Abstract
The genomes of two phenotypically denitrifying type strains of the genus Bacillus were sequenced and the pathways for dissimilatory nitrate reduction were reconstructed. Results suggest that denitrification proceeds in the periplasmic space and in an analogous fashion as in Gram-negative organisms, yet with the participation of proteins that tend to be membrane-bound or membrane-associated. A considerable degree of functional redundancy was observed with marked differences between B. azotoformans LMG 9581T and B. bataviensis LMG 21833T. In addition to the already characterized menaquinol/cyt c-dependent nitric oxide reductase (Suharti et al., 2001, 2004) of which the encoding genes could be identified now, evidence for another novel nitric oxide reductase (NOR) was found. Also, our analyses confirm earlier findings on branched electron transfer with both menaquinol and cytochrome c as reductants. Quite unexpectedly, both bacilli have the disposal of two parallel pathways for nitrite reduction enabling a life style as a denitrifier and as an ammonifying bacterium.
Keywords: dissimilatory nitrate reduction to ammonium (DNRA), denitrification, ammonification, nitric oxide reductase
Introduction
Nitrogen is an essential element of all forms of life, but most of it on Earth is only available in an inaccessible form as dinitrogen gas (N2). Nitrogen-fixing microorganisms, however, are able to bind N2 into ammonium, which serves as a nutrient for other organisms. Nitrifiers use ammonium as the electron donor in their energy metabolism by oxidizing it to nitrite and nitrate. The latter two compounds are utilized by other microbial species for anaerobic respiration, reducing nitrite and nitrate back to dinitrogen gas in a process called denitrification or dissimilatory nitrate/nitrite reduction, thus closing the biogeochemical nitrogen cycle (N-cycle). These three basic processes have been known for over hundred years, but in the last decades it has become clear that the N-cycle is definitely more complex. It now appears that two major processes have been overlooked for a long time: anaerobic ammonium oxidation (anammox) and dissimilatory reduction of nitrite/nitrate into ammonium (DNRA), also known as nitrate/nitrite ammonification [see for recent reviews: Jetten (2008); Kraft et al. (2011); Lam and Kuypers (2011); Martínez-Espinosa et al. (2011)]. Like classical denitrifiers, DNRA bacteria employ nitrite and nitrate as electron acceptors for respiration (Einsle et al., 2002). They, however, reduce these compounds to ammonium as the name suggests. Hitherto, no species are known that carry out both denitrification and DNRA.
In its canonical form, dissimilatory nitrate reduction consists of four consecutive steps: (1) reduction of nitrate to nitrite, (2) nitrite reduction to nitric oxide (NO), (3) NO reduction to produce nitrous oxide (N2O), and (4) reduction of the latter to N2. The specific enzyme for N2O reduction, N2O reductase (NOS, N2OR) is not always present and microorganisms lacking this enzyme make N2O as their end product. In practice, a “true denitrifier” converts at least 80% of the consumed nitrate or nitrite to either N2O or N2 (Mahne and Tiedje, 1995). Also DNRA bacteria are capable of N2O formation, albeit in non-stoichiometric amounts that don't exceed 2–36% of consumed nitrate (Bleakley and Tiedje, 1982; Streminíska et al., 2012). Moreover, the production occurs only during the stationary phase, suggesting emission to be the result of a secondary metabolism (Smith, 1983).
The process of dissimilatory nitrate reduction has been extensively investigated at the genetic, enzymological, and regulatory levels using, among others, Paracoccus denitrificans, Pseudomonas stutzeri, Eschericha coli, and Wolinella succinogenes as model organisms [see for example: Tucker et al. (2005); van Wonderen et al. (2008); Bergaust et al. (2010); Pomowski et al. (2011); Peña et al. (2012), and references in Zumft (1997); Kern and Simon (2009); Simon and Klotz (2012)], of which most belong to Gram-negative Proteobacteria. In the Gram-negative bacteria, the enzymatic reactions involved in denitrification reside at the periplasm, except for nitrate reduction by the Nar-type nitrate reducase, and are catalyzed either by soluble enzymes (periplasmic nitrate-, nitrite-, and N2O reductases) or enzymes having their catalytic site embedded in the membrane (NO reductase). Similarly, electron transfer processes that are related to the different reduction reactions are mediated by a broad variety of soluble cytochrome c type or cupredoxin-like copper proteins localized in the periplasm.
Dissimilatory nitrate reduction is not restricted to Gram-negative species. In fact, denitrification and DNRA activities have been observed in a wide species and niche diversity among Gram-positive microorganisms as well, in particular Bacillus species (de Barjac and Bonnefoi, 1972; Garcia, 1977; Pichinoty et al., 1978, 1983; Tiedje, 1988; Denariaz et al., 1989). However, our knowledge at the molecular and genomic levels on this taxonomic lineage is rather fragmentary. Whereas many genomes of the representatives of the genus Bacillus are available, only four genomes have been reported to contain genes encoding denitrification or ammonification key enzymes. The genome of Bacillus selenitireducens was found to harbor a homolog of the gene coding for dissimilatory nitrite reductase making ammonium (nrfA), while qnorB homologs, encoding the quinol-dependent nitric oxide reductase (NOR), were detected in Bacillus coagulans XZL4 (Su et al., 2011), Bacillus licheniformis ATCC 14580T (Rey et al., 2004; Veith et al., 2004), and Bacillus sp. BTB_CT2. None of these species can be unequivocally designated as denitrifiers. Unlike Gram-negative bacteria, Gram-positive microorganisms only have a very limited periplasmic space. In this respect, it is remarkable that activities of denitrifying enzymes were consistently associated with membrane fractions (Denariaz et al., 1991; Urata and Satoh, 1991; Suharti et al., 2001; Suharti and de Vries, 2005; Fukuda et al., 2011; Matsumoto et al., 2012). Similarly, the limited number of such enzymes that have been purified to date from bacilli or close relatives are all membrane proteins (Denariaz et al., 1991; Urata and Satoh, 1991; Suharti et al., 2001; Suharti and de Vries, 2005; Fukuda et al., 2011; Matsumoto et al., 2012).
Considering the wide-spread occurrence of dissimilatory nitrate reduction and DNRA among Bacillus species (Verbaendert et al., 2011), their ecological relevance, niche, and species differentiation, possibly particular demands that are posed to the localization and organization of enzymes involved in these processes, and an overall limited knowledge regarding their molecular mechanisms, we now sequenced the genomes of two well-known denitrifying, publicly available representatives, Bacillus azotoformans LMG 9581T (Pichinoty et al., 1983) and Bacillus bataviensis LMG 21833T (Heyrman et al., 2004), able to produce dinitrogen and nitrous oxide respectively as end-products (Verbaendert et al., unpublished data). In order to come to a more comprehensive understanding of nitrate respiration by these Gram-positive microorganisms, we focused on their denitrifying inventories. Using bio-informatic tools, all gene products were analyzed for their homology with well studied enzymes from other organisms, their cellular localization and, in case of membrane proteins, for their putative topology and orientation. The results of our study enabled an unprecedented insight into the genomic basis underlying denitrification in both Bacillus species.
Materials and methods
Strains and DNA extraction
Bacillus azotoformans LMG 9581T and Bacillus bataviensis LMG 21833T were obtained from the BCCM/LMG bacteria collection. Strains were aerobically grown on trypticase soy broth at 28°C. Cells were harvested after overnight growth and DNA was extracted by the method of Pitcher et al. (1989), slightly modified as described previously (Heyndrickx et al., 1996).
Genome sequencing
Library preparation and genome sequencing was performed by Baseclear B.V. For sequencing, a paired-end strategy on the Illumina Genome Analyzer IIx was used that yielded reads of an average length of 74 bp. Automatic trimming (based on a threshold of Q = 20) and assembly was performed using CLC Genomics Workbench v4. The k-mer parameter was varied to maximize the N50 of the resulting assembly for each genome. Genome statistics are listed in Table 1.
Table 1.
Genome characteristics of both analyzed genomes.
| B. azotoformans LMG 9581T | B. bataviensis LMG 21833T | |
|---|---|---|
| # contigs | 169 | 197 |
| Size (Mb) | 4,3 MB | 5,4 Mb |
| Av. read coverage | 86x | 79,5x |
| N50 (Kb) | 94,5 | 82,1 |
| % G+C | 39,7 | 39,6 |
| # RNA calls | 8 rRNA | 6 rRNA |
| 25 tRNA | 23 tRNA | |
| # CDS calls | 4226 | 5207 |
| NCBI accession n° | AJLR00000000 | AJLS00000000 |
| NCBI BioProject | PRJNA80827 | PRJNA77725 |
Calls for RNA and CDS were deduced from PGAAP.
Genome annotation
Functional annotation and metabolic reconstruction was performed with (1) the Rapid Annotation Subsystem Technology (RAST) server (Aziz et al., 2008), using Glimmer (Salzberg et al., 1998) for gene calling and allowing frameshift correction, backfilling of gaps, and automatic fixing of errors, (2) KEGG Automatic Annotation Server (KAAS) (Moriya et al., 2007), using Glimmer gene calls from RAST and total prokaryotic genes data set for annotation, and (3) NCBI's Prokaryotic Genome Automatic Annotation Pipeline (PGAAP) (http://www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html) which uses GeneMark and GeneMark.HMM for gene calling (Borodovsky and McIninch, 1993; Lukashin and Borodovsky, 1998). Assigned functions were checked with pBLAST (Altschul et al., 1997) and InterProScan (Zdobnov and Apweiler, 2001). An inventory of genes involved in denitrification and ammonification for both genomes are listed in Tables 2, 3. Missing genes were searched for in the genome with PSI-BLAST using homologous amino acid sequences of closely related Bacillus or Geobacillus species. Possible frameshifts mentioned in the text were not corrected in the submitted genome.
Table 2.
Overview of gene inventory involved in nitrogen assimilation, denitrification and ammonification of B. azotoformans LMG 9581T.
| Function | Gene | Gene coordinates | Size (bp) | pBLAST best hit | ORF identifier | ||
|---|---|---|---|---|---|---|---|
| Genbank identifier | % ID | Rel. gene length | |||||
| Cytoplasmic dissimilatory nitrate reduction | narG1 | contig48_8658_4975 | 3684 | ABO66033 Geobacillus thermodenitrificans NG80-2 | 73 | 100,1 | BAZO_08891 |
| narH1 | contig48_4985_3435 | 1551 | ABO66034.1 Geobacillus thermodenitrificans NG80-2 | 73 | 101,4 | BAZO_08886 | |
| narJ1 | contig48_3463_2900 | 564 | ABO66035 Geobacillus thermodenitrificans NG80-2 | 46 | 101,1 | BAZO_08881 | |
| narI1 | contig48_2856_2140 | 717 | YP_001124781.1 Geobacillus thermodenitrificans NG80-2 | 65 | 103,9 | BAZO_08876 | |
| narG2 | contig69_15227_11544 | 3684 | EEL50916 Bacillus cereus Rock3-44 | 80 | 98,0 | BAZO_10677 | |
| narH2 | contig69_11554_10088 | 1467 | EID44195 Geobacillus thermoglucosidans TNO-09.020 | 84 | 99,8 | BAZO_10672 | |
| narJ2 | contig69_10009_9455 | 555 | YP_005422967 Bacillus amyloliquefaciens subsp. plantarum YAU B9601-Y2 | 46 | 100,0 | BAZO_10667 | |
| narI2 | contig69_9458_8769 | 690 | YP_002949597.1 Geobacillus sp. WCH70 | 69 | 98,3 | BAZO_10662 | |
| Periplasmic dissimilatory nitrate reduction | napG1 | contig124_32161_32757 | 597 | ZP_07949106.1 Eggerthella sp. 1_3_56FAA | 57 | 97,5 | BAZO_15064 |
| napA | contig124_32833_35370 | 2538 | YP_004710008.1 Eggerthella sp. YY7918 | 61 | 99,3 | BAZO_15069 | |
| napB | contig124_35383_35802 | 420 | YP_003303796.1 Sulfurospirillum deleyianum DSM 6946 | 28 | 80,9 | BAZO_15074 | |
| napD | contig124_35847_36104 | 258 | ZP_07949109.1 Eggerthella sp. 1_3_56FAA | 38 | 101,2 | BAZO_15079 | |
| napH | contig121_9516_10421 | 906 | ZP_07949110.1 Eggerthella sp. 1_3_56FAA | 40 | 101,3 | BAZO_14359 | |
| napG2 | contig124_36110_36679 | 570 | ZP_09635522 Desulfitobacterium dehalogenans ATCC 51507 | 36 | 101,6 | BAZO_15084 | |
| Assimilatory nitrate reduction | nasC | Not present | |||||
| Assimilatory nitrite reduction | nirB | Not present | |||||
| nirD | Not present | ||||||
| Nitrate transport | narK | contig69_15531_17030 | 1500 | ZP_08006147.1 Bacillus sp. 2_A_57_CT2 | 66 | 99,6 | BAZO_10682 |
| Nitrite transport | nirC1 | contig05_16758_15988 | 768 | ZP_09351718.1 Bacillus smithii 7_3_47FAA | 62 | BAZO_00505 | |
| nirC2 | contig107_<1_505 | 505 | YP_005876144.1 Lactococcus lactis subsp. cremoris A76 | 97 | BAZO_11644 | ||
| Ammonium transport | Not present | ||||||
| Dissimilatory nitrite reduction to ammonium | nrfH | contig37_9421_9936 | 516 | YP_002435907.1 Desulfovibrio vulgaris str. 'Miyazaki F' | 46 | 108,2 | BAZO_03250 |
| nrfA | contig37_9926_11395 | 1469 | ZP_09602448.1 Bacillus sp. 1NLA3E | 65 | 103,1 | BAZO_03255 | |
| Dissimilatory nitrite reduction to NO | nirK | contig38_30517_31578 | 1062 | ZP_08007035.1 Bacillus sp. 2_A_57_CT2 | 74 | 100,3 | BAZO_03565 |
| Dissimilatory quinol-dependent nitric oxide reduction | qnorB1 | contig04_38763_36502 | 2262 | ZP_09601292.1 Bacillus sp. 1NLA3E | 82 | 99,3 | BAZO_00190 |
| qnorB2 | contig48_12741_10378 | 2364 | ZP_09599319.1 Bacillus sp. 1NLA3E | 77 | 100,5 | BAZO_08916 | |
| norD/yojO | contig15_10303_8381 | 1923 | AEN89960.1 Bacillus megaterium WSH-002 | 58 | 100,5 | BAZO_01537 | |
| norQ/yojN | contig15_11194_10313 | 882 | YP_003597326.1 Bacillus megaterium DSM 319 | 73 | 96,7 | BAZO_01542 | |
| dnrN/norA | contig39_94731_95435 | 705 | ZP_08532570.1 Caldalkalibacillus thermarum TA2.A1 | 56 | 98,7 | BAZO_04395 | |
| Dissimilatory menaquinol/cyt c-dependent nitric oxide reduction (Type I) (qCuANOR, sNOR) | cbaC1 | contig42_104893_105033 | 138 | ZP_08784194.1 Ornithinibacillus scapharcae TW25 | 38 | 102,2 | – |
| cbaB1 | contig42_105051_105512 | 462 | YP_004095308.1 Bacillus cellulosilyticus DSM 2522 | 70 | 99,4 | BAZO_06394 | |
| cbaA1 | contig42_105527_107233 | 1707 | ZP_08532586.1 Caldalkalibacillus thermarum TA2.A1 | 71 | 100,9 | BAZO_06399 | |
| senC1 | contig42_107320_107955 | 636 | YP_004095310.1 Bacillus cellulosilyticus DSM | 62 | 100,5 | BAZO_06404 | |
| cbaD1 | contig42_107955_108527 | 570 | ZP_08008054.1 Bacillus sp. 2_A_57_CT2 | 69 | 102.1 | BAZO_06409 | |
| Putative dissimilatory menaquinol/cyt c-dependent nitric oxide reduction (Type II) | cbaC2 | contig40_60717_60133 | 582 | ZP_03146936.1 Geobacillus sp. G11MC16 | 44 | 101.0 | BAZO_04695 |
| senC2 | contig40_61349_60744 | 606 | ABO66889.1 Geobacillus thermodenitrificans NG80-2 | 44 | 95,7 | BAZO_04700 | |
| cbaA2 | contig40_62823_61342 | 1482 | ABO66888.1 Geobacillus thermodenitrificans NG80-2 | 65 | 102,5 | BAZO_04705 | |
| cbaB2 | contig40_63368_62850 | 519 | ZP_03146939.1 Geobacillus sp. G11MC16 | 58 | 96,6 | BAZO_04710 | |
| Dissimilatory nitrous oxide reduction | nosF0 | contig04_11581_12357 | 777 | ZP_07028021.1 Afipia sp. 1NLS2 | 52 | 101,2 | BAZO_00080 |
| nosC1 | contig04_19964_20425 | 459 | ZP_08982140.1 Desulfosporosinus meridiei DSM 13257 | 36 | 104,1 | BAZO_00115 | |
| nosZ1 | contig04_20450_22300 | 1851 | YP_00454079.1 Desulfotomaculum ruminis DSM 2154 | 66 | 97,3 | BAZO_00120 | |
| nosD1 | contig04_22977_24377 | 1401 | AEG59795.1 Desulfotomaculum ruminis DSM 2154 | 36 | 95,3 | BAZO_00130 | |
| nosL1 | contig04_24398_24883 | 485 | ZP_09074493.1 Paenibacillus elgii B69 | 36 | 83,8 | BAZO_00135 | |
| nosY1 | contig04_24858_25700 | 843 | YP_002456717.1 Desulfitobacterium hafniense DCB-2 | 45 | 101,1 | BAZO_00140 | |
| nosF1 | contig04_25697_26614 | 918 | ZP_09636992.1 Desulfitobacterium dehalogenans | 43 | 100,0 | BAZO_00145 | |
| nosC2 | contig41_6372_6797 | 423 | YP_001125844.1 Geobacillus thermodenitrificans NG80-2 | 48 | 93,4 | BAZO_05335 | |
| nosZ2 | contig41_6822_8702 | 1881 | YP_001125843.1 Geobacillus thermodenitrificans NG80-2 | 76 | 100,8 | BAZO_05340 | |
| nosD2 | contig41_9319_10644 | 1326 | YP_001125841.1 Geobacillus thermodenitrificans NG80-2 | 48 | 102,1 | BAZO_05350 | |
| nosC3 | contig147_57854_58288 | 432 | YP_001125844.1 Geobacillus thermodenitrificans NG80-2 | 51 | 95,4 | BAZO_18221 | |
| nosZ3 | contig147_58304_60181 | 1878 | YP_001125843.1 Geobacillus thermodenitrificans NG80-2 | 78 | 100,6 | BAZO_18226 | |
| nosD3 | contig147_60810_62126 | 1317 | YP_001125841.1 Geobacillus thermodenitrificans NG80-2 | 55 | 101,4 | BAZO_18236 | |
| nosY3 | contig147_62126_62923 | 798 | YP_001125840.1 Geobacillus thermodenitrificans NG80-2 | 48 | 100,4 | BAZO_18241 | |
| nosF3 | contig147_62920_63591 | 672 | YP_001125839.1 Geobacillus thermodenitrificans NG80-2 | 65 | 101,8 | BAZO_18246 | |
| nosD4 | contig147_26847_28190 | 1344 | YP_002315691.1 Anoxybacillus flavithermus DSM 21510 / WK1 | 37 | 100,7 | BAZO_18066 | |
| nosL4 | contig147_28190_28672 | 482 | YP_002315692.1 Anoxybacillus flavithermus DSM 21510 / WK1 | 45 | 108,6 | BAZO_18071 | |
| nosY4 | contig147_28713_29543 | 831 | YP_002315693.1 Anoxybacillus flavithermus DSM 21510 / WK1 | 43 | 103,0 | BAZO_18076 | |
| nosF4 | contig147_29548_30279 | 732 | YP_002315694.1 Anoxybacillus flavithermus DSM 21510 / WK1 | 53 | 104,7 | BAZO_18081 | |
| nosD5 | contig120_85053_86486 | 1434 | YP_003570259.1 Salinibacter ruber M8 | 28 | 102,8 | BAZO_14249 | |
| nosL5 | contig120_86476_86982 | 506 | ZP_08007037.1 Bacillus sp. 2_A_57_CT2 | 41 | 107,4 | BAZO_14254 | |
| nosY5 | contig120_87020_87985 | 966 | ZP_08007038.1 Bacillus sp. 2_A_57_CT2 | 52 | 115,4 | BAZO_14259 | |
| SenC/SCO1-type membrane-bound protein | senC3 | contig09_37800_37213 | 522 | YP_002316585.1 Anoxybacillus flavithermus WK1 | 55 | 88,8 | BAZO_01207 |
| senC4 | contig147_56968_57558 | 591 | ZP_01173458.1 Bacillus sp. NRRL B-14911 | 53 | 101,5 | BAZO_18216 | |
Genes are ordered according to their location in the operon when applicable. Gene size is given in absolute numbers as well as relative to that of best pBLAST hit. No ORF identifier is given when ORF was too small to be recognized in initial annotation.
