Abstract
Recent studies suggest that circulating LDL (low-density lipoproteins) play a central role in the pathogenesis of atherosclerosis, and the oxidized form (ox-LDL) is highly atherogenic. Deposits of ox-LDL have been found in atherosclerotic plaques, and ox-LDL has been shown to promote monocyte recruitment, foam cell formation and the transition of quiescent and contractile vascular SMCs (smooth muscle cells) to the migratory and proliferative phenotype. SMC phenotype transition and hyperplasia are the pivotal events in the pathogenesis of atherosclerosis. To comprehend the complex molecular mechanisms involved in ox-LDL-mediated SMC phenotype transition, we have compared the differential gene expression profiles of cultured quiescent human coronary artery SMCs with cells induced with ox-LDL for 3 and 21 h using Affymetrix HG-133UA cDNA microarray chips. Assignment of the regulated genes into functional groups indicated that several genes involved in metabolism, membrane transport, cell–cell interactions, signal transduction, transcription, translation, cell migration, proliferation and apoptosis were differentially expressed. Our data suggests that the interaction of ox-LDL with its cognate receptors on SMCs modulates the induction of several growth factors and cytokines, which activate a variety of intracellular signalling mechanisms (including PI3K, MAPK, Jak/STAT, sphingosine, Rho kinase pathways) that contribute to SMC transition from the quiescent and contractile phenotype to the proliferative and migratory phenotype. Our study has also identified several genes (including CDC27, cyclin A1, cyclin G2, glypican 1, MINOR, p15 and apolipoprotein) not previously implicated in ox-LDL-induced SMC phenotype transition and substantially extends the list of potential candidate genes involved in atherogenesis.
Keywords: microarray, oxidized low-density lipoprotein, quantitative PCR, transcriptome, vascular smooth muscle cell
Abbreviations: hr, human recombinant; LDL, low-density lipoproteins; MDA, malondialdehyde; n-LDL, non-oxidized LDL; ox-LDL, oxidized low-density lipoproteins; SmBM, SMC basal medium; SMCs, smooth muscle cells; TBARS, thiobarbituric acid-reactive substance
1. Introduction
Atherosclerosis and the subsequent development of occlusive vascular disease is the principal cause of coronary heart disease and cerebral stroke, the most common cause of death and morbidity in industrialized and developing nations (http://www.americanheart.org/presenter.jhtml?identifier = 4478). Thus, the understanding of the cellular and molecular mechanism of atherogenesis should provide insight into pharmacological strategies for limiting the initiation and progression of atherosclerosis prior to the development of clinical consequences. Atherosclerosis is a chronic inflammatory disease during which endothelial and SMCs (smooth muscle cells) of the arterial vessel wall are activated by proinflammatory stimuli such as IL-1 and TNFα elaborated by activated macrophages and T cells. It is characterized by complex interactions between a variety of lipids, mononuclear phagocytes and their soluble mediators in the intima and by intimal hyperplasia. Multiple local and systemic risk factors including mechanical shear stress due to haemodynamic changes, hypercholesterolaemia, hypertension and high plasma levels of inflammatory markers may initiate atherosclerosis by inducing endothelial dysfunction and vascular injury (Ross, 1999).
The ox-LDL (oxidized form of low-density lipoprotein) is a major component of cholesterol involved in hypercholesterolaemia, which is a major risk factor. ECs (endothelial cells), vascular SMCs and infiltrating immune cells have been reported to produce superoxide anion and/or hydrogen peroxide, which mediate the oxidation of a lipid component of LDL (Morel et al., 1984; Navab et al., 2004). ox-LDL has been detected in atherosclerosis plaques as well as plasma of atherosclerosis patients, and several lines of evidence have suggested that ox-LDL may play important roles in the pathogenesis and progression of atherosclerosis and the destabilization of the atherosclerotic plaque (Steinberg et al., 1989; Ross, 1999). ox-LDL can bind to scavenger receptors and the LOX-1 (lectin-like ox-LDL receptor-1), and the accumulation of excess cholesterol and cholesteryl esters by macrophages and vascular SMCs leads to the formation of foam cells that are the hallmarks of early fatty streak lesions and atheroma development (Witztum and Steinberg, 1991; Sawamura et al., 1997; Kataoka et al., 2001). ox-LDL has been shown to induce a wide range of biological effects such as SMC proliferation, monocyte chemotaxis and apoptosis/necrosis of vascular ECs and SMCs, depending on the degree of oxidation and the extracellular concentration. Particularly, minimally oxidized LDL has been shown to induce SMC proliferation and migration, which are pivotal events in intimal hyperplasia and atherogenesis. ox-LDL and its lipid constituents may also cause EC dysfunction by inducing the transcription of proatherogenic genes (Kume and Gimbrone, 1994).
ox-LDL-induced proliferation of quiescent SMCs has been associated with the ability of ox-LDL to simultaneously (i) increase the expression and nuclear localization of specific cell cycle-activating proteins (e.g. CDC2, Cdk2, cdk4, cyclin B1, D1 and PCNA1) and cell cycle-inhibiting proteins (e.g. p21 and p27), and (ii) augment intracellular signalling pathways (e.g. PI3K and PLC pathways) involved in the mitogenic response (Zettler et al., 2003). At higher concentrations, ox-LDL has also been shown to be cytotoxic, inducing apoptosis in intimal vascular SMCs and to increase plaque instability and rapture in acute coronary syndromes (Thorne et al., 1996; Okura et al., 2000). ox-LDL-induced apoptosis has been reported to involve both Fas and TNF receptors I and II signalling pathways leading to (i) down-regulation of antiapoptotic proteins of the Bcl-2 family, (ii) up-regulation of apoptotic proteins including caspase 3, and (iii) activation of MAP and Jun kinase-dependent transcription factors (e.g. STAT, NFkB, p53, ATF-2, ELK-1, CREB and AP-1), which may promote apoptosis or growth and survival (Napoli et al., 2000). ox-LDL may thus play an important role in the pathogenesis and development of atherosclerosis by its effect on vascular SMC proliferation, phenotype modulation and apoptosis (Zhao et al., 2005).
The ability of vascular SMCs in the media of arteries to undergo phenotype modulation from the quiescent and contractile state to the proliferative, migratory and synthetic state underlies their crucial role in the development and progression of vascular pathology, such as atherosclerosis and restenosis. Phenotype modulation involves a cascade of events in which different genes are turned on or off in a regulated manner. To gain insight into the early molecular events associated with ox-LDL-mediated SMC phenotype modulation, we used microarray analysis to compare the gene expression profiles of quiescent human coronary artery SMC stimulated with ox-LDL with control cells treated with n-LDL (non-oxidized LDL). Our results show that the 3 and 21 h transcriptional effects of ox-LDL on SMCs were particularly far-reaching, and a number of genes that are involved in various biological mechanisms were differentially regulated. The ox-LDL effect appeared to be mediated via the transcriptional induction of proinflammatory cytokines and growth factors, and these, in turn, initiated multiple signal transduction pathways that induced effector genes of cell proliferation, migration and extracellular matrix formation. Of particular interest is the induced expression of several nuclear receptor transcription factors. We believe that such a comprehensive analysis of the early events of SMC phenotype transition may identify novel targets for drug discovery for the intervention of the progression of atherosclerosis and the development of occlusive vascular complications.
2. Materials and methods
2.1. Oxidation of LDL
LDL (SIGMA Chemical Company) was dialysed in the dark at 4°C for 24 h against three changes of 100 volumes of PBS, pH 7.4, and then sterilized by filtration through 0.45 μm Millipore. n-LDL control and ox-LDL (100 μg/ml) were prepared as described by Steinbrecher et al. (1984) (Morel et al., 1984) by incubation with PBS or freshly prepared CuSO4 solution in PBS (at a final concentration of 5 μM) respectively for 3 and 12 h at 37°C. Then, LDL samples were extensively dialysed against PBS containing 0.1 mM EDTA and sterilized by filtration through 0.22-μm Millipore filters. The samples were stored at 4°C and used within 6 h of preparation.
Lipoprotein concentration was expressed as protein content and was determined using a BCA kit (Pierce) with albumin as standard. The extent of LDL oxidation was assessed by measuring TBARS (thiobarbituric acid-reactive substance), lipid peroxides and conjugated dienes as described by Morel et al. (1984) using MDA (malondialdehyde) as standard, and then, the values were expressed as nmol MDA equivalents per mg or μg of LDL protein. ox-LDL preparations with TBARS ≥20 nmol MDA/μg were used for SMC treatments. The extent of LDL oxidation was also monitored by the increase in electrophoretic mobility on 0.5% agarose gel in barbital buffer, pH 8.6, relative to n-LDL control.
2.2. Smooth muscle cells
Human coronary artery SMCs were purchased from Clonetics and cultured in SmBM (SMC basal medium) containing SmBM-3 growth supplements [FBS (fetal bovine serum) (5%), bovine insulin (50 ng/ml), hr (human recombinant)-EGF (epidermal growth factor) (5.0 ng/ml), hr-FGF-B (20 ng/ml) and GA-1000 (Gentamicin, Amphotericin B)] supplied by BioWhittaker Inc. The culture medium and FBS contained less than 50 pg of LPS per ml, as measured by the Limulus amoebocyte assay (BioWhittaker Inc.). SMCs were characterized by (i) their typical ‘hill and valley’ growth pattern, (ii) positive staining with anti-SM-α-actin antibody (Dako Diagnostics) and (iii) negative staining of Factor VIII-related antigen, an endothelial cell marker, using anti-factor VIII antibody (Dako Diagnostics). Cells cultures were used between passages 4 and 7 and in accordance with our institutional guidelines for research on human tissues and cells.
2.3. Isolation of total RNA
Confluent SMC cultures in 10 cm diameter Petri dishes were synchronized to quiescence by incubation for 48 h in SmBM+0.5% FBS. The cells were washed and incubated in SmBM+0.5% FBS in the absence or presence of n-LDL or ox-LDL (2 μg/ml) for 3 and 21 h. These two time points were used for the analysis of the regulation of early- and late-response genes, respectively. The reactions were set up in quadruplicates. Total RNA was extracted from the cells using TRIzol reagent (Invitrogen Life Technologies Inc.), and RNA samples from corresponding cell cultures were pooled.
2.4. Microarray analysis of differential gene expression in control n-LDL and ox-LDL-treated SMCs
Total RNA samples were treated with RNase-free DNase, and mRNA was isolated using Oligotex according to the manufacturer’s instructions (Qiagen Inc.). Biotinylated cRNA (complementary RNA) samples for chip hybridization were prepared according to protocols supplied by Affymetrix (Affymetrix) and then hybridized to HG-U133A oligonucleotide array Gene Chip (Affymetrix) following the manufacturer’s protocol. The arrays were washed, stained with streptavidin–phycoerythrin and scanned. Data files were analysed using Affymetrix GeneChip® Operating Software (GCOS) version 1.0 (Affymetrix).
