Abstract
hMSCs (human mesenchymal stem cells) express two isoforms of DNA topo II (topoisomerase II). Although both isoforms have the same catalytic activity, they are specialized for different functions in the cell: while topo IIα is essential for chromosome segregation in mitotic cells, topo IIβ is involved in more specific cellular functions. A number of inhibitors are available that inhibit the catalytic activity of both topo II isoforms. However, in order to investigate the isoform-specific inhibition of these two enzymes, it is necessary to use other techniques such as siRNA (small interfering RNA) interference to selectively silence either one of the isoforms individually. Depending on the lipid charge densities and protein varieties of the cell membrane, previous studies have demonstrated that transfection efficiencies of siRNAs to hMSCs are very low. In the study reported here, we demonstrate the use of Lipofectamine RNAiMAX as an efficient transfection reagent to introduce siRNAs into human mesenchymal stem cells with significantly great efficiency to silence topo IIβ selectively. A high level of transfection efficiency (80%) was achieved by using unlabelled topo IIβ-specific siRNA oligos. Specifically, it was confirmed repeatedly that green labelled siRNAs interfere with the transfection of siRNAs. The reagent induced minimal cytotoxicity (3.5–4.5%), and cell viability of the transfected hMSCs decreased 20–30% compared with untreated cells, depending on the concentration of the reagent.
Keywords: DNA topoisomerase IIβ, human mesenchymal stem cell, RNAiMAX, siRNA transfection
Abbreviations: DMEM, Dulbecco's modified Eagle's medium; GFP, green fluorescent protein; HEK, human embryonic kidney; hMSC, human mesenchymal stem cell; LDH, lactate dehydrogenase; MSC, mesenchymal stem cell; MSC-FBS, MSC-qualified fetal bovine serum; PE, phycoerythrin; RNAi, RNA interference; siRNA, small interfering RNA; topo II, topoisomerase II
1. Introduction
In mammalian cells, two isoforms of topoisomerase II have been identified; a 170-kDa topo IIα (topoisomerase IIα) and the 180-kDa topo IIβ (topoisomerase IIβ). These isoforms are encoded by separate genes, but they are similar in primary structure (72%), and they have similar catalytic properties in vitro (Sakaguchi and Kikuchi, 2004). However, they are regulated very differently. Since they differ in the cell cycle dependency and tissue specificity, it can be assumed that they play separate roles in cellular physiology. Topo IIα is mainly involved in mitotic processes and is only present in proliferating tissues (Drake et al., 1989; Austin and Marsh, 1998). However, topo IIβ is present in all tissues including terminally differentiated ones, and its level is not cell cycle dependent. This indicates that it may play a role in DNA metabolism, especially in the transcriptional activation of some inducible genes, rather than being involved in DNA replication and chromosome condensation/segregation. This possibility is supported by the finding that topo IIβ has been found to be involved in gene induction during neural differentiation (Tsutsui et al., 2001; Sano et al., 2008).
Topoisomerase inhibitors are designed to interfere with the action of topoisomerases. Commonly used topo II inhibitors are 2,6-dioxopiperazines such as ICRF-159, ICRF-187 and ICRF-193 and epipodophyllotoxins such as VP-16 and VM-26. However, these inhibitors are not selective, and they inhibit the enzymatic activity of both topo IIβ and topo IIα. In order to investigate the cellular roles of either topo IIβ or topo IIα isoforms individually, topo IIβ-specific inhibition could be provided by RNA interference with the use of topo IIβ-specific siRNAs (small interfering RNAs).
RNAi (RNA interference) is a form of PTGS (posttranscriptional gene silencing) in most eukaryotic cells, mediated by siRNAs. siRNAs are short double-stranded DNA sequences, which trigger degradation of mRNAs in a sequence-specific manner so that the homologous genes are silenced (Hannon, 2002; Kawasaki et al., 2004). Because of its high efficiency and specificity, RNAi has become an important research tool for analysing gene functions in eukaryotes via the introduction of siRNAs.
Lipid-based transfections are carried out by using commercially available cationic lipids. These cationic liposomes are also known as lipoplexes, and they are capable of delivering both siRNA and siRNA-encoding plasmids through the cell membrane (Sioud and Sorensen, 2003; Spagnou et al., 2004).
MSCs (mesenchymal stem cells) are one of the most promising adult stem cell types due to their availability and relatively simple requirements for in vitro expansion and genetic manipulation. They may be isolated from a number of tissues such as blood, placenta, amniotic fluid, heart, skeletal muscle, adipose tissue, synovial tissue and pancreas; in addition, under appropriate conditions, they can differentiate into several lineages including bone, cartilage, fat, muscle, connective tissue and tendon (Javazon et al., 2004; Kubo et al., 2009). It is difficult to transfect many primary cells, especially stem cells, to achieve gene knockdown by conventional methods. This could be because of a structural variation in lipid molecules of cell membrane that is able to mediate transfection (Dalby et al., 2004). Many groups have developed techniques to deliver plasmid DNA or siRNA into hMSCs with varying efficiencies. Other groups have managed to transfer genes into hMSCs by viral or electroporation techniques. Although cell viability was reduced (16.5%) with Nucleofector technology in hMSCs, an efficiency of 45% was achieved, which is a high transient transfection efficiency compared with other non-viral methods (Lee et al., 2001; Peister et al., 2004; Zhang et al., 2004). Another research group managed non-viral nucleic acid (DNA and siRNA) delivery in hMSCs by using Lipofectamine 2000 with a 50% efficiency (Hoelters et al., 2005). This was the first demonstration of efficient non-viral, liposomal delivery of both nucleic acids (DNA and siRNA) into hMSCs.
