Abstract
XBP1 is a key regulator of the unfolded protein response (UPR), which is involved in a wide range of physiological and pathological processes. XBP1 ablation in liver causes profound hypolipidemia in mice, highlighting its critical role in lipid metabolism. XBP1 deficiency triggers feedback activation of its upstream enzyme IRE1α, instigating regulated IRE1-dependent decay (RIDD) of cytosolic mRNAs. Here, we identify RIDD as a crucial control mechanism of lipid homeostasis. Suppression of RIDD by RNA interference or genetic ablation of IRE1α reversed hypolipidemia in XBP1 deficient mice. Comprehensive microarray analysis of XBP1 and/or IRE1α deficient liver identified genes involved in lipogenesis and lipoprotein metabolism as RIDD substrates, which might contribute to the suppression of plasma lipid levels by activated IRE1α. Ablation of XBP1 ameliorated hepatosteatosis, liver damage and hypercholesterolemia in dyslipidemic animal models, suggesting that direct targeting of either IRE1α or XBP1 might be a feasible strategy to treat dyslipidemias.
Introduction
The unfolded protein response (UPR) was originally identified as a signaling system that promotes the transcription of endoplasmic reticulum (ER) chaperone genes in response to stresses that burden the ER with increased client proteins for folding (Ron and Walter, 2007; Schroder and Kaufman, 2005). In mammals, UPR is initiated by three families of unique ER transmembrane proteins, PERK, IRE1 (IRE1α and IRE1β), and ATF6 (ATF6α and ATF6β). IRE1 is evolutionarily well conserved in all eukaryotes from unicellular organisms to mammals, while the other UPR branches are present only in higher eukaryotes (Cox et al., 1993; Mori et al., 1993; Ron and Walter, 2007; Wang et al., 1998).
XBP1 is the only known transcription factor downstream of IRE1α that is activated through an unconventional mRNA splicing reaction. XBP1 activates the transcription of a variety of genes involved in protein secretory pathways (Acosta-Alvear et al., 2007; Lee et al., 2003; Shaffer et al., 2004). In line with this, IRE1α and XBP1 are required for the development, survival and the protein secretory function of some professional secretory cells (Huh et al., 2010; Iwawaki et al., 2010; Kaser et al., 2008; Lee et al., 2005; Reimold et al., 2001; Zhang et al., 2005).
In addition to activating XBP1, IRE1α can also activate Jun N-terminal kinase (JNK) (Urano et al., 2000) and induce the degradation of certain mRNAs, a process known as regulated IRE1-dependent decay (RIDD) (Han et al., 2009; Hollien et al., 2009; Lee et al., 2011; Lipson et al., 2008; Oikawa et al., 2010). The physiological significance of RIDD was first explored in insect cells, where it was postulated to be a mechanism to reduce ER stress by limiting the entry of cargo proteins to the ER, given the preferential degradation of mRNAs encoding secretory proteins by RIDD (Hollien and Weissman, 2006). Interestingly, in mammalian cells, IRE1α appears to cleave mRNAs encoding not only secretory cargo proteins, but also ER resident proteins that serve in protein folding and secretory pathways. This has led to the hypothesis that IRE1α might promote apoptosis under severe ER stress conditions by diminishing ER capacity to handle stress (Han et al., 2009). The in vivo functions of RIDD are only beginning to be explored. We and others have demonstrated that IRE1α degrades insulin mRNA as well as proinsulin-processing enzyme mRNAs, uncovering an important function of RIDD in insulin secretion from β cells (Han et al., 2009; Lee et al., 2011; Lipson et al., 2008). IRE1β, which is specifically expressed in the epithelial cells of the gastrointestinal tract, was shown to degrade the mRNA encoding microsomal triglyceride transfer protein, and hence to suppress chylomicron production (Iqbal et al., 2008).
We previously reported that XBP1 ablation in the liver profoundly decreased plasma triglyceride (TG) and cholesterol levels in mice, revealing an important role for this factor in hepatic lipid metabolism (Lee et al., 2008). Contrary to our speculation that XBP1 deficiency might induce ER stress in hepatocytes, leading to decreased very-low-density lipoprotein (VLDL) secretion, XBP1 deficient hepatocytes did not exhibit morphological signs of ER dysfunction, defects in apoB100 secretion, TG accumulation, increased apoptosis, or activation of XBP1 independent UPR markers, arguing against the contribution of ER stress to the hypolipidemic phenotype of the mutant mice. Instead, we found that the expression of key lipogenic enzyme genes was reduced in XBP1 deficient liver. Some but not all of these genes were directly induced by XBP1s overexpression, indicating that XBP1 acts as a pro-lipogenic transcription factor.
While XBP1 plays an important role in hepatic lipid metabolism, several studies have also reported UPR activation in alcoholic and non-alcoholic fatty liver diseases (Ji and Kaplowitz, 2003; Ozcan et al., 2004; Puri et al., 2008), suggesting the presence of ER stress in these metabolic abnormalities (Hotamisligil, 2008). Although it remains unclear how lipids activate the UPR (or cause ER stress), the idea that ER stress might contribute to the pathogenesis of dyslipidemic metabolic diseases remains an interesting one.
Although XBP1 ablation did not activate PERK or ATF6, it strongly activated its upstream enzyme, IRE1α, indicating feedback regulation of IRE1α activity by the abundance of its substrate XBP1s (Lee et al., 2008). Hyperactivated IRE1α possesses ribonuclease activity to induce the degradation of certain mRNAs by RIDD, such as those encoding cytochrome P450 enzymes that carry out detoxification of xenobiotics (Hur et al., 2012). Here, we demonstrate that RIDD also plays an important role in hepatic lipid metabolism. Suppression of RIDD by IRE1α siRNA markedly restored plasma TG and cholesterol levels in XBP1 knock out mice, along with the induction of lipid metabolism genes. Gene expression profiling revealed a group of lipid metabolism genes regulated by RIDD, which included Angptl3 and the carboxylesterase 1 (Ces1) gene family, whose suppression decreases plasma lipids (Koishi et al., 2002; Musunuru et al., 2010; Quiroga and Lehner, 2011; Romeo et al., 2009). Contrary to the notion that XBP1 mitigates ER stress induced by fat accumulation, XBP1 ablation ameliorated hepatic steatosis in ob/ob mice, and liver damage induced by long-term high fat diet feeding, consistent with the distinct role of IRE1α/XBP1 in lipid metabolism. We also demonstrate that siRNA-mediated silencing of XBP1 mRNA in the liver effectively lowers plasma lipids in mice, providing “proof of principle” that targeting XBP1 may be a viable approach to the treatment of dyslipidemias.
Results
Partial restoration of plasma lipid levels by suppression of IRE1α in Xbp1Δ mice
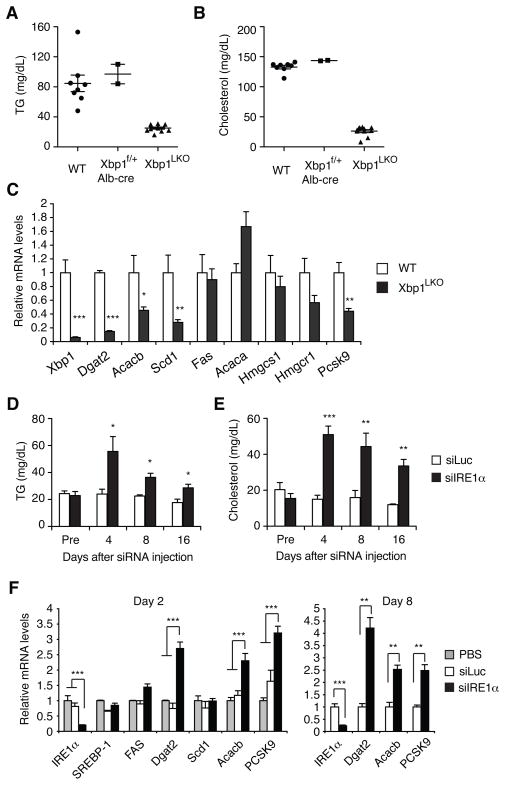
We previously demonstrated that an inducible deletion of Xbp1 in the liver decreased plasma cholesterol and TG levels in mice, while preserving normal hepatic lipid contents and composition (Lee et al., 2008). As XBP1 deficiency also resulted in constitutive activation of IRE1α, which could potentially mediate XBP1-independent functions, we sought to determine the contribution of hyperactivated IRE1α to the hypolipidemic phenotype of XBP1 deficient mice. Xbp1f/f;Alb-cre (Xbp1LKO) mice which expressed Cre recombinase in postnatal liver under the control of the mouse albumin enhancer/promoter displayed markedly lower plasma TG and cholesterol levels compared to their Xbp1f/f littermate controls (Figure 1A and 1B). Plasma lipid levels were not altered in heterozygous Xbp1f/+;Alb-cre mice. Similar results were obtained in females (Figure S1A and S1B). Lipogenic enzyme genes such as Dgat2, Acacb, and Scd1, were suppressed in the liver of Xbp1LKO mice (Figure 1C), similar to what we observed in the inducible XBP1 deficient mouse strain (Lee et al., 2008).
