Abstract
The understanding that oncogenes can have profound effects on cellular metabolism and the discovery of mutations and alterations in several metabolism-related enzymes (IDH1, IDH2, SDH, FH, PKM2) has renewed interest in cancer metabolism and renewed hope of taking therapeutic advantage of cancer metabolism. Otto Warburg observed that aerobic glycolysis was a characteristic of cancer cells. More than 50-years later, we understand that aerobic glycolysis and uptake of glutamine and glycine allow cancer cells to produce energy (ATP) and the nucleotides, amino acids and lipids required for proliferation. Expression of the MYC oncogene drives the increase in cellular biomass facilitating proliferation. PKM2 expression in cancer cells stimulates aerobic glycolysis. Amongst intermediary metabolism enzyme, mutations in succinate dehydrogenase (SDH) occur in gastointestinal stromal tumors and result in a pseudohypoxic metabolic milieu. Fumarate hydratase (FH) mutations lead to a characteristic renal cell carcinoma. Isocitrate dehydrogenase (IDH1/2) mutations have been found in leukemias, gliomas, prostate cancer, colon cancer, thyroid cancer and sarcomas. These recently recognized oncogenic metabolic lesions may be selective targets for new anticancer therapeutics.
Keywords: Cancer metabolism, IDH1/2, PKM2, MYC, succinate dehydrogenase, fumarate hydratase
INTRODUCTION
Cancer requires Specific Metabolic Programming for Proliferation
Even in ancient times, cancer was recognized to be a problem of abnormal growth. The earliest anticancer drugs focused on attacking cancer through selective toxicity toward proliferating cells. Several of the earliest anticancer drugs are the purine and pyrimidine antimetabolites which directly target critical metabolic pathways required for proliferation (1). Taking advantage of the differential expression of specific enzymes by malignant cells led to the drug cyclophosphamide which was designed to be activated selectively in tumor cells which over-express phosphoramidase by a sequence of metabolic reactions (2). The Warburg effect, glycolysis under aerobic conditions, is one of the most highly validated observations in cancer research and continues to be of interest (3). The current wave of interest in cancer metabolism was stimulated by new knowledge regarding mutations in intermediary metabolism enzymes and by an understanding that `oncogenes' can have profound effects on cellular metabolic processes (4). This CCR Focus section highlights aspects of cancer metabolism and renewed hope of taking advantage of altered cancer metabolism therapeutically.
Cellular metabolic regulation is critically linked to availability of nutrients. Cellular proliferation requires active nutrient uptake, synthesis of DNA, RNA, proteins and lipids and energy (ATP). Thus, oncogenes that drive cells to proliferate, also directly or indirectly drive metabolic changes (5, 6). Inhibitors of cellular proliferation which block or terminate DNA synthesis or which cross-link or fragment DNA, characterize many classic anticancer agents. These drugs are directed toward normal cellular proliferation processes and therefore lack selectivity for neoplastic cells over proliferating normal cells. The same is true of classic inhibitors of intermediary metabolism, many of which were enzyme transition state analogs. Mutant enzymes in tumors provide the potential for discovery of selective, drug-like inhibitors of the mutant enzyme. The development of drug-like compounds which selectively block certain protein:protein interactions may allow targeting of amplified or over-expressed metabolism-controlling oncogenic proteins. The systematic characterization of the metabolic pathways active in cancer cells is an on-going endeavor. Jain et al. measured the consumption and release profiles of 219 small molecule metabolites from cell culture media in the NCI-60 cancer cell line panel, integrated these data with gene expression, and found that glycine consumption and the mitochondrial glycine biosynthetic pathway correlated with cellular proliferation rate (7, 8).