Lipofectamine RNAiMAX Transfection Reagent is a cationic lipid-based reagent, which is specially produced for the delivery of siRNA. Compared with other reagents, Lipofectamine RNAiMAX requires lower concentrations of siRNAs, has minimal cytotoxicity and gives maximal cell viability. The use of this reagent in human embryonic stem cells has yielded a 90% transfection efficiency (Zhao et al., 2008). Lipofectamine RNAiMAX is also used in hard-to-transfect MSCs for the delivery of siRNAs (De Becker et al., 2007).
This study has shown for the first time that Lipofectamine RNAiMAX is a suitable transfection reagent, thereby justifying it as the correct preference for selectively silencing topo IIβ in hMSCs.
2. Materials and methods
2.1. Isolation and expansion of hMSCs
This work was performed under the Ethics Committee of Karadeniz Technical University (KTU), process number 2007/26. Human bone marrow aspirates were obtained from KTU, Faculty of Medicine, Department of Hematology. In brief, mononuclear cells were collected from bone marrow aspirates by Ficoll (Biochrom) density gradient centrifugation, and collected cells were suspended in DMEM (Dulbecco's modified Eagle's medium)-LG (Gibco) culture media containing 20% MSC-FBS (qualified fetal bovine serum, Gibco) and 0.2% primocin (InVivoGen). These cells were named as (P0)' (passage 0) and incubated at 37°C in a humidified atmosphere containing 5% CO2. After 72 h, non-adherent cells were removed by culture medium refreshment. Cells were detached using 0.25% trypsin–EDTA solution (Gibco) 10–14 days after isolation and replated and termed as ‘P1’ following this first subculture. Cell expansion was carried out in complete medium containing DMEM-LG and 15% MSC-FBS. After three subcultures, cells selected by plastic adherence were screened by flow cytometry for CD73, CD105, CD45, CD34 and HLA-DR cell surface markers.
2.2. Flow cytometry
To confirm that hMSCs maintain their phenotypic characteristics after growth in culture, undifferentiated hMSCs were subjected to flow cytometry analysis. Fifteen surface markers of hBM (human bone marrow)-MSCs at passage 3 were assayed. After each passage, stem cells were harvested and suspended in their culture medium at a concentration of 1×106 cells/ml. After a brief centrifugation, cells were resuspended in wash buffer (BD Biosciences), and 100 μl of cell suspension was incubated with antigen-specific antibodies for 45 min at room temperature. Flow cytometry was performed using a FACSCalibur (BD Biosciences). The data were analysed with Cell Quest software (BD Biosciences), and the forward and side scatter profile gated out debris and dead cells. Immunophenotyping of hBM-MSCs was performed with antibodies against the following human antigens: CD13, CD29, CD44, CD73, CD90, CD146, CD166, HLA-ABC, CD11b, CD14, CD15, CD34, CD45, CD117, HLA-DR, and their isotype controls [IgG1 FITC, IgG2a PE (phycoerythrin) and IgG1 PE)]. All of the antibodies were supplied by Becton Dickinson.
2.3. LDH (lactate dehydrogenase) assay
The day before the LDH assay (Roche), cells were seeded into 24-well culture plates with complete medium at 30–50% confluency (1×104 hMSCs/well). After siRNA transfection of hMSCs with Lipofectamine RNAiMAX (Invitrogen), cells were incubated at 37°C in a CO2 incubator for 72 h. To determine the LDH activity, 100 μl of cell culture medium was transferred into a 96-well plate at 12, 24, 36, 48, 60 and 72 h. Then, 100 μl of the reaction mixture was added to each well, and the plate was incubated for 30 min at room temperature, protected from light. The absorbance was then measured in a microplate reader (BioTek) at 490/690 nm.
2.4. Cell viability and proliferation assay
First, 2×103 cells/well were seeded into 96-well culture plates with complete medium at 30–50% confluency. In brief, siRNA transfection was carried out on the next day. After siRNA transfection of hMSCs with Lipofectamine RNAiMAX, cells were incubated at 37°C in a 5% CO2 incubator for 72 h. The WST-1 assay was performed according to the manufacturer's instructions after 24, 48 and 72 h of transfection. On the day of WST-1 assay, 10 μl of cell proliferation reagent WST-1 (Roche) was added to each well and incubated at 37°C in 5% CO2 incubator for 4 h. The absorbance was measured in a microplate reader (BioTek) at 450/690 nm.