Figure 1.
Partial restoration of plasma lipid levels by IRE1α silencing in XBP1 deficient mice. (A) Plasma TG and (B) cholesterol levels of male mice with the indicated genotypes measured at fed state. (C) Expression of lipid metabolism genes in male Xbp1LKO mice determined by qRT-PCR. n = 8–9 per group. (D) Xbp1LKO mice were i.v. injected with siRNAs targeting luciferase or IRE1α mRNA. Plasma TG and (E) cholesterol levels were measured at indicated time points. n = 8–9 per group. (F) Xbp1LKO mice were sacrificed 2 or 8 days after siRNA or PBS injection. Hepatic mRNA levels were measured by qRT-PCR. Values represent mean ± SEM. n = 2 (PBS), or 3–6 (siRNA). *P <0.05, **P <0.01, ***P <0.001.
To investigate the function of hyperactivated IRE1α in hepatic lipid metabolism, we silenced IRE1α expression in the liver of Xbp1LKO mice using siRNA, as described elsewhere (Hur et al., 2012). IRE1α siRNA markedly increased plasma TG and cholesterol levels in Xbp1LKO mice, indicating that hyperactivated IRE1α contributed to the reduction of plasma lipids in the setting of XBP1 deficiency (Figure 1D and 1E). Injection of IRE1α siRNA into Xbp1LKO mice significantly increased Dgat2 and Acacb mRNAs, which play crucial roles in fatty acid metabolism and were suppressed in Xbp1 deficient liver (Figure 1F). Pcsk9, which promotes the degradation of LDL receptor (Horton et al., 2007), was also induced by IRE1α siRNA, suggesting that these mRNAs are degraded after cleavage by hyperactivated IRE1α in XBP1 deficient liver. In contrast, wt C57BL/6 mice injected with IRE1α siRNA exhibited slightly lower plasma TG and cholesterol levels compared with the control siRNA injected group (Figure S1C), consistent with the phenotype of genetically IRE1α ablated mice as described below.
Modest hypolipidemia in IRE1α deficient mice
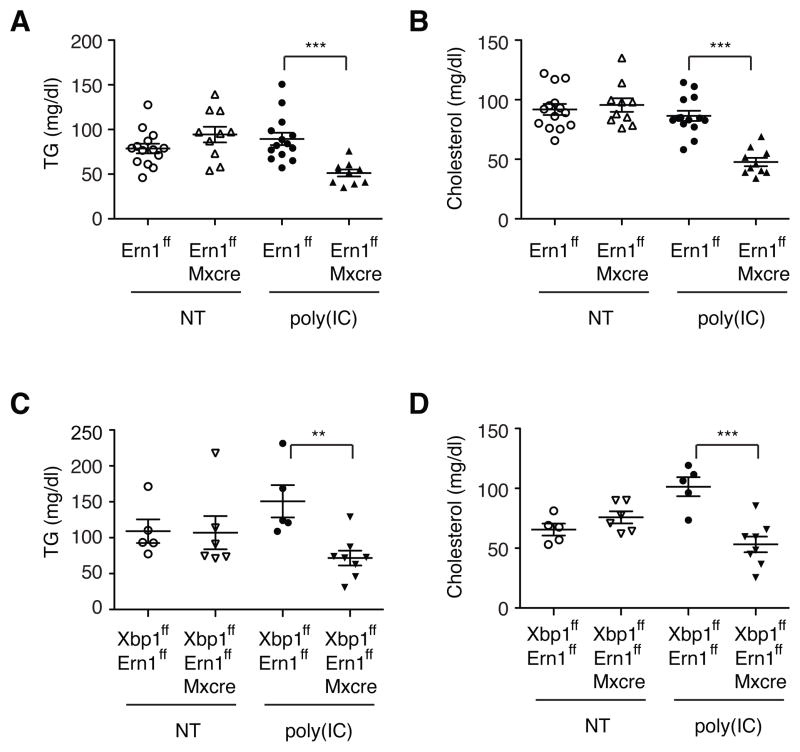
Since IRE1α silencing restored the plasma lipid concentrations to near normal levels in Xbp1 deficient mice, we asked whether XBP1 regulates plasma lipid levels solely by modulating IRE1α activity or has a separate function in hepatic lipid metabolism. We sought to determine the relative contributions of XBP1 deficiency and IRE1α hyperactivation to the dyslipidemia phenotype of Xbp1LKO mice. It is notable that IRE1α siRNA only partially restored plasma lipid levels in Xbp1LKO mice (Figure 1), suggesting that XBP1 itself may also play a role in lipid metabolism independent of RIDD. XBP1 deficient mice with normal IRE1α activity would reveal the direct role of XBP1 in hepatic lipid metabolism, but this was not a feasible approach. As an alternative, we examined IRE1α deficient mice that do not produce the active XBP1s protein. IRE1α deficient mice were generated by crossing Ern1flox mice with Mx1-cre mice, which allowed inducible deletion of IRE1α in the liver upon poly (I:C) administration. Without poly (I:C) administration, plasma TG and cholesterol levels were comparable between Ern1f/f and Ern1f/f;Mx1-cre mice (Figure 2A and 2B). Notably, poly (I:C) administration significantly decreased both plasma TG and cholesterol levels in Ern1f/f;Mx1-cre mice (Ire1Δ), but not in the control Ern1flox/flox mice, indicating that XBP1s is required to maintain normal plasma lipid levels. Moreover, IRE1α and XBP1 double-deficient mice exhibited modestly decreased plasma TG and cholesterol levels compared with WT littermates (Figure 2C and 2D), consistent with the results from IRE1α siRNA-injected Xbp1LKO mice (Figure 1C and 1D). XBP1 and/or IRE1α mRNA levels were decreased by >90% in Ire1Δ/Xbp1Δ and Ire1Δ mice compared to controls (Figure S2). These data suggest that the lack of XBP1s and the hyperactivation of IRE1α both contribute to the dramatic decrease of plasma lipids observed in the setting of XBP1 deficiency and do so via separate mechanisms.
Figure 2.
Plasma lipid levels in IRE1α, and IRE1α/XBP1 double deficient mice. (A) Plasma TG and (B) cholesterol levels were measured before and 3 wks after poly (I:C) injection of male Ern1f/f and Ern1f/f;Mx1-cre mice. Bloods were drawn at fed state. (C–D) Plasma lipid levels of Ern1f/f;Xbp1f/f and Ern1f/f;Xbp1f/f;Mx1-cre mice before and 3 wks after poly(I:C) injection. Values represent mean ± SEM. **P <0.01, ***P <0.001.