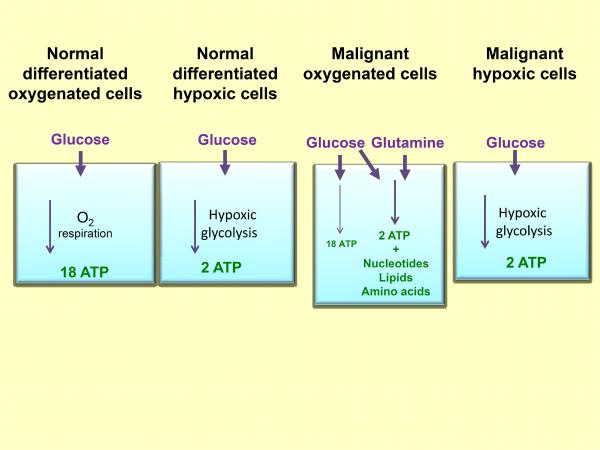
Normal differentiated cells generate the energy mainly by mitochondrial oxidative phosphorylation, while normal proliferating cells and cancer cells favor aerobic glycolysis, a less efficient process for generating ATP but a process that allows production of nucleotides, amino acids and lipids in addition to ATP, thus preparing the cell for division (Figures 1, 2)(9, 10). Cancer cells obtain energy and carbon intermediates from glycolysis without accumulation of reactive oxygen species which can damage DNA and other cellular components. Glucose glycolytic metabolism along with glutamine and glycine uptake provides the small carbon compounds, NADPH and ATP. Glucose glycolytic metabolism is dominant under hypoxic conditions (Figures 1, 2) (11). Understanding metabolism requires knowledge of metabolite profiles, concentrations and fluxes and of enzyme kinetics and networking of intracellular metabolite fluxes. Cascante et al. identified metabolite fluxes through the oxidative and nonoxidative pentose phosphate pathway, and routing of glucose to anaerobic glycolysis and the tricarboxylic acid (TCA) cycle (Figure 2) (12). Stable isotope resolved metabolomics was applied to analysis of gene dysregulations by infusing uniformly labeled 13C-glucose into patients followed by tumor resection. NMR and gas chromatography/mass spectroscopy (GCMS) were used for 13C -based metabolomics analysis. Lactate, alanine, succinate, glutamate, aspartate, and citrate in tumors were 13C-enriched, suggesting glycolysis and TCA cycle activity. Enhanced 13C-aspartate production with three 13C-labeled carbons and the buildup of 13C-2,3-glutamate was consistent with the incorporation of glucose into aspartate or glutamate via glycolysis, pyruvate carboxylation, and the TCA cycle (Figure 2). Pyruvate carboxylation may be important for replenishing TCA cycle intermediates which can be diverted to lipid, protein and nucleic acid biosynthesis (13). Several signaling pathways that incorporate nutrients into cellular components and some cancer-associated mutations facilitate nutrient metabolism to proliferation over ATP production (14).
Figure 1.
Schematic illustrating the sources of ATP energy used by normal and malignant cells under varied conditions of oxygenation.
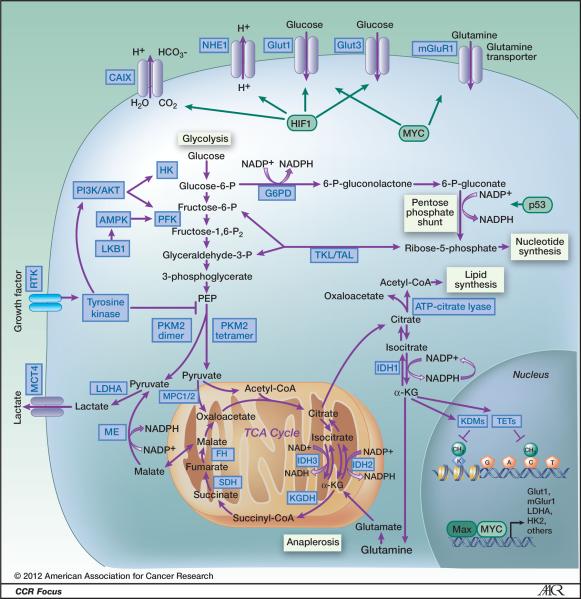
Figure 2.
Schematic illustrating metabolic pathways prominent in malignant cells. Blue boxes indicate enzymes and transporters that may be useful therapeutic targets in cancer. Green boxes represent transcription factors that alter metabolic pathways. Glycolysis is the ten-step metabolic pathway that converts glucose into pyruvate. The free energy released in this process is used to form the high-energy compounds ATP and NADH. The pentose phosphate pathway is a process that generates NADPH and 5-carbon sugars as alternative to glycolysis. Nucleotide synthesis provides molecules that make up the individual structural units of the nucleic acids RNA and DNA. In addition, nucleotides participate in cell signaling (cGMP and cAMP), are cofactors of enzymatic reactions, and nucleoside triphosphates are sources of chemical energy. Fatty acid synthesis is the synthesis of fatty acids from acetyl-CoA and malonyl-CoA precursors via fatty acid synthases. Anaplerotic reactions are those that replenish and maintain intermediates of a metabolic pathway. The abbreviations are: CAIX, Carbonic anhydrase IX; NHE1, Na+/H+ exchanger; Glut1, glucose transporter 1; Glut3, Glucose transporter 3; mGluR1, metabotropic glutamate receptor 1; PI3K/AKT, phophotidylinositol-3-kinase/Protein kinase B; G6PD, glucose-6-phosphate dehydrogenase; AMPK, AMP-activated protein kinase; PFK, phosphofructokinase; LKB1, serine/threonine kinase 11 (STK11), liver kinase B1; TKL/TAL, transketolase-transaldolase; RTK, receptor tyrosine kinase; PKM2, pyruvate kinase M2; MCT4, monocarboxylate transporter 4; IDH1 isocitrate dehydrogenase 1; IDH2, isocitrate dehydrogenase 2; IDH3, isocitrate dehydrogenase 3; LDH-A, lactate dehydrogenase A; MPC1/2, mitochondrial pyruvate carrier 1/2; ME, malic enzyme, malate dehydrogenase; FH, fumarate hydratase; SDH, succinate dehydrogenase; KGDH, 2-keto-glutarate dehydrogenase; TETs, DNA hydroxylases; KDMs, lysine histone demethylases; HK, hexokinase; HIF1, hypoxia-inducible factor 1; α-KG, α-ketoglutarate; TCA cycle, tricarboxylic acid cycle.