2.5. Real-time PCR
Quantitative real-time PCR was performed with an ABI Prism 7900HT Sequence Analyzer using the manufacturer’s recommended protocol (PerkinElmer Applied Biosystems) to validate differential expression of selected genes. Two different primer sets were designed and synthesized for each investigated gene using Primer Express version 2.0 (PerkinElmer Applied Biosystems). Each reaction was run in triplicate in 10 μl volumes containing 4 μl of diluted first-strand cDNA template, 5 μl of SYBR Green PCR Master Mix, 0.1 μl (50 μM) of each forward and reverse primer and 0.8 μl of H2O. Samples were incubated at 95°C for 3 min to activate Taq polymerase, and 40 cycles were performed at 95°C for 10 s, at 65°C for 15 s and at 70°C for 20 s. Sequences for the primers used in this study are available upon request.
3. Results and discussion
3.1. Effect of n-LDL and ox-LDL on the proliferation of SMC
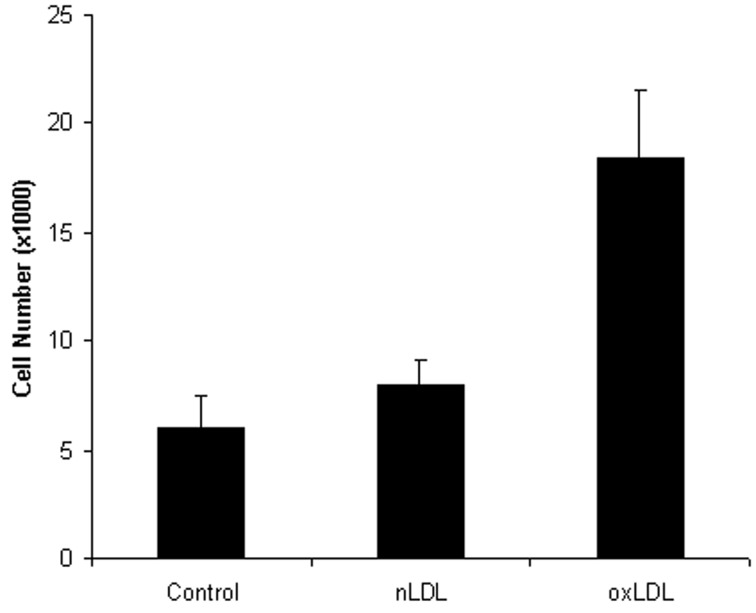
Confluent cultures of human coronary artery SMC were grown in the presence of 0.5% FBS for 48 h to induce quiescence. The cultures were then exposed to 2 μg/ml n-LDL or ox-LDL. The culture medium was replaced at day 2, and the cells were detached at day 5 to assess proliferation by cell counts. The mean and S.E.M. were determined for three separate experiments, each performed in quadruplicate. The increase in cell number in cultures containing n-LDL was only 1.3-fold greater than in cells cultured in the presence of 0.5% FBS. However, in the presence of ox-LDL, SMC proliferation was increased 3.1-fold relative to cells grown in 0.5% FBS alone (Figure 1).
Figure 1. Effect of n (normal) and ox (oxidized) LDL (2 μg/ml) on the proliferation of human coronary artery SMCs.
3.2. Microarray analysis of differential gene expression in ox-LDL-treated SMCs
The differential gene expression responses of SMCs treated with n-LDL and ox-LDL were analysed using Affymetrix oligonucleotide arrays (HG-U133A). Gene regulation in SMCs by ox-LDL was measured relative to n-LDL-treated SMCs and expressed as NC (no change), fold increase or decrease. A total of 1005 genes was found to be differentially regulated by ox-LDL at 3 h (218 genes) and 21 h (833 genes). One hundred and twenty-nine genes were induced, and 89 were suppressed at 3 h, and 311 were induced and 522 were suppressed at 21 h.
3.3. Quantitative real-time PCR validation of microarray analysis
To validate the gene array results, the expression of 24 regulated genes was analysed by quantitative real-time PCR using the expression levels of human GAPDH and beta-2-microglobulin as internal housekeeping gene controls to normalize technical variability between samples (Table 1). These genes were randomly selected from the pool of 80 genes composed of the top 20 genes that were up- or down-regulated at 3 and 21 h. The expression of 16 genes was shown to correlate well in microarray and real-time PCR, whereas the magnitudes of differential expression were somewhat different for eight genes.
Table 1. Microarray compared with real-time PCR.
| Gene symbol | Microarray | Real-time PCR | Gene title | |||
|---|---|---|---|---|---|---|
| Accession number | 3 h | 21 h | 3 h | 21 h | ||
| U12767 | NR4A3 | 37.9 | NC | 30.6 | 0.6 | Mitogen-induced nuclear orphan receptor (MINOR) |
| NM_002546 | TNFRSF11B | 15.1 | NC | 3.1 | 0.9 | Osteoprotegerin (TNFRSF11B) |
| NM_004591 | SCYA20 | 14.2 | NC | 3.8 | 0.3 | Chemokine (cc motif) ligand 20 (CCL20) |
| N32859 | NR1D2 | 8 | NC | 3.3 | 2.2 | Nuclear receptor subfamily 1, group D, member 2 |
| U66838 | CCNA1 | 7.6 | NC | 3.4 | 1.6 | Cyclin A1 (CCNA1) |
| NM_004405 | DLX2 | 5.1 | −1.6 | 4.8 | 1.0 | Distal-less homoeobox 2 (DLX2) |
| NM_004904 | CREB1 | 2.9 | −1.5 | 1.0 | 0.4 | cAMP response element-binding protein CRE-Bpa |
| NM_005544 | IRS1 | 2.8 | NC | 2.1 | 0.9 | Insulin receptor substrate 1 (IRS1) |
| NM_002607 | PDGFA | 2.5 | NC | 1.8 | 0.7 | Platelet-derived growth factor alpha (PDGFA) |
| NM_030751 | TCF8 | −3.3 | −4.3 | 0.9 | 0.7 | Transcription factor 8 (TCF8) |
| NM_004527 | MEOX1 | −4.4 | −3.2 | 0.3 | 0.4 | Mesenchyme homoeobox 1 (MEOX1), transcript variant 1 |
| NM_002224 | ITPR3 | NC | 16.8 | 1.7 | 2.3 | Inositol 1,4,5-triphosphate receptor, type 3 (ITPR3) |
| NM_005526 | HSF1 | NC | 11 | 1.6 | 1.8 | Heat shock transcription factor 1 (HSF1) |
| NM_004672 | MAP3K6 | NC | 8.4 | 4.5 | 4.1 | MAP 3 kinase 6 (MAP3K6) |
| NM_003646 | DGKZ | NC | 6.8 | 24.8 | 22.8 | Diacylglycerol kinase, zeta (DGKZ) |
| NM_005483 | CHAF1A | NC | 5.5 | 9.2 | 9.6 | Chromatin assembly factor 1, subunit A (CHAF1A) |
| M64497 | NR2F2 | NC | 3.5 | 1.3 | 1.2 | Nuclear receptor subfamily 2, group F, member 2 |
| NM_002010 | FGF9 | NC | 2.5 | 112.5 | 91.0 | Fibroblast growth factor 9 (FGF9) |
| NM_006166 | NFYB | NC | −5.5 | 1.2 | 0.9 | nuclear transcription factor Y, beta (NFYB) |
| M68891 | GATA2 | NC | −7.3 | 4.3 | 4.0 | GATA-binding protein 2 |
| NM_004622 | TSN | NC | −9 | 1.0 | 0.5 | Translin |
| NM_001356 | DDX3X | NC | −9.5 | 1.3 | 1.0 | DEAD/H box 3, X-linked (DDX3) |
| NM_003831 | RIOK3 | NC | −12.5 | 0.8 | 0.5 | SudD (suppressor of bimD6 homologue) (SUDD) |
| NM_000618 | IGF1 | NC | −12.5 | 0.4 | 0.1 | Insulin-like growth factor I (somatomedin C) |
3.4. Top 20 SMC genes differentially regulated following 3 and 21 h treatment with ox-LDL
Since it is possible that SMC genes regulated by ox-LDL may be implicated in (i) maintenance of the quiescent phenotype, or (ii) phenotype modulation to the proliferative and synthetic phenotype, we have displayed the top 20 SMC genes regulated by ox-LDL treatments at 3 and 21 h (Table 2). Two genes overexpressed at 3 h, MINOR and NR1D2, are members of nuclear hormone receptor family that function as transcriptional regulators. MINOR is known to exhibit pleiotropic physiological functions including regulation of SMC proliferation (Nomiyama et al., 2006). DLX2 is a transcription regulator that modulates neuron development in ventral embryonic forebrain, and CCL20 is involved in recruitment of activated T cells. RAI3, a G protein-coupled receptor, is implicated in many fundamental cellular processes including embryogenesis, cell growth, differentiation and apoptosis (Cheng and Lotan, 1998), and fjx1 is a protein important for growth and differentiation (Ashery-Padan et al., 1999). Growth-promoting genes such as DKK1, CCNA1, HB-EGF (Davis-Fleischer and Besner, 1998), BMP2, BST1 and CDC27 were also up-regulated at 3 h. The majority of the genes up-regulated at 3 h were early response genes, and their induced expression was transient, declining to levels similar to controls in 21 h.