Identification of direct XBP1 targets and RIDD substrates in liver
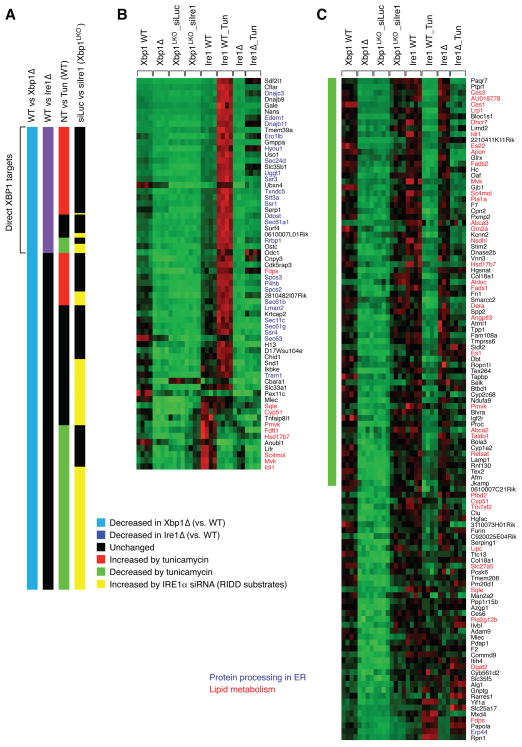
We next sought to identify lipid metabolism genes that are directly regulated by XBP1 and those regulated by RIDD. We reasoned that genes down-regulated in XBP1 deficient liver would fall into two groups: RIDD substrates and direct XBP1 targets. To identify RIDD substrate mRNAs and direct XBP1 targets in the liver, we performed a comprehensive comparative microarray analysis of three groups of RNA samples: WT and XBP1 deficient mice, WT and IRE1α deficient mice untreated or injected with tunicamycin, and XBP1 deficient mice injected with luciferase or IRE1α siRNA. We identified 237 genes whose mRNA levels were significantly lower in XBP1 knockout than in the WT littermate controls (Figure 3 and Table S1). Among these XBP1- dependent genes, 64 genes were also down regulated in IRE1α deficient liver, hence representing likely direct XBP1 target genes. A majority of these genes were induced by tunicamycin treatment in WT, but not in Ire1Δ mice, indicating that XBP1s directly activates their transcription (Figure 3A and 3B). Gene ontology analysis revealed that genes involved in “Protein processing in endoplasmic reticulum (KEGG mmu04141)” were highly enriched in this group, consistent with the critical function of XBP1 in protein secretory pathways (Lee et al., 2003; Shaffer et al., 2004). Notably, the expression of a number of sterol biosynthesis-related genes such as Fdps, Sqle, Cyp51, Pmvk, Fdft1, Hsd17b7, Sc4mol, Mvk, and Idi1 was also reduced in both XBP1 and IRE1α deficient mice, and hence might contribute to the decreased lipid levels in these mutant mice.
Figure 3.
Identification of direct XBP1 targets and RIDD substrates by microarray analysis. (A) Comparison of gene expression profiles between WT and Xbp1Δ; WT and Ire1Δ; untreated (NT) and tunicamycin-treated; siLuc and siIre1 injection to Xbp1LKO. Shown are data for 237 genes that were decreased in Xbp1Δ liver. (B) mRNA levels of 64 genes that were suppressed in both Xbp1Δ and Ire1Δ liver in individual liver samples. (C) mRNA levels of 112 genes that were induced by IRE1α siRNA in Xbp1LKO liver. Genes involved in protein folding processes in the ER are shown in blue. Lipid metabolism genes are highlighted in red.
Among the 237 genes that were suppressed in XBP1 deficient liver, 112 genes were induced by IRE1α siRNA treatment in XBP1 deficient mice- these genes represent RIDD substrates (Figure 3C). Notably, a number of genes involved in lipid metabolism in addition to Dgat2, Acacb and Pcsk9 were identified as RIDD substrates, likely contributing to the suppression of plasma lipid levels by activated IRE1α. Acacb and Pcsk9 were not included in the microarray analysis because of weak signals. Tunicamycin activates IRE1α and hence would decrease the expression of RIDD substrates. Indeed, most of the genes that were suppressed by tunicamycin were induced by IRE1α siRNA in XBP1 deficient liver. (Figure 3A). Among RIDD substrates, Hsd17b7, Idi1, Mvk, Sc4mol and Pmvk, which participate in sterol biosynthesis were suppressed in IRE1α deficient mice, suggesting that XBP1 regulates these genes in both RIDD-dependent and –independent manners.
Identification of Ces1 and Angptl3 mRNAs as RIDD substrates
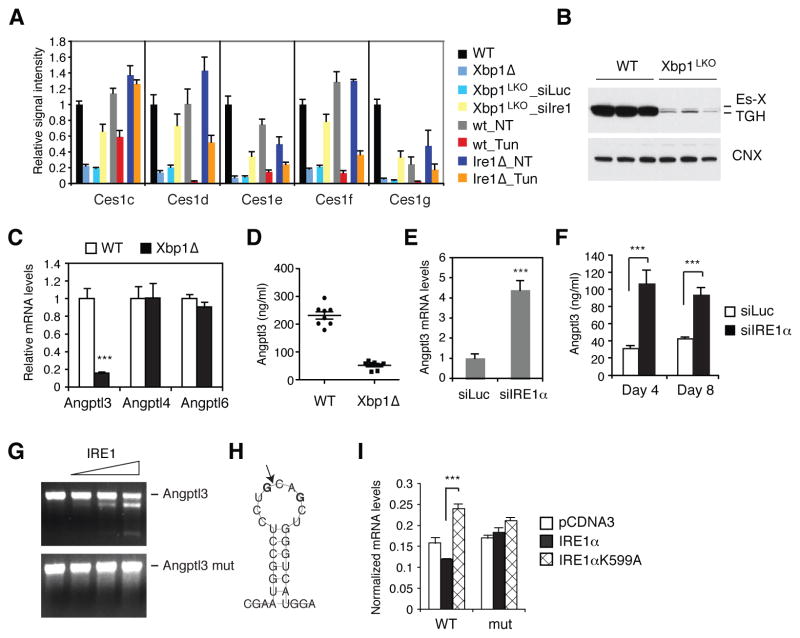
A notable group of RIDD substrates was the carboxylesterase 1 gene family. The mouse genome contains eight highly homologous CES1-like genes in tandem, generated by gene duplication events during evolution (Figure S3A and S3B) (Holmes et al., 2010). Ces1 genes encode enzymes that possess TG and cholesterol-ester hydrolase activities, and have been implicated in the hydrolysis of neutral lipids stored in lipid droplet and VLDL formation (Parathath et al., 2011; Quiroga and Lehner, 2011; Wei et al., 2010). Notably, ablation of Ces3/TGH encoded by the Ces1d gene decreased plasma TG and cholesterol levels without causing steatosis (Wei et al., 2010). Ces1c, Ces1d, Ces1e, Ces1f and Ces1g mRNA levels were strikingly lower in XBP1 deficient, but not in IRE1α deficient liver compared with WT controls (Figure 4A). Further, these mRNAs were induced by IRE1α siRNA in Xbp1LKO liver, and suppressed by tunicamycin treatment in WT mice. Western blot analysis of liver lysates revealed marked reduction of TGH (Ces1d) and Es-X (Ces1g) proteins in XBP1 deficient liver (Figure 4B). IRE1α-induced degradation of Ces1 mRNAs may contribute to the hypolipidemic phenotype of XBP1 deficient mice.
Figure 4.
Identification of Ces1 and Angptl3 mRNAs as RIDD substrates. (A) Microarray signals for Ces1 genes. (B) Western blot analysis of liver lysates of WT and Xbp1LKO mice using TGH (Ces1d) antibody which also detects Es-X (Ces1g). (C) Angptl3, -4, and -6 mRNA levels were measured in the liver of WT and XBP1 deleted male mice. n = 4 per group. (D) Plasma Angptl3 protein levels in WT and Xbp1Δ mice measured by ELISA. (E) Hepatic Angptl3 mRNA levels measured 8 days after siRNA injection of Xbp1LKO mice. n = 3–5 per group. (F) Plasma Angptl3 protein levels measured 4 or 8 days after siRNA injection to Xbp1LKO mice. n = 3–5 per group. (G) In vitro transcribed Angptl3 mRNA was incubated with recombinant IRE1α and then resolved on an agarose gel. Angptl3mut was generated by changing the two G residues to C shown in bold face in (H). (H) Predicted secondary structure of Angptl3 mRNA with a potential IRE1α cleavage site that is depicted by an arrow. (I) WT and mutant Angptl3 constructs were transfected into 293T cells together with WT or mutant IRE1α constructs. EGFP plasmid was also included in the transfection cocktail and served as a normalization control. Angptl3 mRNA levels in the transfected cells were measured by qRT-PCR. Values represent mean ± SEM. ***P <0.001.