2.5. GFP (green fluorescent protein) plasmid and siRNA transfection
HEK (human embryonic kidney) 293 cells, which are originally derived from human embryonic kidney and hMSCs, were used. HEK 293 cells were cultured in DMEM-LG (Gibco) culture media containing 10% FBS (Biochrom) and 1% penicillin/streptomycin (Biochrom). Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2 and subcultured twice in a week. For GFP plasmids and Alexa Fluor 488-labelled siRNA transfections, both cell types, HEK 293 and hMSCs, were seeded into 96-well culture plates at a confluency of 90–95%, 24 h before transfection. Reagents at different concentrations used in these experiments were FuGENE for GFP plasmids, Lipofectamine RNAiMAX for labelled siRNAs and Lipofectamine 2000 for both GFP plasmids and labelled siRNAs. In the case of transfection with unlabelled validated siRNA transfections, cells were seeded into six-well culture plates with complete medium at 30–50% confluency (5×104 hMSCs/well) the day before transfection. Lipofectamine RNAiMAX was diluted with Opti-MEM® in a 1:50 ratio. For the silencing of topo IIβ, two different validated siRNAs (TOP2B_5 and TOP2B_6 from Qiagen) were used for transfection. Sequences of TOP2B_5 and TOP2B_6 used in the experiment were TCGGGCTAGGAAAGAAGTAAA and CAGCCGAAAGACCTAAATACA, respectively. Opti-MEM was used to dilute siRNAs at a ratio of 1:100. Diluted siRNA and reagent were mixed in one tube and incubated for 15 min at room temperature to allow the siRNA–Lipofectamine RNAiMAX complexes to form. Five hundred microlitres of siRNA–Lipofectamine RNAiMAX complexes were added into the wells containing cells and medium, by rocking the plate back and forth. Cells were incubated at 37°C in a 5% CO2 incubator for 24–48 h. After 24 h incubation, the medium containing complex was removed and replaced by the complete medium.
2.6. RT-PCR
RNA samples of hMSCs were extracted by using RNeasy kit (Qiagen). Of total RNA, 0.5 μg was reverse transcribed to obtain cDNA using the Quantitect Reverse Transcription kit (Qiagen). A cDNA library was obtained after 35 cycles of amplification (PCR core kit, Qiagen). Forward and reverse primers were ACAGGTGGTCGTAATG and GTTTCACTGATACACC, yielding a 508-bp PCR product for TOP2β, ACCATTGCAGCCTGTA and GCTCTTCCCATATTATCC yielding a 596-bp PCR product for TOP2α, and CGCACCACTGGCATTGTCAT and GTGGCCATCTCCTGCTCGAA yielding a 208-bp PCR product for actin. PCR was performed 35 cycles for TOP2β (60°C) and TOP2α (54°C), 30 cycles for actin (60°C).
2.7. Western blot
hMSCs were seeded into 10-cm culture Petri dishes (Falcon) for Western blotting with the frequency of 1×106 cells as a final outcome in cell number. After 24–96 h, cells were harvested in lysis buffer (50 mM Tris/HCl pH 6.8, 2 mM EDTA pH 8.0, 1% SDS, 1% 2-mercaptoethanol, 8% glycerol and 2% protease inhibitor cocktail), then protein concentrations were determined by a microassay procedure in accordance with Bradford Method (Protein Assay Kit; Bio-Rad). Cell lysates were analysed via either 6 or 7.5% (w/v) SDS/PAGE gels. Western blot was conducted using a dry blot system (iBlot; Invitrogen). The blots were incubated in blocking solution [5% skimmed milk, 0.1% Tween 20 in 1 M Tris/HCl (pH 7.5), 5 M NaCl] for 2 h at room temperature; then, they were incubated with primary antibodies specific to topo IIβ (1:1000; mouse; BD), and actin (1:500; rabbit; Santa Cruz) for 1 h at 30°C. PVDF membranes were treated with HRP-conjugated either goat-anti-mouse (1:5000; Chemicon) or goat-anti-rabbit (1:25000; Chemicon) secondary antibodies for 1 h at room temperature. Binding was detected on autoradiography film via chemiluminescence substrate (Vector Lab) addition.