Aerobic Glycolysis Supports ATP Production and Macromolecular Synthesis
Uncoupling glycolysis from oxidative phosphorylation in the TCA cycle fulfills requirements for energy production and biosynthesis of cellular components. In glycolysis, the six-carbon sugar glucose is oxidized and split to make 2 molecules of pyruvate and 2 ATP molecules (Figures 1, 2). The pyruvate is further metabolized to acetyl-CoA, ethanol, or lactate. In normal oxygenated conditions, the pyruvate can be converted to acetyl-CoA and via the TCA cycle, and through oxidative phosphorylation to 18 ATP molecules (15).
Glutamine is an important tumor nutrient that contributes to nearly every metabolic function required of proliferating tumor cells (Figure 2). Glutamine is involved in bioenergetics, defense against oxidative stress and along with glucose metabolism in macromolecule precursor biosynthesis. The oncoprotein myc regulates glutamine uptake and degradation (16). Metabotropic glutamate receptors (gene: GRM1; protein: mGluR1) are G-protein-coupled protein receptors expressed in the normal central nervous system and by some cancers including melanoma and triple negative breast cancer. . mGluR1 inhibitors (Riluzole or BAY36-7620) or mGluR1 knockdown can inhibit growth of melanoma and triple negative breast cancer xenografts (17). mGluR1 has been implicated as an oncogene in melanomagenesis. Wangari-Talbot et al. developed a transgenic mouse expressing ectopic GRM1 in melanocytes which was sufficient to induce melanoma (18). Combining the kinase inhibitor sorafenib or the mutant BRaf inhibitor vemurafenib with riluzole had improved antitumor activity in GRM1-expressing melanoma cells harboring either wild-type or mutant B-RAF (19). Clinical trials with riluzole are underway in melanoma and other solid tumors (20, 21).
Myc and Hedgehog in Cancer Bioenergetics
The MYC oncogene is a driver in many human cancers stimulating cellular biomass accumulation (see review by Miller and colleagues in this issue; ref 22). The Myc transcription factor regulates genes involved in cell growth including those regulating ribosome and mitochondrial biogenesis, intermediary metabolism and glycolytic genes, enhancing glycolysis, glutamine uptake and metabolism and lactate production. In normal cells, the MYC gene is regulated by intracellular and extracellular cues such as nutrient and oxygen availability. When nutrients and/or oxygen are low, MYC is down-regulated. In many cancers, MYC is activated or altered resulting in dysregulated ribosome biogenesis and cell mass accumulation; thus, addicting cancer cells to nutrients. In the absence of glucose or glutamine cells with MYC over-expression die (23). Le et al. developed a MYC-inducible human Burkitt lymphoma P493 cell line and examined metabolic response under aerobic and hypoxic conditions, or glucose deprivation, using 13C-based metabolomics. 13C-glucose consumption and lactate production were increased by myc expression and hypoxia. 13C-glutamine uptake and metabolism by the TCA cycle persisted under hypoxia, and contributed to citrate carbons. In the absence of glucose, glutamine-derived fumarate, malate, and citrate increased. Glutamine is essential for cell survival and proliferation in hypoxia and/or low glucose which makes these cells susceptible to glutaminase inhibition (24). The relationship between oncogene signaling and glycolytic activity was explored using a Myc-driven liver cancer model to visualize glycolysis. Lactate dehydrogenase A (LDHA) abundance in tumors correlated with pyruvate conversion to lactate and decreased as tumors regressed (Figure 2) (25, 26). Targeting myc-max dimerization or myc-induced microRNA expression may also be effective therapeutically (27). Cancer cells require glucose and glutamine catabolism as well as other small molecules utilized for macromolecular synthesis to proliferate. Myc transcriptional regulation coordinates glutamine catabolism for protein and nucleotide biosynthesis. Myc-dependent glutaminolysis reprograms mitochondrial metabolism to glutamine catabolism to sustain TCA cycle activity. Mitochondrial glutamine metabolism decreases glucose carbon entering the TCA cycle and glucose mitochondrial-dependent synthesis of phospholipids. Thus, oncogenic MYC induces a transcriptional program that promotes glutaminolysis and cellular addiction to glutamine as a bioenergetic substrate (28).
