Abstract
Photinus pyralis (firefly) luciferase is widely used as a reporter system to monitor alterations in gene promoter and/or signalling pathway activities in vitro. The enzyme catalyses the formation of oxyluciferin from D-luciferin in an ATP-consuming reaction involving photon emission. The purpose of the present study was to characterize the luciferase-inhibiting potential of (E)-2-fluoro-4′-methoxystilbene, which is known as a potent inhibitor of the NF-κB (nuclear factor κB) signalling pathway that is used to modulate the NF-κB signalling pathway in vitro. Results show that (E)-2-fluoro-4′-methoxystilbene effectively inhibits firefly luciferase activity in cell lysates and living cells in a non-competitive manner with respect to the luciferase substrates D-luciferin and ATP. By contrast, the compound has no effect on Renilla and Gaussia luciferases. The mechanism of firefly luciferase inhibition by (E)-2-fluoro-4′-methoxystilbene, as well as its potency is comparable to its structure analogue resveratrol. The in vitro use of trans-stilbenes such as (E)-2-fluoro-4′-methoxystilbene or resveratrol compromises firefly luciferase reporter assays as well as ATP/luciferase-based cell viability assays.
Keywords: firefly, luciferase inhibitor, nuclear factor κB, reporter gene assay, resveratrol
Abbreviations: CCD, charge-coupled device; CMV, cytomegalovirus; NF-κB, nuclear factor κB; NFκBAI4, NF-κB activation inhibitor 4; RT–PCR, reverse transcription–PCR; STF, SuperTopflash
INTRODUCTION
Luciferase reporter systems are widely used reporter genes that allow for an easy luminescence detection of the activity of gene promoters and/or transcriptionally relevant signalling pathways in response to a variety of stimuli and modulators. Among the different luciferases known, Photinus pyralis (firefly) luciferase was the first to be cloned in 1985 [1] and is still most widely used. In a two-step reaction, the second of which is coupled to photon emission, firefly luciferase converts its substrate D-luciferin into oxyluciferin in an ATP- and oxygen-consuming reaction. For a detailed description of the chemistry of luciferase-catalysed reactions, see [2–5]. The luciferase reaction is also used in cytotoxicity/cell viability assays based on assessing cellular ATP levels. Often, coenzyme A is also present in firefly luciferase assay buffers as a light stabilizer due to its ability to perform thiolysis of dehydroluciferyl-AMP, a product of the luciferase reaction capable of inhibiting the enzymatic reaction; e.g. see [5] and references therein.
A number of chemical substances have been described which inhibit firefly luciferase activity by either competitive or non-competitive action. For example, different classes of firefly luciferase inhibitors and their mechanisms of action are discussed in a recent review by Leitao and Esteves da Silva [3], and additional firefly luciferase inhibitors were identified by Auld et al. [6]; as one might expect, various analogues of the substrate luciferin as well as structurally related benzothiazoles inhibit the reaction in a competitive manner [3,6,7]. ATP analogues are also capable of inhibiting firefly luciferase activity [6,8]. The luciferase-inhibitory potential of other classes of chemicals is less self-evident: among others, a number of alcohols, small alkanes, fatty acids, quinoline analogues, substituted benzylamides, different halogenated compounds used as general anaesthetics and certain ionic liquids also interfere with the activity of firefly luciferase [3,6,9,10], as well as the p53 inhibitor pifithrin-α [11] and the widely used antioxidant 3,5,4′-trihydroxy-trans-stilbene, better known as resveratrol [12]. Resveratrol is a potent non-competitive inhibitor of firefly luciferase with a reported Ki value of ~2 μM [12]. The authors of the latter paper stress that luciferase-based analyses of resveratrol-treated cells (e.g. resveratrol is often used for the assessment of antioxidative effects in cell culture) might be biased by the direct luciferase-inhibiting action of the compound. Of course, the same holds true for other luciferase inhibitors if used in cell culture for a purpose different from luciferase inhibition.
In the present study, we demonstrate that (E)-2-fluoro-4′-methoxystilbene {also known as NFκBAI4 [NF-κB (nuclear factor κB) activation inhibitor 4]; Figure 1A}, a resveratrol analogue without antioxidative properties used as a specific inhibitor of NF-κB activation [13], inhibits firefly luciferase, but not other luciferases, in vitro with a potency comparable with resveratrol. In contrast with resveratrol, inhibition of firefly luciferase by NFκBAI4 is sustained for >24 h in living cells, much longer than inhibition by resveratrol.