Table 2A. Top 20 up-regulated genes at 3 h.
| Accession number | Gene title | 3 h | 21 h | Function |
|---|---|---|---|---|
| U12767 | Mitogen-induced nuclear orphan receptor (MINOR) | 37.9 | NC | Regulation of transcription, DNA-dependent |
| NM_002546 | Osteoprotegerin (TNFRSF11B) | 15.1 | NC | Regulates bone resorption |
| NM_004591 | Chemokine (cc motif) ligand 20 (CCL20) | 14.2 | NC | Recruitment of activated T cells |
| NM_012242 | dickkopf (Xenopus laevis) homologue 1 (DKK1) | 13.8 | 3.8 | Growth factor |
| NM_003979 | Retinoic acid-induced 3 (RAI3) | 9.8 | NC | Metabotropic glutamate, GABA-B-like receptor |
| NM_017856 | Hypothetical protein FLJ20514 | 9.3 | NC | |
| N32859 | Nuclear receptor subfamily 1, group D, member 2 (NR1D2) | 8 | NC | Regulation of transcription, DNA-dependent |
| U66838 | Cyclin A1 (CCNA1) | 7.6 | NC | Regulates cell cycle CDK2 and CDC2 |
| AB007938 | KIAA0469 | 7.5 | 6.7 | |
| NM_014344 | Putative secreted ligand homologous to fjx1 | 7 | 7.8 | |
| NM_001945 | Heparin-binding EGF-like growth factor | 7 | −1.8 | Binds EGFR; positive regulation of cell proliferation |
| NM_001200 | Bone morphogenetic protein 2 (BMP2) | 6.7 | NC | Skeletal development |
| NM_004817 | Tight junction protein 2 (zona occludens 2) (TJP2) | 6.7 | NC | Links junctional membrane proteins to actin |
| BC000737 | Regulator of G-protein signalling 4 | 6.6 | NC | Regulates G-protein-coupled receptor signalling |
| NM_004334 | Bone marrow stromal cell antigen 1 (BST1) | 5.4 | NC | Facilitates pre-B-cell growth |
| J00146 | Dihydrofolate reductase pseudogene (psi-hd1) | 5.4 | NC | Converts dihydrofolate into tetrahydrofolate |
| AA166684 | Cell division cycle 27 (CDC27) | 5.4 | 8.2 | Mitotic metaphase/anaphase transition |
| NM_004405 | Distal-less homoeobox 2 (DLX2) | 5.1 | −1.6 | Regulation of transcription, DNA-dependent |
| NM_018039 | Hypothetical gene FLJ10251 | 4.8 | NC | |
| BC001051 | ADP-ribosylation factor-like 7 | 4.7 | 4.6 | Small GTPase-mediated signalling |
Table 2B. Top 20 down-regulated genes at 3 h.
| Accession number | Gene title | 3 h | 21 h | Function |
|---|---|---|---|---|
| AF263928 | Papillomavirus regulatory factor PRF-1 (LOC55893) | −19.7 | NC | |
| NM_017869 | BTG3-associated nuclear protein (BANP) | −9.8 | NC | |
| NM_019058 | Hypothetical protein (FLJ20500) | −9.8 | NC | |
| AK023365 | Liprin-alpha4 | −8.6 | NC | Regulation of cell–matrix interactions |
| BC000069 | Retinoic acid receptor responder 2 | −7.4 | NC | Retinoid metabolism |
| R72286 | Microfibrillar-associated protein 4 | −6.4 | NC | Cell adhesion |
| NM_001873 | Carboxypeptidase E | −6 | −10.8 | Protein catabolism |
| NM_000759 | Colony-stimulating factor 3 (granulocyte) (CSF3) | −5.8 | NC | Positive regulation of cell proliferation |
| NM_031220 | PYK2 N-terminal domain-interacting receptor 1 (NIR1) | −5.6 | NC | Receptor PTK; phosphoinositide transporter |
| M12529 | Apolipoprotein E (APOE) | −5.5 | −6.2 | Lipid metabolism and transport |
| NM_017606 | Hypothetical protein DKFZp434K1210 | −5.3 | NC | |
| NM_000312 | Protein C (PROC) | −5 | NC | Anticoagulant |
| AF056209 | PAM COOH-terminal interactor protein 1 (PCIP1) | −4.9 | NC | Neuropeptide signalling pathway |
| NM_004659 | Matrix metalloproteinase 23A (MMP23A) | −4.7 | NC | Peptidase |
| AK023792 | Hypothetical protein FLJ13074 | −4.5 | −3.1 | |
| NM_004527 | Mesenchyme homoeobox 1 (MEOX1), transcript variant 1 | −4.4 | −3.2 | Homoeobox; transcription factor activity |
| NM_005518 | 3-OH-3-methylglutaryl-CoA synthase 2 (HMGCS2) | −4.3 | NC | Cholesterol biosynthesis |
| NM_030776 | Z-DNA-binding protein 1 (ZBP1) | −4.3 | NC | Binds left-handed Z-DNA |
| AL044326 | Phosphoribosylformylglycinamidine synthase | −4.2 | NC | Purine nucleotide biosynthesis |
| AA621558 | Methionine-tRNA synthetase | −4 | NC | Protein biosynthesis |
| NM_003745 | Suppressor of cytokine signalling-1 (SOCS-1) | −4 | NC | JAK-STAT cascade inhibitor |
| NM_001077 | UDP glycosyltransferase 2, polypeptide B17 (UGT2B17) | −4 | NC | Transferase activity |
Table 2C. Top 20 up-regulated genes at 21 h.
| Accession number | Gene title | 3 h | 21 h | Function |
|---|---|---|---|---|
| D13287 | Glucosidase, beta; acid (GBA) | NC | 27.9 | Sphingoglycolipid metabolism |
| AC005943 | Chromosome 19, cosmid R30538 | NC | 24.3 | |
| BE885926 | KIAA0706 | NC | 17.1 | |
| NM_002224 | Inositol 1,4,5-triphosphate receptor, type 3 (ITPR3) | NC | 16.8 | IP3-sensitive calcium-release channel activity |
| NM_022772 | Hypothetical gene FLJ21935 | NC | 15.4 | |
| NM_024874 | Hypothetical protein FLJ14225 | NC | 14.7 | |
| NM_017585 | Solute carrier family 2 member 6 (SLC2A6) | NC | 11.9 | Facilitates glucose transport |
| NM_014786 | KIAA0337 | NC | 11.7 | |
| U93181 | Nuclear dual-specificity phosphatase (SBF1) | NC | 11.4 | Protein dephosphorylation |
| NM_005526 | Heat shock transcription factor 1 (HSF1) | NC | 11 | Regulation of transcription, DNA-dependent |
| D64109 | Tob family; transducer of ERBB2 | NC | 10.5 | Negative regulation of cell proliferation |
| AF026030 | Mitochondrial inner membrane protein import receptor (hTIM44) | NC | 10.1 | Import of mitochondrial proteins into mitochondria |
| BE305165 | Phospholipase C, beta 3, neighbour pseudogene | NC | 9.5 | |
| NM_002081 | Glypican 1 (GPC1) | NC | 9.4 | Important in endostatin-mediated inhibition of angiogenesis |
| NM_002861 | Phosphate cytidylyltransferase 2, ethanolamine (PCT2E) | NC | 8.8 | Converts ethanolamine into CDP-ethanolamine |
| NM_001492 | Growth differentiation factor 1 (GDF1) | NC | 8.5 | Growth factor |
| NM_004672 | MAP 3 kinase 6 (MAP3K6) | NC | 8.4 | Protein kinase activity |
| NM_002555 | Solute carrier family 22 member 1-like (SLC22A1L) | NC | 8.4 | Organic cation transporter |
| N30649 | Truncated calcium-binding protein (LOC51149) | 2.7 | 8.4 | |
| AA166684 | Cell division cycle 27 (CDC27) | 5.4 | 8.2 | Mitotic metaphase/anaphase transition |
Table 2D. Top 20 down-regulated genes at 21 h.
| Accession number | Gene title | 3 h | 21 h | Function |
|---|---|---|---|---|
| BG257762 | Hypothetical protein | NC | −15.6 | |
| NM_005570 | Lectin, mannose-binding, 1 (LMAN1) | NC | −15.4 | Transport of mannose glycans from ER to Golgi |
| L19161 | Eukaryotic translation initiation factor 2, subunit 3 (EIF2S3) | NC | −13.9 | Translational elongation |
| AK021846 | Sec23-interacting protein p125 | NC | −13.6 | Golgi organization and biogenesis |
| NM_003831 | SudD (suppressor of bimD6 homologue) (SUDD) | NC | −12.5 | Chromosome segregation |
| NM_000618 | Insulin-like growth factor I (somatomedin C) | NC | −12.5 | RAS signal transduction; regulation of proliferation |
| NM_004779 | Transcription complex, subunit 8 (TRC8) | NC | −12.4 | Regulation of transcription, DNA-dependent |
| NM_001873 | Carboxypeptidase E (CPE) | −6 | −10.8 | Protein catabolism |
| AI652662 | Branched-chain aminotransferase 1, cytosolic (BCAT1) | NC | −10.3 | Branched-chain family amino acid biosynthesis |
| NM_001356 | DEAD/H box 3, X-linked (DDX3) | NC | −9.5 | ATP-dependent RNA helicase |
| NM_004622 | Translin | NC | −9 | A recombination hotspot binding protein |
| AF130055 | Translocating chain-associating membrane protein | NC | −8.7 | Protein targeting; co-translational membrane targeting |
| AJ276395 | Migration stimulation factor FN70 (MSF70) | NC | −8.4 | Cell motility |
| NM_003246 | Thrombospondin 1 | NC | −8.2 | Angiogenic activity |
| AF043337 | Interleukin 8 C-terminal variant (IL8) | −1.8 | −8.1 | Cell motility; intracellular signalling cascade |
| BG166705 | Small inducible cytokine subfamily B (CXC), member 5 (SCYB5/CXCL5) | NC | −8 | Chemotaxis; positive regulation of cell proliferation |
| AF021233 | TRAIL-R4-B TNFR superfamily, member 10d | NC | −8 | Decoy with truncated death domain; apoptosis |
| NM_018243 | Hypothetical protein FLJ10849 | NC | −7.9 | |
| AF191653 | Nucleoside diphosphate-linked moiety X-type motif 4 (NUDT4) | NC | −7.9 | Cyclic nucleotide metabolism |
| BE256969 | PAF acetylhydrolase, isoform Ib, alpha subunit (PAFAH1B1) | NC | −7.8 | Lipid metabolism |
The genes overexpressed at 21 h can be grouped into those involved in metabolism (GBA, PCT2E, SBF1), solute transport (SLC2A6, hTIM44, SLC22A1L), regulation of cell proliferation and differentiation (CDC27, GDF1), transcription regulation (HSF1), inhibition of angiogenesis (GPC1) and signalling (ITPR3, MAP3K6). Several novel genes were also up-regulated at 21 h (KIAA0706, FLJ21935, FLJ14225, KIAA0337).
The transcript with the greatest fold decrease in expression at 3 h was the PBF-1 (papillomavirus regulatory/binding factor), which is a nuclear shuttling transcription factor that mediates inhibition of cell growth (Sichtig et al., 2007). The homoeobox transcription factor, MEOX1, that plays a role in the commitment of mesodermal cells in the developing somite to the skeletal muscle lineage, was also down-regulated (Petropoulos et al., 2004). Genes involved in cell–matrix interactions (liprin-alpha4) and biosynthesis of cholesterol (HMGCS2) were down-regulated. Several metabolic enzymes such as CPE, BCAT1, NUDT4 and PAFAH1B1 were also down-regulated by ox-LDL at 21 h. The genes highly repressed at 21 h included a membrane transporter (LMAN1), a transcriptional regulator (TRC8), a translational elongator (EIF2S3), and positive regulators of proliferation (somatomedin C) and cell motility (MSF70, IL8, CXCL5).