Table 3.
Overview of gene inventory involved in nitrogen assimilation, denitrification and ammonification of B. bataviensis LMG 21833T.
| Function | Gene | Gene coordinates | Size (bp) | pBLAST best hit | ORF identifier | ||
|---|---|---|---|---|---|---|---|
| Genbank identifier | % ID | Rel. gene length | |||||
| Cytoplasmic dissimilatory nitrate reduction | narG | contig126_61985_58326a | 3660 | YP_001124778.1 Geobacillus thermodenitrificans NG80-2 | 73 | 99,4 | BABA_18597 |
| narH | contig126_58336_56810 | 1527 | YP_001124779.1 Geobacillus thermodenitrificans NG80-2 | 82 | 99,8 | BABA_18592 | |
| narJ | contig126_56817_56287 | 531 | YP_001124780.1 Geobacillus thermodenitrificans NG80-2 | 44 | 95,2 | BABA_18587 | |
| narI | contig126_56275_55547 | 729 | YP_001124781.1 Geobacillus thermodenitrificans NG80-2 | 66 | 105,7 | BABA_18582 | |
| Periplasmic dissimilatory nitrate reduction | napA | not present | |||||
| napB | not present | ||||||
| napD | not present | ||||||
| napH | contig114_85875_86333 | 459 | AEN91893.1 Bacillus megaterium WSH-002 | 41 | 90,0 | BABA_14547 | |
| napG | not present | ||||||
| Assimilatory nitrate reduction | nasC | contig121_7018_4829 | 2190 | YP_005311789.1 Paenibacillus mucilaginosus 3016 | 80 | 99,5 | BABA_16882 |
| Assimilatory nitrite reduction | nirB | contig05_23966_21540 | 2427 | ZP_09602179.1 Bacillus sp. 1NLA3E | 75 | 100,1 | BABA_00915 |
| nirD | contig05_21543_21223 | 321 | ZP_09602178.1 Bacillus sp. 1NLA3E | 60 | 99,1 | BABA_00910 | |
| Nitrate transport | narK1 | contig115_127063_125732 | 1332 | YP_003562112.1 Bacillus megaterium QM B1551 | 84 | 98,4 | BABA_15587 |
| narK2 | contig126_45914_44613 | 1302 | YP_004643043.1 Paenibacillus mucilaginosus KNP414 | 78 | 99,5 | BABA_18532 | |
| narK3 | contig126_47642_46140 | 1503 | YP_002774204.1 Brevibacillus brevis NBRC 100599 | 65 | 100,4 | BABA_18537 | |
| narK4 | contig135_17368_16046 | 1323 | ZP_08680547.1 Sporosarcina newyorkensis 2681 | 68 | 99,8 | BABA_20711 | |
| Nitrite transport | nirC | contig74_20117_19245 | 816 | ZP_09600582.1 Bacillus sp. 1NLA3E | 73 | 98,6 | BABA_12510 |
| Ammonium transport | amt | contig121_78501_79763 | 561 | ZP_10130407.1 Bacillus methanolicus PB1 | 87 | 97,4 | BABA_17227 |
| Dissimilatory nitrite reduction to ammonium | nrfA | contig123_39386_40837 | 1452 | ZP_09602448.1 Bacillus sp. 1NLA3E | 74 | 102,1 | BABA_17657 |
| nrfH | contig123_38870_39382 | 513 | ZP_09602447.1 Bacillus sp. 1NLA3E | 55 | 98,8 | BABA_17652 | |
| Dissimilatory nitrite reduction to NO | nirK | contig42_38340_38029; contig42_38025_37288 | 1053 | ZP_08007035.1 Bacillus sp. 2_A_57_CT2 | 71 | 99,4 | BABA_p06582 |
| Dissimilatory quinol-dependent nitric oxide reduction | qnorB1 | contig121_76393_77409 | 1017 | ZP_09601292.1 Bacillus sp. 1NLA3E | 79 | 44,7 | BABA_17212 |
| qnorB2 | contig55_76161_75334; contig55_77043_76291 | 1707 | ZP_09599319.1 Bacillus sp. 1NLA3E | 77 | 72,5 | BABA_p08977 | |
| norD | not present | ||||||
| norQ | not present | ||||||
| dnrN/norA | contig134_9390_8692 | 699 | ZP_09599666.1 Bacillus sp. 1NLA3E | 65 | 100,9 | BABA_20471 | |
| Dissimilatory menaquinol/cyt c-dependent nitric oxide reduction (Type I) (qCuANOR, sNOR) | cbaD1 | contig02_44305_43748 | 570 | ZP_08532584.1 Caldalkalibacillus thermarum TA2.A1 | 62 | 95,9 | BABA_00215 |
| senC1 | contig02_44940_44308 | 633 | YP_004095310.1 Bacillus cellulosilyticus DSM 2522 | 65 | 100,0 | BABA_00220 | |
| cbaA1 | contig02_46749_45067 | 1683 | ZP_08008056.1 Bacillus sp. 2_A_57_CT2 | 82 | 99,5 | BABA_00225 | |
| cbaB1 | contig02_47231_46764 | 468 | ZP_08008055.1 Bacillus sp. 2_A_57_CT2 | 66 | 74,3 | BABA_00230 | |
| cbaC1 | contig02_47384_47244 | 138 | YP_004095307.1 Bacillus cellulosilyticus DSM 2522 | 46 | 92,0 | – | |
| ctaB | contig02_48671_47742 | 927 | ZP_9916061.1 Lentibacillus sp. Grbi | 64 | 99,4 | BABA_00235 | |
| Putative dissimilatory menaquinol/cyt c-dependent nitric oxide reduction (Type II) | cbaB2 | contig126_34202_34621 | 420 | ZP_08007586.1 Bacillus sp. 2_A_57_CT2 | 66 | 76,5 | BABA_18497 |
| ccaA2 | contig126_34739_36205 | 1467 | ZP_08007585.1 Bacillus sp. 2_A_57_CT2 | 74 | 100,2 | BABA_18502 | |
| SenC/SCO1-type membrane-bound protein | senC2 | contig36_3182_4066 | 885 | YP_823233.1 bacterium Ellin514 | 28 | 96,1 | BABA_04124 |
| senC3 | contig55_18362_17802 | 561 | EIJ83190.1 Bacillus methanolicus MGA3 | 54 | 97,4 | BABA_08661 | |
| senC4 | contig126_63835_64437 | 603 | ZP_10131691.1 Bacillus methanolicus PB1 | 78 | 58,9 | BABA_18617 | |
Genes are ordered according to their location in the operon when applicable. Gene size is given in absolute numbers as well as relative to that of best pBLAST hit. No ORF identifier is given when ORF was too small to be recognized in initial annotation.
The startcodon of BABA_18597 as deposited at DDBJ/EMBL/GenBank, contains a false start codon with an extra 30-bp sequence at the 5′ terminus.
Location of proteins in cells was initially predicted with sequence-based tools as described by Emanuelsson et al. (2007): SignalP 4.0 (Petersen et al., 2011) was used for prediction of secretory signal proteins, TatP 1.0 for twin-arginine translocation signal proteins (Bendtsen et al., 2005), TMHMM 2.0 for transmembrane α-helices (Krogh et al., 2001), LipoP 1.0 (Rahman et al., 2008) for lipoprotein signal proteins, and SecretomeP 2.0 for signal peptide-less secretion (Bendtsen et al., 2004). If a transmembrane helix was predicted in the same region as a signal peptide, results were verified with Phobius (Käll et al., 2004). Whenever available, predictions were checked by comparison to homologous proteins with validated function and resolved crystal structure.
Alignment and phylogenetic analysis were performed with ClustalW 2.0 (Larkin et al., 2007) and MEGA 5.0 (Tamura et al., 2011).
Accession numbers
The Whole Genome Shotgun projects of Bacillus azotoformans LMG 9581T and Bacillus bataviensis LMG 21833T have been deposited at DDBJ/EMBL/GenBank under the accession numbers AJLR00000000 and AJLS00000000, respectively. The versions described in this paper are the first versions, AJLR01000000 and AJLS01000000.
Results and discussion
Bacillus azotoformans was originally isolated from garden soil and the organism has been known for decades to be a vigorous denitrifier (Pichinoty et al., 1983). Several biochemical studies investigating the enzymes involved in denitrification were conducted but no genomic data was available to aid interpretation of observations. Therefore, the genome of the type strain of the species was sequenced and the genes associated with dissimilatory nitrate reduction were analyzed. Unexpectedly, the genome contained the genes encoding both the complete denitrification pathway and nitrite ammonification (Table 2). No other organism is known to possess both pathways and also no experimental data has been reported that suggested their concurrence in one organism. This, however, may not be exceptional. Next to Bacillus azotoformans LMG 9581T we sequenced the genome of the type strain of B. bataviensis LMG 21833T, originally isolated from soil (Heyrman et al., 2004). This strain was arbitrarily chosen, but it also contained the gene inventory for both processes (Table 3). Hereafter, we will discuss denitrification and ammonification in B. azotoformans step by step, after which both processes will be described for B. bataviensis with an emphasis on the differences between both species.
Nitrate reduction
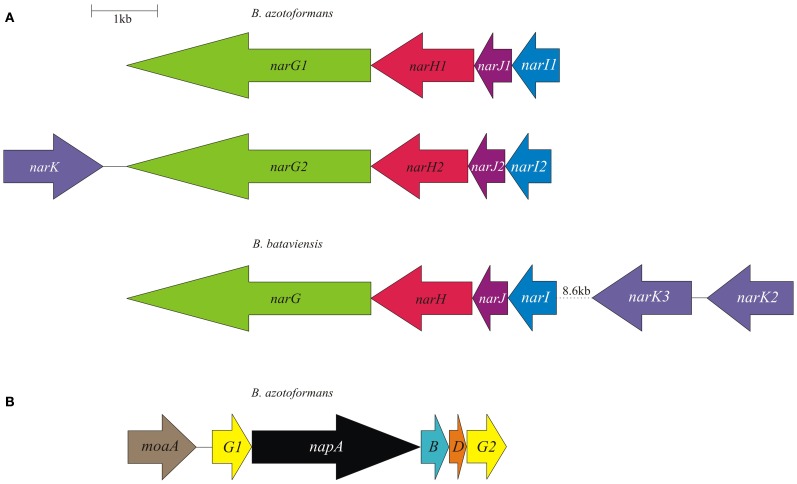
The reduction of nitrate to nitrite in Bacteria is catalyzed by three different types of enzymes, that all bind a molybdenum bis molybdopterin guanine dinucleotide (Mo-bis-MGD) cofactor at the catalytic subunit together with a 4Fe-4S cluster for electron transfer (Rothery et al., 2008). Two of these nitrate reductases (NARs) are involved in respiration, the cytoplasmic Nar, and the periplasmic Nap. The third one, Nas, acts in nitrogen assimilation and is localized in the cytoplasm. In the genome of B. azotoformans a gene coding for Nas is absent, but the organism contains the inventory for two functional Nar systems as well as one Nap protein complex (Figure 1). As yet, the presence of a periplasmic NAR has not been reported for a Gram-positive bacterium.
Figure 1.
Physical map of the B. azotoformans LMG 9581T and B. bataviensis LMG 21833Tnar (A) and nap (B) gene clusters. Arrows show the direction of transcription. Open reading frames are drawn to scale. Homologous genes are shown in identical colors.