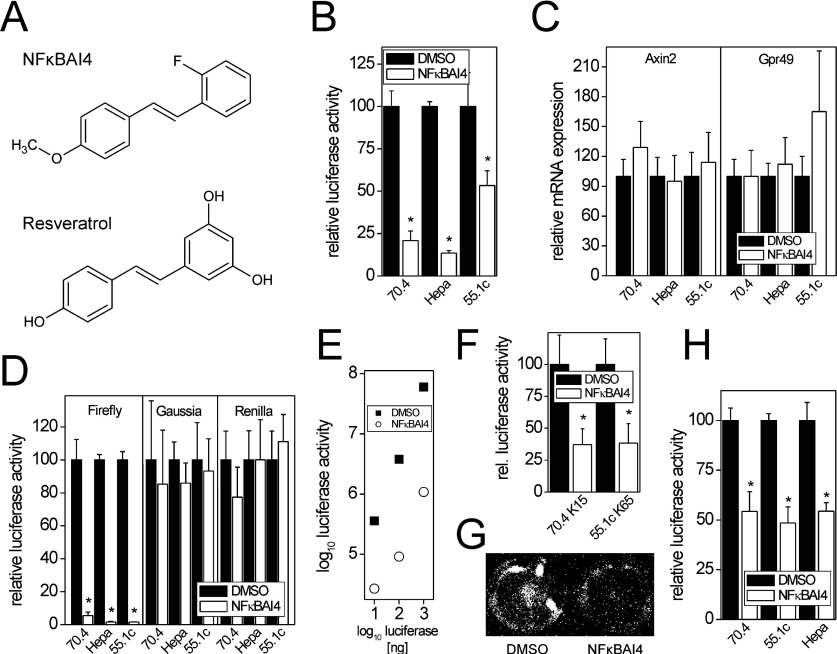
Figure 1. Inhibition of firefly luciferase by the NF-κB inhibitor NFκBAI4.
(A) Chemical structures of NFκBAI4 and its analogue resveratrol. (B) Inhibition of firefly luciferase activity is observed in mouse hepatoma cells transiently transfected with the β-catenin-driven firefly luciferase reporter STF after 24 h of incubation of the cells with 20 μM NFκBAI4. Luciferase signals were normalized to cell vitality, as determined by the Alamar Blue assay. Means±S.D. (n=4) are given; *P<0.05. (C) Lack of inhibition of the known β-catenin target genes Axin2 and Gpr49 by 24 h treatment of cells with 20 μM NFκBAI4. Means±S.D. (n=6) are given. (D) Inhibition of firefly luciferase signals after addition of 20 μM NFκBAI4 [1% (v/v) of 2 mM NFκBAI4 solution in DMSO] to lysates of untreated, transiently luciferase-expressing Hepa1c1c7, 70.4 and 55.1c cells 5 min prior to measurement. No inhibition of Gaussia or Renilla luciferases is observed. Means±S.D. (n=3 and 4) are given; *P<0.05. (E) Inhibition of commercially available firefly luciferase by 10 μM NFκBAI4 at different concentrations of the enzyme. The means of two experiments are shown. (F) Inhibition of firefly luciferase activity in living stably luciferase-expressing cells (cell lines 70.4 K15 and 55.1c K65) by NFκBAI4. Cells were pre-incubated with 20 μM NFκBAI4 for 30 min prior to analysis. Means±S.D. (n=4) are given; *P<0.05. (G) Luciferase inhibition in living cells was also monitored using a CCD camera system. A representative image from 55.1c K65 cells is shown. (H) Inhibition of luminescence signals from the CellTiter-Glo cell viability assay kit, which is based on luciferase-dependent detection of cellular ATP, by addition of 200 μM NFκBAI4. Please note that 200 μM NFκBAI4 is needed for ~50% inhibition of the Ultra-Glo luciferase used in this assay. Means±S.D. (n=3–5, each experiment performed in eight determinations) are given; *P<0.05.
MATERIALS AND METHODS
Chemicals and reagents
NFκBAI4 (catalogue no. 481412; Merck) and resveratrol (catalogue no. R5010; Sigma) were dissolved in DMSO in concentrations up to 20 mM and stored at −20°C for no longer than 4 weeks before use. Cell culture media, supplements and transfection reagents were purchased from Invitrogen. Chemicals for the preparation of luciferase assay buffers were purchased from PJK. D-Luciferin-ethylester was from Gentaur and firefly luciferase was from Roche; passive lysis buffer for luciferase assays was purchased from Promega. All other chemicals were purchased from Merck, if not otherwise indicated.
Cell culture and treatment
Mouse hepatoma cell lines Hepa1c1c7, 70.4 and 55.1c were grown in DMEM (Dulbecco's modified Eagle's medium)/F-12 medium supplemented with 10% fetal calf serum and antibiotics at 37°C and 5% CO2 in a humidified atmosphere. Cells were seeded at a concentration of 50000 cells/cm2 24 h prior to transfection and treated with the indicated concentrations of the compounds 24 h later. Concentration of the solvent DMSO was limited to 0.1% in all assays except for the in vivo monitoring of firefly luciferase activity, in which 1% DMSO was present.