3.5. Functional characterization of the ox-LDL-regulated genes
In order to study the effect of ox-LDL treatment on SMC phenotype modulation further, ox-LDL-regulated genes were clustered into functional groups/subgroups using gene annotation information from the Affymetrix database. The functional groups that we believe to be important for SMC phenotype modulation are shown (Table 3). The complete Tables of the functional categories of the regulated SMC genes are available in the online publication (Supplementary Material at http://www.cellbiolintrep.org/cbr/017/cbr0170033add.htm). For most functional groups, it is difficult to speculate on the overall effect of ox-LDL treatment, since a number of genes with various functional effects were modulated at the same time. However, genes categorized under apoptosis (overall inhibition) and cell proliferation (overall induction) predominantly support the proliferative SMC phenotype induced by ox-LDL treatment. Also, the regulation of many cytokines/chemokines (CCL20, CCL7, CSF3, IL6, IL12B, TGFB2, IL11 and CXCL5) and growth factors [PDGFA, GDF1, GDF11, FGF9 and VEGF (vascular endothelial growth factor)] probably also contributes to the induction of proliferation in SMC. Our data show that (i) the majority of the genes were regulated at 21 h compared with 3 h, (ii) very little overlap exists between genes that are differentially regulated at 3 and 21 h, indicating that the early response (3 h) is distinct from the late response (21 h) and subsides by 21 h, and (iii) modulation of genes that are involved in apoptosis, cell proliferation and cytokines/growth factors could support the induction of proliferation by ox-LDL. The data suggest that the induction of the early response cytokine and growth factor genes may be involved in the induction of the late-response apoptosis, proliferation and structural and ECM (extracellular matrix) genes that are characteristic of the transition of the quiescent SMC to the proliferative and synthetic phenotype.
Table 3. Functional categories.
Numbers in bold indicate increases/decreases greater than 2-fold
| Accession number | Gene title | 3 h | 21 h | Function |
|---|---|---|---|---|
| Apoptosis | ||||
| AF083421 | Immediate early response 3 (IER3) | −2.1 | NC | Apoptosis inhibitor activity |
| NM_005178 | B-cell CLL/lymphoma 3 (BCL3) | −2.4 | NC | Cell cycle regulation |
| AF069073 | p8 protein homologue (COM1) | NC | 2.9 | Induction of apoptosis |
| NM_002342 | Lymphotoxin beta receptor (LTBR) (TNFRSF3) | NC | 2.8 | TNFR-related protein |
| NM_005380 | Neuroblastoma, suppression of tumourigenicity 1 (NBL1) | NC | 2.2 | Negative regulation of cell cycle |
| NM_022121 | P53-induced protein (PIGPC1) | NC | −2 | |
| Z70519 | FASApo 1 protein (TNFRSF6) | NC | −2.2 | Induction of apoptosis |
| NM_014452 | death receptor 6 | NC | −2.4 | Induction of apoptosis |
| NM_002583 | PRKC, apoptosis WT1 regulator (PAWR) | NC | −2.6 | Negative regulation of proliferation |
| NM_003842 | TNFR superfamily, member 10b (TNFRSF10B) | NC | −3.4 | Induction of apoptosis |
| NM_013437 | Potential tumour suppressor (ST7) | NC | −3.6 | Tumour suppressor |
| NM_021960 | Myeloid cell leukaemia sequence 1 (BCL2-related) | NC | −6 | Apoptotic program |
| Cell adhesion and cell–cell signalling | ||||
| NM_001200 | Bone morphogenetic protein 2 (BMP2) | 6.7 | NC | Skeletal development |
| NM_013372 | Cysteine knot superfamily 1 (CKTSF1B1) | 2.8 | NC | Block BMP signalling |
| AF154054 | DRM; cysteine knot superfamily 1 | 2.4 | NC | Antagonist of bone morphogenetic protein |
| NM_016157 | Trophinin (TRO) | −3.7 | NC | Embryo implantation |
| NM_016223 | PKC and casein kinase substrate in neurons 3 (PACSIN3) | NC | 7.3 | Kinesin complex; focal adhesion |
| NM_004952 | Ephrin-A3 | NC | 7 | Cell–cell signalling |
| AI692180 | Liprin beta 2 | NC | 6.3 | Cell adhesion |
| NM_002587 | Protocadherin 1 (cadherin-like 1) (PCDH1) | NC | 3.9 | Calcium-dependent cell–cell adhesion |
| NM_002204 | Integrin, alpha 3; CD49C (ITGA3) | NC | 2.2 | Cell matrix adhesion |
| NM_002087 | Granulin (GRN) | NC | 2.1 | Cell–cell signalling; signal transduction |
| BC004542 | Plexin B2 | NC | 2.1 | Cell adhesion molecule |
| NM_013231 | Fibronectin leucine-rich transmembrane protein 2 | NC | 2 | Cell adhesion |
| NM_001792 | Cadherin 2, type 1 (CDH2) | NC | −2.1 | Cell adhesion |
| NM_001078 | Vascular cell adhesion molecule 1 (VCAM1) | NC | −2.1 | Adhesion of monocytes and lymphocytes |
| NM_000885 | Alpha 4 subunit of VLA-4 receptor CD49D (ITGA4) | NC | −2.6 | Cell–matrix adhesion; integrin-mediated signalling pathway |
| AF152501 | Protocadherin beta 8 (PCDHB8) | NC | −2.6 | Homophilic cell adhesion; cell adhesion |
| NM_005506 | CD36 | NC | −2.8 | Cell adhesion |
| AF263279 | Sialomucin CD164 | NC | −3.3 | Regulation of haematopoiesis; cell adhesion |
| AU135154 | A disintegrin and metalloproteinase domain 10 | NC | −3.4 | Cell–cell signalling |
| Cell motility and cytoskeleton | ||||
| NM_004817 | Tight junction protein 2 (zona occludens 2) (TJP2) | 6.7 | NC | Links junctional membrane proteins to actin |
| NM_012134 | Leiomodin 1 (LMOD1) | 3.1 | NC | Tropomyosin binding |
| AF043337 | interleukin 8 C-terminal variant (IL8) | −1.8 | −8.1 | Cell motility; intracellular signalling cascade |
| D49372 | Eotaxin | −2.2 | −2.5 | Chemokine; signal transduction |
| U88321 | Beta chemokine Exodus-3 | −3 | −4.1 | Chemotaxis; cell communication |
| NM_004999 | Myosin VI | NC | 6.1 | Myosin ATPase activity; motor activity |
| NM_006709 | HLA-B-associated transcript 8 (BATS8) | NC | 5 | Histone-lysine N-methyltransferase activity |
| M13452 | Lamin A | NC | 4.8 | Interacts with intermediate filaments |
| BG475299 | ems1 (cortactin) p8085 src substrate | NC | 3.1 | Actin-binding protein |
| NM_020987 | Ankyrin 3 (ANK3) | NC | 2.7 | Cytoskeletal anchoring |
| NM_004395 | Drebrin 1 (DBN1) | NC | 2.4 | Actin binding |
| NM_006848 | Hepatitis delta antigen-interacting protein A (DIPA) | NC | 2.4 | Kinesin complex |
| M86406 | Skeletal muscle alpha 2 actinin (ACTN2) | NC | 2.4 | Anchor myofibrillar actin filaments |
| NM_002373 | Microtubule-associated protein | NC | 2.1 | Modulate the assembly of microtubules |
| NM_002480 | Myosin phosphatase, target subunit 1 (MYPT1) | NC | −2.1 | Regulation of muscle contraction |
| NM_000366 | Tropomyosin 1 (alpha) (TPM1) | NC | −2.5 | Regulation of muscle contraction |
| AI214061 | Tropomyosin 4 | NC | −2.9 | Constituent of muscle |
| NM_005722 | Actin-related protein 2, yeast homologue (ACTR2) | NC | −3 | Cell motility |
| BE675337 | Gelsolin | NC | −6.4 | Actin filament polymerization |
| BC001352 | Tubulin, beta polypeptide | NC | −6.5 | Microtubule-based movement |
| AJ276395 | Migration stimulation factor FN70 | NC | −8.4 | Cell motility |
| Cell proliferation | ||||
| U66838 | Cyclin A1 (CCNA1) | 7.6 | NC | Regulates cell cycle CDK2 and CDC2 |
| AA166684 | Cell division cycle 27 (CDC27) | 5.4 | 8.2 | Mitotic metaphase/anaphase transition |
| NM_021120 | Discs, large homologue 3 (DLG3) | 4.6 | NC | Negative regulation of cell proliferation |
| AI770084 | Dihydropyrimidinase-like 2 | 3.8 | NC | Regulates axonal growth and branching |
| AW189518 | Piwi (Drosophila)-like 1 | 3.2 | NC | Oogenesis; spermatogenesis |
| D84212 | Serine/threonine kinase 6 (STK6) | 2.9 | NC | Cell growth |
| NM_001423 | Epithelial membrane protein 1 (EMP1) | 2.3 | NC | Cell proliferation; epidermal differentiation |
| AF188298 | Disabled 2 p93 (DAB2) | 2.2 | NC | Cell proliferation; tumour suppressor |
| BF514079 | Gut-enriched Kruppel-like factor (Gklf) | 2.1 | NC | Inhibition of DNA synthesis |
| NM_002510 | Glycoprotein transmembrane nmb (GPNMB) | 2 | 4.9 | Negative regulation of cell proliferation |
| NM_002048 | Growth arrest-specific 1 (GAS1) | −3.3 | NC | Cell cycle arrest |
| D64109 | Tob family; transducer of ERBB2 | NC | 10.5 | Negative regulation of cell proliferation |
| NM_003308 | Testis specific protein, Y-linked (TSPY) | NC | 5.5 | Spermatogenesis |
| NM_016195 | M-phase phosphoprotein 1 (MPHOSPH1) | NC | 3.4 | Microtubule disassembly at G2- to M-phase |
| NM_000820 | Growth arrest-specific 6 (GAS6), mRNA. | NC | 2.7 | Negative regulation of cell proliferation |
| M73554 | Bcl-1; cyclin D1 (PRAD1) | NC | 2.5 | G1/S transition of mitosis |
| L13720 | Growth-arrest-specific protein (gas) | NC | 2.5 | Negative regulation of cell proliferation |
| NM_021873 | Cell division cycle 25B (CDC25B) | NC | 2.3 | Positive regulation of cell cycle |
| BC000076 | Cyclin D1 (PRAD1) | NC | 2.3 | Activates cdc2 (p34) |
| NM_004864 | Prostate differentiation factor | NC | 2.2 | |
| AK023348 | Clone 24720 epithelin 1 and 2; granulin | NC | 2 | Growth modulatory activity |
| NM_015392 | Neural proliferation differentiation and control 1 (NPDC1) | NC | 2 | |
| L49506 | Cyclin G2 | NC | −2 | Regulates specific cell cycle CDKs |
| AV700514 | Ceroid-lipofuscinosis, neuronal 5 | NC | −2.4 | Cell growth and/or maintenance |