Sonic hedgehog (Shh) signaling is aberrantly active in a spectrum of human malignancies. High lipid synthesis is a hallmark of Shh-driven medulloblastoma. Hedgehog signaling inactivates the Rb/E2F tumor suppressor complex to promote lipogenesis. Shh-metabolic activity facilitates tumor progression in an E2F1- and FASN-dependent manner. The nutrient sensor PPARγ, a component of the Shh metabolic network, regulates glycolysis and is elevated in Shh-driven medulloblastoma. PPARγ inhibition and/or Rb inactivation inhibits cell proliferation, and drives medulloblastoma cell death. Mitogenic Shh coupling to a nutrient sensor and metabolic transcriptional regulator defines the mechanism through which Shh drives cancer, controls the cell cycle, and regulates the glycolysis (29).
Addiction to Glucose and Glutamine
Targeting glucose metabolism may be a selective way to kill cancer cells. Several glycolytic enzymes are required to maintain a high glucose metabolism (30). Transcriptomic, proteomic, functional and structural properties of cancer cell mitochondria indicate impaired biogenesis and organelle activity. Cancer has a bioenergetics signature described by the ratio of oxidative phosphorylation and glycolysis that reflects metabolic activity and may correlate with response to chemotherapy (31). Some human carcinomas over-express mitochondrial ATPase Inhibitory Factor 1(IF1) which blocks the activity of mitochondrial H+-ATP synthase and facilitates metabolic adaptation to aerobic glycolysis. IF1 up-regulation in colon cancer cells triggers mitochondrial hyperpolarization and promotes proliferation (32).
Cancer cells are resistant to mitochondrial apoptosis perhaps due to up-regulation of glycolysis, accumulation of mitochondrial genome mutations, switch from respiration to glycolysis or the metabolic reprogramming from mutant enzymes like fumarate hydratase (FH) and succinate dehydrogenase (SDH)(33). In tumor cell mitochondria, oxidative metabolism of glucose- and glutamine-derived carbon produces citrate and acetyl-coenzyme A for lipid synthesis (Figure 2). Tumor cells with defective mitochondria use glutamine-dependent reductive carboxylation via mitochondrial and cytosolic NADP/NADPH-dependent isocitrate dehydrogenase (IDH) as the major citrate forming pathway. Metabolism of glutamine-derived citrate provides both the acetyl coenzyme A for lipid synthesis and the four-carbon intermediates needed to produce TCA cycle metabolites and related macromolecular precursors. The glutamine-dependent pathway is the dominant mode of metabolism in malignant cells with mutations in electron transport chain complex I or complex III, and in renal carcinoma cells with mutant FH. The glutamine-dependent pathway reverses many canonical TCA cycle reactions, supports tumor cell growth, and explains generation of TCA cycle intermediates by cells with impaired mitochondria (34).
Mitochondrial membrane permeabilization, a critical event in physiologic or stress induced apoptosis, is controlled by the permeability transition pore complex. Oncoproteins from the Bcl-2 family and tumor suppressor proteins from the Bax family interact with the permeability transition pore complex to inhibit or facilitate membrane permeabilization, respectively. Experimental agents including lonidamine, arsenite, betulinic acid, CD437, and several amphipathic cationic α-helical peptides, act on mitochondrial membranes and/or on the permeability transition pore complex to induce apoptosis. Mitochondrial membrane stabilization by anti-apoptotic Bcl-2-like proteins reduces the cytotoxicity of these agents (35, 36). In HCT116 colon cancer cells treated with oligomycin repression of nuclear-encoded mitochondrial protein synthesis occurs, but glycolytic protein synthesis is not affected. Translational control of mitochondrial proteins is controlled by AMP-activated protein kinase (AMPK), general control non-derepressible 2 (GCN2) kinase and silencing of activating transcription factor 4. Thus, glycolysis and oxidative phosphorylation are controlled at different levels of gene expression, providing the cell a mechanistic strategy for metabolic adaptation under stressful conditions (37). Metformin inhibits gluconeogenesis gene transcription in the liver, increases glucose uptake in skeletal muscle, and decreases circulating insulin. Thereby, metformin reduces circulating glucose, increases insulin sensitivity, reduces insulin resistance-associated hyperinsulinemia and activates AMPK. Preclinical data showing potential metformin anticancer activity along with epidemiological and retrospective data supporting metformin antineoplastic effects provided a rationale to evaluate metformin in clinical trials (38).