Interestingly, angiopoietin-like protein 3 (Angptl3) mRNA was also identified as a RIDD substrate in the microarray analysis. ANGPTL3 protein is produced and secreted mainly by liver both in humans and mice, and possesses inhibitory activity toward lipoprotein lipase and endothelial lipase (Oike et al., 2005). Accordingly, genetic loss of ANGPTL3 resulted in decreased plasma TG and cholesterol levels in both species (Fujimoto et al., 2006; Koishi et al., 2002; Musunuru et al., 2010; Shimamura et al., 2007). qRT-PCR confirmed that Angptl3 mRNA levels were markedly lower in XBP1 deficient liver compared with WT (Figure 4C). Angptl4 and Angptl6 mRNAs that are also abundantly produced in liver were not altered by XBP1 deficiency (Figure 4C). Consistent with the reduction of Angptl3 mRNA by XBP1 deficiency, plasma Angptl3 protein concentration was also markedly decreased in Xbp1Δ mice (Figure 4D). Administration of IRE1α siRNA into Xbp1LKO mice significantly induced hepatic Angptl3 mRNA and plasma Angptl3 protein levels, indicating that hyperactivated IRE1α reduces Angptl3 expression in the liver (Figure 4E and 4F). We used Angpt13 siRNA to reduce Angptl3 expression in WT mice to levels comparable to what we observed in Xbp1Δ mice. This strategy markedly decreased both plasma TG and cholesterol levels, suggesting that IRE1α-induced degradation of Angplt3 mRNA contributes to the hypolipidemia phenotype of XBP1 deficient mice (Figure S3C-S3F).
To directly demonstrate that Angptl3 mRNA is cleaved by IRE1α, we performed in vitro cleavage assays using recombinant IRE1α. Angptl3 RNA synthesized in vitro was efficiently cleaved by IRE1α into two smaller fragments (Figure 4G). Analysis of the mRNA secondary structure predicted the presence of a hairpin structure in Angptl3 mRNA, which was similar to the IRE1α cleavage site in XBP1 mRNA (Figure 4H). Mutation of the potential cleavage site abolished the cleavage of Angptl3 mRNA by IRE1α, demonstrating that IRE1α recognizes the hairpin structure and cleaves Angptl3 mRNA (Figure 4G). To further validate the IRE1α cleavage site in Angptl3 mRNA, we transfected 293T cells with WT or cleavage site mutant Angptl3 expression plasmids together with IRE1α, and measured the mRNA levels. IRE1α is activated by overexpression in 293T cells and reduced the expression of WT, but not the mutant Angptl3 mRNA (Figure 4I), indicating that IRE1α degraded only the WT Angptl3 mRNA.
Ablation of XBP1 ameliorates hepatic steatosis and liver damage in dietary and genetic animal disease models
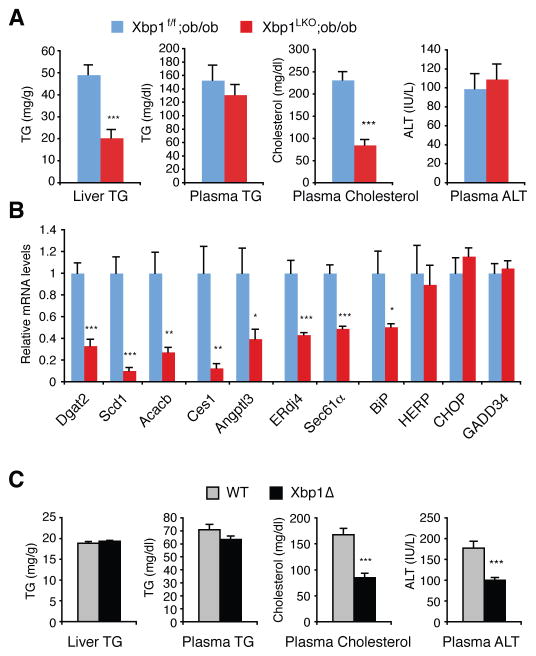
IRE1α and XBP1 activate hepatic lipid metabolism via transcriptional and post-transcriptional regulation of genes in lipid metabolic pathways. However, the IRE1α/XBP1 signaling pathway is also considered to have a cytoprotective role against ER stress, which has been implicated in metabolic abnormalities in various organs (Cnop et al., 2011; Ron and Walter, 2007). Several reports have demonstrated that fat accumulation causes ER stress in liver (Nakatani et al., 2005; Ozcan et al., 2004; Puri et al., 2008; Sha et al., 2011). We asked whether XBP1 deficiency would ameliorate fat accumulation in liver and blood under metabolic stress conditions, given its lipid-lowering effect, or would instead heighten ER stress and lipotoxicity, leading to increased cell death. We investigated the effects of XBP1 deficiency in two dyslipidemic animal models: leptin deficient ob/ob mice, and mice fed a high-fat diet.
Ablation of Xbp1 in the liver of ob/ob mice was achieved by generating compound mutant mice (Xbp1LKO;ob/ob) harboring a homozygous Xbp1 flox allele, Alb-cre transgene, and ob/ob mutation. Compared with the Xbp1f/f;ob/ob mice littermate control, Xbp1LKO;ob/ob mice displayed markedly lower hepatic TG and plasma cholesterol levels (Figure 5A and S4A), associated with decreased lipogenic gene mRNA levels (Figure 5B), similar to Xbp1LKO mice. Plasma ALT levels were comparable between Xbp1f/f;ob/ob and Xbp1LKO;ob/ob mice, indicating that the ablation of Xbp1 did not augment liver damage in ob/ob mice (Figure 5A). CHOP, GADD34, Herpud1 and BiP mRNAs which are strongly induced under ER stress conditions by the PERK and ATF6 pathways were not induced in Xbp1LKO;ob/ob mice (Figure 5B), suggesting that XBP1 ablation did not induce measurable ER stress in ob/ob mouse liver. Furthermore, contrary to the notion that fat accumulation induces ER stress in liver, we failed to detect any measurable induction of UPR markers such as phospho-IRE1α, phospho-PERK, phospho-eIF2α, processed nuclear ATF6α and XBP1s proteins, and BiP, CHOP and ERdj4 mRNAs in ob/ob mouse liver (Figure S4B and S4C), suggesting that the causal relationship between fat accumulation and ER stress induction in liver should be reassessed.
Figure 5.
Effects of XBP1 ablation in dyslipidemic mouse models. (A) Metabolic parameters of 8-week old Xbp1f/f;ob/ob and Xbp1LKO;ob/ob mice measured after a 4-h fasting. n = 4–6 female mice per group. (B) Hepatic mRNA levels were measured by qRT-PCR. (C) WT and Xbp1Δ mice were fed a high fat diet for 5 months, and then sacrificed to measure metabolic parameters. n = 9–10 male mice per group. Values represent mean ± SEM. *P <0.05, **P <0.01, ***P <0.001.
Feeding a high-fat diet increased plasma and hepatic TG in Xbp1Δ mice to a level similar to WT mice (Figure 5C and S4D). However, plasma cholesterol levels were lower in HFD fed Xbp1Δ mice than in WT mice. Interestingly, the plasma ALT level was significantly lower in Xbp1Δ compared to WT mice (Figure 5C), indicating decreased hepatotoxicity in the absence of XBP1. We did not observe any differences in glucose tolerance and insulin tolerance tests between WT and Xbp1Δ mice under HFD conditions (Figure S4E and S4F).