| NM_012325 | Microtubule-associated protein, RPEB family, member 1 | NC | −2.5 | Regulation of cell cycle |
| NM_004404 | Neural precursor cell expressed, developmentally down-regulated 5 | NC | −2.8 | Cell cycle; cytokinesis |
| NM_006431 | Chaperonin-containing TCP1, subunit 2 (beta) | NC | −3.2 | Cyclin E maturation |
| M27281 | Vascular endothelial growth factor (VEGF) | NC | −3.9 | Mitogen that specifically acts on endothelial cells |
| NM_078487 | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) (CDKN2B) | NC | −4.1 | Inhibits CDK4 and induces G1-phase cell cycle arrest |
| Extracellular matrix | ||||
| NM_004659 | Matrix metalloproteinase 23A (MMP23A) | −4.7 | NC | Peptidase |
| R72286 | Microfibrillar-associated protein 4 | −6.4 | NC | Cell adhesion |
| AK023365 | Liprin-alpha4 | −8.6 | NC | Regulation of cell–matrix interactions |
| U48734 | Non-muscle alpha-actinin | NC | 2.3 | Attachment of microfilament bundles to adherens-type junctions |
| NM_002421 | Matrix metalloproteinase 1 (MMP1) | NC | 2.1 | Collagen I, II and III catabolism |
| NM_000362 | Tissue inhibitor of metalloproteinase-3 | NC | −2 | Metalloendopeptidase inhibitor |
| BE350145 | Collagen, type VI, alpha 1 | NC | −2.6 | Component of microfibrillar structures |
| NM_000138 | Fibrillin 1 | NC | −3 | Component of extracellular microfibrils |
| U77706 | Laminin alpha 4 chain (LAMA4) | NC | −4 | Non-collagenous constituent of basement membranes |
| AV721177 | Phosphatidylinositol-binding clathrin assembly protein | NC | −4.5 | Protein complex assembly; vesicle-mediated transport |
| NM_003246 | Thrombospondin 1 | NC | −8.2 | Angiogenic activity |
| Receptors and membrane proteins | ||||
| NM_004334 | Bone marrow stromal cell antigen 1 (BST1) | 5.4 | NC | Facilitates pre-B-cell growth |
| U01157 | Glucagon-like peptide-1 receptor | 3.4 | NC | Stimulator of glucose-induced insulin secretion |
| M90657 | Transmembrane 4 superfamily member 1 (TM4SF1) | 2.8 | 2.1 | Protein complex assembly; tumour metastasis |
| AF043498 | Prostate stem cell antigen (PSCA) | 2.7 | NC | Prostrate cancer progression |
| NM_031220 | PYK2 N-terminal domain-interacting receptor 1 (NIR1) | −5.6 | NC | Receptor PTK; phosphoinositide transporter |
| BC000069 | Retinoic acid receptor responder 2 | −7.4 | NC | Retinoid metabolism |
| NM_002081 | Glypican 1 (GPC1) | NC | 9.4 | Important in endostatin mediated inhibition of angiogenesis |
| AF020314 | Leucocyte membrane antigen (CMRF-35H) | NC | 4.7 | May play a regulatory role in leukocyte function |
| U72069 | Karyopherin (importin) beta 2 | NC | 3.7 | Targets cytoplasmic proteins to the nucleus |
| NM_000319 | Peroxisome receptor 1 (PXR1) | NC | 3.4 | Protein-peroxisome targeting |
| AK022910 | Nuclear transport receptor; transportin-SR | NC | 2.8 | Nucleocytoplasmic transport |
| NM_004616 | Transmembrane 4 superfamily member 3 (TM4SF3) | NC | 2.5 | Protein complex assembly |
| AI859060 | Cholinergic receptor, epsilon polypeptide | NC | 2.1 | Synaptic transmission |
| NM_003801 | GPI anchor attachment protein 1 (GPAA1) | NC | 2.1 | Links proteins to cell membrane |
| NM_014045 | Low-density lipoprotein receptor-related protein 10 (LRP10) | NC | 2 | Lipoprotein metabolism |
| BC000389 | Transmembrane 4 superfamily member 7 (TM4SF7) | NC | 2 | Protein complex assembly |
| NM_003999 | Oncostatin M receptor (OSMR) | NC | −2.4 | IL6 cell surface receptor linked signal transduction |
| NM_003144 | Signal sequence receptor, alpha (SSR1) | NC | −2.7 | Co-translational membrane targeting |
| U50748 | Leptin receptor short form (db) | NC | −2.8 | Gene transcription via activation of STAT |
| U52914 | Leptin receptor | NC | −5.3 | Gene transcription via activation of STAT |
| NM_002888 | Retinoic acid receptor responder 1 (RARRES1) | NC | −5.3 | Negative regulation of cell proliferation |
| Signal transduction | ||||
| NM_003979 | Retinoic acid induced 3 (RAI3) | 9.8 | NC | Metabotropic glutamate, GABA-B-like receptor |
| BC000737 | Regulator of G-protein signalling 4 | 6.6 | NC | Regulates G-protein-coupled receptor signalling |
| BC001051 | ADP-ribosylation factor-like 7 | 4.7 | 4.6 | Small GTPase-mediated signalling |
| AF091395 | Triple functional domain (PTPRF interacting) | 3.2 | NC | Receptor protein tyrosine phosphatase signalling |
| AY00716 | EH domain-containing 1 (EHD1) | 3 | NC | Endocytosis of IGF1 receptors |
| M16591 | Haemopoietic cell kinase (HCK) | 2.9 | NC | Protein tyrosine kinase activity |
| BE737620 | Myosin phosphatase, target subunit 1 | 2.9 | NC | Regulates phosphatidylinositol signalling system |
| BE466525 | Ecotropic viral integration site 1(EVI1) | 2.8 | NC | JUN kinase binding; protein kinase inhibitor |
| NM_005544 | Insulin receptor substrate 1 (IRS1) | 2.8 | NC | Stimulates mitogenesis |
| NM_006482 | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 | 2.7 | NC | Protein phosphorylation |
| AF238083 | Sphingosine kinase 1 (SPHK1) | 2.7 | NC | Sphingosine metabolism |
| AB026436 | Dual-specificity phosphatase 10 | 2.3 | NC | Inactivate MAPKs |
| AA780381 | MAP2 kinase 3 (ERK kinase 3) | 2.3 | NC | Protein phosphorylation |
| NM_003749 | Insulin receptor substrate-2 (IRS2) | 2.2 | NC | Stimulates mitogenesis |
| AB023137 | A kinase (PRKA) anchor protein 2 (AKAP2) | 2.2 | 1.8 | Activates adenylate cyclase |
| AF015043 | SH3-domain-binding protein 4 (SH3BP4) | 2.1 | NC | Signal transducer activity |
| NM_005104 | Bromodomain-containing 2 (BRD2) | 2 | NC | Protein serine/threonine kinase activity |
| AB003476 | Gravin; A kinase (PRKA) anchor protein 12 | 2 | NC | G-protein-coupled receptor protein signalling |
| U77914 | Jagged 1 (JAG1) | 2 | NC | G-protein-coupled receptor protein signalling, ligand in the Notch signalling pathway |
| NM_002648 | Protein kinase-related oncogene (PIM1) | −2.9 | NC | Haematopoietic development |
| L37882 | Frizzled homology 2 (FZD2) | −2.9 | 2.2 | Signal transduction; intracellular Ca release |
| NM_005204 | MAP3 kinase 8 (MAP3K8) | −3 | NC | Ser/thr kinase; regulation of cellular transformation |
| NM_004842 | A kinase (PRKA) anchor protein 7 (AKAP7) | −3.9 | NC | Protein kinase A anchor protein activity |
| NM_003745 | Suppressor of cytokine signalling-1 (SOCS-1) | −4 | NC | JAK-STAT cascade inhibitor |
| NM_002224 | Inositol 1,4,5-triphosphate receptor, type 3 (ITPR3) | NC | 16.8 | IP3-sensitive calcium-release channel activity |
| U93181 | Nuclear dual-specificity phosphatase (SBF1) | NC | 11.4 | Protein dephosphorylation |
| NM_004672 | MAP 3 kinase 6 (MAP3K6) | NC | 8.4 | Protein kinase activity |
| NM_003646 | Diacylglycerol kinase, zeta (DGKZ) | NC | 6.8 | Phosphatidylinositol signalling |
| NM_030981 | Small GTP-binding protein (RAB1B) | NC | 4.3 | Ras; small monomeric GTPase activity |
| AF272035 | Rag C protein (GTR2) | NC | 3.4 | Small monomeric GTPase activity |
| U96922 | Inositol polyphosphate-4-phosphatase, type II alpha | NC | 3.3 | Regulates phosphatidylinositol signalling system |
| BF062886 | Vaccinia-related kinase 3 (VRK3) | NC | 3.3 | Protein phosphorylation |
| AV700224 | Casein kinase 1, delta | NC | 3.1 | Protein phosphorylation |
| NM_017572 | G-protein-coupled receptor kinase 7 (GPRK7) | NC | 2.7 | Protein phosphorylation |
| BE138888 | GTP-binding protein Rac2 | NC | 2.6 | Small GTPase-mediated signal transduction |
| AF022212 | Rho GTPase-activating protein 6 isoform 2 | NC | 2.6 | Actin filament polymerization |
| NM_002547 | Oligophrenin 1 (OPHN1) | NC | 2.5 | Rho GTPase-activating protein; cell migration |
| M55268 | Casein kinase 2, alpha prime polypeptide (CSNK2A2) | NC | 2.3 | Protein phosphorylation |
| NM_016602 | CC chemokine receptor 10 (CCR10) | NC | 2.3 | G-protein-coupled receptor protein signalling |
| NM_006182 | Discoidin domain receptor family, member 2 (DDR2) | NC | 2.3 | Receptor protein tyrosine kinase |
| NM_002712 | Protein phosphatase 1, regulatory subunit 7 (PPP1R7) | NC | 2.2 | Protein dephosphorylation |
| AB009358 | MAP2 kinase 7; JNK-activating kinase 2 | NC | −2 | Specific activator of JNK1 and JNK2 |
| NM_002356 | Myristoylated alanine-rich protein kinase C substrate (MARCKS) | NC | −2 | A filamentous actin cross-linking protein |
| NM_012250 | Oncogene TC21 (TC21) | NC | −2 | Small GTPase-mediated signal transduction |
| NM_000291 | Phosphoglycerate kinase 1 (PGK1) | NC | −2 | Phosphoglycerate kinase activity |
| NM_005607 | PTK2 protein tyrosine kinase 2 (PTK2) | NC | −2 | Protein kinase activity |
| NM_003022 | SH3 domain-binding glutamic acid-rich protein like (SH3BGRL) | NC | −2 | SH3/SH2 adaptor protein activity |
| NM_022650 | GTPase-activating protein (GAP) | NC | −2.1 | Bind activated Rho GTPases and stimulate GTP hydrolysis |
| NM_005261 | GTP-binding protein overexpressed in skeletal muscle (GEM) | NC | −2.1 | Small GTPase-mediated signal transduction |
| NM_004578 | Ras-associated protein (RAB4) | NC | −2.1 | Rho small monomeric GTPase activity |