70.4- and 55.1c-derived subclones stably transfected with the artificial β-catenin-driven firefly luciferase reporter plasmid STF (SuperTopflash) [14] and a plasmid-mediating resistance against G418 (pSV2neo; BD Biosciences) were routinely grown in medium additionally supplemented with 400 μg/ml G418 [15]. The selection antibiotic was removed from the cultures when plating them for experiments. Stably transfected cell lines are referred to as 70.4STF K15, 70.4STF K31 (both 70.4-derived), and 55.1cSTF K16, K60, K53 and K65 (55.1c-derived).
Cell viability assays
Cell viability/cytotoxicity of all compounds was analysed by the Neutral Red uptake and Alamar Blue assays using standard methodology. All concentrations used for cell treatment in the presented experiments did not cause any significant alterations in cell viability. Analyses with the CellTiter-Glo Luminescent Cell Viability Assay Kit (Promega) based on the detection of cellular ATP levels by a firefly luciferase-dependent reaction (modified Ultra-Glo recombinant luciferase) were performed as recommended by the manufacturer.
Transfections
Cells were transfected with the STF reporter plasmid (see above), the CMV (cytomegalovirus) promoter-driven Renilla luciferase expression plasmid pRL-CMV (Promega), or an AP-1-responsive Gaussia luciferase expression vector [16] using Lipofectamine™ 2000 according to the manufacturer's instructions. Stably transfected cells derived from the 55.1c cell line were established as recently described [15].
Luciferase activity assays
Firefly luciferase activity was determined in a 96-well plate reader (Victor3V; PerkinElmer) as described previously [17] using a reaction buffer containing 20 mM tricine, 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM DTT (dithiothreitol), 270 μM co-enzyme A, 470 μM D-luciferin and 530 μM ATP at pH 7.8 [18]. Renilla luciferase reaction buffer contained 220 mM K3PO4, 1.1 M NaCl, 2.2 mM EDTA, 0.44 g/l BSA, 1.3 mM NaN3 and 1.43 μM coelenterazine at pH 5.0 [19]. The same buffer was used for measurement of Gaussia luciferase with the only modification that it contained 5.72 μM coelenterazine. Then 10 μl of cell lysate (firefly, Renilla; prepared in 1× passive lysis buffer) or cell culture medium supernatant (Gaussia) was mixed with 50 μl of the respective reaction buffer. Renilla buffer was added to the lysate/firefly reaction buffer mix after measurement of firefly luciferase activity. Luminescence was measured for a period of 10 s. For analysis of decay rates of the enzyme reaction, luminescence was continuously monitored for 9 min after addition of the reaction buffer. If luminescence counts are presented in the Figures instead of relative luciferase activity, counts/s are shown. In the case of the addition of substances dissolved in DMSO to one of the luciferase assay buffers, DMSO concentration was limited to 5%. The firefly luciferase stock solution was prepared by dissolving 1 mg of the protein in 1 ml luciferase assay buffer, without ATP and D-luciferin, supplemented with 0.1% BSA.
Monitoring of luciferase in living cells was performed similar to [20] using a buffer containing 25 mM Tris/HCl, pH 7.5, 150 mM NaCl, 100 μM D-luciferin-ethylester and 1% DMSO. Cells were pre-incubated with 20 μM NFκBAI4 in culture medium for 30 min at 37°C and 5% CO2 followed by washing with PBS. Then, pre-warmed (37°C) assay buffer was added to the cells and luciferase signals were assessed after an additional 5 min of incubation in the plate reader or by the use of a CCD (charge-coupled device) camera (Raytest) using a time frame of 4 min (plate reader) or 8 min (CCD camera).
For the calculation of relative luciferase activity values, luminescence counts for each well were normalized to its corresponding cell vitality, as determined by the Alamar Blue assay, prior to cell lysis. Cell vitality-normalized luciferase activities are given relative to cell vitality-normalized values of untreated cells (percentage of control).
RNA isolation and real-time RT–PCR (reverse transcription–PCR)
Isolation of total RNA, RT by avian myeloblastosis reverse transcriptase (Promega), and real-time RT–PCR on a LightCycler instrument by the use of the FastStart DNA Master SYBR Green I kit (Roche) have been described recently [21]. Target gene expression was normalized to 18S rRNA expression according to [22]. PCR primers were as follows: Axin2_fwd, 5′-CGACGCACTGACCGACGATT-3′; Axin2_rev, 5′-TCCAGACTATGGCGGCTTTCC-3′; 18S rRNA_fwd, 5′-CGGCTACCACATCCAAGGAA-3′; 18S rRNA_rev, 5′-GCTGGAATTACCGCGGCT-3′.