3. Results
3.1. Immunophenotype of isolated hMSCs from BM (bone marrow)
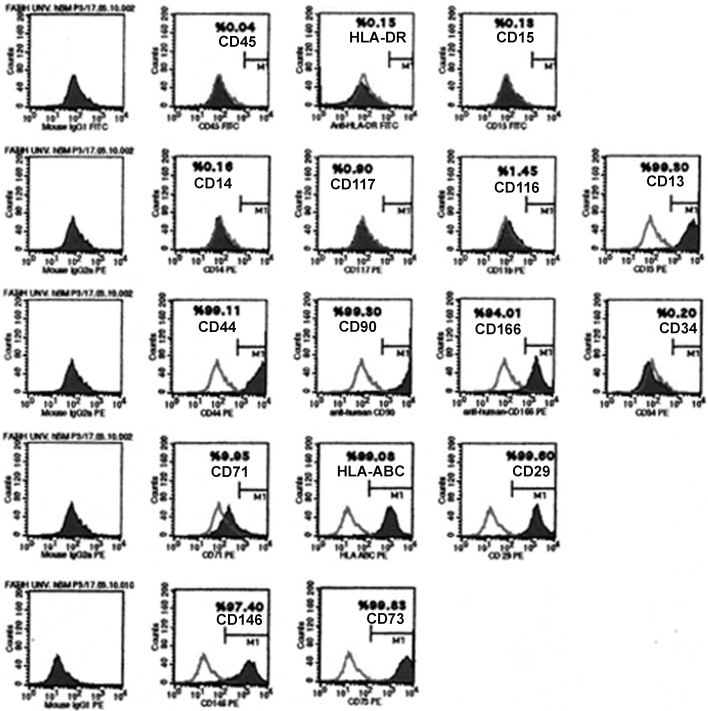
Flow cytometry results showed that after isolation by differential centrifugation, hMSCs at passage 3 expressed CD13, CD29, CD44, CD73, CD90, CD146, CD166, and HLA ABC, whereas they showed a lack of CD11b, CD14, CD15, CD34, CD45, CD117 or HLA-DR antigens (Figure 1, Table 1). This data indicated that hMSCs used in this study had the same characteristics of hMSCs reported in previous studies (Kemp et al., 2005; Motaln et al., 2010).
Figure 1. Representative flow cytometry analysis of cell surface markers in hBM-MSCs at passage 3.
Table 1. List of cell surface markers in hBM-MSCs at passage 3 indicated in Figure 1.
| Cell surface antigen | Presence in hMSCs at passage 3 (%) |
|---|---|
| CD13 | 99.3 |
| CD14 | 0.16 |
| CD15 | 0.13 |
| CD29 | 99.6 |
| CD44 | 99.11 |
| CD45 | 0.04 |
| CD71 | 9.95 |
| CD73 | 99.83 |
| CD90 | 99.3 |
| CD116 | 1.45 |
| CD117 | 0.90 |
| CD146 | 97.4 |
| CD166 | 94.01 |
| HLA-DR | 0.15 |
| HLA-ABC | 99.08 |
3.2. GFP plasmid and siRNA transfection of hMSCs
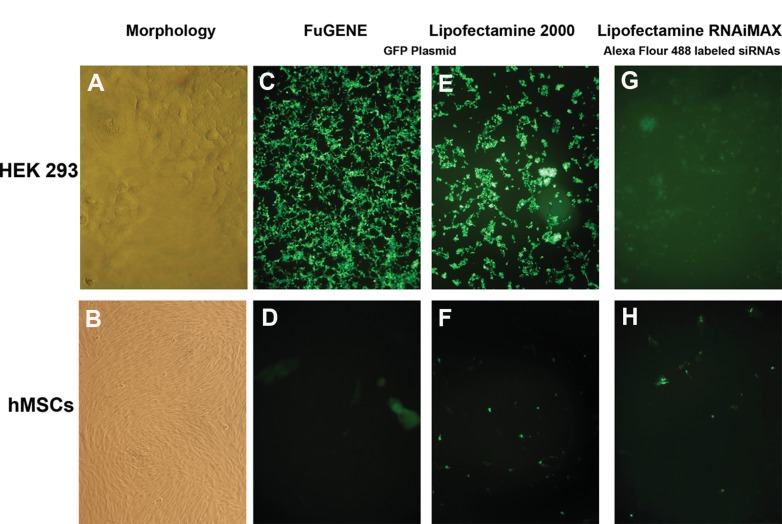
In this study, hMSCs derived from bone marrow were chosen due to their high abundance in the bone marrow and also easiness to isolate and expand in the culture. After flow cytometry analysis, cells at third passage were subjected to the GFP–plasmid and siRNA transfection experiments in hMSCs. HEK 293 cell line was used as a positive control. Approximately 100% efficiency was observed for GFP plasmid transfection of HEK 293 with Lipofectamine 2000 and FuGENE HD transfection reagents. Increasing concentrations of reagent and lack of the serum resulted in higher cytotoxicity for HEK 293 cells (data not shown). However, for MSCs, efficiency did not exceed 25%. Alexa Fluor 488-labelled control siRNAs were delivered with Lipofectamine RNAiMAX and Lipofectamine 2000 to HEK 293 cells with about 95–100% efficiency. Unfortunately, this method was also insufficient for hMSCs. The possibility that the label at the end of oligo might be preventing efficient delivery of the siRNA into the cell led us to use unlabelled siRNAs (Figure 2).
Figure 2. GFP plasmid and siRNA transfection of HEK-293 and hMSCs.
Morphology of HEK-293 and hMSCs at third passage (A, B). GFP plasmid transfection of HEK-293 and hMSCs with Lipofectamine 2000 and FuGENE transfection reagents. While approximately 100% transfection efficiency was observed for HEK-293 cells, it was about 25% for Lipofectamine 2000 and 15% for FuGENE (C, D, E, F). Transfection with RNAiMAX transfection reagent by using Alexa Fluor 488-labelled siRNA oligos in HEK-293 and hMSCs with approximately 100% and 15% transfection efficiencies, respectively. All pictures were taken following 24 h of transfection (G, H).