| AI571798 | Rho GDP dissociation inhibitor (GDI) alpha | NC | −2.1 | Negative regulation of cell adhesion |
| NM_006241 | Protein phosphatase 1, regulatory (inhibitor) subunit 2 (PPP1R2) | NC | −2.2 | Ser-/thr-specific protein phosphatase inhibitor |
| AL136139 | Enhancer of filamentation 1 (HEF1) | NC | −2.3 | Integrin-initiated cytoskeleton-linked signalling |
| NM_016322 | GTPase Rab14 | NC | −2.3 | Ras; small monomeric GTPase activity |
| BC005122 | ADP-ribosylation factor GTPase activating protein 1 | NC | −2.4 | Regulation of signalling, growth by hydrolysis of GTP |
| AF218074 | MAP3 kinase 7 | NC | −2.4 | Phosphorylates MKK6 to stimulate JNK; NFKB translocation |
| AF001362 | Jak2 kinase (JAK2) | NC | −2.5 | Protein tyrosine kinase activity; JAK-STAT cascade |
| NM_002731 | Protein kinase, cAMP-dependent, catalytic, beta (PRKACB) | NC | −2.6 | Protein kinase activity |
| NM_002716 | Protein phosphatase 2 regulatory subunit A beta isoform | NC | −2.6 | Regulation of proliferation, contraction, transcription |
| AF002280 | Alpha-actinin-2 associated LIM protein alternatively spliced | NC | −2.7 | Interacts with alpha-actinin-2 in cytoskeletal assembly |
| NM_003507 | Frizzled (Drosophila) homologue 7 (FZD7) | NC | −2.7 | Fz7-mediated signalling controls cell sorting in mesoderm |
| NM_004161 | RAB1, member RAS oncogene family | NC | −2.7 | Small GTPase-mediated signalling; vesicle-mediated transport |
| J03005 | G-protein alpha-inhibiting activity polypeptide 3 (GNAI3) | NC | −2.8 | G-protein-coupled receptor protein signalling pathway |
| NM_003463 | Protein tyrosine phosphatase type IVA, member 1 | NC | −2.9 | Protein dephosphorylation; oncogenesis |
| NM_006575 | MAP 4 kinase 5 | NC | −3 | Activates JNK but not ERK1 |
| M18468 | Protein kinase, cAMP-dependent, regulatory, type I, alpha | NC | −3.1 | cAMP-dependent protein kinase |
| NM_001506 | G-protein-coupled receptor 32 (GPR32) | NC | −3.2 | G-protein-coupled receptor protein signalling |
| NM_005242 | Coagulation factor II (thrombin) receptor-like 1 (F2RL1) | NC | −3.3 | G-protein-coupled receptor protein signalling pathway |
| AF092132 | p21 (CDKN1A)-activated kinase 2 (PAK2) | NC | −3.5 | Negative regulation of protein kinase activity |
| NM_000945 | Calcineurin B, type I (CNB1) | NC | −3.6 | Ca-dependent ser/thr phosphatase activity; calcium binding |
| NM_004162 | RAB5A, member RAS oncogene family | NC | −3.6 | Bind GTP and exhibits GTPase activity; regulation of endocytosis |
| X75208 | HEK2 protein tyrosine kinase receptor | NC | −3.7 | Receptor tyrosine kinase signalling |
| NM_006654 | FGF receptor substrate 2 (FRS2) | NC | −3.8 | FGF signalling; cell growth and differentiation |
| NM_003688 | Calcium Calmodulin-dependent serine protein kinase (CASK) | NC | −4.2 | Cytoskeletal membrane scaffold; cortical cytoskeleton signalling |
| AF127481 | Dual-specificity phosphatase 1 (DUSP1) | NC | −4.4 | Dephosphorylate and inactivates p44MAPK (ERK1) |
| AW665024 | Protein tyrosine kinase 9 | NC | −4.7 | Protein phosphorylation |
| NM_016277 | RAB23, member RAS oncogene family | NC | −4.9 | Small GTPase-mediated signalling; intracellular protein transport |
| S69182 | Protein tyrosine phosphatase (PTPG1); non-receptor type 12 | NC | −5.5 | Protein dephosphorylation |
| NM_001346 | Diacylglycerol kinase, gamma (DGKG) | NC | −5.6 | PKC activation |
| Z25435 | Protein-ser/thr kinase gene | NC | −5.8 | Protein phosphorylation |
| AF051311 | Ras-GTPase activating protein SH3 domain-binding protein 2 | NC | −6.1 | RAS protein signal transduction |
| NM_002184 | gp130, oncostatin M receptor | NC | −7 | Cell surface receptor-linked signal transduction |
| NM_002869 | RAB6, member RAS oncogene family | NC | −7.4 | Small GTPase-mediated signalling; non-selective vesicle transport |
| AF021233 | TRAIL-R4-B TNFR superfamily, member 10d | NC | −8 | Decoy with truncated death domain; apoptosis |
| Transcription and translation | ||||
| U12767 | Mitogen-induced nuclear orphan receptor (MINOR) | 37.9 | NC | Regulation of transcription, DNA-dependent |
| N32859 | Nuclear receptor subfamily 1, group D, member 2 | 8 | NC | Regulation of transcription, DNA-dependent |
| NM_004405 | Distal-less homoeobox 2 (DLX2) | 5.1 | −1.6 | Regulation of transcription, DNA-dependent |
| S77154 | Beta-type transcription factor homologue human | 3.2 | NC | Regulation of transcription, DNA-dependent |
| NM_004904 | cAMP response element-binding protein CRE-Bpa | 2.9 | −1.5 | Transcription factor activated by translocation of PKC |
| NM_006981 | Nuclear receptor subfamily 4, group A, member 3 (NR4A3) | 2.8 | NC | Regulation of transcription, DNA-dependent |
| NM_004472 | Forkhead box D1 (FOXD1) | 2.6 | NC | Regulation of transcription, DNA-dependent |
| AL021977 | V-maf musculoaponeurotic fibrosarcoma oncogene homologue F | 2.2 | 2.2 | Regulation of transcription, DNA-dependent |
| NM_003201 | Transcription factor 6-like 1 (TCF6L1) | 2.1 | NC | Mitochondrial DNA transcriptional activator |
| NM_022898 | B-cell lymphoma/leukaemia 11B (BCL11B) | 2 | NC | Regulation of transcription, zinc finger protein |
| M83667 | CCAAT-enhancer-binding protein (CEBP), delta (CEBPD) | −2.1 | NC | Regulation of transcription, DNA-dependent |
| L07648 | MAX-interacting protein 1 (MXI1) | −2.1 | NC | Transcription factor |
| NM_005384 | Nuclear factor, interleukin 3 regulated (NFIL3) | −2.2 | NC | Transcriptional co-repressor |
| BE542323 | TONDU | −2.2 | NC | Regulation of transcription, DNA-dependent |
| NM_014112 | Trichorhinophalangeal syndrome I gene (TRPS1) | −2.2 | NC | Zn finger transcription factor |
| NM_003670 | Basic helix-loop-helix domain containing, class B, 2 | −2.4 | −3.9 | Regulation of transcription, DNA-dependent |
| BG250310 | Zinc finger protein 36, C3H type-like 1 (ZFP36L1) | −2.9 | NC | Transcription factor |
| AF055993 | Sin3-associated polypeptide (SAP30) | −3.2 | NC | Transcription co-repressor activity |
| NM_020529 | NFk light polypeptide gene enhancer in B-cells inhibitor, alpha | −3.3 | NC | Cytoplasmic sequestering of NF-kappaB |
| NM_030751 | Transcription factor 8 (TCF8) | −3.3 | −4.3 | Represses interleukin 2 expression |
| NM_004527 | Mesenchyme homoeobox 1 (MEOX1), transcript variant 1 | −4.4 | −3.2 | Homoeobox; transcription factor activity |
| NM_005526 | Heat shock transcription factor 1 (HSF1) | NC | 11 | Regulation of transcription, DNA-dependent |
| AB015332 | Neighbour of A-kinase-anchoring protein 95 | NC | 6.1 | DEAD/H-box RNA helicase binding |
| NM_005483 | Chromatin assembly factor 1, subunit A (CHAF1A) | NC | 5.5 | Assembles histone octamers onto replicating DNA |
| NM_003597 | TGFB-inducible early growth response 2 | NC | 5.5 | Transcriptional regulator |
| NM_004083 | DNA-damage-inducible transcript 3 | NC | 4.3 | A dominant-negative inhibitor of C/EBP and LAP |
| AL161985 | Transcription factor binding to IGHM enhancer 3 | NC | 4.2 | Regulation of transcription, DNA-dependent |
| AF106934 | Thyroid hormone receptor-associated protein (TRAP95) | NC | 3.7 | Transcription co-activator |
| M64497 | Nuclear receptor subfamily 2, group F, member 2 | NC | 3.5 | Transcription co-repressor activity |
| AB019219 | Similar to yeast pre-mRNA splicing factors, Prp1Zer1 and Prp6 | NC | 3.5 | Spliceosome assembly |
| U67734 | HIV-1 Tat interactive protein | NC | 3.2 | Transcriptional co-activator |
| NM_005859 | Purine-rich element-binding protein A (PURA) | NC | 3.2 | Regulation of transcription, binds to GAGA boxes |
| X72631 | Rev-ErbA alpha | NC | 3 | Regulation of transcription, DNA-dependent |
| U19769 | Centromere protein F (mitosin) (CENPF) | NC | 2.8 | Regulation of mitosis |
| NM_014140 | HepA-related protein (HARP) | NC | 2.8 | ATP-dependent helicase |
| AC004908 | Ribosomal protein L23a | NC | 2.8 | Protein biosynthesis |
| NM_006943 | SRY (sex-determining region Y)-box 22 (SOX22) | NC | 2.8 | Regulation of transcription from Pol II promoter |
| NM_020310 | MAX-binding protein (MNT) | NC | 2.7 | Regulation of transcription; negative regulation of cell proliferation |
| BC002704 | Signal transducer and activator of transcription 1 (STAT1) | NC | 2.7 | Transcription from Pol II promoter |
| AF295773 | Ral guanine nucleotide dissociation stimulator (RALGDS) | NC | 2.6 | RAS protein signal transduction |
| AF055078 | Zinc finger protein 42 (ZNF42) | NC | 2.6 | Regulation of transcription, DNA-dependent |
| U79283 | Albumin D-box-binding protein | NC | 2.5 | Regulation of transcription |
| AI884867 | Ribosomal protein L26 | NC | 2.4 | Protein biosynthesis |
| NM_006736 | Hsp, neuronal DNAJ-like 1 (HSJ1) subfamily B, member 2 | NC | 2.2 | Co-chaperone activity |
| NM_004176 | Sterol regulatory element binding transcription factor 1 (SREBF1) | NC | 2.2 | Regulation of transcription; LDL metabolism regulation |
| NM_014292 | Chromobox homologue 6 | NC | 2.1 | Regulation of transcription; chromatin modification |
| AW517464 | Ribosomal protein L3 | NC | 2.1 | Protein biosynthesis |