RESULTS AND DISCUSSION
Inhibition of firefly but not Renilla or Gaussia luciferases by NFκBAI4
In a series of experiments aimed at analysing a possible interplay of β-catenin signalling and other cellular signalling pathways, the murine hepatoma cell lines Hepa1c1c7, 70.4 and 55.1c were transiently transfected with the β-catenin-driven firefly luciferase reporter STF and treated with 20 μM NFκBAI4 for 24 h. As shown in Figure 1(B), NFκBAI4 treatment caused an unexpected strong decrease of luminescence in all three cell systems. A similar inhibitory effect of NFκBAI4 was observed when a number of stably STF-transfected cell clones derived from the 70.4 or 55.1c cell lines were treated with the compound (Figure 3B). This was, however, not accompanied by a decrease in mRNA levels of the known β-catenin target genes Axin2 and Gpr49 (G-protein-coupled receptor 49) [15], thus casting doubt on a true inhibition of the pathway (Figure 1C), especially as a considerable concomitant reduction of target gene expression and reporter activity can be achieved by transfection of siRNA (small interfering RNA) directed against β-catenin mRNA (results not shown). One possible explanation for this discrepancy of β-catenin-dependent firefly luciferase reporter and target mRNA data was that NFκBAI4 causes an inhibition of the firefly luciferase reaction. To test this hypothesis, 20 μM NFκBAI4 was added directly into lysates of untreated Hepa1c1c7, 70.4 and 55.1c cells which had been transiently transfected with expression vectors for the firefly, Renilla or Gaussia luciferase 24 h before. NFκBAI4 strongly inhibited firefly luciferase signals in all cell lines, whereas the other luciferases, i.e. Gaussia and Renilla luciferase, were not affected (Figure 1D), demonstrating the specificity of NFκBAI4 for firefly luciferase. Pure commercially available firefly luciferase was also inhibited by NFκBAI4, demonstrating that the observed effects are not due to other components present in cell lysates (Figure 1E). Inhibition of firefly luciferase activity was further monitored in living luciferase-expressing cells incubated with a buffer containing the cell-permeable D-luciferin derivative D-luciferin-ethylester [20] following exposure of the cells to NFκBAI4 (Figures 1F and 1G).
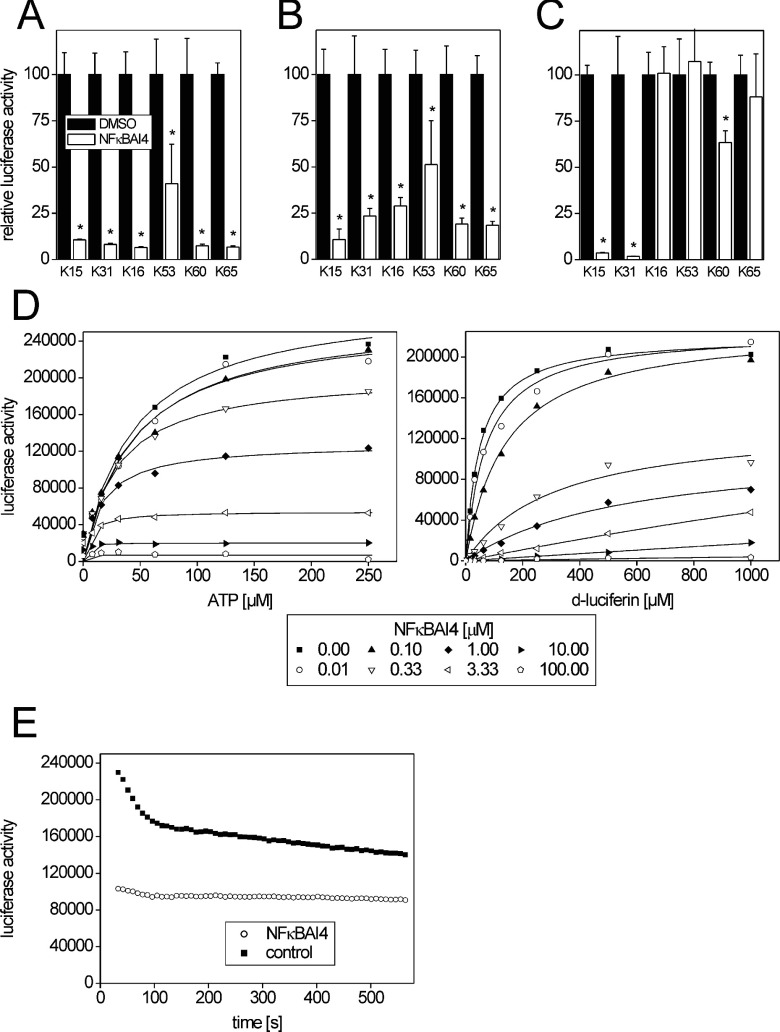
Figure 3. Time-dependent firefly luciferase inhibition by NFκBAI4 in cell culture and analysis of the inhibition mechanism.