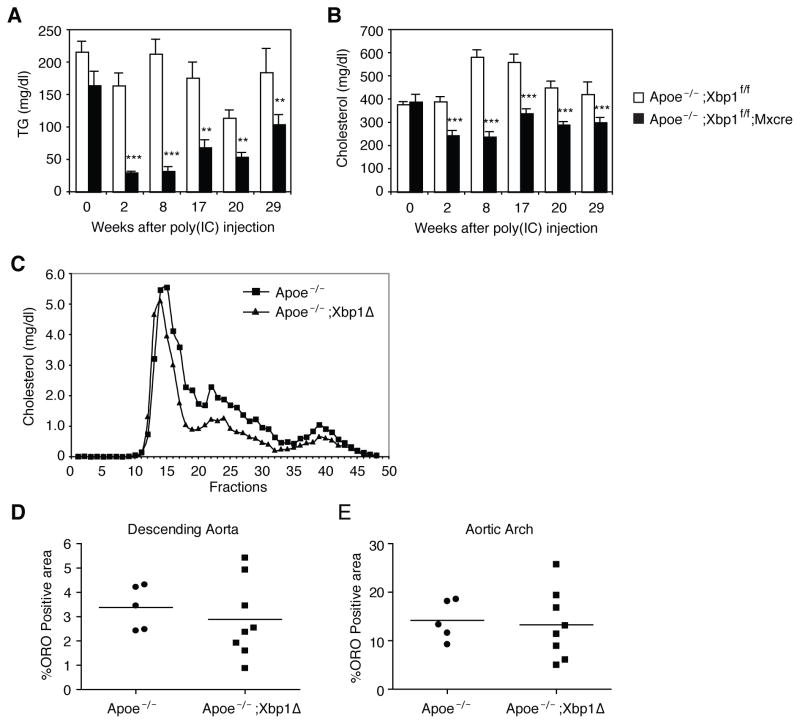
Ablation of XBP1 in ApoE deficient mice can significantly decrease plasma cholesterol
ApoE deficient mice display high levels of plasma cholesterol and are widely used as a model for atherosclerosis (Zadelaar et al., 2007). To test whether XBP1 ablation could lower cholesterol levels and reduce atherosclerotic lesion development under such extreme conditions, we generated ApoE−/−;Xbp1f/f;Mxcre mice that harbored a poly (I:C)-inducible cre recombinase transgene. Poly (I:C) administration activated the Cre and ablated XBP1 to generate ApoE and XBP1 double knock out (ApoE−/−;Xbp1Δ) mice. Inducible XBP1 deletion in ApoE−/− mice significantly decreased plasma TG and cholesterol levels, although the latter still remained substantially elevated at >200 mg/dL (Figure 6A and 6B). FPLC analysis of the lipoprotein profile demonstrated that the cholesterol distribution was not altered by XBP1 ablation in ApoE−/− mice (Figure 6C). Not surprisingly, despite reductions in plasma cholesterol and TG levels in ApoE−/−;Xbp1Δ compared with ApoE−/− mice, atherosclerotic lesion formation was not significantly different between these two groups, as measured by the oil red O stained areas in the descending aorta and aortic arch (Figure 6D, 6E and S5). Thus the 30–40% reduction in plasma cholesterol levels achieved by XBP1 ablation, while very significant, was not sufficient to prevent lesion formation in the setting of ApoE deficiency.
Figure 6.
Ablation of XBP1 in apoE knock out mice reduces plasma lipids. (A) Apoe−/−;Xbp1f/f and Apoe−/−;Xbp1f/f;Mx-cre mice were injected with poly(I:C) on day 0, day 4, week 12 and week 20. Bloods were drawn at fed state to measure plasma TG and (B) cholesterol levels. n = 8–9 per group. Values represent mean ± SEM. **P <0.01, ***P <0.001 compared with the control group (Apoe−/−;Xbp1f/f mice). (C) Plasma collected at weeks 8 and 17 were pooled and subjected to FPLC analysis. (D–E) Descending aorta and aortic arch were stained with oil red O (ORO) and ORO positive areas were quantified.
Effect of XBP1 siRNA on plasma lipids
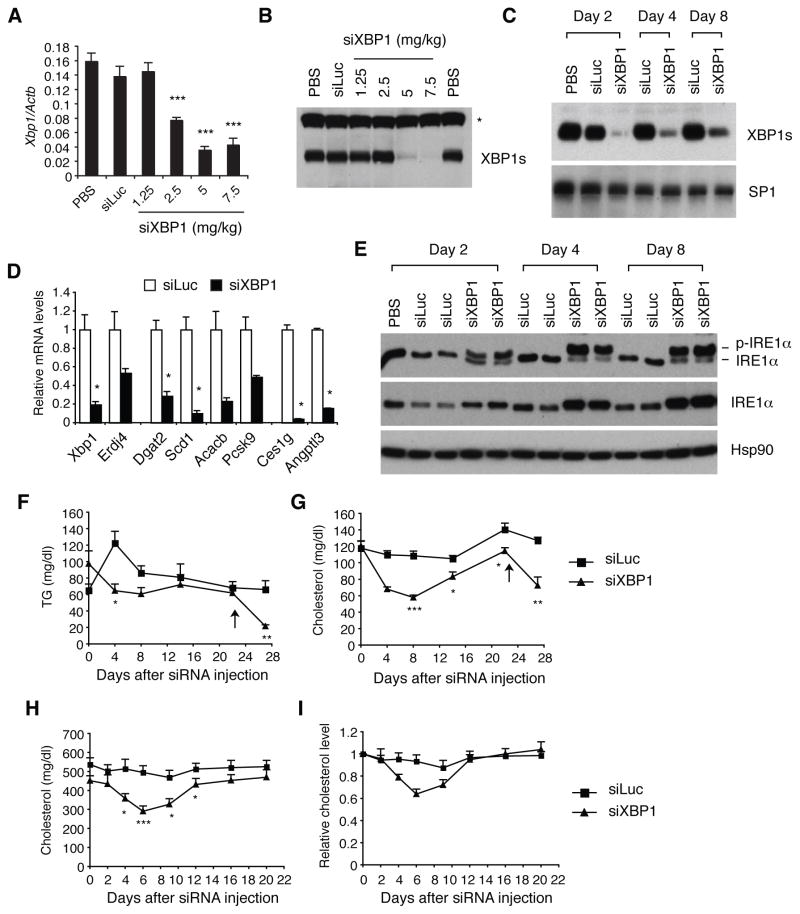
In recent years, RNA interference has received considerable attention as a novel therapeutic strategy, as sequence specific siRNA molecules can target virtually any disease-associated gene with high specificity (Vaishnaw et al., 2010). XBP1 is an attractive target for RNA therapeutics, given the profound reduction of plasma TG and cholesterol by XBP1 ablation. We investigated if the targeted delivery of XBP1 siRNA to the liver would decrease plasma lipid levels. Intravenous injection of lipidoid formulated XBP1 siRNA decreased XBP1 mRNA and protein levels in the liver in a dose-dependent manner, resulting in near complete suppression of XBP1s protein at doses > 5 mg/kg (Figure 7A and 7B). It is notable that a intermediate dose of 2.5 mg/kg siRNA had no effect on XBP1s protein level, despite ~50% reduction of XBP1 mRNA, indicating precise regulation of IRE1α activation to maintain constant XBP1s protein levels in the liver. Silencing of XBP1s expression was stably maintained for more than a week after a single injection of XBP1 siRNA at 7.5 mg/kg (Figure 7C). Similar to the effect of genetic ablation of XBP1, XBP1 siRNA induced IRE1α hyperactivation and decreased the expression of genes involved in lipid metabolism, similar to what we observed in XBP1 knock-out mice (Figure 7D and 7E). XBP1 siRNA transiently decreased plasma TG and cholesterol levels not only in C57BL/6 mice (Figure 7F and 7G), but also in apoE deficient mice (Figure 7H and 7I), indicating that XBP1 siRNA can efficiently reduce even very elevated plasma lipid levels.
Figure 7.
Silencing of XBP1 mRNA in vivo. (A) Female C57BL/6 mice were i.v. injected with PBS, luciferase siRNA (5 mg/kg), or varying doses of XBP1 siRNA. Mice were sacrificed 48 hrs later. Hepatic XBP1 mRNA levels were measured by qRT-PCR. n = 4 mice per group. Values represent mean ± SEM. ***P <0.001 compared with the control groups (PBS and luciferase siRNA groups). (B) Western blot to measure XBP1s protein levels in pooled nuclear extracts. *non-specific band. (C) Mice were injected with siRNAs at 7.5 mg/kg, and sacrificed after indicated days to measure hepatic XBP1s protein levels by western blot. SP1 serves as a loading control. n = 3–4 mice per group. (D) qRT-PCR analysis of hepatic gene expression. n = 3 per group. *P <0.05 (E) Western blot analysis of IRE1α in liver lysates. The panel shows IRE1α phosphorylation measured by Phos-Tag western blot. (F–G) Male C57BL/6, or (H–I) Apoe−/− mice were i.v. injected with siRNAs (7.5 mg/kg), and bled to measure plasma lipid levels. n = 5 –8 per group.. Values represent mean ± SEM. *P <0.05, **P <0.01, ***P <0.001 compared with the control group (siLuc).
Discussion
We have previously demonstrated that ablation of XBP1 in the liver resulted in a profound reduction of plasma TG and cholesterol unaccompanied by hepatic steatosis (Lee et al., 2008). Here, we demonstrate that IRE1α hyperactivation and the consequent RIDD substantially contributes to the hypolipidemia phenotype of XBP1 deficient mice. XBP1 deficiency results in a feedback activation of IRE1α, inducing the degradation of mRNAs of a cohort of lipid metabolism genes, such as Dgat2, Acacb, Pcsk9, Angptl3 and Ces1, which regulate TG and cholesterol metabolism at multiple levels. Dgat2 and Acacb encode key lipogenic enzymes; Pcsk9 is involved in LDL clearance; Angptl3 suppresses LPL mediated TG clearance; Ces1 possesses TG hydrolyzing activity and is implicated in fatty acids mobilization from lipid droplets to nascent VLDL. Hence, suppression of these genes by IRE1α is likely to contribute to the striking hypolipidemic phenotype mediated by liver-specific loss of XBP1.