| NM_003925 | Methyl-CpG binding endonuclease (MED1) | NC | −2 | DNA repair |
| NM_000176.1 | Nuclear receptor subfamily 3, group C, member 1 (NR3C1) | NC | −2 | Regulation of transcription, DNA-dependent |
| AF098483 | PC4- and SFRS1-interacting protein 2 (PSIP2) | NC | −2 | Transcriptional co-activators |
| AK026426.1 | SWI-/SNF-related, matrix associated, subfamily a, member 1 | NC | −2 | Chromatin modelling |
| NM_004500 | Heterogeneous nuclear ribonucleoprotein C (C1C2) | NC | −2.1 | mRNA Splicing |
| NM_014319 | Integral inner nuclear membrane protein (MAN1) | NC | −2.1 | Nuclear membrane localization |
| NM_005324 | H3 histone, family 3B (H3.3B) | NC | −2.2 | Nucleosome assembly |
| D13889 | Inhibitor of DNA-binding 1 (ID1) | NC | −2.2 | Regulation of transcription from Pol II promoter |
| AK021418 | Putative RNA helicase | NC | −2.2 | rRNA processing |
| BC000451 | Splicing factor, arginine-/serine-rich 10 | NC | −2.2 | mRNA splicing |
| AL136621 | Zinc finger protein 198 | NC | −2.2 | Protein binding |
| NM_004379 | cAMP-responsive element-binding protein 1 | NC | −2.3 | Regulation of transcription, DNA-dependent |
| M62829 | Early growth response 1 (EGR1) | NC | −2.3 | Regulation of transcription |
| AA679988 | Polypyrimidine tract-binding protein 1(PTPB1) | NC | −2.3 | mRNA splicing |
| BC000627 | Signal transducer and activator of transcription 3 (STAT 3) | NC | −2.3 | JAK-STAT cascade; transcription factor activity |
| NM_012266 | DnaJ (Hsp40) homologue, subfamily B, member 5 | NC | −2.4 | Response to stress |
| BC000806 | Polymerase (RNA) II (DNA directed) polypeptide K (POLR2K) | NC | −2.4 | Transcription from Pol III promoter |
| NM_003016 | Splicing factor, arginine-/serine-rich 2 | NC | −2.4 | mRNA splicing |
| BC000997 | Splicing factor, arginine-/serine-rich 7 (SFRS7) | NC | −2.5 | mRNA processing |
| AF309553 | Meiotic recombination protein REC14 | NC | −2.6 | Meiotic recombination |
| NM_006902 | Paired mesoderm homoeobox 1a (PMX1a) | NC | −2.6 | Regulation of transcription, DNA-dependent |
| AL117487 | Transcriptional adaptor 3-like (ADA3) | NC | −2.6 | Transcriptional activity |
| M94630 | Heterogeneous nuclear ribonucleoprotein D (HNRPD) | NC | −2.9 | RNA processing |
| D13891 | Inhibitor of DNA-binding 2 (ID2) | NC | −2.9 | Transcriptional repressor |
| AL553320 | Stress-induced phosphoprotein 1 (STIP1) | NC | −2.9 | Association of molecular chaperones HSP70 and HSP90 |
| L23959 | Transcription factor Dp-1 (TFDP1) | NC | −2.9 | Regulation of transcription from Pol II promoter |
| NM_006265 | RAD21 (S. pombe) homologue | NC | −3 | Chromosome segregation |
| NM_006924 | Splicing factor, arginine-/serine-rich 1 (SFRS1) | NC | −3 | mRNA splice site selection |
| NM_004779 | CCR4-NOT transcription complex, subunit 8 (CNOT8) | NC | −3.1 | Regulation of transcription, DNA-dependent |
| AF039942 | HCF-binding transcription factor Zhangfei (ZF) | NC | −3.2 | Regulation of transcription, DNA-dependent |
| AB009023 | RNA guanylyltransferase and 5-phosphatase (RNGTT) | NC | −3.2 | mRNA capping |
| NM_003017 | Splicing factor, arginine-/serine-rich 3 (SFRS3) | NC | −3.3 | mRNA splicing |
| M97935 | Transcription factor ISGF-3 | NC | −3.3 | IRF; transcription factor activity |
| BC001255 | Nuclear cap-binding protein subunit 2 (NCBP2) | NC | −3.5 | snRNA-nucleus export |
| U12170 | Zinc finger homoeodomain protein; transcription factor 8 | NC | −3.7 | Represses interleukin 2 expression |
| AF061261 | Muscleblind-like 2 (Drosophila) (MBNL2) | NC | −4.2 | Transcription factor activity |
| AF072814 | PHD finger DNA-binding protein isoform 1 (M96) | NC | −4.3 | regulation of transcription, DNA-dependent |
| BF983406 | heterogeneous nuclear ribonucleoprotein H1 | NC | −4.4 | RNA processing |
| NM_021038 | Muscleblind (Drosophila)-like | NC | −4.8 | Nucleic acid-binding activity |
| AI217362 | Trinucleotide repeat containing 11 (THR-associated protein) | NC | −5 | Regulation of transcription, DNA-dependent |
| NM_006166 | Nuclear transcription factor Y, beta (NFYB) | NC | −5.5 | Regulation of transcription, DNA-dependent |
| U71300 | snRNA activating protein complex subunit (SNAP50) | NC | −5.5 | snRNA transcription |
| NM_007034 | DnaJ (Hsp40) homologue, subfamily B, member 4 | NC | −6.3 | Heat shock protein activity |
| M68891 | GATA-binding protein 2 | NC | −7.3 | Regulation of transcription, DNA-dependent |
| NM_004622 | Translin | NC | −9 | A recombination hotspot-binding protein |
| NM_001356 | DEAD/H box 3, X-linked (DDX3) | NC | −9.5 | ATP-dependent RNA helicase |
| NM_004779 | Transcription complex, subunit 8 | NC | −12.4 | Regulation of transcription, DNA-dependent |
| NM_003831 | SudD (suppressor of bimD6 homologue) (SUDD) | NC | −12.5 | Chromosome segregation |
| Cytokine and growth factor | ||||
| NM_002546 | Osteoprotegerin (TNFRSF11B) | 15.1 | NC | Regulates bone resorption |
| NM_004591 | Chemokine (cc motif) ligand 20 (CCL20) | 14.2 | NC | Recruitment of activated T cells |
| NM_012242 | dickkopf (Xenopus laevis) homologue 1 (DKK1) | 13.8 | 3.8 | Growth factor |
| NM_001945 | Heparin-binding EGF-like growth factor | 7 | −1.8 | Binds EGFR; positive regulation of cell proliferation |
| NM_000417 | Interleukin 2 receptor, alpha (IL2RA) | 3.8 | NC | T-cell proliferation |
| NM_002506 | Nerve growth factor, beta (NGFB) | 2.8 | 1.6 | Survival of nerve cells |
| NM_002607 | Platelet-derived growth factor alpha (PDGFA) | 2.5 | NC | Cell proliferation |
| NM_013246 | Cardiotrophin-like cytokine (CLC) | 2 | NC | IL-6 family of cytokines |
| AF229253 | FGF2-interacting factor (API5) | −1.5 | −2.9 | Apoptosis inhibitor |
| S69738 | Monocyte chemotactic protein human (MCP-1) | −1.8 | −2 | Recruitment of monocytes |
| AF125392 | Insulin-induced protein 2 | −2 | −2.3 | |
| NM_006273 | Chemokine (C–C motif) ligand 7 (CCL7) | −2 | −2.6 | Monocytes/macrophages recruitment |
| X16323 | Hepatocyte growth factor (HGF) | −2.3 | −4.1 | Cell proliferation |
| M59465 | TNF alpha-induced protein 3 (TNFAIP3) | −2.7 | −1.9 | Inhibits NFk-B and TNF-mediated apoptosis |
| M57731 | Gro-beta; GRO2 oncogene | −3.1 | NC | G-protein-coupled receptor protein signalling pathway |
| NM_000759 | Colony-stimulating factor 3 (granulocyte) (CSF3) | −5.8 | NC | Positive regulation of cell proliferation |
| NM_001492 | Growth differentiation factor 1 (GDF1) | NC | 8.5 | Growth factor |
| AF028333 | Growth differentiation factor-11 (GDF11) | NC | 4.8 | Neurogenesis; skeletal development |
| NM_021805 | Single Ig IL-1R-related molecule (SIGIRR) | NC | 4.7 | Subtype of the IL-1R superfamily |
| NM_002010 | Fibroblast growth factor 9 (FGF9) | NC | 2.5 | Cell proliferation |
| NM_000599 | Insulin-like growth factor-binding protein 5 (IGFBP5) | NC | 2.1 | IGF binding; regulation of cell growth |
| NM_000600 | Interleukin 6 (IL6) | NC | −2 | Acute-phase response; cell proliferation |
| AF214570 | Vascular endothelial growth factor (VEGF) | NC | −2.4 | Positive regulation of cell proliferation; angiogenesis |
| NM_000598 | Insulin-like growth factor binding protein 3 | NC | −2.5 | Regulation of cell growth |
| U81380 | Interleukin-13 receptor soluble form | NC | −2.6 | IL-13 regulation |
| NM_002187 | Interleukin 12B (IL12B) | NC | −2.7 | Positive regulation of activated T-cell proliferation |
| BC001281 | TNF receptor superfamily, member 10b | NC | −2.8 | Induction of apoptosis via death domain receptors |
| D78132 | Ras homologue enriched in brain 2 (RHEB2) | NC | −2.9 | Ras-related growth factor |
| U19495 | Intercrine-alpha stromal cell-derived factor 1 | NC | −3.1 | Regulation of actin polymerization; cell–cell signalling |
| AW770896 | Insulin-like growth factor-binding protein 7 | NC | −3.7 | Negative regulation of cell proliferation |
| M19154 | Transforming growth factor, beta 2 (TGFB2) | NC | −4 | Regulation of proliferation |
| NM_005711 | EGF-like repeats and discoidin I-like domains 3 | NC | −5.3 | Integrin binding; cell adhesion |
| NM_000641 | Interleukin 11 (IL11) | NC | −6.4 | Positive regulation of cell proliferation |
| BG166705 | Small inducible cytokine subfamily B (CXC), member 5 (SCYB5) | NC | −8 | Chemotaxis; positive regulation of cell proliferation |
| NM_000618 | Insulin-like growth factor I (somatomedin C) | NC | −12.5 | RAS signal transduction; regulation of proliferation |
The profile of the regulated genes observed in this study showed several similarities to the recent report by Deng et al. (2006) on differentially expressed genes in human coronary artery SMCs treated with 40 μg/ml ox-LDL for 24 h. Among the top 50 up- and down-regulated genes, GPC1, DGKZ, DDR2, HMOX1 and FOXD1 were up-regulated in both studies, and thrombospondin 1 and VCAM1 were down-regulated. Whereas COL6A1, PTGF1, CD36, GPR32 and DCN were shown to be down-regulated in our study, these genes were reported to be up-regulated by Deng et al. We believe that the observed differences may be due to the differences in the time of exposure of the cells and the concentrations of ox-LDL used.