Luciferase activity was assessed in lysates from various cell lines, derived from 70.4 or 55.1c mouse hepatoma cells, with stable expression of firefly luciferase. Then 20 μM NFκBAI4 was added to cell cultures and incubated for 1 h (A), 24 h (B) or 48 h (C) prior to lysis and measurement. Luciferase signals were normalized to cell vitality, as determined by the Alamar Blue assay. Means±S.D. (n=3 and 4) are given; *P<0.05. For comparison see data for firefly luciferase inhibition by resveratrol in Supplementary Figure S2 at http://www.bioscirep.org/bsr/032/bsr0320531add.htm. (D) Dose-response analysis of firefly luciferase activity for the substrates D-luciferin and ATP in the presence of different amounts of NFκBAI4. When varying D-luciferin content of the reaction mixture, ATP was kept constant at 500 μM; when varying ATP levels, D-luciferin was kept constant at 1000 μM. Solid lines represent a global fit of the mixed hyperbolic equation and show the expected non-competitive inhibition of firefly luciferase activity by NFκBAI4. For comparison, see results obtained with resveratrol in [12]. (E) Decay of the firefly luciferase reaction during a 9 min time frame after addition of coenzyme A-free reaction buffer in the presence or absence of 1 μM NFκBAI4. The means of two experiments are given.
Using a commercially available cell viability assay based on the detection of cellular ATP levels by a modified firefly luciferase (Ultra-Glo recombinant luciferase, derived from the luciferase of the firefly Photinus pennsylvanica; Promega), the inhibition of luciferase activity by NFκBAI4 was also detectable (Figure 1H). However, much higher concentrations of NFκBAI4 were needed for the inhibition of Ultra-Glo luciferase (IC50≈200 μM), as compared with conventional firefly luciferase.
Comparison with the structurally related firefly luciferase inhibitor resveratrol
It has been reported that resveratrol, structurally related to NFκBAI4, inhibits firefly luciferase [6,12]. We thus compared the ability of both compounds to inhibit firefly luciferase activity derived from lysates of untreated firefly luciferase-expressing cells (Figure 2, and Supplementary Figure S1 at http://www.bioscirep.org/bsr/032/bsr0320531add.htm). The inhibitory potency of both substances was very similar in all four cell lines analysed, with IC50 values of ~1 μM (Table 1). Almost identical concentration-dependent inhibition (IC50≈1 μM) of firefly luciferase by NFκBAI4 was detected when a reaction buffer without coenzyme A was used (results not shown; for comparison, see also results in Figure 3E). Values obtained with resveratrol were comparable with a previously reported IC50 value of ~2 μM [12].
Figure 2. Dose–dependency of firefly luciferase inhibition by NFκBAI4 and resveratrol.
Dose-effect curves of luciferase inhibition by NFκBAI4 (A) or resveratrol (B) are shown in lysates from untreated 70.4STF K15 and 55.1cSTF K65 cells with stable expression of firefly luciferase. Then 20 μM NFκBAI4 was added to lysates of untreated cells 5 min prior to measurement. Means±S.D. (n=4) are given. Inhibition of firefly luciferase activity in lysates derived from other cell lines is depicted in Supplementary Figure S1 at http://www.bioscirep.org/bsr/032/bsr0320531add.htm.
Table 1. IC50 values for firefly luciferase inhibition by NFκBAI4 and resveratrol.
Values were obtained with lysates from four stably luciferase-expressing mouse hepatoma cell lines. Underlying data are shown in Figure 2(A) and Supplementary Figure S1 at http://www.bioscirep.org/bsr/032/bsr0320531add.htm.
| IC50 (μM) | ||
|---|---|---|
| Cell line | NFκBAI4 | Resveratrol |
| 70.4STF K15 | 0.85 | 1.11 |
| 70.4STF K31 | 0.88 | 0.77 |
| 55.1cSTF K53 | 0.95 | 0.90 |
| 55.1cSTF K65 | 1.42 | 1.30 |
We further compared the luciferase-inhibiting ability of NFκBAI4 and resveratrol in cells after different periods of incubation with 20 μM of the respective substance. NFκBAI4 strongly inhibited firefly luciferase after 1 h of incubation and the effect was still pronounced after 24 h (Figures 3A and 3B). Following 48 h of incubation with NFκBAI4, the inhibitory effect was still visible in 70.4-derived, but not in 55.1c-derived, cell lines (Figure 3C). By contrast, inhibition of firefly luciferase activity by resveratrol was weaker after 1 h and disappeared in all cell lines already at 24 h after addition of the compound to the cells (see Supplementary Figure S2 at http://www.bioscirep.org/bsr/032/bsr0320531add.htm). A possible explanation for this dissimilar behaviour of the two substances which share an equal potency of firefly luciferase inhibition directly after addition to the cell lysates can be taken from the chemical structure of both molecules: resveratrol possesses hydroxy groups that allow for an easy and rapid cellular metabolism via conjugation with glucuronic acid or sulfate [23,24]. The lack of hydroxy groups in the NFκBAI4 molecule makes it necessary to chemically modify the molecule prior to further metabolism, which is a type of reaction that is typically carried out by xenobiotic-metabolizing enzymes from the cytochrome P450 family. The 55.1c and 70.4 hepatoma cell lines exhibit much lower levels of most cytochromes P450 than normal liver (results not shown), meaning that one would expect a rather slow metabolism of NFκBAI4.