The Nar enzyme is composed of three subunits (NarGHI). In the genome, the genes coding for the three subunits are usually linked with the narJ gene encoding a maturation protein. NarGHI is well characterized by the resolution of the atomic structure of the enzyme from E. coli (Bertero et al., 2003, 2005). In the complex, NarG is the catalytic subunit housing the Mo-bis-MGD cofactor and a 4Fe-4S cluster. In Proteobacteria, NarG and NarH are localized in the cytoplasm, despite the presence in the former of a TAT signal for protein export, but the TAT sequence is non-functional (Ize et al., 2009). NarH binds three 4Fe-4S clusters and one 3Fe-4S cluster in tandem and mediates electron transfer between NarG and NarI. NarI is a membrane protein with five transmembrane helices (TMHs) that sandwich two cytochrome b molecules, one near the cytoplasmic and the other one near the periplasmic face. In addition, NarI interacts with the electron donor for nitrate reduction, menaquinol, notably at the periplasmic side (Bertero et al., 2005). The complex is organized such that the uptake of two protons in the cytoplasm during nitrate reduction is associated with the release of two protons at the periplasm upon quinol oxidation, thus contributing to the building of proton-motive force (pmf) by a redox loop mechanism (Rothery et al., 2008; Simon et al., 2008).
As mentioned, the genome of B. azotoformans codes for two different Nar systems (NarG1H1J1I1, BAZO_08891-08876; NarG2H2J2I2, BAZO_10677-10662) (Figure 1A). In both NarGs (with 74.9% aa sequence identity), all structurally relevant amino acids are fully conserved with respect to those in E. coli NarG (data not shown). Similarly, all structurally and functionally relevant amino acids are conserved in both NarH and NarI copies with respect to those of the E. coli enzyme. These observations indicate that the two B. azotoformans Nars are functional. NarG1 and NarG2 are devoid of a TAT signal sequence, indicative of their cytoplasmic localization. In agreement herewith, the narG2H2J2I2 gene cluster is linked to a gene coding for a protein (BAZO_10682) of the NarK family of nitrate transporters (Moir and Wood, 2001). More specifically, the pertinent protein is a member of the NarK2 subfamily of nitrate–nitrite antiporters. In other parts of the genome two genes (BAZO_00505, nirC1; BAZO_11644, nirC2) are found coding for members of the NirC/FocA Major Facilitator Superfamily (MFS). Proteins belonging to this superfamily facilitate the translocation across the cytoplasmic membrane of nitrite (NirC), formate (FocA), or even both (Moir and Wood, 2001; Falke et al., 2010). BAZO_11644 shows 97% sequence identity to fdhC gene product of Lactococcus lactis subsp. cremoris (YP_001032837). FdhC is a formate transporter associated with the formate dehydrogenase, which highly suggests a same function for BAZO_11644. A BlastP search indicates BAZO_00505 to be most closely related to putative formate/nitrite transporters from other Bacillus species including B. subtilis (NP_390598, 61% aa identity). As compared to FocA and NirC proteins of which the function has been established, sequence similarity of BAZO_00505 is higher with respect to the NirC members, which might imply that this B. azotoformans protein favors nitrite export.
The presence of a periplasmic NAR system comes as a surprise. Nature has invented several variations on the Nap theme (Simon and Klotz, 2012) and B. azotoformans adds one more. First of all, the nap gene cluster organization (napG1ABDG2-moaA BAZO_15084-15059) is unusual with two copies of the napG gene (napG1, BAZO_15064; napG2, BAZO_15084) and the presence of a gene (moaA; BAZO_15069) coding for one of the enzymes involved in Mo-bis-MGD biosynthesis (Table 2; Figure 1B). The nap gene cluster is devoid of genes coding for NapH, which is found in another part of the genome (BAZO_14359), whereas coding sequences for the cytoplasmic maturation factors NapF, a 4Fe-4S protein, and NapL are completely absent. Furthermore, no gene accounting for quinol-oxidizing membrane-bound NapC is found on the genome. These genes are also missing in Campylobacter jejuni, where nrfH seems to replace napC (Pittman et al., 2007). Still, the available genes would suffice for an operational Nap system (González et al., 2006). In this system, NapA represents the catalytic subunit harboring a Mo-bis-MGD cofactor and a 4Fe-4S cluster like in NarG. NapD is involved in the posttranslational assembly of NapA. NapA receives its electrons for nitrate reduction from the companion diheme c protein NapB. NapAB are particularly well characterized by resolution of the crystal structures from at least four different species (Arnoux et al., 2003; Jepson et al., 2007; Kern and Simon, 2008; Najmudin et al., 2008; Coelho et al., 2011). The comparison of the amino acid sequence of B. azotoformans NapA with those of known atomic structures reveals the conservation of all amino acids related with the binding of the molybdopterin cofactor and of the iron sulfur cluster. However, NapB contains some specific insertions and deletions with respect to NapB from Rhodobacter sphaeroides (Arnoux et al., 2003) and Cupriavidus necator (Coelho et al., 2011), possibly indicating somewhat different interactions with other subunits within the enzyme complex, but with conservation of the two heme c binding sites (data not shown). B. azotoformans NapA contains a distinct TAT signal, whereas NapB has an N-terminal signal sequence, indicating that both proteins are exported to the periplasm, as expected. NapH and one or both NapG proteins most likely constitute a membrane-bound electron-transfer module, as has been established for other organisms (Richardson et al., 2001; Simon et al., 2003; Kern and Simon, 2008). NapH is a membrane-bound enzyme (with four TMHs) that specifically oxidizes menaquinol while NapG is a periplasmic adapter protein that is thought to deliver electrons from menaquinol oxidation (Kern and Simon, 2008). In agreement herewith, NapH from B. azotoformans is predicted to contain 4 TMHs, whereas two Cx3CP motifs and the cysteines binding two 4Fe-4S clusters in the periplasmic domain are fully conserved with respect to the NapH proteins from E. coli and P. denitrificans. Both NapG's are shorter than NapG from W. succinogenes (Kern and Simon, 2008), E. coli (YP_002403484.1), or Campylobacter upsaliensis (ZP_00370550.1) (198 for NapG1 and 189 for NapG2 vs 232-266 aa), lacking a C-terminal part. However, they still contain a 4Fe-4S binding motif (Kern and Simon, 2008) suggesting functionality of both copies. As expected for the periplasmic localization of this iron-sulfur protein, NapG2 (BAZO_15084) has a clear TAT signal sequence, unlike NapG1 (BAZO_15064). Its absence would localize NapG1 in the cytoplasm. It is conceivable that the protein substitutes at this side for NapF found in other organisms. By the presence of the NapAB and NapGH modules, possibly assembled as one membrane-bound complex, B. azotoformans has the disposal of a second quinol-dependent NAR system. Presently, it is not understood why B. azotoformans as well as many other microorganisms harbor two dissimilatory reductases. By its topology and architecture, Nap is not expected to contribute to the pmf. It has been suggested that a main function of Nap would be nitrate-dependent regeneration of quinone from quinol that is produced in concert with the oxidation of the (organic) substrates (Richardson, 2000). Alternatively, the Nap module and the dissimilatory nitrite reductase (NrfHA) discussed next, may provide B. azotoformans with a high-affinity system for N assimilation under low nitrate conditions (Pittman et al., 2007; Kim et al., 2012).
Nitrite reduction
In general, the one-electron reduction of nitrite to NO is catalyzed by two genetically and biochemically distinct nitrite reductases, the cytochrome cd1 protein (NirS) and copper-containing NirK. B. azotoformans contains the latter representative (Table 2). DNRA bacteria possess NrfA as their key enzyme. Assisted by its redox partner NrfH, NrfA catalyzes the six-electron reduction of nitrite to ammonium (Simon, 2002; Einsle, 2011; Simon and Klotz, 2012). NrfHA can also be involved in stress response to nitric oxide, hydroxylamine, and hydrogen peroxide (Kern et al., 2011). B. azotoformans contains both NrfA and NrfH. In a similar fashion, the NirB and NirD proteins catalyze the NAD(P)H-dependent reduction of nitrite to ammonium for nitrogen assimilation (Luque-Almagro et al., 2011), but homologs of these cannot be detected in the genome of B. azotoformans.
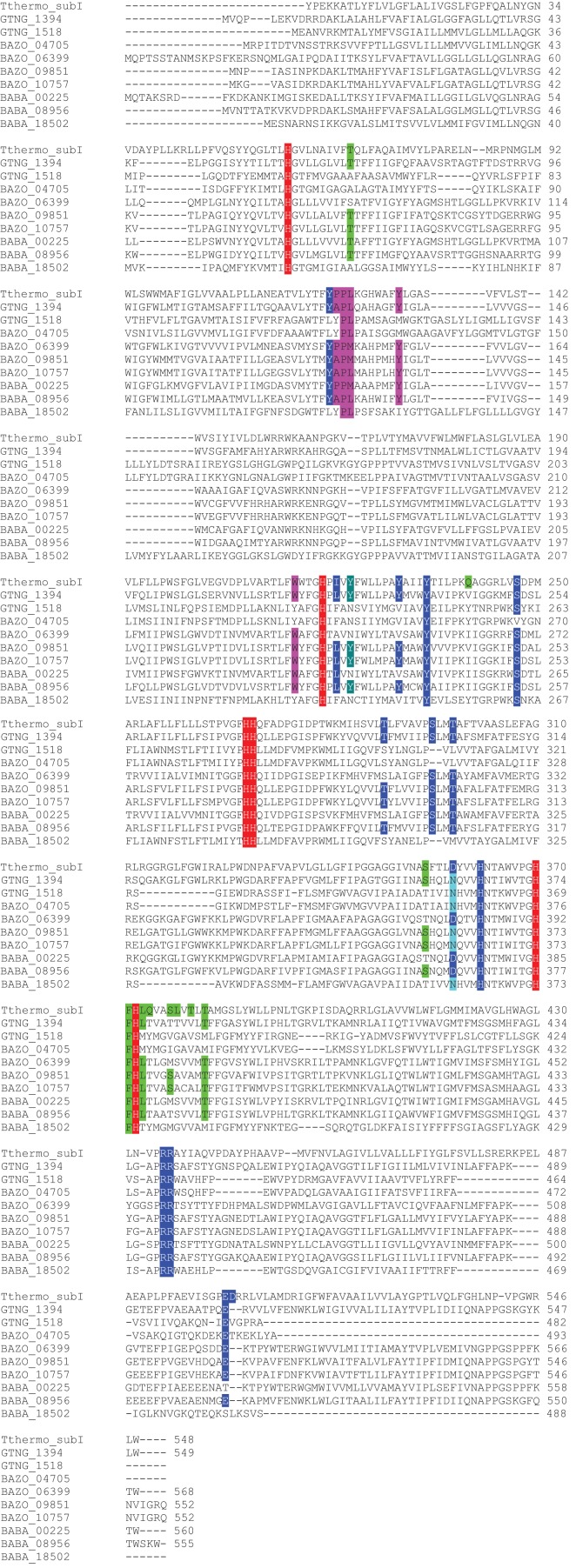
NirK is encoded by BAZO_03565 and the protein shows a high degree of identity with known NirKs of which crystal structures are available (Ellis et al., 2001; Tocheva et al., 2004; Jacobson et al., 2005; Fukuda et al., 2011). Among the structurally well characterized proteins, sequence identity (80%) is highest with NirK (GK0767; YP_146620) from Gram-positive Geobacillus kaustophilus (Fukuda et al., 2011). BAZO_03565 shares three characteristic loop regions, with deletions in the “linker loop” and “tower loop,” as well as an “extra loop.” These features are typical for the NirK2 family (Boulanger and Murphy, 2002). The tower loop was suggested to “facilitate a more intimate interaction with the lipid membrane” (Boulanger and Murphy, 2002). BAZO_03565 is preceded by a Sec signal for protein export, indicative of a periplasmic localization of the processed protein. Such localization holds for all NirKs known to date. Quite interestingly, the LipoP program predicts the B. azotoformans protein to be a lipoprotein. Such covalent binding confirms results by Suharti and de Vries (2005), who found that nitrite reductase activity of B. azotoformans NCCB 10003 was exclusively associated with the membrane fraction. We may note that NirK from randomly chosen Gram-positive bacteria [Geobacillus thermodenitrificans NG80-2 (GTNG_0650), Geobacillus kaustophilus HTA-426 (GK0767), and Geobacillus thermoglucosidasius C56-YS93 (Geoth_3084)] are also identified as putative lipoproteins. Suharti and de Vries (2005) proposed NirK from B. azotoformans to be a dual-function enzyme: it could use both menaquinol and reduced cytochrome c as electron donors. However, it remained unclear whether menaquinol acted in a direct way, via a menaquinol oxidizing enzyme, or indirectly via the action of menaquinol: cytochrome c oxidase (b6f, complex III; see below). Unfortunately, BAZO_03565 takes an isolated position within the genome and the gene context does not give a clue about its redox partners. This is in contrast with observations in the genomes of closely related denitrifiers Geobacillus thermodenitrificans (Feng et al., 2007) and G. kaustophilus, in which the nirK is imbedded in a gene cluster with genes encoding NAR and associated proteins, NOR genes, a nitrate transporter, and several regulatory genes.
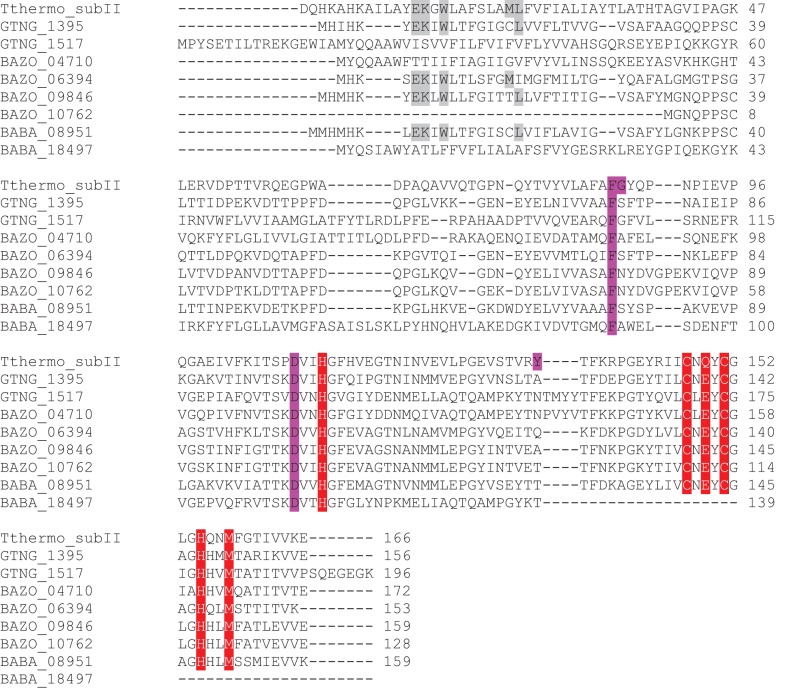
Ammonium-forming nitrite-reducing NrfHA are encoded by BAZO_03250-03255 (Table 2). The nrfHA operon organization is similar to that described for several bacteria capable of DNRA such as Campylobacter jejuni, Desulfovibrio vulgaris, Geobacter sulfurreducens (Simon, 2002), and Desulfitobacterium hafniense DCB-2 (Kim et al., 2012). NrfA is a periplasmic pentaheme c protein in which four cytochromes c are involved in electron transfer while the fifth one has a catalytic function. Their presence in the amino acid sequence is characterized by CXXCH (electron transfer) and CXXCK (catalysis) motifs. BAZO_03255, indeed, shows these motifs and it displays also all other sequence features of NrfA proteins of which crystal structures are available (Bamford et al., 2002; Cunha et al., 2003; Rodrigues et al., 2006a) (Figure A1). Consistent with the localization of all known NrfA's, BAZO_03255 contains an N-terminal signal sequence for protein export. Known NrfA's are soluble proteins, but the LipoP program predicts BAZO_03255 to be a lipoprotein. As far as could be checked, the lipoprotein nature might also hold for NrfA's from other Firmicutes, namely Bacillus selenitireducens MLS10 (Bsel_1305) and Desulfitobacterium hafniense DCB-2 (Dhaf_4234). NrfH is a membrane-bound tetraheme cytochrome c belonging to the NapC/NirT family of menaquinol oxidases (Simon et al., 2000). The comparison with NrfH from the Deltaproteobacterium Desulfovibrio vulgaris, of which the structure has been resolved (Rodrigues et al., 2006b, 2008), establishes the conservation in BAZO_03250 of all relevant amino acids implemented with structuring the N-terminal TMH, quinol binding, and ligation of the four hemes c. Taken together, our observations indicate that both NrfA and NrfH are functional proteins in B. azotoformans. As a membrane-bound complex, NrfAH would facilitate in B. azotoformans menaquinol oxidation coupled with the reduction of nitrite making ammonium. One may note that both proteins are rich in heme c molecules. In this respect it is interesting that nrfAH is linked to a gene cluster (ccmEFHABC; BAZO_03265-BAZO_03300) encoding six out of eight proteins of the System I cytochrome c biogenesis machinery (Kranz et al., 2009).