Recently Reeve et al. (2007) have reported that treatment of human coronary artery SMCs with ox-LDL induced a gene regulation profile comparable with the gene expression pattern in the aorta of apoE−/− mice. An analysis of expression of antioxidant genes in this study indicated that ox-LDL induced an oxidative stress response in coronary artery SMCs with increased expression of Hsp70, HSF-1, MnSOD, HO-1 (haem oxygenase-1) and ferritin that induced coronary artery SMC death in a caspase-independent manner. In agreement with Reeve et al, our data also document that (i) ox-LDL induced expression of HSF-1, chromobox homologue 6, NQO1 [NAD(P)H dehydrogenase quinone 1], truncated calcium-binding protein and dual-specificity tyrosine-phosphorylation-regulated kinase 2 and (ii) down-regulation of VEGFA (vascular endothelial growth factor A) precursor and API5. This agreement further supports the idea that the effect of ox-LDL on SMC bears relevance to the development of atherosclerosis.
In our study, several transcription and chromatin-remodelling genes were found to be differentially regulated, which have not been reported previously. Thirty-seven genes encoding transcription and chromatin-remodelling factors were induced more than 2-fold (11 at 3 h and 26 at 21 h), whereas 47 genes were down-regulated (12 at 3 h and 35 at 21 h). In particular, NR4A3, NR1D2, NR2F2, Tes1, CREB1 and FOXD1, which were identified as transient immediate early TF genes, were highly up-regulated (3- to 38-fold) within 3 h, whereas HSF1, CHAF1A, TIEG2, GADD153, TRAP95, NR2F2, PURA, SMARCAL1, MNT and SREBF1 were induced 3- to 11-fold at 21 h. Thus, our data are consistent with the notion that the expression of genes that mediate SMC phenotype modulation is regulated by a variety of transcription factors. Furthermore, the rapid, abundant and transient induction of CREB1 and several genes belonging to the nuclear hormone receptor superfamily transcription factors (including NR4A3, NR1D2, NR2F2 and TRAP59) suggest that these transcription factors may play a fundamental role in the induction of genes required for ox-LDL-induced activation and proliferation of quiescent SMC. We believe that our study contributes further to (i) the understanding of the molecular mechanism of ox-LDL-mediated vascular SMC phenotype modulation, and (ii) the identification of several potential biomarker genes for targeted disruption and overexpression for the strategic development of treatment for various vascular diseases.
Online data
Footnotes
This work was supported by a grant from the University of Toronto, Faculty of Medicine Dean’s Research Fund.
Author contribution
Joe Minta designed and directed the experiments, discussed the results and wrote the manuscript. James Jungwon Yun performed the quantitative real-time PCR experiments and the classification of regulated genes into functional groups. Rosanne St-Bernard took part in cell growth assays, isolation of total RNA from oxidized and non-oxidized-LDL-treated cells and the initial assignment of biological functions to genes regulated in the microarray data.
References
- Ashery-Padan R, Alvarez-Bolado G, Klamt B, Gessler M, Gruss P. Fjx1, the murine homologue of the Drosophila four-jointed gene, codes for a putative secreted protein expressed in restricted domains of the developing and adult brain. Mech Dev. 1999;80:213–7. doi: 10.1016/s0925-4773(98)00218-4. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Lotan R. Molecular cloning and characterization of a novel retinoic acid-inducible gene that encodes a putative G protein-coupled receptor. J Biol Chem. 1998;273:35008–15. doi: 10.1074/jbc.273.52.35008. [DOI] [PubMed] [Google Scholar]
- Davis-Fleischer KM, Besner GE. Structure and function of heparin-binding EGF-like growth factor (HB-EGF). Front Biosci. 1998;3:d288–99. doi: 10.2741/a241. [DOI] [PubMed] [Google Scholar]
- Deng DX, Spin JM, Tsalenko A, Vailaya A, Ben-Dor A, Yakhini Z. Molecular signatures determining coronary artery and saphenous vein smooth muscle cell phenotypes: distinct responses to stimuli. Arterioscler Thromb Vasc Biol. 2006;26:1058–65. doi: 10.1161/01.ATV.0000208185.16371.97. [DOI] [PubMed] [Google Scholar]
- Kataoka H, Kume N, Miyamoto S, Minami M, Morimoto M, Hayashida K. Oxidized LDL modulates Bax/Bcl-2 through the lectinlike Ox-LDL receptor-1 in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2001;21:955–60. doi: 10.1161/01.atv.21.6.955. [DOI] [PubMed] [Google Scholar]
- Kume N, Gimbrone MA Lysophosphatidylcholine transcriptionally induces growth factor gene expression in cultured human endothelial cells. J Clin Invest. 1994;93:907–11. doi: 10.1172/JCI117047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel DW, DiCorleto PE, Chisolm GM. Endothelial and smooth muscle cells alter low density lipoprotein in vitro by free radical oxidation. Arteriosclerosis. 1984;4:357–64. doi: 10.1161/01.atv.4.4.357. [DOI] [PubMed] [Google Scholar]
- Napoli C, Quehenberger O, De Nigris F, Abete P, Glass CK, Palinski W. Mildly oxidized low density lipoprotein activates multiple apoptotic signaling pathways in human coronary cells. FASEB J. 2000;14:1996–2007. doi: 10.1096/fj.99-0986com. [DOI] [PubMed] [Google Scholar]
- Navab M, Ananthramaiah GM, Reddy ST, Van Lenten BJ, Ansell BJ, Fonarow GC. The oxidation hypothesis of atherogenesis: the role of oxidized phospholipids and HDL. J Lipid Res. 2004;45:993–1007. doi: 10.1194/jlr.R400001-JLR200. [DOI] [PubMed] [Google Scholar]
- Nomiyama T, Nakamachi T, Gizard F, Heywood EB, Jones KL, Ohkura N. The NR4A orphan nuclear receptor NOR1 is induced by platelet-derived growth factor and mediates vascular smooth muscle cell proliferation. J Biol Chem. 2006;281:33467–76. doi: 10.1074/jbc.M603436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura Y, Brink M, Itabe H, Scheidegger KJ, Kalangos A, Delafontaine P. Oxidized low-density lipoprotein is associated with apoptosis of vascular smooth muscle cells in human atherosclerotic plaques. Circulation. 2000;102:2680–6. doi: 10.1161/01.cir.102.22.2680. [DOI] [PubMed] [Google Scholar]
- Petropoulos H, Gianakopoulos PJ, Ridgeway AG, Skerjanc IS. Disruption of Meox or Gli activity ablates skeletal myogenesis in P19 cells. J Biol Chem. 2004;279:23874–81. doi: 10.1074/jbc.M312612200. [DOI] [PubMed] [Google Scholar]
- Reeve JL, Stenson-Cox C, O’Doherty A, Porn-Ares I, Ares M, O’Brien T. OxLDL-induced gene expression patterns in CASMC are mimicked in apoE−/− mice aortas. Biochem Biophys Res Commun. 2007;356:681–6. doi: 10.1016/j.bbrc.2007.03.027. [DOI] [PubMed] [Google Scholar]
- Ross R. Atherosclerosis is an inflammatory disease. Am Heart J. 1999;138:S419–20. doi: 10.1016/s0002-8703(99)70266-8. [DOI] [PubMed] [Google Scholar]
- Sawamura T, Kume N, Aoyama T, Moriwaki H, Hoshikawa H, Aiba Y. An endothelial receptor for oxidized low-density lipoprotein. Nature. 1997;386:73–7. doi: 10.1038/386073a0. [DOI] [PubMed] [Google Scholar]
- Sichtig N, Silling S, Steger G. Papillomavirus binding factor (PBF)-mediated inhibition of cell growth is regulated by 14-3-3beta. Arch Biochem Biophys. 2007;464:90–9. doi: 10.1016/j.abb.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Steinberg D, Parthasarathy S, Carew TE, Khoo JC, Witztum JL. Beyond cholesterol. Modifications of low-density lipoprotein that increase its atherogenicity. N Engl J Med. 1989;320:915–24. doi: 10.1056/NEJM198904063201407. [DOI] [PubMed] [Google Scholar]
- Steinbrecher UP, Parthasarathy S, Leake DS, Witztum JL, Steinberg D. Modification of low density lipoprotein by endothelial cells involves lipid peroxidation and degradation of low density lipoprotein phospholipids. Proc Natl Acad Sci U S A. 1984;81:3883–7. doi: 10.1073/pnas.81.12.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne SA, Abbot SE, Winyard PG, Blake DR, Mills PG. Extent of oxidative modification of low density lipoprotein determines the degree of cytotoxicity to human coronary artery cells. Heart. 1996;75:11–6. doi: 10.1136/hrt.75.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witztum JL, Steinberg D. Role of oxidized low density lipoprotein in atherogenesis. J Clin Invest. 1991;88:1785–92. doi: 10.1172/JCI115499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettler ME, Prociuk MA, Austria JA, Massaeli H, Zhong G, Pierce GN. OxLDL stimulates cell proliferation through a general induction of cell cycle proteins. Am J Physiol Heart Circ Physiol. 2003;284:H644–53. doi: 10.1152/ajpheart.00494.2001. [DOI] [PubMed] [Google Scholar]
- Zhao GF, Seng JJ, Zhang H, She MP. Effects of oxidized low density lipoprotein on the growth of human artery smooth muscle cells. Chin Med J (Engl) 2005;118:1973–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.