Using a synthetic lethal screen, Chan et al. identified compounds that exploit the loss of the von Hippel-Lindau tumor suppressor gene in renal cell carcinoma. Renal cell carcinoma is dependent on aerobic glycolysis for ATP production which is facilitated by induction of glucose transporter 1 (GLUT1) (Figure 2). Exposure to GLUT1 antagonists inhibits renal cell carcinoma cell proliferation by blocking glucose uptake (39). The spatial distribution of GLUT1 and monocarboxylate transporter (MCT) was examined in relation to vasculature and survival in NSCLC fresh frozen biopsies. Squamous cell carcinomas had the highest GLUT1 expression. In squamous cell carcinomas, GLUT1 and MCT expression increased with increasing distance from vasculature. In adenocarcinomas, high GLUT1 expression correlated with poor differentiation and high GLUT1 plus high MCT expression was associated with poor survival (40).
PKM2 and Glycolysis
Pyruvate kinase has two isoforms: M1 (PKM1) and M2 (PKM2). PKM2 expression in cancer cells stimulates aerobic glycolysis (Figure 2) (see review by Tamada and colleagues in this issue; ref 41). Several siRNAs targeting PKM2 decreased viability in multiple cancer cell lines. In vivo, siPKM2 delivery caused regression of established xenografts (42). Control of intracellular reactive oxygen species (ROS) is critical for cell survival. In human lung cancer cells, acute increases in intracellular ROS inhibited PKM2. PKM2 inhibition is required to direct glucose into the pentose phosphate pathway to generate reducing potential for ROS detoxification. Lung cancer cells transfected with an oxidation-resistant mutant PKM2 had increased sensitivity to oxidative stress. PKM2 may protect cancer cells from oxidative stress (43).
The mitochondrial pyruvate carrier (MPC) is a heterocomplex of MPC1 and MPC2on the inner mitochondrial membrane which carries pyruvate into mitochondria, (44). The absence of MPC1 or MPC2 impairs pyruvate oxidation. Human genetic defects in three families with children suffering from lactic acidosis and hyperpyruvatemia mapped to the MPC1 locus (45). MCT inhibition is a potential metabolic therapeutic target in cancer. Cancer cells proliferate more rapidly in the presence of pyruvate supplementation which stimulates mitochondrial oxygen consumption and increases reserve respiratory capacity. A MCT inhibitor decreased mitochondrial respiration and cell growth (46). PKM2 regulates the serine synthetic pathway. Tumor cells use serine-dependent regulation of PKM2 and GCN2 to modulate glycolytic intermediate flux in support of cell proliferation (47).
A proteomic screen for phosphotyrosine-binding proteins found that PKM2 binds directly to tyrosine-phosphorylated peptides resulting in release of fructose-1,6-bisphosphate and in PKM2 inhibition. PKM2 regulation by phosphotyrosine signaling diverts glucose metabolites from energy production to anabolic processes for cell growth (48). CD44 interacts with PKM2 and enhances the glycolytic phenotype of cancer cells that are either deficient in p53 or exposed to hypoxia. RNAi CD44 ablation increases metabolic flux to mitochondrial respiration and inhibits glycolysis and the pentose phosphate pathway (49).
Mutant Succinate Dehydrogenase (SDH) in Cancer
The frequent mutations of several metabolic enzymes in cancer support a driver role for altered metabolic enzymes in tumorigenesis (50). A mutation in a conserved SDH domain in two siblings with complex II deficiency presenting as Leigh syndrome, a rare neurometabolic disorder that affects the central nervous system, caused a mitochondrial respiratory chain deficiency (51). In cancer cells with mutant SDH, succinate can accumulate in mitochondria and cytosol, and inhibit prolyl hydroxylase enzymes. Depending on the specific prolyl hydroxylase inhibited, cells become resistant to apoptotic signals and/or activate a pseudohypoxic response that enhances glycolysis via hypoxia-inducible factor (HIF) (Figure 2) (52). Carney-Stratakis syndrome, an inherited condition predisposing individuals to gastrointestinal stromal tumor and paraganglioma, is caused by SDH germline mutations, leading to electron transport chain complex II dysfunction. When sporadic gastrointestinal stromal tumor patients were tested 4 of 34 patients (12%) had germline mutations in SDH (53, 54). SDH germline mutations are responsible for sporadic paragangliomas (6%), sporadic phaeochromocytomas (9%) and >80% of familial paraganglioma and phaeochromocytoma (55). The loss of SDH leads to accumulation of succinate that acts as a competitor to α-KG to inhibit TET catalyzed hydroxylation of 5-methylcytosine leading to DNA hypermethylation (56).