3.3. Selective silencing of topo IIβ with Lipofectamine RNAiMAX reagent
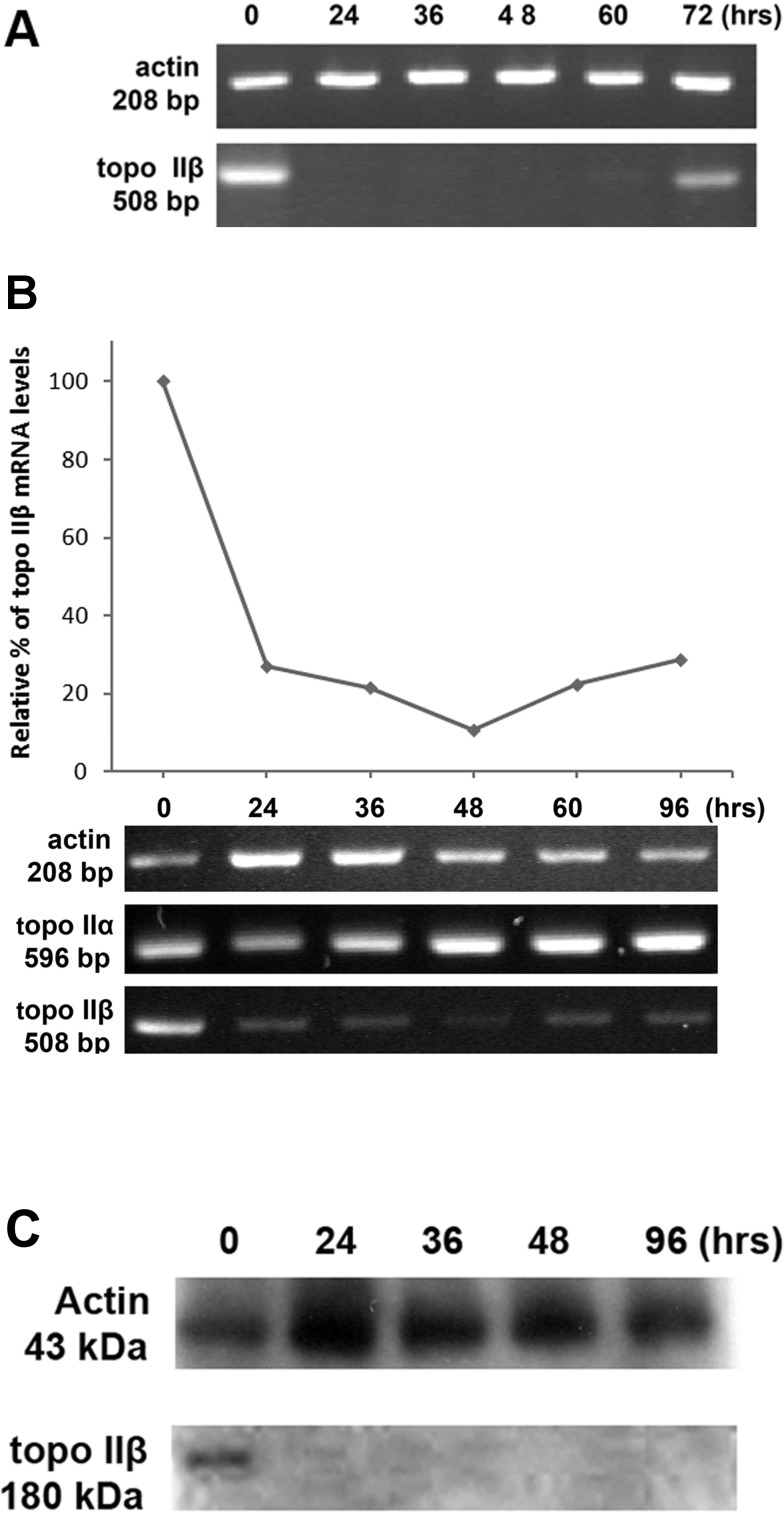
Two RNA duplexes of 21 nucleotides in length for topo IIβ (validated unlabelled siRNAs) were obtained from a commercial source, and hMSCs were transfected with a combination of these two siRNAs in Lipofectamine RNAiMAX reagent. Cells were assayed 24, 36, 48, 60 and 72 h after transfection by RT-PCR (Figure 3A). Specific human DNA primers were used for actin (as a control) and topo IIβ genes. Cells transfected with topo IIβ-specific siRNAs resulted in a significant reduction in topo IIβ at mRNA level. Most of the endogenous topo IIβ mRNA disappeared between 24 and 60 h after transfection. At 60 h of transfection, endogenous topo IIβ mRNA reappeared slightly, and after 72 h, almost more than half of the mRNA was retrieved (Figure 3A). A cytoskeleton protein, actin, was used to show that transfection is not non-specific. Actin mRNA was not affected by transfection. To extend the silencing period of the topo IIβ gene, a second transfection was performed after 48 h. Then, silencing of the topo IIβ was reanalysed between 24 and 96 h after transfection by RT-PCR (Figure 3B). At this time, almost 80% silencing was reached after 24 h of the transfection, and this low level of topo IIβ was observed even after 96 h after transfection. Also, since topo IIβ has its isoform, topo IIα, the mRNA level of topo IIα was also checked during 96 h of siRNA transfection to investigate whether topo IIβ is silenced selectively. The results clarified that topo IIα expression at mRNA level is not changed by the siRNA transfection (Figure 3B). Topo IIβ gene silencing was also confirmed by Western blotting at the protein level (Figure 3C). In a Western blot, monoclonal antibody raised against topo IIβ, clone 40, specifically recognized a 180-kDa polypeptide in hMSCs. On the first day after treatment, the topo IIβ protein level was approximately 10–15%, and no protein bands were observed in the later days of the transfection.