It now appears that B. azotoformans has the disposal of two parallel pathways for nitrite reduction enabling a life style as a denitrifier and as a DNRA bacterium. It has been argued that denitrification is more favorable under carbon limitation, whereas as nitrate/nitrite ammonification is more attractive under electron acceptor limitation (Tiedje et al., 1982; Tiedje, 1988). By the presence of the two, B. azotoformans may benefit from the best of two worlds. Still, the metabolism leaves us with one puzzle. One may note that the organism is devoid of the assimilatory nitrate and nitrite reductases. This would make B. azotoformans dependent on ammonium as the nitrogen source. Obviously, ammonium can be produced by the action of the Nar, Nap, and Nrf systems and even more so, since nrfA in E. coli is known to be expressed under low nitrate conditions, while NirB operates at high nitrate concentrations (Wang et al., 2000). So, B. azotoformans might have adapted to low nitrate/nitrite conditions. The problem, however, is that no gene is found in the genome of B. azotoformans coding for a known (AmtB-type) transporter to take periplasmically produced ammonium into the cell.
Nitric oxide reduction to nitrous oxide
The product of nitrite reductase NirK, NO, is a very reactive and toxic free radical compound and a range of known or predicted but still to be validated enzymes exists to convert it into N2O (Richardson, 2000; de Vries and Schröder, 2002; Tavares et al., 2006; Hemp and Gennis, 2008; Watmough et al., 2009; Kraft et al., 2011; Martínez-Espinosa et al., 2011; Stein, 2011). NORs fall into two different classes: (1) NorVW flavorubredoxin that is employed by many organisms for NO detoxification in response to nitrosative stress from the environment and (2) NORs belonging to the heme-copper oxidase (HCO) superfamily. The enzymes combine two NO molecules to make N2O by the input of two electrons. Presently, three types of NORs have been studied in some detail: (1) cNOR (or NorBC) that uses reduced cytochrome c as the reductant, (2) quinol-dependent qNOR (qNorB), and (3) qCuANOR that takes both as electron donors. For cNOR and qNOR atomic structures are available (Hino et al., 2012; Matsumoto et al., 2012). Presently, only one qCuANOR has been purified, notably from Bacillus azotoformans NCCB 10003 that is closely related to the strain (LMG 9581T) discussed here (Suharti et al., 2001, 2004; Lu et al., 2004). The enzyme was reported to be composed of two subunits, a small CuA-type subunit and a large one with two heme b molecules. Although quite well characterized, the encoding genes remained elusive.
Besides NORs, the HCO superfamily comprises a broad variety of terminal oxidases (Hemp and Gennis, 2008; Sousa et al., 2012). The common property is a membrane-bound catalytic subunit with 12–14 TMHs that bind a heme b (or a) for electron transfer and a second heme (b3, a3, or o3) constituting the catalytic center together with an iron (FeB in NOR) or a copper ion (CuB in oxidases). Both FeB and CuB are ligated by three conserved histidines. Next, two histidines coordinate the electron-transferring heme, whereas one more histidine serves as the proximal ligand to the catalytic heme. This histidine sextet is a signature for HCOs. Oxidases are distinguished by the presence of a tyrosine near the catalytic side that makes a covalent bond with one of the histidines binding CuB. In NORs the pertinent tyrosine is replaced by a glutamate, glutamine, aspartate, or asparagine (Hemp and Gennis, 2008). Moreover, in certain NOR types one of the CuB- or FeB-ligating histidines is substituted by an aspartate (Hemp and Gennis, 2008; Sievert et al., 2008). Besides the catalytic subunit, heme copper oxidases may contain one or more additional subunits for electron transfer as well as membrane-spanning polypeptides for structural integrity. Electron transfer subunits have heme c or copper (CuA)-containing cupredoxins as redox components. In addition, heme copper oxidases are distinguished on the basis of their use of the electron donors for O2 or NO reduction, which can be either reduced cyt c or quinol.
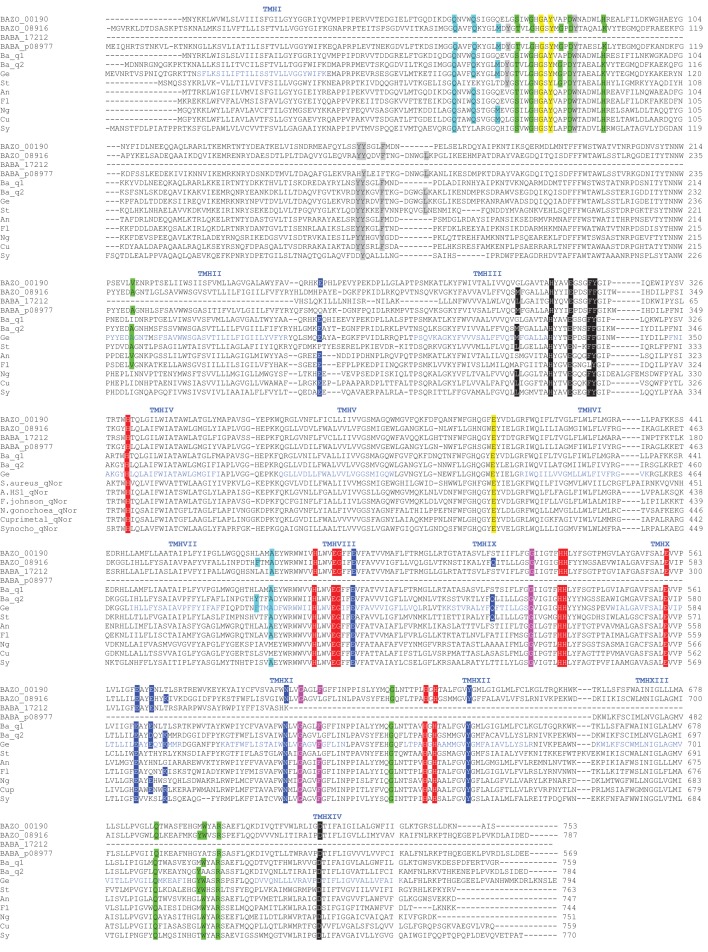
While cNORs are absent in the genome of B. azotoformans, we could identify two genes coding for qNORs, BAZO_00190, and BAZO_08916. The preference for quinol-dependent NOR seems to be a common property of Gram-positive microorganisms. Although BAZO_00190 and BAZO_08916 share only 38% sequence identity, the comparison of their amino acid sequences with those of which the functions have been established, including qNOR from Geobacillus stearothermophilus having a known crystal structure (Matsumoto et al., 2012) (Figure A2), suggests functionality of both B. azotoformans proteins. Briefly, BAZO_00190 and BAZO_08916 are composed of one subunit in which 13 TMHs surround the catalytic module. A 14th (N-terminal) TMH precedes a soluble domain facing the periplasm with a heme c fold, but heme c itself is absent. In both proteins amino acids are conserved binding both heme b molecules, non-heme iron, a specific calcium atom, the quinol substrate as well as the amino acids lining a proposed water/proton channel down from cytoplasm to the catalytic side, hydrophobic amino acids along two other putative water channels, and aromatic amino acids that stabilize that heme c fold of the soluble domain (Figure A2).
cNOR (NorBC) activity depends on ancillary proteins (NorDEFQ) that tend to be encoded on the same operon as the structural proteins (Zumft, 2005b), but qNOR can do without these. Nevertheless, genes norDQ (BAZO_01537; BAZO_01542) are present in the genome, although not directly linked to BAZO_00190 and BAZO_08916. This is also the case in genomes of other Gram-positives harboring the qnorB gene, including G. thermodenitrificans NG80-2, G. kaustophilus HTA-426, Anoxybacillus flavithermus WK1, Bacillus licheniformis ATCC 14580, Oceanobacillus iheyensis HTE831, making it a common observation among Gram-positives. However, it is difficult to speculate on their necessity for qNOR functionality in these organisms, as the precise functions of NorD and NorQ remain to be established. In addition, the genome of B. azotoformans contains the norA gene (BAZO_04395), also termed dnrA or scdA), which codes for a putative iron-sulfur cluster repair protein in response to NO damage (Overton et al., 2008).
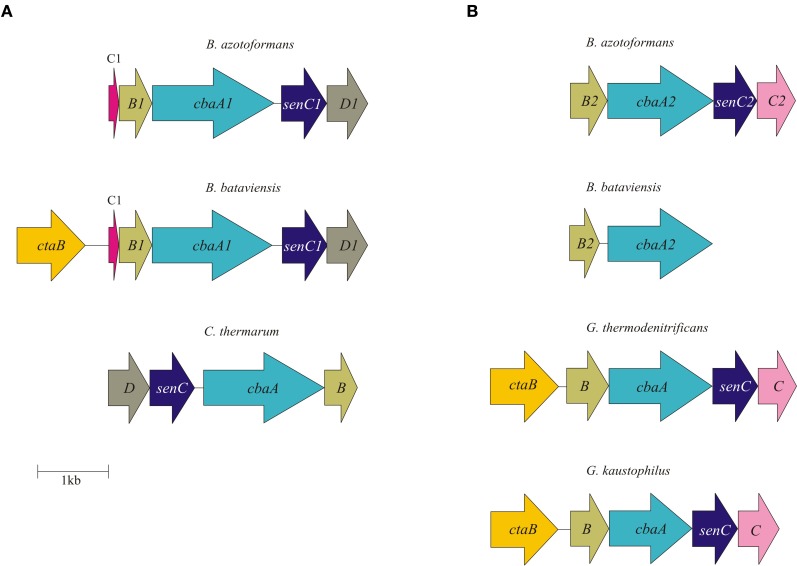
Besides the two qNORs, no less than four gene clusters are found in the genome of B. azotoformans encoding members of the HCO superfamily that share significant sequence homology with cytochrome c-dependent ba3 oxidase (CbaAB) from Thermus thermophilus (Figures A3, A4). This is a terminal oxidase that is structurally and functionally quite well investigated [see amongst others: Soulimane et al. (2000); Fee et al. (2008); Smirnova et al. (2008); Liu et al. (2012); von Ballmoos et al. (2012)]. ba3-type oxidase receives its electrons for O2 reduction from cyt c. The electrons are transferred via CuA in subunit II (CbaB), and cyt b in subunit I (CbaA) to the a3-CuA catalytic center in this subunit. In agreement herewith, all amino acids that had been structurally assigned to CuA binding and to electron transfer were conserved in the B. azotoformans subunits II (Figure A4). Importantly, the N-terminal sequence of BAZO_06394 (MHKSEKIWLTLSFGMIMGFM) is identical to the one of subunit II of qCuANOR published by Suharti et al. (2001). These authors (Suharti et al., 2004) also presented the N-terminal sequence of the large subunit (MTKKNTQEVVKEGREGIGTFIGVGIVGAV), but this was not found in the corresponding subunit I (BAZO_06399). Rather, the almost identical sequence—MATTKNTQEVVKEGREGIGTFIGVGIVGAV—was retrieved in the adjacent gene (contig42_104893_105033) (Figure 2A). This gene encodes a small (46 amino acids) membrane-bound peptide. Hence, it is very well conceivable that this peptide formed part of the enzyme preparation purified by Suharti et al. (2001, 2004). The comparison of the amino acid sequence of subunit I (BAZO_06399) with that of ba3 from T. thermophilus revealed that the tyrosine (Y223, T. thermophilus numbering in Figure A3) covalently binding a CuB-associated histidine (H219) was substituted by an asparagine in the B. azotoformans protein, in agreement with predictions made by Hemp and Gennis (2008) for NOR functionality (Figure A3). These findings identify BAZO_06394-BAZO_06399 as the dual-function quinol and cyt c-dependent qCuANOR described by Suharti et al. (2001, 2004). We may note that the genes coding for these three subunits are linked to two other ones, one (BAZO_06404) coding for a SenC/SCO1-type membrane-bound protein that has been implemented with the insertion of copper (CuA) into subunit I, and BAZO_06409 that is predicted to comprise six TMHs (Figure 2A). In protein databases, homologs of the latter are found being annotated as cyt c oxidase-associated membrane proteins. Furthermore, the presence of BAZO_06394-06399 qCuANOR is not restricted to B. azotoformans and B. bataviensis (see below), but close homologs sharing the same sequential features and the same cluster organization are found in various other Gram-positive bacteria, including Caldalkalibacillus thermarum TA2, but also in Nitrosomonas eutropha and other nitrifiers (Figure 2A and data not shown). In N. eutropha the particular NOR was designated sNOR (Stein et al., 2007). Transcriptome analysis suggested sNOR as a suitable candidate for aerobic NO reduction, next to NorBC (Cho et al., 2006).
Figure 2.
Physical map of qCuANOR type I (A) and qCuANOR type II (B) gene cluster in B. azotoformans LMG 9581T, B. bataviensis LMG 21833T, Caldalkalibacillus thermarum TA2.A1, Geobacillus thermodenitrificans NG80-2, and G. kaustophilus HTA426. Arrows show the direction of transcription. Open reading frames are drawn to scale. Homologous genes are shown in identical colors.
B. azotoformans likely contains one more novel type of qCuANOR (type II) of which the subunits I and II are encoded by BAZO_04705 and BAZO_04710, respectively (Figure 2B). Again, subunit II is of the CuA-type and in subunit I the cross-linking tyrosine is substituted by an asparagine (Figures A3, A4). At the C-terminal part, the large subunit I is substantially shorter than in the previous case and in T. thermophilus ba3 oxidase, yet with the conservation of all general HCO characteristics. Being shorter, subunit I is predicted to be structured by 12 TMHs, instead of 13. In contrast, subunit II contains two N-terminal TMHs, one more than in BAZO_06394, and subunit II of T. thermophilus ba3 oxidase. As above, BAZO_04705-04710 are linked to genes encoding a SenC/SCO1 paralog (BAZO_04700) and a polypeptide (BAZO_04695) with 4–5 predicted TMHs that might constitute a third subunit. Again, the presence of the BAZO_04695-4710 is not restricted to B. azotoformans. Close homologs, both at the sequence and cluster organization levels, are found in other Gram-positive bacteria, including G. thermodenitrificans NG80-2 (GTNT_1517-1520) and G. kaustophilus HTA-426 (GK1670-1673) (Figure 2B and data not shown). Quite remarkably, the gene clusters in both Geobacillus species are associated with ones coding for heme o oxygenase (GTNG_1516, GK1690). This enzyme converts heme o into heme a. This might imply that that the Geobacillus enzymes would contain heme a instead of heme b, thereby inventing one more variation on the NOR theme.
The two remaining ba3 oxidase-related HCO members (BAZO_09851-09846, BAZO_10757-10762) are highly related to one and another. Their large (BAZO_09851, BAZO_10757) and small subunits (BAZO_09846, BAZO_10762) are 83 and 90% identical, respectively. However lacking the first 31 N-terminal amino acids, BAZO_10762 is markedly shorter than BAZO_09846 (128 vs 159 aa). Sequence analyses suggest these HCOs to be genuine (O2-reducing) oxidases, having a tyrosine for histidine cross linking. At the sequence levels, the large (I) and small (II) subunits of both B. azotoformans proteins are 40 and 45% identical to the ones of ba3 oxidase from T. thermophilus (Figures A3, A4). The differences in amino acid sequences might suggest that the large subunits could bind other hemes and/or differ in the nature of the proton channels. Indeed, sequence identities of BAZO_09851 and BAZO_10757 are significantly higher (67%) with respect to the large subunit of bo3 cytochrome c oxidase isolated from Geobacillus (previously Bacillus) stearothermophilus having heme o3 at the catalytic side (Sakamoto et al., 1997; Nikaido et al., 1998). The latter protein catalyzes O2 reduction with reduced cyt c551, a lipoprotein, as the specific reductant (Km = 0.15 μM). The bo3 cytochrome c oxidase actively pumps protons across the cell membrane during the reaction. It is not known if quinol can substitute for cyt c551 as electron donor. Also here, the genes coding for both B. azotoformans oxidases are linked to ones (BAZO_09841, BAZO_10767) encoding small membrane-bound polypeptides (46 amino acids) possibly serving as a third subunit. We may note that close homologs to BAZO_09851-09841 and BAZO_10757-10767 are present in G. thermodenitrificans NG80-2 (GTNT_1394-1396) and G. kaustophilus HTA-426 (GK1546-1548), as well as in many other Bacillus-related species.