Mutant Fumarate Hydratase (FH) in Cancer
FH is an enzyme of the TCA cycle that catalyzes the hydration of fumarate into malate (Figure 2). Hereditary leiomyomatosis renal cell carcinoma, an aggressive form of renal cell carcinoma characterized by FH germline mutation, is highly metastatic and often lethal. FH absence leads to fumarate accumulation, which impairs HIF degradation at normal oxygen tensions. Cell survival without a functional TCA cycle was explored using Fh1-deleted mouse kidney cells and a computer model of metabolism in these cells. A metabolic pathway beginning with glutamine uptake and ending with bilirubin excretion from Fh1-deleted cells involving the biosynthesis and degradation of heme enabled Fh1-deleted cells to use accumulated TCA cycle metabolites and permitted some mitochondrial NADH production. Targeting this pathway was lethal to Fh1-deleted cells, but not Fh1 wild-type cells. Therefore, inhibition of heme oxygenation is synthetically lethal when combined with Fh1-deletion (57, 58). FH inactivation drives a metabolic shift to aerobic glycolysis in FH-deficient kidney tumors resulting in decreased AMPK and p53 tumor suppressor, and activation of the anabolic factors, acetyl-CoA carboxylase and ribosomal protein S6. Reduced AMPK leads to diminished iron transport. The consequent iron deficiency activates iron regulatory proteins, and increases expression of HIF-1α. HIF-1α silencing or AMPK activation decreases invasion, suggesting that HIF-1α and AMPK alterations contribute to FH-deficient cell growth (59). FH tumor suppressor gene loss results in tumor development that can progress to aggressive, metastatic cancers. Fh1 knockdown in mouse embryo fibroblasts results in increased c-met through HIF stabilization. Restoration of Fh1 expression abrogated the mitogenic and transformed phenotype of Fh1-deleted cells (60). Von Hippel-Lindau (VHL) or FH gene mutations produce morphologically distinct renal cell carcinomas with altered HIF-1αexpression resulting in a pseudohypoxic state (61). The UOK 262 cell line, derived from a patient with an aggressive, FH deficient renal cell carcinoma, was characterized for gene expression, chromosome profiles, bioenergetics, mitochondrial ultrastructure, FH catabolic activity, invasiveness, and glucose requirements. UOK262 cells have compromised oxidative phosphorylation, are dependent on aerobic glycolysis and glucose, have high lactate efflux, and over-express GLUT1 and LDHA (Figure 2). UOK262 xenografts retain the characteristics of FH-deficient renal cell carcinoma (62). Inactivating FH mutations in UOK262 cells result in glucose-mediated ROS generation and ROS-dependent HIF-1α stabilization (63). The germline FH missense mutation UOK268 cell line has a mutant FH protein which localizes to the mitochondria, but has almost no catalytic activity resulting in compromised oxidative phosphorylation and increased glycolytic flux (64). Fh1-deficient mouse embryonic fibroblast lines have up-regulated HIF-1α and HIF target genes, dependence on glycolysis, and an elevated lactate efflux, associated with glycolytic enzyme up-regulation. In a variety of FH-deficient tissues, there was increased PKM2 and LDHA (65).
IDH1 and IDH2 Mutants in Cancer
Frequent IDH1 and IDH2 heterozygous somatic mutations in acute myeloid leukemia (33%), glioblastoma (70% of low-grade gliomas and secondary glioblastomas) and chondrosarcomas (56%) have stimulated interest in targeting the mutant enzymes (see review by Yang and colleagues in this issue; ref 66). Mutant IDH1 and IDH2 activity changes in cell metabolic profile may tie to cancer epigenetics (67–69). Glioblastoma genome wide mutational analysis showed somatic IDH1 mutations occur frequently in tumors that evolved from lower-grade gliomas. Ichimura et al. screened IDH1 gene exon 4 for mutations in 596 primary intracranial tumors. Codon 132 mutation occurred in 54% of astrocytomas, 65% of oligodendroglial tumors and in 6% of glioblastomas (3% of primary and 50% of secondary glioblastomas). TP53 gene mutations and total 1p/19q deletions were mutually exclusive, and IDH1 mutation along with TP53 mutation or total 1p/19q loss was a frequent, early change in oligodendroglial tumors, diffuse astrocytomas, anaplastic astrocytomas, and secondary glioblastomas (70, 71). In 445 CNS tumors and 494 non-CNS tumors, mutations that affect IDH1 amino acid 132 were found in >70% of astrocytomas and oligodendrogliomas and in glioblastomas developed from lower-grade lesions. Tumors without IDH1mutations often had mutations affecting IDH2 amino acid R172. Patients with IDH1 or IDH2 mutant tumors had better outcomes than those with wild-type IDH genes (72, 73). When IDH1 mutant diffuse glioma infiltration patterns were determined by single tumor cell identification using an anti-mutant IDH1 R132H antibody, tumor cells appeared throughout the brain; thus, diffuse glioma is a whole brain disease (74, 75).
The frequency of IDH R132 mutations in 1200 mesenchymal tumors (220 cartilaginous tumors, 222 osteosarcomas and 750 bone and soft tissue tumors) was determined. Cartilaginous tumors and chondroblastic osteosarcomas, wild-type for IDH1 R132, were analyzed for IDH2 (R172, R140) mutations. Heterozygous somatic IDH1/IDH2 mutations were detected in 56% of central and periosteal cartilaginous tumors (40% R132C; 60% R132H). The ratio of IDH1:IDH2 mutations was 10.6:1. IDH1 and IDH2 mutations are the first common genetic abnormalities identified in conventional central and periosteal cartilaginous tumors (76).