IRE1α regulates hepatic lipid metabolism via two distinct mechanisms. First, IRE1α promotes the degradation of mRNAs encoding lipid metabolism genes such as Dgat2, Acacb, Ces1, Angptl3, and Pcsk9, which play important roles in de novo lipogenesis, hydrolysis of cholesterol ester and TG, and lipoprotein catabolism. Second, IRE1α activates its downstream transcription factor XBP1, which can directly activate certain lipid metabolism genes. Hence, the combined effects of XBP1 deficiency coupled with IRE1α hyperactivation causes a profound reduction of plasma lipids in XBP1 deficient mice. IRE1α deficient mice are defective in both XBP1s-mediated activation of lipogenic genes and IRE1α-mediated mRNA degradation. Under regular chow-fed conditions, IRE1α activity in liver is low, exerting minimal effect on the levels of RIDD substrates, but producing measurable amounts of XBP1s protein, suggesting that the modest hypolipidemia of IRE1α deficient mice is caused mainly by the lack of XBP1s.
In that case, how does XBP1s deficiency cause hypolipidemia independently of RIDD in IRE1α deficient mice? We previously demonstrated that XBP1 directly activated Dgat2 and Acacb lipogenic genes, which might contribute to the hypolipidemia observed in the absence of XBP1. However, these genes were not suppressed in IRE1α deficient liver, but were induced by IRE1α siRNA in XBP1 deficient liver, indicating that these genes are primarily regulated by RIDD. In contrast, microarray analysis identified various lipid metabolism genes such as Fdps, Sqle, Cyp51, Pmvk, Fdft1, Hsd17b7, Sc4mol, Mvk, and Idi1 that were suppressed in both XBP1 and IRE1α deficient liver, hence representing direct XBP1 targets. We speculate that XBP1 directly regulates the expression of a subset of lipid metabolism genes including those listed above, contributing to the hypolipidemia phenotype observed in IRE1α deficient mice. It is also possible that the alteration of ER protein folding homeostasis caused by XBP1 deficiency indirectly affected lipid metabolism. For example, VLDL assembly occurs in the ER lumen and involves the folding and lipidation of apoB-100. Although apoB turnover was not substantially altered in XBP1 or IRE1α deficient primary hepatocytes (Lee et al., 2008; Zhang et al., 2011), it is possible that XBP1 deficiency impairs VLDL assembly and/or secretion in vivo. Supporting this scenario, IRE1α deficient mice were reported to be more susceptible to tunicamycin-induced hepatic steatosis (Zhang et al., 2011), which is likely to be caused by the inhibition of hepatic VLDL secretion (Lee and Glimcher, 2009; Liao and Chan, 2001).
It is notable that XBP1 ablation did not cause any deleterious effect on the viability of hepatocytes. On the contrary, XBP1 ablation in the liver protected mice from hepatic steatosis and liver damage caused by prolonged HFD feeding or by leptin deficiency (ob/ob mice). XBP1 ablation in the liver of ob/ob mice markedly ameliorated hepatic steatosis, paralleled by the suppression of the expression of lipogenic genes. Feeding a high-fat diet caused liver damage in WT mice, which was ameliorated in XBP1 deficient mice. Oxidative stress has been implicated in HFD-induced liver cell death (Anstee and Goldin, 2006). It remains to be determined if XBP1 deficiency altered the generation of ROS, or the sensitivity of hepatocytes to oxidative damage.
The ER is a crucial subcellular compartment for many physiological functions of the liver, including lipogenesis, lipoprotein production, protein secretion, and detoxification of xenobiotics. One might speculate that an increased burden of lipids, proteins, and xenobiotics on the ER would impair ER function, evoking a stress response. Supporting this hypothesis, several papers have reported UPR activation in various liver diseases as a marker of ER stress (Hotamisligil, 2008; Ozcan et al., 2004; Puri et al., 2008). Notably, it has been shown that TG accumulation in the liver in obesity causes ER stress, and the disruption of the UPR exacerbates the consequences of metabolic abnormalities such as insulin resistance (Hotamisligil, 2008; Ozcan et al., 2004; Park et al., 2010; Winnay et al., 2010; Zhou et al., 2011). In stark contrast, we failed to detect any measurable UPR activation (IRE1α and PERK phosphorylation; ATF6α processing; CHOP and BiP mRNA induction) in the liver of ob/ob mice, arguing against the involvement of the UPR in the metabolic abnormalities in this animal model. Our data are consistent with a recent report that demonstrated increased hepatic insulin sensitivity in XBP1 deficient mice, which was associated with decreased TG accumulation in the liver and plasma (Jurczak et al., 2011). It is important to note that our data does not disprove the involvement of ER dysfunction in metabolic stress, given that we actually did not measure “ER stress”, defined as the increase of misfolded or unfolded protein species in the ER, in ob/ob mice, which is currently not feasible. Although UPR activation has been used as a marker of ER stress, the immediate upstream stimuli activating UPR in normal and diseased liver is not defined. It is possible that metabolic stresses compromise the ER function, without UPR activation.
XBP1 deficient liver displays a qualitatively and quantitatively normal lipid profile with no hepatic steatosis. This is in contrast to ApoB siRNA treated or Mttp mutant mice where lipid accumulates in the liver due to impaired VLDL assembly/secretion (Raabe et al., 1999; Tadin-Strapps et al., 2011). Dramatic reduction of plasma lipids coupled with preservation of the normal hepatic lipid profile in XBP1 deficient mice suggests that XBP1 is a promising target for drug development to treat dyslipidemias. We demonstrated that XBP1 siRNA lowered plasma lipid levels both in WT C57BL/6 and apoE deficient mice, providing proof of principle for targeting XBP1 to treat dyslipidemias. Alternatively, small molecule compounds modulating IRE1α could be useful to lower plasma lipid levels. Since genetic ablation of IRE1α in the liver decreased plasma lipid levels, compounds that inhibit IRE1α activity are expected to have similar lipid-lowering effects. IRE1α activators could also decrease plasma lipid levels by promoting the degradation of mRNAs of lipid metabolism genes. IRE1α activation would also induce XBP1s, which can promote lipogenesis. The effects of IRE1α activation coupled with XBP1s induction on lipid homeostasis remain to be further investigated.
Methods
Mice
Xbp1flox mice were crossed with interferon inducible B6.Cg-Tg(Mx1-cre)1Cgn/J or C57BL/6-Tg(Alb-cre)21Mgn/J strains of mice (Jackson Laboratory) that produce cre recombinase under the control of the mouse albumin promoter and efficiently delete the floxed gene in the liver as previously described (Lee et al., 2008). Ern1flox mice have been previously described (Iwawaki et al., 2009), and crossed with Mx1-cre mice to inducibly delete IRE1α in the liver. Mice were housed in a specific pathogen free facility at the Harvard School of Public Health and had free access to water and standard chow diet (PicoLab Rodent diet 20, #5058, Lab diet), which consisted of 12% fat, 23.5% protein, and 64.5% carbohydrate, or a high-fat diet (45% fat, 18.6% protein, and 36.4% carbohydrate; TestDiet, #58G8). Mice heterozygous for a leptin mutation (B6.V-Lepob/J) were obtained from The Jackson Laboratory, and intercrossed to produce homozygous ob/ob mice, or bred onto Xbp1LKO mice to generate Xbp1LKO;ob/ob mice. Xbp1f/f;Mx1-cre mice were crossed to ApoE−/− mice (Jackson laboratory, B6.129P2-Apoetm1Unc/J), and injected with poly (I:C) to generate XBP1 and ApoE double deficient mice. Animal studies and experiments were approved and carried out according to the guidelines of the Animal Care and Use Committee of Harvard University.