For oxygen respiration, model organism Bacillus subtilis strain 168 has the disposal of four different oxidases, cyt c551-dependent caa3 oxidase, which is preferentially used at high oxygen concentrations, and three types that rely on quinol: aa3-600 oxidase, bd oxidase, and a bb' oxidase (Lauraeus et al., 1991; Azarkina et al., 1999; Winstedt and von Wachenfeldt, 2000). Apart from the b(a/o)3-type oxidase discussed before, which seems to be absent in B. subtilis, the genome of B. azotoformans has the inventory of two of these. Genes coding for bd oxidase are not found in B. azotoformans, unlike many other Bacillus species including B. bataviensis discussed below. Four-subunit quinol-dependent aa3-600 oxidase encoded by the qoxBACD genes are represented by the gene products of BAZO_10131-10146 (subunit II, BAZO_10131; subunit I, BAZO_10136; subunit III, BAZO_10141; subunit IV, BAZO_10146). Amino acid sequences of the B. azotoformans gene products readily compare those of established functions. Cyt c-dependent caa3 oxidase is also composed of four subunits (CtaCDEF). The structural genes are usually clustered with genes coding for accessory proteins, including CtaB and CtaA that catalyze the subsequent conversion of heme b into heme o and heme a, and CtaG, a cytochrome c oxidase assembly factor. The gene cluster organization (ctaABCDEFG) is conserved in B. azotoformans (BAZO_04065-BAZO_04035). Furthermore, the individual gene products are well conserved with respect to the ones of which the function has been established. These observations support functionality of both quinol-dependent aa3-600 (QoxBACD) and of cyt c-dependent caa3 oxidase (CtaCDEF) in B. azotoformans.
By exploiting variations on the HCO theme, B. azotoformans harbors a repertoire of four different NO reductases and four different oxidases that utilize reduced cyt c and/or quinol as reductants. Obviously, this repertoire and the branching in electron transfer may lend the organism the metabolic versatility to cope with large changes in respiratory conditions in the environment.
Nitrous oxide reduction
The last step in the denitrification pathway to dinitrogen gas is the two-electron reduction of N2O to produce, with the input of two protons, N2 and water. While the previous steps can be catalyzed by different types of enzymes, only one exists for N2O reduction, nitrous oxide reductase (NOS, N2OR, NosZ) [see for reviews Zumft (1997, 2005a,b); Zumft and Kroneck (2007)]. NOS is a homodimeric protein with two multinuclear copper centers. In all organisms investigated thus far, it is located in the periplasm and in Gram-negative species it is a soluble protein. For its assembly, NOS depends on a number of accessory proteins (NosADLYF). Assembly is thought to take place in the cytoplasm after which NOS is exported using the TAT translocon. In Gram-negative bacteria, the TAT signal sequence is unusual long counting approximately fifty amino acids.
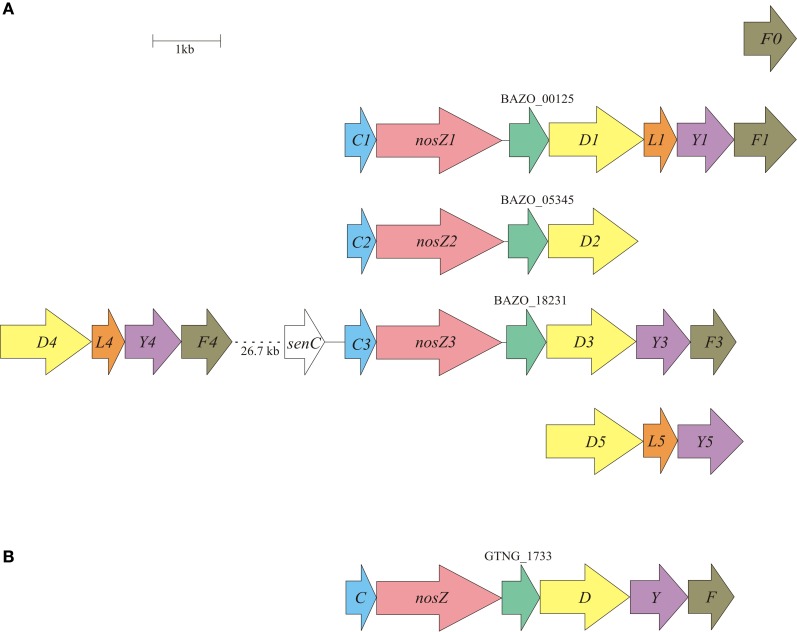
The genome of B. azotoformans encodes no less than five nos gene clusters, with three clusters including a nosZ gene, and one orphan nosF (Table 2, Figure 3A). Gene arrangements differ in every cluster (Figure 3A), but they are most similar to the atypical nos gene cluster (nosCZ-ORF-nosDYF-ORF) found in the three other Gram-positives available in the databases with nos genes: Geobacillus thermodenitrificans NG80-2 (Figure 3B), Desulfitobacterium hafniense T51 (Liu et al., 2008), and Desulfotomaculum ruminis DSM 2154. Each nosZ gene is consistently preceded by nosC, coding for a cytochrome c, which is predicted (LipoP) to be a lipoprotein with the heme c part facing the periplasm. NosZ is followed by a conserved hypothetical open reading frame encoding a membrane protein with four predicted TMHs. Interestingly, the latter is also the case in W. succinogenes (Ws0918) (Simon et al., 2004). In W. succinogenes, the two copies of nosC are located downstream of nosZ and are flanked by nosG and nosH genes, which are absent in B. azotoformans. NosGH represent a membrane-bound quinone-oxidizing electron transfer module, resembling NapGH in the periplasmic NAR (Nap) system described above. B. azotoformans also lacks a nosA gene that codes for a periplasmic accessory protein involved in Cu binding. However, we note the presence of one of the four senC/SCO1 paralogs upstream of nosC3. As mentioned before, SenC has been implemented with the insertion of copper (CuA); genes coding for the other three paralogs are associated with the alternative NORs. NosL, which is present in three copies in the nos gene clusters (Figure 3A), is a lipoprotein that stoichiometrically binds Cu. This property suggests NosL to be a copper chaperone for metal-center assembly (Zumft, 2005a). NosDYF represent a Cu-ABC transport system with a periplasmic Cu-binding protein (NosD), a membrane-bound permease (NosY), and a cytoplasmic ATPase (NosF) that provides the driving force for Cu translocation (Zumft, 2005a). As can be seen from Figure 3A, nosDYF come in different arrangements in the genome of B. azotoformans, either with or not associated with NosZ. Moreover, sequence analysis reveals marked differences among the individual gene products. All five nosD gene products have a putative transmembrane helix at the C-terminus, but only three of these have a predicted Sec-signal (NosD1, BAZO_00130; NosD4, BAZO_18066; NosD5, BAZO_14249), as expected for periplasmic proteins. NosY3 contains a seventh TMH, one more than usual, while significant insertions without any known domain are found in NosY4 (39 AA at the N-terminus) and NosF1 (49 AA at the C-terminus). Regardless of these differences, B. azotoformans has the disposal of a multicopy factory that could supply NosZ, and possibly also other copper proteins like NirK, with the high demand on copper (Richardson et al., 2009).
Figure 3.
Physical map of the B. azotoformans LMG 9581T (A) and Geobacillus thermodenitrificans NG80-2 (B) nos gene clusters. Arrows show the direction of transcription. Open reading frames are drawn to scale. Homologous genes are shown in identical colors.
The deduced primary structure of the three B. azotoformans NosZ proteins (between 76.5 and 83.1% sequence identity) show all conserved ligands of CuA and CuZ centers that have been identified in the crystal structures (Figure A5) (Brown et al., 2000; Paraskevopoulos et al., 2006). Nevertheless, the NosZ sequences contain insertions and deletions that are shared with nitrous oxide reductases from other Gram-positive species, likely placing these in a distinct family. The differences could be related with domain-specific interactions in Gram-positives and Gram-negatives with other components of the NosZ system. Another feature is that the N-terminal sequence in Gram-positive NosZ's is shorter (Figure A5). Moreover, SignalP and TatP prediction programs are somewhat ambiguous regarding the presence of N-terminal leader and TAT signal sequences. For instance, whereas NosZ1 from B. azotoformans is predicted to have a TAT leader, yet lacking an N-terminal cleavage side, the opposite holds for NosZ2 and NosZ3. These differences might point to, even protein-specific, differences in assembly, and transport. It is conceivable that transport proceeds not only by the TAT—but also by the Sec translocon. Indeed, it has been described for W. succinogenes that transport of non-folded NosZ and subsequent Cu insertion results in a fully functional protein (Heikkilä et al., 2001).
NosZ from Gram-negative bacteria is, as mentioned, a soluble protein in the periplasm where it receives its electrons for N2O reduction from reduced cytochrome c. In Gram-positive microorganisms, including B. azotoformans strain NCCB 10003, nitrous oxide reductase activity appears to be associated with the membrane fraction only [(Suharti and de Vries, 2005), and references herein]. Considering that the B. azotoformans NosZ gene products do not possess any TMH other than an N-terminal Sec-or TAT-signal, the conserved ORF encoding the polypeptide with four TMHs might provide a platform for membrane association. In addition, B. azotoformans nitrous oxide reductase(s) employ both cyt c and menaquinol as electron donors (Suharti and de Vries, 2005). Clearly, the particular lipoprotein cytochromes c that are encoded by the nosC genes might assist in cyt c-dependent electron transfer, but the role of menaquinol is more puzzling. Menaquinol could act directly or indirectly, viz. via menaquinol: cytochrome c oxidoreductase (bc1/b6f complex; see hereafter). A more direct role presumes the presence of a membrane-bound quinol oxidase in contact with NosZ. Here, the conserved membrane protein would come to mind. Alternatively, NapGH might serve such function. Experimental research has to decide between these or other alternatives.
bc1/b6f complex of B. azotoformans
In nitrate/nitrite-respiring Gram-negative bacteria, (mena)quinol: cytochrome c oxidoreductase (bc1 complex) plays a central role in the energy metabolism. By the complex, quinol that is produced concomitant to the oxidation of (organic) substrates is re-oxidized with cyt c as electron acceptor. Taking advantage of the proton-motive Q cycle, quinol:cyt c oxidoreduction is associated with the pumping of protons (4H+/2e−) across the cell membrane. Reduced cyt c serves as the reductant of the nitrate-reducing steps that, in case of a localization in the periplasm, do not contribute to pmf generation themselves. The work by the group of de Vries (Suharti et al., 2004, Suharti and de Vries, 2005) demonstrates that B. azotoformans opts for branched electron transfer, using both quinols and reduced cyt c for denitrification (see also above). While the cyt c-dependent pathway is energetically more favorable, a direct coupling of the denitrification steps with quinol oxidation could be kinetically more attractive (Suharti et al., 2004; Suharti and de Vries, 2005). The former pathway calls for the presence of a bc1/b6f complex in B. azotoformans, as well as suitable cytochromes c for electron transfer.
In its simple, canonical form the bc1 complex comprises three subunits: a Rieske-type 2Fe-2S protein and a membrane protein that binds one heme b and one quinone at the cytoplasmic face and another heme b and a quinone near the periplasm. The third component is cyt c. In Bacillus subtilis, the organization of the complex is different, resembling b6f complexes of Cyanobacteria and chloroplasts (Yu et al., 1995; Yu and Le Brun, 1998). The Rieske iron sulfur protein (QcrA) is of a general type, but the cytochrome b subunit is split into two parts (QcrA and QcrB). QcrB is homologous to the N-terminal part and QcrC is to the C-terminal part of common cytochrome b and of subunit IV of the b6f complex. At its N-terminus, QcrC is fused to a c-type cytochrome localized in the periplasm. The particular organization and the related amino acid sequences are conserved in QcrA (BAZO_09251), QcrB (BAZO_09246), and QcrC (BAZO_09241) from B. azotoformans (data not shown). We note the presence in the qcrABC gene cluster of a set of flanking genes coding for membrane-bound proteins (BAZO_09231; BAZO_09236; BAZO_09261) and for proteins with tetratrico peptide repeats (TPR) (BAZO_09256; BAZO_09266). Their presence is conserved in many Bacillus species, including B. bataviensis. This observation might suggest b6f of these organisms to be more “complex.”
For electron transfer, B. subtilis has the disposal of only two small membrane-bound c-type cytochromes: CccB, a cyt c551 lipoprotein, and CccA, a cyt c550 that is anchored to the membrane by a single C-terminal TMH (Bengtsson et al., 1999). De Vries and coworkers (Suharti et al., 2004; Suharti and de Vries, 2005) were able to isolate and characterize three different types of monoheme cytochrome c-type lipoproteins from B. azotoformans, namely cyt c550, cyt c551, and cyt c552; their encoding genes were not identified. All three could be reduced in concert with menaquinol oxidation by the b6f complex. Cyt c551 functioned as the specific electron donor to qCuANOR, but it was inactive in nitrite and in nitrous oxide reduction. In addition, pseudoazurin or other cupredoxins, the principal electron donors for copper-containing NirK, appeared to be absent (Suharti and de Vries, 2005). Now looking at the genome of B. azotoformans, we note the presence of four genes coding for monoheme cyt c-type lipoproteins, three of which being linked to the nosZ genes (see above). This would leave the gene product of the fourth candidate (BAZO_07449)—indeed annotated as cyt c551—as the electron carrier for qCuANOR. Furthermore, diheme cyt c5 is specifically required for NirK activity in Neisseria meningitides (Deeudom et al., 2008). The genome of B. azotoformans houses only one gene encoding a diheme cytochrome c lipoprotein (BAZO_12419) that consequently might serve nitrite reduction by NirK in this organism.
Gene inventory of B. bataviensis
B. bataviensis is capable of denitrification, albeit not to N2 but with the stoichiometric production of N2O from nitrate [(Verbaendert et al., 2011); Verbaendert et al., unpublished data]. This presumes the presence of the complete denitrification pathway lacking nitrous oxide reductase (NosZ). A first glance at the genome of B. bataviensis shows this to be the case. NosZ and its accessory proteins are absent, but the gene inventory for reduction of nitrate to N2O is available (Table 3). The more close inspection reveals a more complicated situation with sets of truncated or degenerated (pseudo)genes that are difficult to reconcile with full functionality of their gene products. In addition, we note quite interesting differences in the denitrification potentials between B. bataviensis and B. azotoformans.
For cytoplasmic dissimilatory nitrate reduction to nitrite, the genome of B. bataviensis contains one narGHJI gene cluster (BABA_18582-18597) with two linked NarK copies (BABA_18532, BABA_18537) in close vicinity (Figure 1A). The NarG, H, J, and I proteins display all conserved features, indicating that NAR is the active NAR in B. bataviensis. This has to be the case since the organism is devoid of the alternative, periplasmic NAR system (NAP). Indeed, none of the nap genes are found in the genome, except a putative napH (BABA_14547). Its gene product (153 aa) is significantly shorter than NapH (~300 AA) with an established function (Simon et al., 2003; Kern and Simon, 2008), thereby lacking all cysteines for the binding of iron sulfur clusters. Rather, InterProScan suggests BABA_14547 to be an FMN-binding protein.
For dissimilatory reduction of nitrite to NO, again only one candidate is present, a nirK gene that is designated as a pseudogene (BABA_p06582) by the presence of an apparent interspersing stop codon. However, by the correction for a single non-sense mutation (A–C at position 38028 of contig 42), the DNA sequence translates into an amino acid sequence that is 79.8% identical to NirK from B. azotoformans, differing only three amino acids in length. Moreover, by this substitution all features would be conserved that are related with the binding of the catalytic copper atoms and with the characteristic secondary structural elements of NirK2 family proteins (see above). Again bearing in mind that B. bataviensis is capable of nitrite reduction to NO, we anticipate that this pseudogene, caused by a single non-sense mutation, is probably the result of a sequencing artifact (although base calling was unambiguous). Proteome analysis should confirm this expectation. Like B. azotoformans, B. bataviensis is proposed to carry out also dissimilatory nitrate reduction to ammonia (Table 3). The nrfHA gene products (BAZO_17652-17657) meet all criteria described above for functionality (see Figure A1). The question is under which conditions the genes are expressed, taking into account that during standard denitrifying growth nitrate is quantitatively reduced to N2O.