The mutant IDH enzymes produce 2-hydroxyglutarate (2HG) from α-KG (Figure 2). 2HG-producing IDH mutants can prevent histone demethylation required for lineage-specific progenitor cells to differentiate. In glioma samples, IDH mutations associated with gene expression profiles enriched for genes expressed in neural progenitor cells, and increased histone methylation. In adipocytes, mutant IDH transfection or exposure to cell-permeable 2HG repressed differentiation gene expression and increased repressive histone methylation marks. Stable 2HG-producing mutant IDH transfection in immortalized astrocytes produced progressive histone methylation (77). Glioblastomas and other cancers with the CpG island methylator phenotype (CIMP) constitute a subset of tumors with extensive epigenomic aberrations. IDH1 mutation establishes CIMP by remodeling the methylome and transcriptome. Mutant IDH1 epigenomic alterations activate glioma-CIMP-positive proneural gene expression programs (78). Mutant IDH inhibitors in combination with the DNA methylation inhibitors, decitabine and azacitidine, may be useful therapeutically (79).
Human oligodendroglioma metabolites (>200) were profiled to examine mutant IDH1 and IDH2 effects. Amino acids, glutathione metabolites, choline derivatives and TCA cycle intermediates were altered in mutant IDH1- and IDH2-expressing cells. Similar changes occurred after exposure to 2HG. N-acetyl-aspartyl-glutamate (NAAG), a brain dipeptide, was reduced 50-fold in IDH1 mutant cells and 8.3-fold in IDH2 mutant cells (80). 2HG is a competitive inhibitor of α-KG-dependent dioxygenases, including histone demethylases and TET family 5-methlycytosine (5mC) hydroxylases (Figure 2). IDH1 mutations are associated with increased histone methylation and decreased 5-OH-mC (81). In 460 pediatric AML samples, a small number of genes distinguished IDH mutated patients and IDH wildtype patients, including a deregulated tryptophan metabolism pathway in IDH mutated cases (82). Mutational and epigenetic patient profiling showed that IDH1/2-mutant leukemias had DNA hypermethylation. IDH1/2 mutations were mutually exclusive from α-KG-dependent enzyme TET2 mutations, and TET2 loss-of-function mutations were associated with similar epigenetic defects as IDH1/2 mutations. Mutant IDH1/2 expression or TET2 depletion impaired hematopoietic differentiation and increased progenitor cell marker expression (83). Metabolomic studies of IDH1/2 mutant cells indicate alterations in glutamine, fatty acid, and citrate synthesis pathways (84, 85).
Ollier disease and Maffucci syndrome are non-hereditary skeletal disorders characterized by multiple enchondromas (Ollier disease) combined with soft tissue spindle cell hemangiomas (Maffucci syndrome). Somatic heterozygous mutations in IDH1 (R132C and R132H) or IDH2 (R172S) occur in 87% of enchondromas (benign cartilage tumors) and in 70% of spindle cell hemangiomas (benign vascular lesions). In total, 35 of 43 (81%) Ollier disease and 10 of 13 (77%) Maffucci syndrome patients carried IDH1 (98%) or IDH2 (2%) mutations. Immunohistochemistry for mutant IDH1 R132H protein suggested intraneoplastic and somatic mosaicism (86, 87).
Therapeutic Opportunities
Lessons from a century of metabolic research can be explained by a small set of biochemical principles. Glycolysis is a pathway that meets proliferating cellular demands (88). Metabolic modeling allows systematic, genome-scale study of human metabolism by using high-throughput `omics' data (see review by Jerby and Ruppin in this issue; ref 89). The first genome scale network model of cancer metabolism, validated by correctly identifying genes essential for proliferation in cancer cell lines, predicted 52 drug targets of which 40% are targeted by approved or experimental anticancer drugs (90).
Metabolomics is the quantitative assessment of endogenous metabolites within a biological system. Knowing a metabolome may allow early cancer detection and diagnosis or be a predictive and pharmacodynamic marker of drug effect. Metabolic and molecular imaging technologies, such as positron emission tomography (PET) and magnetic resonance spectroscopic imaging (MRI), enable noninvasive metabolic marker detection (91). The HIF1 pathway affects response to radiotherapy by HIF1 protection of vasculature after irradiation and regulation of glycolysis and the pentose phosphate pathway; thus increasing tumor antioxidant capacity (see review by Meijer and colleagues in this issue; ref 92). [18F]-fluorodeoxyglucose-PET images can be used to quantitatively determine glucose metabolic rate and pharmacokinetic rate constants in tissue volumes which is useful for radiotherapy Pharmacokinetic analysis was performed to determine the rate constants of fluorodeoxyglucose metabolism on 41 patients (104 lesions). Highest glucose metabolic rate tumor regions had high cellular uptake and phosphorylation rate constants with relatively low blood volume. In regions with less metabolic activity, the blood volume fraction was higher and cellular uptake and phosphorylation rate constants lower. Thus, tumor glucose phosphorylation rate was not dependent on nutrient transport (93, 94).