Figure 3. RT-PCR and Western blot analysis topo IIβ after siRNA transfections with Lipofectamine RNAiMAX.
Transfections were performed once at 0 h (A) and twice at 0 and 48 h (B, C). Although actin level did not change during transfection, topo IIβ disappeared at 24 h and reappeared at 72 h (A). While the level of topo IIα and actin did not change during 96 h of transfection, 80% of topo IIβ mRNA disappeared after 24 h, and this low level was observed even after 96 h (B). Protein level of topo IIβ decreased 85–90% during transfection after 24 h, but actin did not change.
3.4. Cytotoxicity and proliferation assays
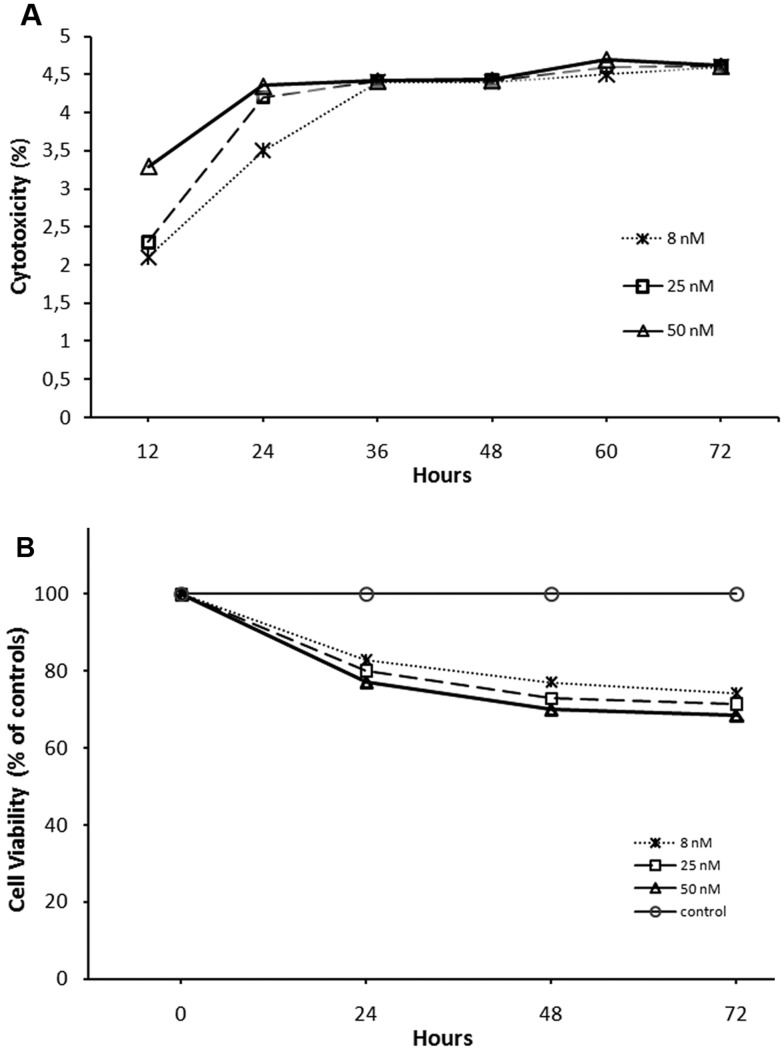
The LDH cytotoxicity detection kit is a colorimetric assay for the quantification of cell death and cell lysis, based on the determination of LDH activity released from the cytosol of damaged cells into the medium, thus indicating cell membrane damage. The LDH cytotoxicity assay kit was used following the manufacturer's instructions. The integrity of the plasma membrane was evaluated by an LDH assay following 12, 24, 36, 48, 60 and 72 h incubating the cells with 8, 25 and 50 nM of topo IIβ-specific siRNAs by Lipofectamine RNAiMAX transfection reagent (Figure 4A). Relative cytotoxicity of the reagent in transfected hMSCs increases in the first 24 h in a dose-dependent manner and then become stabilized in a time-dependent manner. The percentage of the damaged cells by the reagent during this time period was approximately in the range of 3.5–4.5%. These results demonstrated that Lipofectamine RNAiMAX did not cause considerable damage to the plasma membrane of hMSCs. The proliferation assay was performed to elucidate the effect of the same transfection reagent on hMSCs. We established a similar experimental setup as we did for the LDH assay to compare the proliferation rates of the cells treated with 8, 25 and 50 nM of siRNAs after 24, 48 and 72 h of transfection. A graph of the transfection reagent-treated and untreated samples revealed that cell viability of transfected cells decreased about 20% compared with control cells with 8 nM siRNA concentration (Figure 4B). The increased concentration of topo IIβ-specific siRNAs (25 and 50 nM) did not cause a significant difference in the viability of the transfected cells compared with the lower concentration.