Contrary to B. azotoformans, B. bataviensis seems to have the genetic potential for assimilatory nitrate and nitrite reduction to ammonium. The gene inventory is made up by nasC (BABA_16882) and nirDB (nasGB) (BABA_00910-00915) coding for assimilatory nitrate and nitrite reductase, respectively. Mechanistically, the process could be somewhat different from the one investigated thus far. In many microorganims, nasC and nirDB (nasGB) are localized on the same operon together with NarK-type transporters and regulatory proteins (Luque-Almagro et al., 2011). In P. denitrificans NasC and NasGB presumably function as one protein complex (Gates et al., 2011). Herein, NasB (NirB) binds NADH that serves as the reductant for both the six-electron reduction of nitrite to ammonia (by NasB) and the two-electron reduction of nitrate to nitrite (by NasC). The Rieske-type 2Fe-2S protein NasG (NirD) intermediates electron transfer between both catalytic subunits. In B. bataviensis nasC and nirBD are found in different parts of the genome and are not linked to NarK-type transporters. We note, however, the presence in the nirBD cluster of three genes (BABA_00890-00905) encoding key enzymes for the synthesis of sirohydrochlorin, the prosthetic group of NirB. NasC and NirB are highly similar to the respective proteins of which the function has been established, albeit with a notable difference. NasC (729 AA) from B. bataviensis is shorter than the protein from, for instance, P. denitrificans (870 AA) and it lacks at its C-terminal part a number of cysteines that might bind an iron-sulfur cluster. However, this difference is shared with many other Gram-positive microorganisms. Although B. bataviensis NirD is 44–57% identical to NirDs from other Bacillus-related species, there is one important distinction: in the former two key amino acids (cysteine, histidine) are substituted that are involved in the binding of the 2Fe-2S cluster. In contrast, nasC is linked with two genes coding for a hybrid iron sulfur protein (HCP) (BABA_16877) and for another Rieske-type 2Fe-2S protein (BABA_16872). The particular properties are consistently shared with other species, like Bacillus sp. 1NLA3E, suggesting an alternative, yet functional assimilatory nitrate/nitrite reductase system in B. bataviensis. For the uptake of nitrate, the genome of B. bataviensis contains four narK copies, unlike B. azotoformans that has only one. Two of these (NarK2, BABA18537; NarK3, BABA_18532) were already encountered in connection with dissimilatory NAR. They belong to the NarK2 family of nitrate:nitrite antiporters, facilitating the uptake of nitrate and the export of toxic nitrite. NarK1 (BABA_15587) and NarK4 (BABA_18532) are affiliated with the NarK1 family of (high-affinity) nitrate:proton symporters, which would make these proteins attractive candidates to serve N-assimilation. In contrast again to B. azotoformans, a gene is present in the genome of B. bataviensis (BABA_17227) coding for an AmtB-type of ammonium transporter.
Regarding NO reduction to N2O and the role therein of HCO proteins, the genome picture is ambiguous. B. bataviensis contains two copies of qnorB and three HCO copies related to cytochrome c-dependent ba3 oxidase (cbaAB) from Thermus thermophilus (Table 3). One qnorB is a pseudogene (BABA_p08977), with two frame shifts, either resulting from actual point mutations or sequencing errors (although base calling was unambiguous). When the gene sequence was altered manually at both positions (changing T into C at position contig55_76292 and A into G at position contig55_76206), the resulting amino acid sequence became highly similar to qNorB sequences from Geobacillus and Bacillus strains (AA sequence identity, 79–83%). However, it still contained a 217-AA deletion near the C-terminus (from position 464 to 760 in G. thermodenitrificans numbering; see Figure A2) comprising only 8 out of 14 TMHs and lacking five out of six conserved histidine residues. Also the other qnorB gene (BABA_17212) is truncated. It is devoid of 273 N-terminal and approximately 150 C-terminal amino acids, again spanning only 8 TMHs. Thus, it seems that both qNorBs are corrupted and inactive. Sequence analysis addresses two of the three ba3-type oxidases (CbaAB) (BABA_00225-00230, BABA_18502-18497) to the alternative NORs (Figures A3, A4). However, subunit II of the latter one (BABA_18497) lacks approximately 30 amino acids at its C-terminus, including the ones involved in binding of CuA. In addition, BABA_18502-18497 are not linked to genes coding for the proposed membrane-bound subunit III and for a SenC/SCO1 protein for CuA insertion. These observations would overrule a role for cyt c as an electron donor, although a function as such for quinol is still feasible. Subunits I (BABA_00225) and II (BABA_00230) of the second candidate are 70 and 79% identical to the respective subunits of BAZO_06399-006394 that has been identified above as qCuANOR (sNOR) (Figures 2, A3, A4). Furthermore, the N terminus of subunit II is quite similar to the one determined by Suharti et al. (2004), having only three mismatches. In full agreement with BAZO_06399-06394, the genes of the B. bataviensis are linked to ones encoding a small membrane-bound polypeptide (46 amino acids), another membrane protein (BABA_00215) with 6 TMHs and a SenC/SCO1-type protein (BABA_00220) (Table 3; Figure 2). Unlike BAZO_06399-006394 and all its other relatives, the B. bataviensis gene cluster is associated with a gene coding for heme o synthetase (ctaB; BABA_00235). This might imply that the particular B. bataviensis HCO contains cyt o3 at its catalytic side. Nevertheless, the striking resemblance regarding amino acid sequences and the organization of the gene cluster highly suggest BABA_00225-00230 and its associated small membrane subunit to be a menaquinol/cyt c-dependent NOR.
The third ba3-type (CbaAB) HCO (BABA_08951-08956) is most likely an oxidase, as deduced from its amino acid sequence having the crosslinking tyrosine (Figure A3). It is one of the four oxidases that are detected in the genome. Like in B. azotoformans, oxygen respiration may be mediated by quinol-dependent dependent aa3-600 oxidase encoded by the qoxBACD genes (BABA_05106-05091) and by cyt c-dependent caa3 oxidase (CtaCDEF). In agreement with other organisms, structural genes form part of a larger cluster (ctaABCDEFG) (BABA_16147-16177) that includes the ctaA and ctaB genes encoding cyt a and cyt o biosynthesis proteins. A sequence comparison of the gene products just mentioned with those of verified proteins indicates aa3-600 oxidase and cyt c-dependent caa3 oxidase to be functional (data not shown). Besides these three, we find conserved genes encoding the alternative quinol-dependent cytochrome bd oxidase (CydAB, BABA_01890-01895). CydAB is an oxidase that is found in many microorganisms and that, because of its high affinity for oxygen, (Borisov et al., 2011) is expressed under O2 limitation. B. azotoformans does not contain this oxidase. By the availability of the four different oxidases, B. bataviensis would have the potential to aerobically respire under a wide range of oxygen concentrations. However, the organism seems to be more limited in nitrate respiration as compared to B. azotoformans.
Concluding remarks
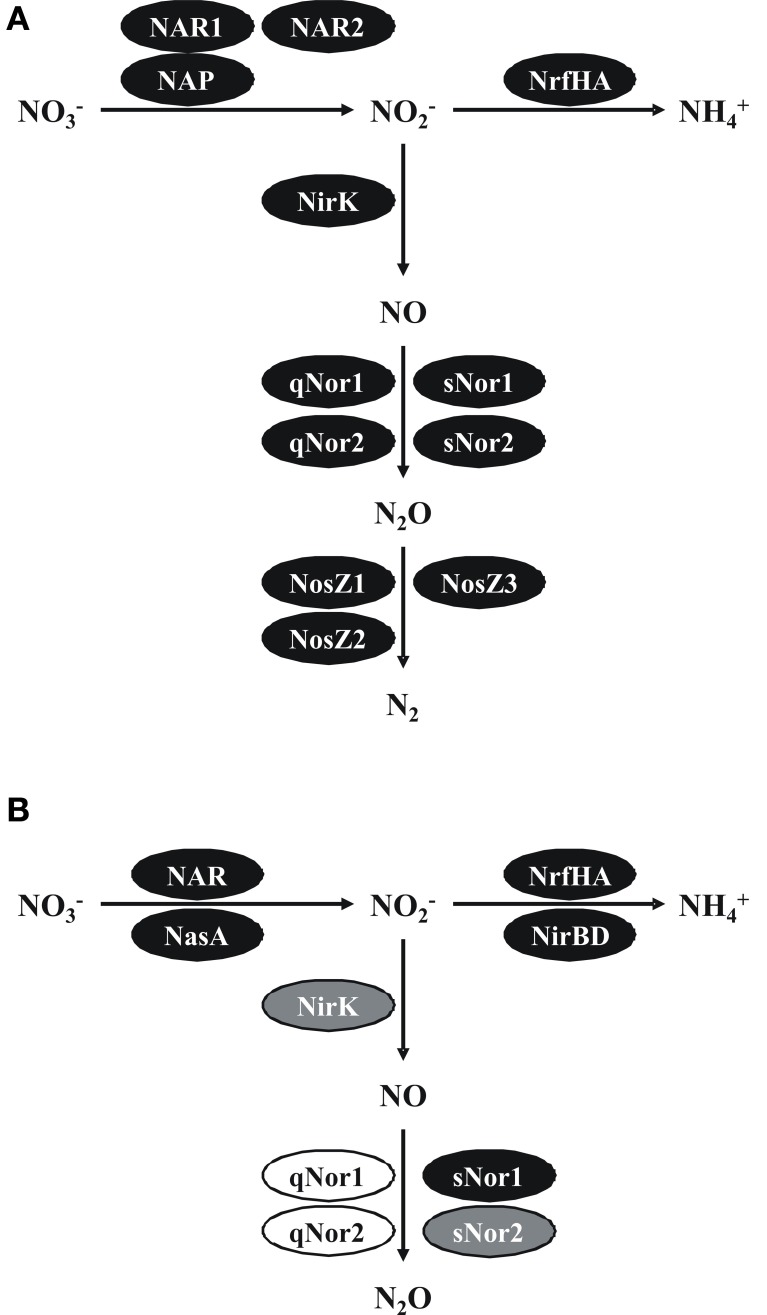
Above, we made a comprehensive analysis of the functional denitrification gene inventory of two Bacillus species. Our results are schematically presented in Figure 4. Our exploration came with quite some surprises, we detected potential new enzymes, supported previous suggestions and raised interesting questions that need experimental answers, which we briefly would like to summarize.
Figure 4.
Pathways and functional gene inventories for denitrification and ammonification in B. azotoformans LMG 9581T (A) and B. bataviensis LMG 21833T (B). Black ovals indicate predicted functional enzymes, gray ovals likely functional enzymes and white ovals pseudogenes or corrupted enzymes. NAR, cytoplasmic dissimilatory nitrate reductase; NAP, periplasmic dissimilatory nitrate reductase; NasC, assimilatory nitrate reductase; NrfHA, dissimilatory nitrite reductase to ammonium; NirBD, assimilatory nitrite reductase; NirK, periplasmic nitrite reductase to nitric oxide; qNor, dissimilatory quinol-dependent nitric oxide reductase; sNor, dissimilatory menaquinol/cyt c-dependent nitric oxide reductase; NosZ, dissimilatory nitrous oxide reductase. See text for more explanations.
A first surprise was that B. azotoformans, a well-known denitrifier, and B. bataviensis that is less well understood in this respect, are both capable of denitrification and dissimilatory ammonification (DNRA). Especially, B. azotoformans that makes N2 as the end product of nitrate denitrification has a large arsenal of apparently redundant enzymes, but that most likely would enable the organism to thrive under highly variable environmental conditions. The N2O-producing B. bataviensis is much more restricted in this. We note the presence of various pseudogenes that could be relicts of earlier times or situations in which this microorganism could be more dependent on anaerobic nitrate respiration. Striking is the way both Bacillus species took advantage of the opportunities of heme copper oxidases for NO and O2 reduction. Our analysis permitted the identification of the genes coding for bi-functional qCuANOR purified and characterized before Suharti et al. (2001, 2004). Yet, we could detect other related novel enzymes that either reduce NO or O2, but that still need to be validated by biochemical research.
Gram-positive microorganisms typically have limited space in their periplasm, which might pose specific demands on the way denitrification processes are structured in the periplasm. Suharti and de Vries (2005) already noticed that virtually all denitrifying partial reactions in B. azotoformans strain NCCB 10003 were associated with the cell membrane. Our genome analyses support their observations. All enzymes appear to be membrane-bound, either by TMHs at catalytic enzymes themselves, through association with membrane-bound partners or by covalent binding to lipids (NirK, NosZ). Membrane association most likely requires specific structural and architectural adaptations that are reflected in amino acid sequences. In agreement herewith, we note that the denitrification enzymes discussed often form part of (sub)families that are specific for Gram-positive microorganisms. Another aspect that relates to the membrane-bound character is energy metabolism. As pointed out again by De Vries and his coworkers (Suharti et al., 2004; Suharti and de Vries, 2005), all reactions tested used menaquinol as the reductant. Our analyses confirm these findings. However, also cyt c could be utilized as the reductant. This branched electron transfer adds to the metabolic flexibility. The use of (mena)quinol may allow reactions to proceed at high rates, also under conditions of substrate limitation. The use of reduced cytochrome(s) produced through quinol oxidation by the bc1/b6f complex is energetically more favorable.
All in all, our analyses suggest an astonishing versatility in denitrification opportunities, especially for B. azotoformans. The important question is, if and under which environmental conditions the different (partial) processes are expressed. An answer to this needs experimental research.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Flemish Fund for Scientific Research (FWO11/KAN/043 and FWO11/PDO/084). We would like to thank H. Op den Camp, K. Ji, and J. van de Vossenberg form the Department of Microbiology of the Radboud University Nijmegen for their helpful suggestions for initial genome assembly and annotation. We would also like to thank the reviewers for their detailed and constructive comments on earlier versions of the manuscript.
Appendix
Figure A1.
Multiple sequence alignment of NrfA from B. azotoformans LMG 9581T, B. bataviensis LMG 21833T, and other bacteria. Heme-binding motifs are highlighted in red (white lettering), the lysine at the catalytic heme is highlighted in green, active site residues are printed in blue, distal heme-ligating histidines for hemes 2–6 are printed in red [according to Einsle et al. (2000)]. Bs, Bacillus selenitireducens MLS10 (Bsel_1305); Dv, Desulfovibrio vulgaris DP4 (PDB 2J7A); Dd, Desulfovibrio desulfuricans (PDB 10AH_A). PDB, protein database accession number.
Figure A2.
Multiple sequence alignment of qNOR from B. azotoformans LMG 9581T, B. bataviensis LMG 21833T, and other bacteria. Two non-sense mutations in BABA_p08977 have been corrected in this figure (see text for further explanation). Amino acids involved in the binding of catalytic centers (cyt b, b3, FeB) are highlighted in red, residues for quinol binding in black, bulky residues in the hydrophilic part in gray, residues for Ca2+ binding in yellow, residues lining the long water channel from the cytoplasm in dark blue, residues at water channel 1 in green and water channel 2 in light blue; transmembrane regions are indicated in blue and numbered on top of the alignment [as described by Matsumoto et al. (2012)]. Ba_q1, Bacillus sp. 1NLA3E (B1NLA3EDRAFT_3340); Ba_q2, Bacillus sp. 1NLA3E (B1NLA3EDRAFT_1567); Ge, Geobacillus stearothermophilus (PDB 3AYF, 3AYG); St, Staphylococcus aureus (SAEMRSA15_02230); An, Anaerophaga sp. HS1 (AnHS1_010100003178); Fl, Flavobacterium johnsoniae UW101 (Fjoh_2413); Ng, Neisseria gonorrhoeae (NgonD_010100007739); Cu, Cupriavidus metallidurans CH34 (Rmet_3167); Sy, Synechocystis sp. PCC 6803 (sll0450). PDB, protein database accession number.
Figure A3.
Multiple sequence alignment of CbaA from B. azotoformans LMG 9581T, B. bataviensis LMG 21833T, Geobacillus thermodenitrificans NG80-2 (GTNG_1394; GTNG_1518), and. T. thermophilus (PDB 3EH3). Amino acids involved in the binding of heme b, heme b3, and CuB/FeB are highlighted in red, residues forming D-water channel in blue, residues forming Q-water channel in green, crosslinking tyrosine in oxidases in dark green, residues related with electron transfer in purple. PDB, protein database accession number.
Figure A4.
Multiple sequence alignment of CbaB from B. azotoformans LMG 9581T, B. bataviensis LMG 21833T, Geobacillus thermodenitrificans NG80-2 (GTNG_1395; GTNG_1517), and T. thermophilus (PDB 3EH3). Contact site residues are highlighted in gray, residues for CuA-binding in red, and residues for electron transport in pink. PDB, protein database accession number.