Mechanism of enzyme inhibition
To exclude an attenuation of luciferase signals caused by absorption of the emitted photons by NFκBAI4, we recorded UV/visual spectra of NFκBAI4 in aqueous solution. An absorption peak at 315 nm was observed, whereas no absorption was detected at higher wavelengths (results not shown). The 315 nm peak is far away from the emission peak of firefly luciferase at 560 nm [25]. Thus the inhibitory effect of NFκBAI4 is not due to light absorption by the compound.
Resveratrol was shown to inhibit firefly luciferase in a non-competitive manner with respect to its substrates ATP and D-luciferin [12]. We assayed firefly luciferase activity in the presence of various amounts of NFκBAI4 and different concentrations of its substrates ATP and D-luciferin, similar to the analyses performed with resveratrol [12]. A mixed hyperbolic equation was fitted to the data (Figure 3D). The results appear almost identical with those reported for resveratrol [12], demonstrating that firefly luciferase is inhibited by NFκBAI4 in a non-competitive manner, comparable with resveratrol (Figure 3D).
We further analysed whether NFκBAI4 would affect the decay of the luminescence signal. For this purpose, lysates were treated with 1 μM NFκBAI4 (equivalent to the IC50 of the compound). Analyses were performed in the absence of coenzyme A. As expected, an overall decreased intensity of the luminescence signal was observed when the inhibitor was present (Figure 3E). After a rapid initial decay of the signal during the first 2 min, signal intensity declined more slowly during the rest of the observation period when the untreated lysate was used, whereas the decrease appeared to be somewhat less pronounced in the presence of NFκBAI4 (Figure 3E).
Improvement of a dual luciferase assay buffer by NFκBAI4
An inhibition of firefly luciferase might be desired under certain conditions: for example, dual luciferase assays normally consist of an analysis of firefly luciferase activity followed by the assessment of Renilla luciferase activity in the same cell lysate. This implies that the activity of firefly luciferase is effectively quenched before measuring luminescence produced by the Renilla enzyme, an issue that is solved by a change of buffer composition and pH in the reaction tube. Using our buffers, residual activity of firefly luciferase is ~0.02% of the initial activity, as determined by measuring lysates containing only firefly but not Renilla luciferase before and after addition of the Renilla buffer (Figure 4A). The addition of 20 μM NFκBAI4 to the Renilla buffer further improved its ability to quench the firefly signal (Figure 4A), but did not influence Renilla luciferase activity, as determined by the addition of 20 μM NFκBAI4 to the Renilla luciferase reaction buffer in a classic dual luciferase assay with transiently transfected Hepa1c1c7 cells (Figure 4B). Thus, in principle, dual luciferase systems might be improved by addition of a firefly luciferase inhibitor to the Renilla luciferase reaction buffer.
Figure 4. Improvement of the ability of a Renilla luciferase reaction buffer to quench firefly luciferase activity.
(A) Remnant activity of firefly luciferase in Renilla buffer is further reduced by addition of 20 μM NFκBAI4. Values are given as percentage of control (i.e. lysate with firefly buffer prior to the addition of Renilla buffer) (B) Renilla luciferase activity is not altered by the presence of NFκBAI4. Renilla luciferase activity was assayed using Renilla buffer with or without 20 μM NFκBAI4 in lysates from transiently transfected Hepa1c1c7 cells. Means±S.D. (n=6) are given; *P<0.05.
Conclusions
The potential of NFκBAI4 to inhibit firefly luciferase is important to know when conducting luciferase reporter analyses in lysates from cells treated with the compound, since these analyses might be compromised by the direct action of NFκBAI4 on the luciferase enzyme. The same applies to cell vitality assays that make use of firefly luciferase to determine the levels of cellular ATP as an indicator of metabolically active intact cells. In accordance with previous work [6,12], we found that the structurally related molecule resveratrol also inhibits firefly luciferase, in a manner comparable with NFκBAI4. Moreover, the glutamate receptor antagonist SIB-1893 [26], a trans-stilbene like NFκBAI4 and resveratrol, has been shown to inhibit firefly luciferase with a somewhat lower efficacy than NFκBAI4 [6] (please note that the compound is erroneously depicted as a cis-stilbene in the latter paper). Thus, it seems likely that trans-stilbenes constitute a structural class of firefly luciferase-inhibiting molecules which, however, has not been clearly identified as such in previous studies.