Figure 4. Cytotoxicity and cell viability assays of hMSCs after siRNA transfections with Lipofectamine RNAiMAX.
Cytotoxicity of transfected hMSCs increases in the first 24 h in a dose-dependent manner and then become stabilized in a time-dependent manner (A). Cell viability assay of siRNA transfected hMSCs with Lipofectamine RNAiMAX reagent. Cell viability was affected by Lipofectamine RNAiMAX 20–30% in hMSCs (B). Transfections were performed twice at 0 and 48 h.
4. Discussion and conclusion
In mammals, topo II has two isoforms called topo IIα and topo IIβ. Since commercial topo II inhibitor drugs inhibit both isoforms of topo IIα and topo IIβ, a selective method such as siRNA interference for specific topo IIα or topo IIβ silencing was used. There are several studies (Sakaguchi and Kikuchi, 2004; Emmons et al., 2006) which have used siRNAs for selective topo IIβ inhibition in cancer lines, but there are none in hMSCs. siRNA duplexes have been widely used to investigate the roles of some critical genes in the self-renewal maintenance and differentiation of hMSCs (Hoelters et al., 2005). But, as with many primary cells and stem cells, hMSCs are very difficult to transfect with respect to cell lines. Although several common transfection reagents and Lipofectamine RNAiMAX have been described to transfect siRNAs into hMSCs (De Becker et al., 2007; Ho et al., 2009) so far, a detailed study about Lipofectamine RNAiMAX should be exploited.
In this study, three reagents were used in order to obtain desired silencing at hMSCs. With Lipofectamine 2000 and FuGENE HD transfection reagents, almost 95–100% efficiency was observed at GFP plasmid transfection of control HEK 293 cell line. However, in MSCs, efficiency did not exceed 25%. Also, carrying out transfections in serum-free medium did not cause any increase in efficiency even if it increased the cytotoxic effect of the reagents. These results led us to try delivering siRNA oligos, since they have smaller sizes, and they do not need to reach the nucleus to induce RNA interference. Alexa Fluor 488-labelled control siRNAs were delivered with Lipofectamine RNAiMAX and Lipofectamine 2000 to HEK 293 with about 100% efficiency. Unfortunately, this method was also ineffective for hMSCs. The possibility that the label at the end of oligo might be preventing efficient delivery of the siRNA into the cell led us to use unlabelled siRNAs. Topo IIβ-specific siRNAs were delivered with Lipofectamine RNAiMAX to hMSCs. A significant silencing (about 80%) was observed at the mRNA level and 85–90% at the protein level of the topo IIβ enzyme during 96 h of siRNA transfection.
Concerning the cytotoxicity, the transfection of hMSCs with Lipofectamine RNAiMAX caused cytotoxicity in only 3.5–4.5% of cells. A significant decrease in cell viability was not observed in relation with the cytotoxicity. Considering the graphical analysis of these two assays, an almost negative correlation between the cytotoxicity and cell viability assays was indicated.
In conclusion, we achieved to transfect hMSCs with topo IIβ-specific siRNA oligos by using Lipofectamine RNAiMAX reagent with high transfection efficiency, low cytotoxicity and high cell viability. In further studies, the results of selective topo IIβ inhibition will lead us to reveal the function of topo IIβ during transdifferentiation of hMSCs into neuronal cells.
Acknowledgements
We thank Professor Serdar Bedii Omay for supplying us bone marrow samples for hMSCs isolation. The results of the study were presented at “The 19th CDB Meeting: RNA Sciences in Cell and Developmental Biology”.
Footnotes
This work was supported by TUBITAK [grant numbers 106S279, SBAG-K-116], and Fatih University Research Project Foundation [grant number P50030706].
Author contribution
All authors participated in planning the study and preparing the manuscript. Sevim Isik designed the study, analysed the data and wrote the manuscript. Nebiyyeh Kamaci, Tuba Emnacar, Nihal Karakas and Gulsum Arikan conducted the experiments.