Figure A5.
A Multiple sequence alignment of NosZ from B. azotoformans LMG 9581T and other bacteria. Conserved histidine ligands of the CuZ center are highlighted in dark blue; cysteine and other ligands of the CuA center in yellow, and turquoise respectively; partially conserved histidine residues in brown; calcium atom and chloride ion ligands in pink and green, respectively [according to Simon et al. (2004)]. TAT sequences are printed in red, N-terminal cleavage in blue. Gt, Geobacillus thermodenitrificans NG80-2 (GTNG_1734); Dh, Desulfitobacterium hafniense T51 (DSY0261); Dr, Desulfotomaculum. ruminis DSM 2154 (Desru_1528), Pd, Paracoccus denitrificans DSM 413 (PDB 1FWX), Ac, Achromobacter cycloclastes 1013 (PDB 21WF); Ps, Pseudomonas stutzeri (PDB 3SBP). PDB, protein database accession number.
References
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoux P., Sabaty M., Alric J., Frangioni B., Guigliarelli B., Adriano J.-M., et al. (2003). Structural and redox plasticity in the heterodimeric periplasmic nitrate reductase. Nat. Struct. Biol. 10, 928–934 10.1038/nsb994 [DOI] [PubMed] [Google Scholar]
- Azarkina N., Siletsky S., Borisov V., Von Wachenfeldt C., Hederstedt L., Konstantinov A. (1999). A cytochrome bb'-type quinol oxidase in Bacillus subtilis strain 168. J. Biol. Chem. 274, 32810–32817 10.1074/jbc.274.46.32810 [DOI] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., Dejongh M., Disz T., Edwards R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford V., Angove H., Seward H., Thomson A., Cole J., Jn B., et al. (2002). Structure and spectroscopy of the periplasmic cytochrome c nitrite reductase from Escherichia coli. Biochemistry 41, 2921–2931 10.1021/bi015765d [DOI] [PubMed] [Google Scholar]
- Bendtsen J. D., Jensen L. J., Blom N., Heijne G. V., Brunak S. (2004). Feature based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17, 349–356 10.1093/protein/gzh037 [DOI] [PubMed] [Google Scholar]
- Bendtsen J. D., Nielsen H., Widdick D., Palmer T., Brunak S. (2005). Prediction of twin-arginine signal peptides. BMC Bioinform. 6:167 10.1186/1471-2105-6-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson J., Rivolta C., Hederstedt L., Karamata D. (1999). Bacillus subtilis contains two small c-type cytochromes with homologous heme domains but different types of membrane anchors. J. Biol. Chem. 274, 26179–21684 10.1074/jbc.274.37.26179 [DOI] [PubMed] [Google Scholar]
- Bergaust L., Mao Y., Bakken L. R., Frostegård Å. (2010). Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal pH on nitrogen oxide reductase in Paracoccus denitrificans. Appl. Environ. Microbiol. 76, 6387–6396 10.1128/AEM.00608-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero M., Rothery R., Boroumand N., Palak M., Blasco F., Ginet N., et al. (2005). Structural and biochemical characterization of a quinol binding site of Escherichia coli nitrate reductase A. J. Biol. Chem. 280, 14836–14843 10.1074/jbc.M410457200 [DOI] [PubMed] [Google Scholar]
- Bertero M., Rothery R., Palak M., Hou C., Lim D., Blasco F., et al. (2003). Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Biol. 10, 681–687 10.1038/nsb969 [DOI] [PubMed] [Google Scholar]
- Bleakley B. H., Tiedje J. M. (1982). Nitrous oxide production by organisms other than nitrifiers or denitrifiers. Appl. Environ. Microbiol. 44, 1342–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov V., Gennis R., Hemp J., Verkhovsky M. (2011). The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 1807, 1398–1413 10.1016/j.bbabio.2011.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky M., McIninch J. (1993). GeneMark: parallel gene recognition for both DNA strands. Comput. Chem. 17, 123–133 [Google Scholar]
- Boulanger M. J., Murphy M. E. P. (2002). Crystal structure of the soluble domain of the major anaerobically induced outer membrane protein (AniA) from pathogenic Neisseria: a new class of copper containing nitrite reductases. J. Mol. Biol. 315, 1111–1127 10.1006/jmbi.2001.5251 [DOI] [PubMed] [Google Scholar]
- Brown K., Djinovic-Carugo K., Haltia T., Cabrito I., Saraste M., Moura J., et al. (2000). Revisiting the catalytic CuZ cluster of nitrous oxide (N2O) reductase. Evidence of a bridging inorganic sulfur. J. Biol. Chem. 275, 41133–41136 10.1074/jbc.M008617200 [DOI] [PubMed] [Google Scholar]
- Cho C. M.-H., Yan T., Liu X., Wu L., Zhou J., Stein L. Y. (2006). Transcriptome of a Nitrosomonas europaea mutant with a disrupted nitrite reductase gene (nirK). Appl. Environ. Microbiol. 72, 4450–4454 10.1128/AEM.02958-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho C., González P. J., Moura J. J. G., Moura I., Trincão J., Romão M. J. (2011). The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states. J. Mol. Biol. 408, 932–948 10.1016/j.jmb.2011.03.016 [DOI] [PubMed] [Google Scholar]
- Cunha C., Macieira S., Dias J., Almeida G., Goncalves L., Costa C., et al. (2003). Cytochrome c nitrite reductase from Desulfovibrio desulfuricans ATCC 27774. The relevance of the two calcium sites in the structure of the catalytic subunit (NrfA). J. Biol. Chem. 278, 17455–17465 10.1074/jbc.M211777200 [DOI] [PubMed] [Google Scholar]
- de Barjac H., Bonnefoi A. (1972). Essai de classification biochimique de 64 “Bacillus” des groupes II et III représentant 11 espèces différentes. Ann. Inst. Pasteur 122, 463–473 [PubMed] [Google Scholar]
- Deeudom M., Koomey M., Moir J. (2008). Roles of c-type cytochromes in respiration in Neisseria meningitidis. Microbiology 154, 2857–2864 10.1099/mic.0.2008/020339-0 [DOI] [PubMed] [Google Scholar]
- Denariaz G., Payne W. J., Legall J. (1989). A halophilic denitrifier, Bacillus halodenitrificans sp. nov. Int. J. Syst. Bacteriol. 39, 145–151 [Google Scholar]
- Denariaz G., Payne W. J., Legall J. (1991). The denitrifying nitrite reductase of Bacillus halodenitrificans. Biochim. Biophys. Acta 1056, 225–232 [Google Scholar]
- de Vries S., Schröder I. (2002). Comparison between the nitric oxide reductase family and its aerobic relatives, the cytochrome oxidases. Biochem. Soc. Trans. 30, 662–667 [DOI] [PubMed] [Google Scholar]
- Einsle O. (2011). Structure and function of formate-dependent cytochrome c nitrite reductase, NrfA. Methods Enzymol. 496, 399–422 10.1016/B978-0-12-386489-5.00016-6 [DOI] [PubMed] [Google Scholar]
- Einsle O., Messerschmidt A., Huber R., Kroneck P. M. H., Neese F. (2002). Mechanism of the six-electron reduction of nitrite to ammonia by cytochrome c nitrite reductase. J. Am. Chem. Soc. 124, 11737–11745 10.1021/ja0206487 [DOI] [PubMed] [Google Scholar]
- Einsle O., Stach P., Messerschmidt A., Simon J., Kröger A., Huber R., et al. (2000). Cytochrome c nitrite reductase from Wolinella succinogenes. J. Biol. Chem. 275, 39608–39616 10.1074/jbc.M006188200 [DOI] [PubMed] [Google Scholar]
- Ellis M. J., Dodd F. E., Strange R. W., Prudêncio M., Sawers G., Eady R. R., et al. (2001). X-ray structure of a blue copper nitrite reductase at high pH and in copper-free form at 1.9 A resolution. Acta Crystallogr. D Biol. Crystallogr. 57, 1110–1118 10.1107/S0907444901008654 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., Heijne G. V., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971 10.1038/nprot.2007.131 [DOI] [PubMed] [Google Scholar]
- Falke D., Schulz K., Doberenz C., Beyer L., Lilie H., Thiemer B., et al. (2010). Unexpected oligomeric structure of the FocA formate channel of Escherichia coli: a paradigm for the formate-nitrite transporter family of integral membrane proteins. FEMS Microbiol. Lett. 303, 69–75 10.1111/j.1574-6968.2009.01862.x [DOI] [PubMed] [Google Scholar]
- Fee J., Case D., Noodleman L. (2008). Toward a chemical mechanism of proton pumping by the B-type cytochrome c oxidases: application of density functional theory to cytochrome ba3 of Thermus thermophilus. J. Am. Chem. Soc. 130, 15002–15021 10.1021/ja803112w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Wang W., Cheng J., Ren Y., Zhao G., Gao C., et al. (2007). Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc. Nat. Acad. Sci. U.S.A. 104, 5602–5607 10.1073/pnas.0609650104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y., Tamada T., Takami H., Suzuki S., Inoue T., Nojiri M. (2011). Cloning, expression, purification, crystallization and preliminary X-ray crystallographic study of GK0767, the copper-containing nitrite reductase from Geobacillus kaustophilus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 692–695 10.1107/S1744309111013297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J.-L. (1977). Etude de la dénitrification chez une bactérie thermophile sporulée. Ann. Microbiol. 128A, 447–458 [PubMed] [Google Scholar]
- Gates A., Luque-Almagro V., Goddard A., Ferguson S., Roldán M., Richardson D. (2011). A composite biochemical system for bacterial nitrate and nitrite assimilation as exemplified by Paracoccus denitrificans. Biochem. J. 435, 743–753 10.1042/BJ20101920 [DOI] [PubMed] [Google Scholar]
- González P. J., Correia C., Moura I., Brondino, Moura J. J. G. (2006). Bacterial nitrate reductases: molecular and biological aspects of nitrate reduction. J. Inorg. Biochem. 100, 1015–1023 10.1016/j.jinorgbio.2005.11.024 [DOI] [PubMed] [Google Scholar]
- Heikkilä M., Honisch U., Wunsch P., Zumft W. (2001). Role of the Tat transport system in nitrous oxided reductase translocation and cytochrome cd1 biosynthesis in Pseudomonas stutzeri. J. Bacteriol. 183, 1663–1671 10.1128/JB.183.5.1663-1671.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemp J., Gennis R. B. (2008). Diversity of the heme–copper superfamily in Archaea: insights from genomics and structural modeling. Results Probl. Cell Differ. 45, 1–31 10.1007/400_2007_046 [DOI] [PubMed] [Google Scholar]
- Heyndrickx M., Vauterin L., Vandamme P., Kersters K., De Vos P. (1996). Applicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J. Microbiol. Meth. 26, 247–259 [Google Scholar]
- Heyrman J., Vanparys B., Logan N. A., Balcaen A., Rodrıguez-Dıaz M., Felske A., et al. (2004). Bacillus novalis sp. nov., Bacillus vireti sp. nov., Bacillus soli sp. nov., Bacillus bataviensis sp. nov. and Bacillus drentensis sp. nov., from the Drentse A grasslands. Int. J. Syst. Evol. Microbiol. 54, 47–57 10.1099/ijs.0.02723-0 [DOI] [PubMed] [Google Scholar]
- Hino T., Nagano S., Sugimoto H., Tosha T., Shiro Y. (2012). Molecular structure and function of bacterial nitric oxide reductase. Biochim. Biophys. Acta 1817, 680–687 10.1016/j.bbabio.2011.09.021 [DOI] [PubMed] [Google Scholar]
- Ize B., Coulthurst S. J., Hatzixanthis K., Caldelari I., Buchanan G., Barclay E. C., et al. (2009). Remnant signal peptides on non-exported enzymes: implications for the evolution of prokaryotic respiratory chains. Microbiology 155, 3992–4004 10.1099/mic.0.033647-0 [DOI] [PubMed] [Google Scholar]
- Jacobson F., Guo H., Olesen K., Okvist M., Neutze R., Sjölin L. (2005). Structures of the oxidized and reduced forms of nitrite reductase from Rhodobacter sphaeroides 2.4.3 at high pH: changes in the interactions of the type 2 copper. Acta Crystallogr. D Biol. Crystallogr. 61, 1190–1198 10.1107/S0907444905017488 [DOI] [PubMed] [Google Scholar]
- Jepson B. J. N., Mohan S., Clarke T. A., Gates A. J., Cole J. A., Butler C. S., et al. (2007). Spectropotentiometric and structural analysis of the periplasmic nitrate reductase from Escherichia coli. J. Biol. Chem. 282, 6425–6437 10.1074/jbc.M607353200 [DOI] [PubMed] [Google Scholar]
- Jetten M. S. (2008). The microbial nitrogen cycle. Environ. Microbiol. 10, 2903–2909 10.1111/j.1462-2920.2008.01786.x [DOI] [PubMed] [Google Scholar]
- Käll L., Krogh A., Sonnhammer E. L. L. (2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Kern M., Simon J. (2008). Characterization of the NapGH quinol dehydrogenase complex involved in Wolinella succinogenes nitrate respiration. Mol. Microbiol. 69, 1137–1152 10.1111/j.1365-2958.2008.06361.x [DOI] [PubMed] [Google Scholar]
- Kern M., Simon J. (2009). Electron transport chains and bioenergetics of respiratory nitrogen metabolism in Wolinella succinogenes and other Epsilonproteobacteria. Biochim. Biophys. Acta 1787, 646–656 10.1016/j.bbabio.2008.12.010 [DOI] [PubMed] [Google Scholar]
- Kern M., Volz J., Simon J. (2011). The oxidative and nitrosative stress defence network of Wolinella succinogenes: cytochrome c nitrite reductase mediates the stress response to nitrite, nitric oxide, hydroxylamine and hydrogen peroxide. Environ. Microbiol. 13, 2478–2494 10.1111/j.1462-2920.2011.02520.x [DOI] [PubMed] [Google Scholar]
- Kim S.-H., Harzman C., Davis J. K., Hutcheson R., Broderick J. B., Marsh T. L., et al. (2012). Genome sequence of Desulfitobacterium hafniense DCB-2, a Gram-positive anaerobe capable of dehalogenation and metal reduction. BMC Microbiol. 12:21 10.1186/1471-2180-12-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft B., Strous M., Tegetmeyer H. E. (2011). Microbial nitrate respiration – genes, enzymes and environmental distribution. J. Biotechnol. 155, 104–117 10.1016/j.jbiotec.2010.12.025 [DOI] [PubMed] [Google Scholar]
- Kranz R., Richard-Fogal C., Taylor J., Frawley E. (2009). Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol. Mol. Biol. Rev. 73, 510–528 10.1128/MMBR.00001-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., Heijne G. V., Sonnhammer E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Lam P., Kuypers M. (2011). Microbial nitrogen cycling processes in oxygen minimum zones. Ann. Rev. Mar. Sci. 3, 317–345 [DOI] [PubMed] [Google Scholar]
- Larkin M., Blackshields G., Brown N., Chenna R., McGettigan P., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lauraeus M., Haltia T., Saraste M., Wikström M. (1991). Bacillus subtilis expresses two kinds of haem-A-containing terminal oxidases. Eur. J. Biochem. 197, 699–705 10.1111/j.1432-1033.1991.tb15961.x [DOI] [PubMed] [Google Scholar]
- Liu B., Zhang Y., Sage J., Soltis S., Doukov T., Chen Y., et al. (2012). Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: a combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B). Biochim. Biophys. Acta 1817, 658–665 10.1016/j.bbabio.2011.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Gao C., Zhang A., Jin P., Wang L., Feng L. (2008). The nos gene cluster from gram-positive bacterium Geobacillus thermodenitrificans NG80-2 and functional characterization of the recombinant NosZ. FEMS Microbiol. Lett. 289, 46–52 10.1111/j.1574-6968.2008.01362.x [DOI] [PubMed] [Google Scholar]
- Lu S., Suharti, Vries S. D., Moënne-Loccoz P. (2004). Two CO molecules can bind concomitantly at the diiron site of NO reductase from Bacillus azotoformans. J. Am. Chem. Soc. 126, 15332–15333 10.1021/ja045233v [DOI] [PubMed] [Google Scholar]
- Lukashin A., Borodovsky M. (1998). GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26, 1107–1115 10.1093/nar/26.4.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque-Almagro V., Gates A., Moreno-Vivián C., Ferguson S., Richardson D., Roldán M. (2011). Bacterial nitrate assimilation: gene distribution and regulation. Biochem. Soc. Trans. 39, 1838–1843 10.1042/BST20110688 [DOI] [PubMed] [Google Scholar]