In vivo siRNA delivery
XBP1 siRNA (sense, 5′-CACCCUGAAUUCAUUGUCU-3′; antisense, 5′-AGACAAUGAAUUCAGGGUG-3′) was formulated with lipidoid 98N12-5 as described previously (Frank-Kamenetsky et al., 2008). XBP1 and luciferase siRNAs (Frank-Kamenetsky et al., 2008) were diluted in PBS and injected into mice via the tail vein at 5 μl/g body weight. IRE1α (sense, 5′-AuGccGAAGuucAGAuGGAdTsdT-3′; antisense, 5′-UCcAUCUGAACUUCGGcAUdTsdT-3′) and control luciferase siRNAs formulated with lipidoid C12–200 were injected via tail vein as described previously (Hur et al., 2012; Love et al., 2010).
Blood chemistry and measurement of liver TG contents
Plasma TG and cholesterol levels were measured by using commercial Triglyceride Reagent (Sigma) and the Amplex® Red Cholesterol Assay Kit (Invitrogen), respectively, following manufacturer’s instructions. Plasma Angplt3 levels were measured using a commercial ELISA kit (R&D Systems). Fast performance liquid chromatography (FPLC) analysis of plasma samples was performed as described previously (Lee et al., 2011). Serum alanine aminotransferase (ALT) activity was measured using a commercial reagent (Bioquant). Lipids were extracted from liver tissue by Folch’s method, and subjected to TG assay.
RNA isolation, qRT-PCR and microarray analysis
Total RNA was extracted from liver using Qiazol lysis reagent (Qiagen), and reverse transcribed into cDNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR was performed as described previously (Lee et al., 2008). RNA samples from individual mice were further purified using RNeasy MinElute Cleanup Kit (Qiagen), and then used for the production of biotin-labeled cRNA followed by hybridization with HT MG-430 PM Array Plate (Affymetrix). 237 XBP1-dependent genes were identified by comparing WT and Xbp1Δ liver samples with minimum fold-change criterion of 1.6 and P-value ≤ 0.005. Relaxed criteria (P-value ≤ 0.05) were used for the comparison between WT and Ire1Δ; untreated and tunicamycin treated; luciferase and IRE1α siRNA. Microarray data have been deposited in the Gene Expression Omnibus (GEO) with accession code GSE40515.
Cell culture and Transfection
HEK293T cells were cultured in DMEM supplemented with 10% fetal bovine serum. Transient transfection was performed using Lipofectamine 2000 (Invitrogen), as described previously (Lee et al., 2011).
Western blot
Liver tissues were homogenized in RIPA buffer (50 mM Tris–Cl pH 8.0, 150 mM NaCl, 1% NP40, 0.5% deoxycholate, 0.1% SDS, 50 mM NaF) supplemented with protease inhibitor tablet (Roche). Homogenates were centrifuged at 12,000g for 10 min at 4°C, and the supernatants were collected. Liver nuclear extracts were prepared as described previously (Lee et al., 2008). Liver lysates were used for western blotting as described previously (Hur et al., 2012).
In vitro IRE1α-mediated mRNA cleavage assays
Cleavage of in vitro transcribed Angptl3 mRNA by recombinant IRE1α was tested as described previously (Lee et al., 2011). Briefly, pCMV-SPORT6-Angptl3 (BC019491, Open Biosystems) plasmid was linearized by BglII digestion and incubated with SP6 polymerase to produce Angptl3 RNA. Angptl3mut construct was generated using QuikChange Site-Directed Mutagenesis Kit (Stratagene), based on the predicted mRNA secondary structure (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). In vitro transcribed RNAs were incubated with the recombinant protein including the cytosolic domain of human IRE1α, resolved on a 1.2% denaturing agarose gel and visualized by ethidium bromide staining.
Quantification of atherosclerosis lesions
The aortas were harvested and fixed in 10% buffered formalin solution overnight. Atherosclerotic lesions were analyzed in the aortic arch, and descending aorta by oil red O staining as previously described (Gotsman et al., 2007; Gotsman et al., 2006).
Statistical Analysis
Values represent mean ± SEM. Statistical significance of differences between groups was determined by student T tests. p-values less than 0.05 were considered to be significant.
Supplementary Material
Research highlights.
XBP1-deficient mice exhibit low plasma TG and cholesterol levels.
Suppression of IRE1α increases plasma lipid level in XBP1-deficient mice.
IRE1α degrades mRNAs encoding lipid metabolism regulators.
XBP1 deficiency in liver ameliorates steatosis and liver damage in dyslipidemic animal models.
Acknowledgments
This study was supported by NIH grants AI32412 (LHG), DK082448 (LHG), DK089211 (A-HL), and grants from the American Heart Association (A-HL) and Bristol-Myers Squibb Company (A-HL). We thank David Cohen for FPLC and helpful discussions, Richard Lehner for TGH (Ces1) antibody, Kirsten Sigrist for help with animal maintenance, and Wen-Pin Yang, Aiqing He, Bo Guan and David Gordon for microarray analysis. We also thank Brian Bettencort and Greg Hinkle (design and bioinformatics), Akin Akinc and Alnylam formulations group, Muthiah Manoharan, Martin Maier, Satya Kumchimanchi, Klaus Charisse, William Querbes and the rest of the Alnylam chemistry team for siRNA synthesis.
L.H. Glimcher is a Member of the Board of Directors of and holds equity in Bristol Myers Squibb.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Anstee QM, Goldin RD. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int J Exp Pathol. 2006;87:1–16. doi: 10.1111/j.0959-9673.2006.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnop M, Foufelle F, Velloso LA. Endoplasmic reticulum stress, obesity and diabetes. Trends Mol Med. 2011 doi: 10.1016/j.molmed.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A, Butler D, Charisse K, Dorkin R, Fan Y, Gamba-Vitalo C, Hadwiger P, Jayaraman M, John M, Jayaprakash KN, Maier M, Nechev L, Rajeev KG, Read T, Rohl I, Soutschek J, Tan P, Wong J, Wang G, Zimmermann T, de Fougerolles A, Vornlocher HP, Langer R, Anderson DG, Manoharan M, Koteliansky V, Horton JD, Fitzgerald K. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc Natl Acad Sci U S A. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto K, Koishi R, Shimizugawa T, Ando Y. Angptl3-null mice show low plasma lipid concentrations by enhanced lipoprotein lipase activity. Exp Anim. 2006;55:27–34. doi: 10.1538/expanim.55.27. [DOI] [PubMed] [Google Scholar]
- Gotsman I, Grabie N, Dacosta R, Sukhova G, Sharpe A, Lichtman AH. Proatherogenic immune responses are regulated by the PD-1/PD-L pathway in mice. J Clin Invest. 2007;117:2974–2982. doi: 10.1172/JCI31344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotsman I, Grabie N, Gupta R, Dacosta R, MacConmara M, Lederer J, Sukhova G, Witztum JL, Sharpe AH, Lichtman AH. Impaired regulatory T-cell response and enhanced atherosclerosis in the absence of inducible costimulatory molecule. Circulation. 2006;114:2047–2055. doi: 10.1161/CIRCULATIONAHA.106.633263. [DOI] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Holmes RS, Wright MW, Laulederkind SJ, Cox LA, Hosokawa M, Imai T, Ishibashi S, Lehner R, Miyazaki M, Perkins EJ, Potter PM, Redinbo MR, Robert J, Satoh T, Yamashita T, Yan B, Yokoi T, Zechner R, Maltais LJ. Recommended nomenclature for five mammalian carboxylesterase gene families: human, mouse, and rat genes and proteins. Mamm Genome. 2010;21:427–441. doi: 10.1007/s00335-010-9284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Cohen JC, Hobbs HH. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem Sci. 2007;32:71–77. doi: 10.1016/j.tibs.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS. Inflammation and endoplasmic reticulum stress in obesity and diabetes. Int J Obes (Lond) 2008;32(Suppl 7):S52–54. doi: 10.1038/ijo.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WJ, Esen E, Geahlen JH, Bredemeyer AJ, Lee AH, Shi G, Konieczny SF, Glimcher LH, Mills JC. XBP1 Controls Maturation of Gastric Zymogenic Cells by Induction of MIST1 and Expansion of the Rough Endoplasmic Reticulum. Gastroenterology. 2010;139:2038–2049. doi: 10.1053/j.gastro.2010.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur KY, So JS, Ruda V, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Iwawaki T, Glimcher LH, Lee AH. IRE1alpha activation protects mice against acetaminophen-induced hepatotoxicity. J Exp Med. 2012;209:307–318. doi: 10.1084/jem.20111298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Dai K, Seimon T, Jungreis R, Oyadomari M, Kuriakose G, Ron D, Tabas I, Hussain MM. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 2008;7:445–455. doi: 10.1016/j.cmet.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Kohno K. IRE1alpha disruption causes histological abnormality of exocrine tissues, increase of blood glucose level, and decrease of serum immunoglobulin level. PLoS One. 2010;5:e13052. doi: 10.1371/journal.pone.0013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Yamanaka S, Kohno K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci U S A. 2009;106:16657–16662. doi: 10.1073/pnas.0903775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C, Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology. 2003;124:1488–1499. doi: 10.1016/s0016-5085(03)00276-2. [DOI] [PubMed] [Google Scholar]