References
- Austin CA, Marsh KL. Eukaryotic DNA topoisomerase II beta. BioEssays. 1998;20:215–26. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Dalby B, Cates S, Harris A, Ohki EC, Tilkins ML. Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods. 2004;33:95–103. doi: 10.1016/j.ymeth.2003.11.023. [DOI] [PubMed] [Google Scholar]
- De Becker A, Van Hummelen P, Bakkus M, Vande Broek I, De Wever J, De Waele M. Migration of culture-expanded human mesenchymal stem cells through bone marrow endothelium is regulated by matrix metalloproteinase-2 and tissue inhibitor of metalloproteinase-3. Haematologica. 2007;92:440–9. doi: 10.3324/haematol.10475. [DOI] [PubMed] [Google Scholar]
- Drake FH, Hofmann GA, Bartus HF, Mattern MR, Crooke ST, Mirabelli CK. Biochemical and pharmacological properties of p170 and p180 forms of topoisomerase II. Biochemistry. 1989;28:8154–60. doi: 10.1021/bi00446a029. [DOI] [PubMed] [Google Scholar]
- Emmons M, Boulware D, Sullivan DM, Hazlehurst LA. Topoisomerase II beta levels are a determinant of melphalan-induced DNA crosslinks and sensitivity to cell death. Biochem Pharmacol. 2006;72:11–8. doi: 10.1016/j.bcp.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–51. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Ho IA, Chan KY, Ng WH, Guo CM, Hui KM, Cheang P. Matrix metalloproteinase 1 is necessary for the migration of human bone marrow-derived mesenchymal stem cells toward human glioma. Stem Cells. 2009;27:1366–75. doi: 10.1002/stem.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelters J, Ciccarella M, Drechsel M, Geissler C, Gulkan H, Bocker W. Nonviral genetic modification mediates effective transgene expression and functional RNA interference in human mesenchymal stem cells. J Gene Med. 2005;7:718–28. doi: 10.1002/jgm.731. [DOI] [PubMed] [Google Scholar]
- Javazon EH, Beggs KJ, Flake AW. Mesenchymal stem cells: paradoxes of passaging. Exp Hematol. 2004;32:414–25. doi: 10.1016/j.exphem.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Wadhwa R, Taira K. World of small RNAs: from ribozymes to siRNA and miRNA. Differentiation. 2004;72:58–64. doi: 10.1111/j.1432-0436.2004.07202006.x. [DOI] [PubMed] [Google Scholar]
- Kemp KC, Hows J, Donaldson C. Bone marrow-derived mesenchymal stem cells. Leukemia Lymphoma. 2005;46:1531–44. doi: 10.1080/10428190500215076. [DOI] [PubMed] [Google Scholar]
- Kubo H, Shimizu M, Taya Y, Kawamoto T, Michida M, Kaneko E. Identification of mesenchymal stem cell (MSC)-transcription factors by microarray and knockdown analyses, and signature molecule-marked MSC in bone marrow by immunohistochemistry. Genes Cells. 2009;14:407–24. doi: 10.1111/j.1365-2443.2009.01281.x. [DOI] [PubMed] [Google Scholar]
- Lee K, Majumdar MK, Buyaner D, Hendricks JK, Pittenger MF, Mosca JD. Human mesenchymal stem cells maintain transgene expression during expansion and differentiation. Mol Ther. 2001;3:857–66. doi: 10.1006/mthe.2001.0327. [DOI] [PubMed] [Google Scholar]
- Motaln H, Schichor C, Lah TT. Human mesenchymal stem cells and their use in cell-based therapies. Cancer. 2010;116:2519–30. doi: 10.1002/cncr.25056. [DOI] [PubMed] [Google Scholar]
- Peister A, Mellad JA, Wang M, Tucker HA, Prockop DJ. Stable transfection of MSCs by electroporation. Gene Ther. 2004;11:224–8. doi: 10.1038/sj.gt.3302163. [DOI] [PubMed] [Google Scholar]
- Sakaguchi A, Kikuchi A. Functional compatibility between isoform alpha and beta of type II DNA topoisomerase. J Cell Sci. 2004;117:1047–54. doi: 10.1242/jcs.00977. [DOI] [PubMed] [Google Scholar]
- Sano K, Miyaji-Yamaguchi M, Tsutsui KM, Tsutsui K. Topoisomerase IIbeta activates a subset of neuronal genes that are repressed in AT-rich genomic environment. PLoS One. 2008;3:e4103. doi: 10.1371/journal.pone.0004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M, Sorensen DR. Cationic liposome-mediated delivery of siRNAs in adult mice. Biochem Biophys Res Commun. 2003;312:1220–5. doi: 10.1016/j.bbrc.2003.11.057. [DOI] [PubMed] [Google Scholar]
- Spagnou S, Miller AD, Keller M. Lipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry. 2004;43:13348–56. doi: 10.1021/bi048950a. [DOI] [PubMed] [Google Scholar]
- Tsutsui K, Sano K, Kikuchi A, Tokunaga A. Involvement of DNA topoisomerase IIbeta in neuronal differentiation. J Biol Chem. 2001;276:5769–78. doi: 10.1074/jbc.M008517200. [DOI] [PubMed] [Google Scholar]
- Zhang XY, La Russa VF, Reiser J. Transduction of bone-marrow-derived mesenchymal stem cells by using lentivirus vectors pseudotyped with modified RD114 envelope glycoproteins. J Virol. 2004;78:1219–29. doi: 10.1128/JVI.78.3.1219-1229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Yang H, Jiang X, Zhou W, Zhu B, Zeng Y. Lipofectamine RNAiMAX: an efficient siRNA transfection reagent in human embryonic stem cells. Mol Biotechnol. 2008;40:19–26. doi: 10.1007/s12033-008-9043-x. [DOI] [PubMed] [Google Scholar]