- Mahne I., Tiedje J. M. (1995). Criteria and methodology for identifying respiratory denitrifiers. Appl. Environ. Microbiol. 61, 1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Espinosa R., Cole J., Richardson D., Watmough N. (2011). Enzymology and ecology of the nitrogen cycle. Biochem. Soc. Trans. 39, 175–178 10.1042/BST0390175 [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Tosha T., Pisliakov A. V., Hino T., Sugimoto H., Nagano S., et al. (2012). Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus. Nat. Struct. Mol. Biol. 19, 238–246 10.1038/nsmb.2213 [DOI] [PubMed] [Google Scholar]
- Moir J. W. B., Wood N. J. (2001). Nitrate and nitrite transport in bacteria. Cell. Mol. Life Sci. 58, 215–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y., Itoh M., Okuda S., Yoshizawa A. C., Kanehisa M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185 10.1093/nar/gkm321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmudin S., González P. J., Trincão J., Coelho C., Mukhopadhyay A., Cerqueira N. M. F. S. A., et al. (2008). Periplasmic nitrate reductase revisited: a sulfur atom completes the sixth coordination of the catalytic molybdenum. J. Biol. Inorg. Chem. 13, 737–753 10.1007/s00775-008-0359-6 [DOI] [PubMed] [Google Scholar]
- Nikaido K., Noguchi S., Sakamoto J., Sone N. (1998). The cbaAB genes for bo3-type cytochrome c oxidase in Bacillus stearothermophilus. Biochim. Biophys. Acta 1397, 262–267 10.1016/S0167-4781(98)00043-8 [DOI] [PubMed] [Google Scholar]
- Overton T. W., Justino M. C., Li Y., Baptista J. M., Melo A. M. P., Cole J. A., et al. (2008). Widespread distribution in pathogenic bacteria of di-iron proteins that repair oxidative and nitrosative damage to iron-sulfur centers. J. Bacteriol. 190, 2004–2013 10.1128/JB.01733-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulos K., Antonyuk S., Sawers R., Eady R., Hasnain S. (2006). Insight into catalysis of nitrous oxide reductase from high-resolution structures of resting and inhibitor-bound enzyme from Achromobacter cycloclastes. J. Mol. Biol. 362, 55–65 10.1016/j.jmb.2006.06.064 [DOI] [PubMed] [Google Scholar]
- Peña A., Busquets A., Gomila M., Bosch R., Nogales B., García-Valdés E., et al. (2012). Draft genome of Pseudomonas stutzeri strain ZoBell (CCUG 16156), a marine isolate and model organism for denitrification studies. J. Bacteriol. 194, 1277–1278 10.1128/JB.06648-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., Heijne G. V., Nielsen H. (2011). SignalP 4.0, discriminating signal peptides from transmembrane regions. Nat. Methods 8, 784–786 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Pichinoty F., de Barjac H., Mandel M., Asselineau J. (1983). Description of Bacillus azotoformans sp. nov. Int. J. Syst. Bacteriol. 33, 660–662 [Google Scholar]
- Pichinoty F., Garcia J. L., Job C., Durand M. (1978). La dénitrification chez Bacillus licheniformis. Can. J. Microbiol. 24, 45–49 [DOI] [PubMed] [Google Scholar]
- Pitcher D. G., Saunders N. A., Owen R. J. (1989). Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8, 151–156 [Google Scholar]
- Pittman M. S., Elvers K. T., Lee L., Jones M. A., Poole R. K., Park S. F., et al. (2007). Growth of Campylobacter jejuni on nitrate and nitrite: electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol. Microbiol. 63, 575–590 10.1111/j.1365-2958.2006.05532.x [DOI] [PubMed] [Google Scholar]
- Pomowski A., Zumft W. G., Kroneck P. M. H., Einsle O. (2011). N2O binding at a [4Cu:2S] copper–sulphur cluster in nitrous oxide reductase. Nature 477, 234–238 10.1038/nature10332 [DOI] [PubMed] [Google Scholar]
- Rahman O., Cummings S. P., Harrington D. J., Sutcliffe I. C. (2008). Methods for the bioinformatic identification of bacterial lipoproteins encoded in the genomes of Gram-positive bacteria. World J. Microbiol. Biotechnol. 24, 2377–2382 [Google Scholar]
- Rey M. W., Ramaiya P., Nelson B. A., Brody-Karpin S. D., Zaretsky E. J., Tang M., et al. (2004). Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome Biol. 5, R77 10.1186/gb-2004-5-10-r77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson D. (2000). Bacterial respiration: a flexible process for a changing environment. Microbiology 146, 551–571 [DOI] [PubMed] [Google Scholar]
- Richardson D., Felgate H., Watmough N., Thomson A., Baggs E. (2009). Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle – could enzymic regulation hold the key? Trends Biotechnol. 27, 388–397 10.1016/j.tibtech.2009.03.009 [DOI] [PubMed] [Google Scholar]
- Richardson D. J., Berks B. C., Russell D. A., Spiro S., Taylor C. J. (2001). Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell. Mol. Life Sci. 58, 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M., Oliveira T., Matias P., Martins I., Valente F., Pereira I., et al. (2006a). Crystallization and preliminary structure determination of the membrane-bound complex cytochrome c nitrite reductase from Desulfovibrio vulgaris Hildenborough. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 565–568 10.1107/S1744309106016629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M. L., Oliveira T. F., Pereira I. A. C., Archer M. (2006b). X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination. EMBO J. 25, 5951–5960 10.1038/sj.emboj.7601439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M. L., Scott K. A., Sansom M. S. P., Pereira I. A. C., Archer M. (2008). Quinol oxidation by c-type cytochromes: structural characterization of the menaquinol binding site of NrfHA. J. Mol. Biol. 381, 341–350 10.1016/j.jmb.2008.05.066 [DOI] [PubMed] [Google Scholar]
- Rothery R. A., Workun G. J., Weiner J. H. (2008). The prokaryotic complex iron–sulfur molybdoenzyme family. Biochim. Biophys. Acta 1778, 1897–1929 10.1016/j.bbamem.2007.09.002 [DOI] [PubMed] [Google Scholar]
- Sakamoto J., Handa Y., Sone N. (1997). A novel cytochrome b(o/a)3-type oxidase from Bacillus stearothermophilus catalyzes cytochrome c-551 oxidation. J. Biochem. 122, 764–771 [DOI] [PubMed] [Google Scholar]
- Salzberg S. L., Delcher A. L., Kasif S., White O. (1998). Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26, 544–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert S. M., Scott K. M., Klotz M. G., Chain P. S. G., Hauser L. J., Hemp J., et al. (2008). Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl. Environ. Microbiol. 74, 1145–1156 10.1128/AEM.01844-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. (2002). Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol. Rev. 26, 285–309 [DOI] [PubMed] [Google Scholar]
- Simon J., Einsle O., Kroneck P. M. H., Zumft W. G. (2004). The unprecedented nos gene cluster of Wolinella succinogenes encodes a novel respiratory electron transfer pathway to cytochrome c nitrous oxide reductase. FEBS Lett. 569, 7–12 10.1016/j.febslet.2004.05.060 [DOI] [PubMed] [Google Scholar]
- Simon J., Gross R., Einsle O., Kroneck P. M. H., Kröger A., Klimmek O. (2000). A NapC/NirT-type cytochrome c (NrfH) is the mediator between the quinone pool and the cytochrome c nitrite reductase of Wolinella succinogenes. Mol. Microbiol. 35, 686–696 10.1046/j.1365-2958.2000.01742.x [DOI] [PubMed] [Google Scholar]
- Simon J., Klotz M. G. (2012). Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochim. Biophys. Acta Available online at: http://dx.doi.org/10.1016/j.bbabio.2012.1007.1005 10.1016/j.bbabio.2012.1007.1005 [DOI] [PubMed] [Google Scholar]
- Simon J., Sänger M., Schuster C., Gross R. (2003). Electron transport to periplasmic nitrate reductase (NapA) of Wolinella succinogenes is independent of a NapC protein. Mol. Microbiol. 49, 69–79 10.1046/j.1365-2958.2003.03544.x [DOI] [PubMed] [Google Scholar]
- Simon J., van Spanning R., Richardson D. (2008). The organisation of proton motive and non-proton motive redox loops in prokaryotic respiratory systems. Biochim. Biophys. Acta 1777, 1480–1490 10.1016/j.bbabio.2008.09.008 [DOI] [PubMed] [Google Scholar]
- Smirnova I., Zaslavsky D., Fee J., Gennis R., Brzezinski P. (2008). Electron and proton transfer in the ba(3) oxidase from Thermus thermophilus. J. Bioenerg. Biomembr. 40, 281–287 10.1007/s10863-008-9157-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. S. (1983). Nitrous oxide production by Escherichia coli is correlated with nitrate reductase activity. Appl. Environ. Microbiol. 45, 1545–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulimane T., Buse G., Bourenkov G., Bartunik H., Huber R., Than M. (2000). Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from Thermus thermophilus. EMBO J. 19, 1766–1776 10.1093/emboj/19.8.1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa F., Alves R., Ribeiro M., Pereira-Leal J., Teixeira M., Pereira M. (2012). The superfamily of heme-copper oxygen reductases: types and evolutionary considerations. Biochim. Biophys. Acta 1817, 629–637 10.1016/j.bbabio.2011.09.020 [DOI] [PubMed] [Google Scholar]
- Stein L. Y., Arp D. A., Berube P. M., Chain P. S. G., Hauser L., Jetten M. S. M., et al. (2007). Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation. Environ. Microbiol. 9, 2993–3007 10.1111/j.1462-2920.2007.01409.x [DOI] [PubMed] [Google Scholar]
- Stein L. Y. (2011). Surveying N2O-producing pathways in bacteria, in Methods in Enzymology: Research on Nitrification and Related Processes, ed Klotz M. G. (San Diego, CA: Elsevier Academic Press; ), 131–152 10.1016/B978-0-12-381294-0.00006-7 [DOI] [PubMed] [Google Scholar]
- Streminíska M. A., Felgate H., Rowley G., Richardson D. J., Baggs E. M. (2012). Nitrous oxide production in soil isolates of nitrate-ammonifying bacteria. Environ. Microbiol. Rep. 4, 66–71 [DOI] [PubMed] [Google Scholar]
- Su F., Xu K., Zhao B., Tai C., Tao F., Tang H., et al. (2011). Genome sequence of the thermophilic strain Bacillus coagulans XZL4, an efficient pentose-utilizing producer of chemicals. J. Bacteriol. 193, 6398–6399 10.1128/JB.06157-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suharti, de Vries S. (2005). Membrane-bound denitrification in the Gram-positive bacterium Bacillus azotoformans. Biochem. Soc. Trans. 33, 130–133 10.1042/BST0330130 [DOI] [PubMed] [Google Scholar]
- Suharti, Heering H. A., de Vries S. (2004). NO reductase from Bacillus azotoformans is a bifunctional enzyme accepting electrons from menaquinol and a specific endogenous membrane-bound cytochrome c551. Biochemistry 43, 13487–13495 10.1021/bi0488101 [DOI] [PubMed] [Google Scholar]
- Suharti, Strampaard M. J. F., Schröder I., de Vries S. (2001). A novel copper A containing menaquinol NO reductase from Bacillus azotoformans. Biochemistry 40, 2632–2639 10.1021/bi0020067 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5, molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares P., Pereira A. S., Moura J. J. G., Moura I. (2006). Metalloenzymes of the denitrification pathway. J. Inorg. Biochem. 100, 2087–2100 10.1016/j.jinorgbio.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Tiedje J. M. (1988). Ecology of denitrification and dissimilatory nitrate reduction to ammonium, in Biology of Anaerobic Microorganisms, ed Zehnder A. (New York, NY: John Wiley and Sons; ). [Google Scholar]
- Tiedje J., Sexstone A., Myrold D., Ja R. (1982). Denitrification: ecological niches, competition and survival. Antonie Van Leeuwenhoek 48, 569–583 [DOI] [PubMed] [Google Scholar]
- Tocheva E. I., Rosell F. I., Mauk A. G., Murphy M. E. P. (2004). Side-on copper-nitrosyl coordination by nitrite reductase. Science 304, 867–870 10.1126/science.1095109 [DOI] [PubMed] [Google Scholar]
- Tucker N. P., Autreaux B. D., Spiro S., Dixon R. (2005). Mechanism of transcriptional regulation by the Escherichia coli nitric oxide sensor NorR. Biochem. Soc. Trans. 34, 191–194 10.1042/BST0340191 [DOI] [PubMed] [Google Scholar]
- Urata K., Satoh T. (1991). Enzyme localization and orientation of the active site of dissimilatory nitrite reductase from Bacillus firmus. Arch. Microbiol. 156, 24–27 [Google Scholar]
- van Wonderen J. H. V., Burlat B., Richardson D. J., Cheesman M. R., Butt J. N. (2008). The nitric oxide reductase activity of cytochrome c nitrite reductase from Escherichia coli. J. Biol. Chem. 283, 9587–9594 10.1074/jbc.M709090200 [DOI] [PubMed] [Google Scholar]
- Veith B., Herzberg C., Steckel S., Feesche J., Maurer K. H., Ehrenreich P., et al. (2004). The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J. Mol. Microbiol. Biotechnol. 7, 204–211 10.1159/000079829 [DOI] [PubMed] [Google Scholar]
- Verbaendert I., Boon N., De Vos P., Heylen K. (2011). Denitrification is a common feature among members of the genus Bacillus. Syst. Appl. Microbiol. 34, 385–391 10.1016/j.syapm.2011.02.003 [DOI] [PubMed] [Google Scholar]
- von Ballmoos C., Adelroth P., Gennis R., Brzezinski P. (2012). Proton transfer in ba(3) cytochrome c oxidase from Thermus thermophilus. Biochim. Biophys. Acta 1817, 650–657 10.1016/j.bbabio.2011.11.015 [DOI] [PubMed] [Google Scholar]
- Wang H., Robert O., Hall J., Gunsalus P. (2000). The nrfA and nirB nitrite reductase operons in Escherichia coli are expressed differently in response to nitrate than to nitrite. J. Bacteriol. 182, 5813–5822 10.1128/JB.182.20.5813-5822.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watmough N., Field S., Hughes R., Richardson D. (2009). The bacterial respiratory nitric oxide reductase. Biochem. Soc. Trans. 37, 392–399 10.1042/BST0370392 [DOI] [PubMed] [Google Scholar]
- Winstedt L., von Wachenfeldt C. (2000). Terminal oxidases of Bacillus subtilis strain 168, one quinol oxidase, cytochrome aa(3) or cytochrome bd, is required for aerobic growth. J. Bacteriol. 182, 6557–6567 10.1128/JB.182.23.6557-6564.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Hederstedt L., Piggot P. (1995). The cytochrome bc complex (menaquinone:cytochrome c reductase) in Bacillus subtilis has a nontraditional subunit organization. J. Bacteriol. 177, 6751–6760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Le Brun N. (1998). Studies of the cytochrome subunits of menaquinone:cytochrome c reductase (bc complex) of Bacillus subtilis. Evidence for the covalent attachment of heme to the cytochrome b subunit. J. Biol. Chem. 273, 8860–8866 10.1074/jbc.273.15.8860 [DOI] [PubMed] [Google Scholar]
- Zdobnov E. M., Apweiler R. (2001). InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848 10.1093/bioinformatics/17.9.847 [DOI] [PubMed] [Google Scholar]
- Zumft W., Kroneck P. (2007). Respiratory transformation of nitrous oxide (N2O) to dinitrogen by bacteria and archaea. Adv. Microb. Physiol. 52, 107–227 10.1016/S0065-2911(06)52003-X [DOI] [PubMed] [Google Scholar]
- Zumft W. G. (1997). Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft W. G. (2005a). Biogenesis of the bacterial respiratory Cu, A, Cu-S enzyme nitrous oxide reductase. J. Mol. Microbiol. Biotechnol. 10, 154–166 10.1159/000091562 [DOI] [PubMed] [Google Scholar]
- Zumft W. G. (2005b). Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme–copper oxidase type. J. Inorg. Biochem. 99, 194–215 10.1016/j.jinorgbio.2004.09.024 [DOI] [PubMed] [Google Scholar]