- Jurczak MJ, Lee AH, Jornayvaz FR, Lee HY, Birkenfeld AL, Guigni BA, Kahn M, Samuel VT, Glimcher LH, Shulman GI. Dissociation of inositol requiring enzyme (IRE1alpha)-mediated JNK activation from hepatic insulin resistance in conditional X-box binding protein-1 (XBP1) knockout mice. J Biol Chem. 2011 doi: 10.1074/jbc.M111.316760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koishi R, Ando Y, Ono M, Shimamura M, Yasumo H, Fujiwara T, Horikoshi H, Furukawa H. Angptl3 regulates lipid metabolism in mice. Nat Genet. 2002;30:151–157. doi: 10.1038/ng814. [DOI] [PubMed] [Google Scholar]
- Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. Embo J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Glimcher LH. Intersection of the unfolded protein response and hepatic lipid metabolism. Cell Mol Life Sci. 2009;66:2835–2850. doi: 10.1007/s00018-009-0049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Heidtman K, Hotamisligil GS, Glimcher LH. Dual and opposing roles of the unfolded protein response regulated by IRE1{alpha} and XBP1 in proinsulin processing and insulin secretion. Proc Natl Acad Sci U S A. 2011;108:8885–8890. doi: 10.1073/pnas.1105564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Chan L. Tunicamycin induces ubiquitination and degradation of apolipoprotein B in HepG2 cells. Biochem J. 2001;353:493–501. doi: 10.1042/0264-6021:3530493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein L, Kersten S. Modulation of plasma TG lipolysis by Angiopoietin-like proteins and GPIHBP1. Biochim Biophys Acta. 2010;1801:415–420. doi: 10.1016/j.bbalip.2009.12.015. [DOI] [PubMed] [Google Scholar]
- Lipson KL, Ghosh R, Urano F. The role of IRE1alpha in the degradation of insulin mRNA in pancreatic beta-cells. PLoS One. 2008;3:e1648. doi: 10.1371/journal.pone.0001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, Frank-Kamenetsky M, Yip KN, Alvarez R, Sah DW, de Fougerolles A, Fitzgerald K, Koteliansky V, Akinc A, Langer R, Anderson DG. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, Fennell T, Banks E, Ambrogio L, Cibulskis K, Kernytsky A, Gonzalez E, Rudzicz N, Engert JC, DePristo MA, Daly MJ, Cohen JC, Hobbs HH, Altshuler D, Schonfeld G, Gabriel SB, Yue P, Kathiresan S. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Kaneto H, Kawamori D, Yoshiuchi K, Hatazaki M, Matsuoka TA, Ozawa K, Ogawa S, Hori M, Yamasaki Y, Matsuhisa M. Involvement of endoplasmic reticulum stress in insulin resistance and diabetes. J Biol Chem. 2005;280:847–851. doi: 10.1074/jbc.M411860200. [DOI] [PubMed] [Google Scholar]
- Oikawa D, Tokuda M, Hosoda A, Iwawaki T. Identification of a consensus element recognized and cleaved by IRE1 alpha. Nucleic Acids Res. 2010;38:6265–6273. doi: 10.1093/nar/gkq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oike Y, Akao M, Kubota Y, Suda T. Angiopoietin-like proteins: potential new targets for metabolic syndrome therapy. Trends Mol Med. 2005;11:473–479. doi: 10.1016/j.molmed.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Parathath S, Dogan S, Joaquin VA, Ghosh S, Guo L, Weibel GL, Rothblat GH, Harrison EH, Fisher EA. Rat carboxylesterase ES-4 enzyme functions as a major hepatic neutral cholesteryl ester hydrolase. J Biol Chem. 2011;286:39683–39692. doi: 10.1074/jbc.M111.258095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Zhou Y, Lee J, Lu A, Sun C, Chung J, Ueki K, Ozcan U. The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat Med. 2010;16:429–437. doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri P, Mirshahi F, Cheung O, Natarajan R, Maher JW, Kellum JM, Sanyal AJ. Activation and dysregulation of the unfolded protein response in nonalcoholic fatty liver disease. Gastroenterology. 2008;134:568–576. doi: 10.1053/j.gastro.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Quiroga AD, Lehner R. Role of endoplasmic reticulum neutral lipid hydrolases. Trends Endocrinol Metab. 2011;22:218–225. doi: 10.1016/j.tem.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Raabe M, Veniant MM, Sullivan MA, Zlot CH, Bjorkegren J, Nielsen LB, Wong JS, Hamilton RL, Young SG. Analysis of the role of microsomal triglyceride transfer protein in the liver of tissue-specific knockout mice. J Clin Invest. 1999;103:1287–1298. doi: 10.1172/JCI6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Romeo S, Yin W, Kozlitina J, Pennacchio LA, Boerwinkle E, Hobbs HH, Cohen JC. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest. 2009;119:70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Sha H, He Y, Yang L, Qi L. Stressed out about obesity: IRE1alpha-XBP1 in metabolic disorders. Trends Endocrinol Metab. 2011;22:374–381. doi: 10.1016/j.tem.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Shimamura M, Matsuda M, Yasumo H, Okazaki M, Fujimoto K, Kono K, Shimizugawa T, Ando Y, Koishi R, Kohama T, Sakai N, Kotani K, Komuro R, Ishida T, Hirata K, Yamashita S, Furukawa H, Shimomura I. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler Thromb Vasc Biol. 2007;27:366–372. doi: 10.1161/01.ATV.0000252827.51626.89. [DOI] [PubMed] [Google Scholar]
- Tadin-Strapps M, Peterson LB, Cumiskey AM, Rosa RL, Mendoza VH, Castro-Perez J, Puig O, Zhang L, Strapps WR, Yendluri S, Andrews L, Pickering V, Rice J, Luo L, Chen Z, Tep S, Ason B, Somers EP, Sachs AB, Bartz SR, Tian J, Chin J, Hubbard BK, Wong KK, Mitnaul LJ. siRNA-induced liver ApoB knockdown lowers serum LDL-cholesterol in a mouse model with human-like serum lipids. Journal of lipid research. 2011;52:1084–1097. doi: 10.1194/jlr.M012872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Vaishnaw AK, Gollob J, Gamba-Vitalo C, Hutabarat R, Sah D, Meyers R, de Fougerolles T, Maraganore J. A status report on RNAi therapeutics. Silence. 2010;1:14. doi: 10.1186/1758-907X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei E, Ben Ali Y, Lyon J, Wang H, Nelson R, Dolinsky VW, Dyck JR, Mitchell G, Korbutt GS, Lehner R. Loss of TGH/Ces3 in mice decreases blood lipids, improves glucose tolerance, and increases energy expenditure. Cell Metab. 2010;11:183–193. doi: 10.1016/j.cmet.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Winnay JN, Boucher J, Mori MA, Ueki K, Kahn CR. A regulatory subunit of phosphoinositide 3-kinase increases the nuclear accumulation of X-box-binding protein-1 to modulate the unfolded protein response. Nat Med. 2010;16:438–445. doi: 10.1038/nm.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadelaar S, Kleemann R, Verschuren L, de Vries-Van der Weij J, van der Hoorn J, Princen HM, Kooistra T. Mouse models for atherosclerosis and pharmaceutical modifiers. Arterioscler Thromb Vasc Biol. 2007;27:1706–1721. doi: 10.1161/ATVBAHA.107.142570. [DOI] [PubMed] [Google Scholar]
- Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G, Chang L, Xu W, Miao H, Leonardi R, Chen YE, Jackowski S, Kaufman RJ. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. EMBO J. 2011;30:1357–1375. doi: 10.1038/emboj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Lee J, Reno CM, Sun C, Park SW, Chung J, Fisher SJ, White MF, Biddinger SB, Ozcan U. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nat Med. 2011;17:356–365. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.