The problems caused by conducting luciferase assays in the presence of NFκBAI4 can be circumvented by using different luciferases (e.g. Gaussia luciferase). Another alternative might be the modified Ultra-Glo luciferase, which is present in Promega's cell viability assay kit. The latter enzyme was inhibited by NFκBAI4 in our analyses, but only at very high concentrations of the compound which exceed the concentrations routinely used in cell culture. Although a direct quantitative comparison with the degree of inhibition obtained with the wild-type is impeded by the different reaction conditions of the assays, it is highly plausible that the modified enzyme is more resistant against inhibition by NFκBAI4, as it is generally more resistant with respect to several other luciferase inhibitors [27]. This idea is supported by data presented on the Promega Corporation web page demonstrating that Ultra-Glo luciferase is more resistant against inhibition by the NFκBAI4 structural analogue resveratrol (http://www.promega.com/de-de/resources/scientific_posters/posters/compound-interference-of-celltiterglo-vs-pe-atplite-1step-poster/).
Online data
AUTHOR CONTRIBUTION
Albert Braeuning designed the experiments, performed the data analysis, conducted the minor part of the experiments and wrote the paper. Silvia Vetter performed the majority of the experimental work. Both authors read and approved the final version of the paper.
ACKNOWLEDGEMENTS
We thank Michael Schwarz (Tübingen, Germany) for helpful discussions and Eva Zeller (Tübingen, Germany) for proofreading of the manuscript.
FUNDING
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
References
- 1.de Wet J. R., Wood K. V., Helinski D. R., DeLuca M. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1985;82:7870–7873. doi: 10.1073/pnas.82.23.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navizet I., Liu Y. J., Ferre N., Roca-Sanjuan D., Lindh R. The chemistry of boluminescence: an analysis of chemical functionalities. ChemPhysChem. 2011;12:3064–3076. doi: 10.1002/cphc.201100504. [DOI] [PubMed] [Google Scholar]
- 3.Leitao J. M., Esteves da Silva J. C. Firefly luciferase inhibition. J. Photochem. Photobiol. B. 2010;101:1–8. doi: 10.1016/j.jphotobiol.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Hosseinkhani S. Molecular enigma of multicolor bioluminescence of firefly luciferase. Cell. Mol. Life Sci. 2011;68:1167–1182. doi: 10.1007/s00018-010-0607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraga H., Fernandes D., Fontes R., Esteves da Silva J. C. Coenzyme A affects firefly luciferase luminescence because it acts as a substrate and not as an allosteric effector. FEBS J. 2005;272:5206–5216. doi: 10.1111/j.1742-4658.2005.04895.x. [DOI] [PubMed] [Google Scholar]
- 6.Auld D. S., Southall N. T., Jadhav A., Johnson R. L., Diller D. J., Simeonov A., Austin C. P., Inglese J. Characterization of chemical libraries for luciferase inhibitory activity. J. Med. Chem. 2008;51:2372–2386. doi: 10.1021/jm701302v. [DOI] [PubMed] [Google Scholar]
- 7.Ribeiro C., Esteves da Silva J. C. Kinetics of inhibition of firefly luciferase by oxyluciferin and dehydroluciferyl-adenylate. Photochem. Photobiol. Sci. 2008;7:1085–1090. doi: 10.1039/b809935a. [DOI] [PubMed] [Google Scholar]
- 8.Filippova N., Ugarova N. N. Inhibition of luciferase from the firefly Luciola mingrelica by ATP analogs. Biokhimiia. 1982;47:1342–1348. [PubMed] [Google Scholar]
- 9.Franks N. P., Jenkins A., Conti E., Lieb W. R., Brick P. Structural basis for the inhibition of firefly luciferase by a general anesthetic. Biophys. J. 1998;75:2205–2211. doi: 10.1016/S0006-3495(98)77664-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebrahimi M., Hosseinkhani S., Heydari A., Khavari-Nejad R. A., Akbari J. Controversial effect of two methylguanidine-based ionic liquids on firefly luciferase. Photochem. Photobiol. Sci. 2012;11:828–834. doi: 10.1039/c2pp05389f. [DOI] [PubMed] [Google Scholar]
