [PubMed] [Google Scholar]
- Wilkie AOM, Buckle VJ, Harris PC, Lamb J, Barton NJ, Reeders ST, Lindenbaum RH, Nicholls RD, Barrow M, Bethlenfalvay NC, et al. 1990a. Clinical features and molecular analysis of the a thalassaemia/mental retardation syndromes. I: Cases due to deletions involving chromosome band 16p13.3. Am J Hum Genet 46: 1112–1126 [PMC free article] [PubMed] [Google Scholar]
- Wilkie AOM, Zeitlin HC, Lindenbaum RH, Buckle VJ, Fischel-Ghodsian N, Chui DHK, Gardner-Medwin D, MacGillivray MH, Weatherall DJ, Higgs DR 1990b. Clinical features and molecular analysis of the α thalassemia/mental retardation syndromes. II: Cases without detectable abnormality of the a globin complex. Am J Hum Genet 46: 1127–1140 [PMC free article] [PubMed] [Google Scholar]
- Wilkie AOM, Pembrey ME, Gibbons RJ, Higgs DR, Porteous MEM, Burn J, Winter RM 1991. The non-deletion type of a thalassaemia/mental retardation: A recognisable dysmorphic syndrome with X-linked inheritance. J Med Genet 28: 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH 2010. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res 20: 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W 2003. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci 100: 10635–10640 [DOI] [PMC free article] [PubMed] [Google Scholar]