- 11.Rocha S., Campbell K. J., Roche K. C., Perkins N. D. The p53-inhibitor pifithrin-alpha inhibits firefly luciferase activity in vivo and in vitro. BMC Mol. Biol. 2003;4:9. doi: 10.1186/1471-2199-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bakhtiarova A., Taslimi P., Elliman S. J., Kosinski P. A., Hubbard B., Kavana M., Kemp D. M. Resveratrol inhibits firefly luciferase. Biochem. Biophys. Res. Commun. 2006;351:481–484. doi: 10.1016/j.bbrc.2006.10.057. [DOI] [PubMed] [Google Scholar]
- 13.Heynekamp J. J., Weber W. M., Hunsaker L. A., Gonzales A. M., Orlando R. A., Deck L. M., Jagt D. L. Substituted trans-stilbenes, including analogues of the natural product resveratrol, inhibit the human tumor necrosis factor α-induced activation of transcription factor nuclear factor κB. J. Med. Chem. 2006;49:7182–7189. doi: 10.1021/jm060630x. [DOI] [PubMed] [Google Scholar]
- 14.Nordeen S. K. Luciferase reporter gene vectors for analysis of promoters and enhancers. Biotechniques. 1988;6:454–458. [PubMed] [Google Scholar]
- 15.Braeuning A., Menzel M., Kleinschnitz E. M., Harada N., Tamai Y., Kohle C., Buchmann A., Schwarz M. Serum components and activated Ha-Ras antagonize expression of perivenous marker genes stimulated by β-catenin signaling in mouse hepatocytes. FEBS J. 2007;274:4766–4777. doi: 10.1111/j.1742-4658.2007.06002.x. [DOI] [PubMed] [Google Scholar]
- 16.Ring L., Perobner I., Karow M., Jochum M., Neth P., Faussner A. Reporter gene HEK-293 cells and WNT/frizzled fusion proteins as tools to study WNT signaling pathways. Biol. Chem. 2011;392:1011–1020. doi: 10.1515/BC.2011.164. [DOI] [PubMed] [Google Scholar]
- 17.Uibel F., Muhleisen A., Kohle C., Weimer M., Stummann T. C., Bremer S., Schwarz M. ReProGlo: a new stem cell-based reporter assay aimed to predict embryotoxic potential of drugs and chemicals. Reprod. Toxicol. 2010;30:103–112. doi: 10.1016/j.reprotox.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Gaunitz F., Papke M. Gene transfer and expression. Methods Mol. Biol. 1998;107:361–370. doi: 10.1385/0-89603-519-0:361. [DOI] [PubMed] [Google Scholar]
- 19.Dyer B. W., Ferrer F. A., Klinedinst D. K., Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 2000;282:158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- 20.Craig F. F., Simmonds A. C., Watmore D., McCapra F., White M. R. Membrane-permeable luciferin esters for assay of firefly luciferase in live intact cells. Biochem. J. 1991;276:637–641. doi: 10.1042/bj2760637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braeuning A., Schwarz M. Zonation of heme synthesis enzymes in mouse liver and their regulation by β-catenin and Ha-ras. Biol. Chem. 2010;391:1305–1313. doi: 10.1515/BC.2010.115. [DOI] [PubMed] [Google Scholar]
- 22.Pfaffl M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walle T., Hsieh F., DeLegge M. H., Oatis J. E., Jr, Walle U. K. High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab. Dispos. 2004;32:1377–1382. doi: 10.1124/dmd.104.000885. [DOI] [PubMed] [Google Scholar]
- 24.Yu C., Shin Y. G., Chow A., Li Y., Kosmeder J. W., Lee Y. S., Hirschelman W. H., Pezzuto J. M., Mehta R. G., van Breemen R. B. Human, rat, and mouse metabolism of resveratrol. Pharm. Res. 2002;19:1907–1914. doi: 10.1023/a:1021414129280. [DOI] [PubMed] [Google Scholar]
- 25.Ugarova N. N. Luciferase of Luciola mingrelica fireflies. Kinetics and regulation mechanism. J. Biolumin. Chemilumin. 1989;4:406–418. doi: 10.1002/bio.1170040155. [DOI] [PubMed] [Google Scholar]
- 26.Varney M. A., Cosford N. D., Jachec C., Rao S. P., Sacaan A., Lin F. F., Bleicher L., Santori E. M., Flor P. J., Allgeier H., et al. SIB-1757 and SIB-1893: selective, noncompetitive antagonists of metabotropic glutamate receptor type 5. J. Pharmacol. Exp. Ther. 1999;290:170–181. [PubMed] [Google Scholar]
- 27.Auld D. S., Zhang Y. Q., Southall N. T., Rai G., Landsman M., MacLure J., Langevin D., Thomas C. J., Austin C. P., Inglese J. A basis for reduced chemical library inhibition of firefly luciferase obtained from directed evolution. J. Med. Chem. 2009;52:1450–1458. doi: 10.1021/jm8014525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.