Understanding the metabolic basis of cancer has the potential to provide the foundation for the development of novel approaches targeting tumor metabolism. Tumors characterized by aerobic glycolysis and/or glucose-dependence could be more sensitive than other tumors to agents targeting the tumor vasculature and glucose transport. Tumors characterized by impaired TCA cycle function and/or respiration that is glutamine-dependent could be sensitive to agents targeting glutamine metabolism (such as glutaminase). Tumors which have impaired mitochondrial/electron transport function could be sensitive to agents which target the reductive carboxylation and fatty acid synthesis pathways. Malignancies which are characterized by IDH1, IDH2, fumarate hydratase or succinate dehydrogenase mutations could affect TET2 function, resulting in a hypermethylation phenotypes (56, 83). Such malignancies could be responsive to hypomethylating agents.
The re-emergence of interest in cancer metabolism stems from discoveries that a number of oncogenes and tumor suppressor genes control cell metabolism and that mutated intermediary metabolism enzymes can act as human tumor suppressors or oncogenes. Targeting cancer metabolism with drugs requires a therapeutic window in which tumor cells have a greater dependence on specific enzymes than normal tissues. Tumors with distinct oncogenic lesions are more likely to respond to drugs that target altered metabolic pathways (95).
REFERENCES
- 1.Parker WB. Enzymology of purine and pyrimidine antimetabolites used in the treatment of cancer. Chem Rev. 2009;109:2880–93. doi: 10.1021/cr900028p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brock N. Oxaphosphorine cytostatics: past-present-future. Cancer Res. 1989;49:1–7. [PubMed] [Google Scholar]
- 3.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 4.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell Death Dis. 2012;3:e248. doi: 10.1038/cddis.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148:1132–44. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–4. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomita M, Kami K. Systems biology, metabolomics, and cancer metabolism. Science. 2012;336:990–1. doi: 10.1126/science.1223066. [DOI] [PubMed] [Google Scholar]
- 9.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annual Rev Cell Dev Biol. 2011;27:441–64. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 11.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–90. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cascante M, Benito A, Zanuy M, Vizan P, Marin S, de Atauri P. Metabolic network adaptations in cancer as targets for novel therapies. Biochem Soc Trans. 2010;38:1302–6. doi: 10.1042/BST0381302. [DOI] [PubMed] [Google Scholar]
- 13.Fan TWM, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM. Altered regulation of metabolic pathways in human lung cancer discerned by 13C stable isotope-resolved metabolomics (SIRM) Molec Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nature Rev Drug Discovery. 2011;10:671–84. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 15.Porporato PE, Dhup S, Dadhich RK, Copetti T, Sonveaux P. Anticancer targets in the glycolytic metabolism of tumors: a comprehensive review. Frontiers Pharm. 2011;2:49. doi: 10.3389/fphar.2011.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeBerardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–24. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Speyer CL, Smith JS, Banda M, DeVries JA, Mekani T, Gorski DH. Metabotropic glutamate receptor-1: a potential therapeutic target for the treatment of breast cancer. Breast Cancer Res Treat. 2012;132:565–73. doi: 10.1007/s10549-011-1624-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wangari-Talbot J, Goydos J, Chen S. Role of the G protein-coupled receptor, mGlu1, in melanoma development. Pharmaceuticals. 2010;3:2821–37. doi: 10.3390/ph3092821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee HJ, Wall BA, Wangari-Talbot J, Shin SS, Rosenberg S, Chan JLK, Namkoong J, Goydos JS, Chen S. Glutamatergic pathway targeting in melanoma: single-agent and combinatorial therapies. Clin Cancer Res. 2011;17:7080–92. doi: 10.1158/1078-0432.CCR-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehnert JM, Lee JH, Shirk J, Bhavsar D, Anosike O, Chan JL, et al. A phase II trial of riluzole, an antagonist of metabotropic glutamate receptor 1 (GRM1) signaling, in metastatic melanoma. J Clin Oncol. 2010;28:15s. abstr TPS309. [Google Scholar]
- 21.Mehnert JM, Semlani N, Wen Y, Tan AR, Moss RA, Adams S, et al. A phase I trial of riluzole and sorafenib in patients with advanced solid tumors and melanoma. J Clin Oncol. 2012;30(suppl) abstr TPS3112. [Google Scholar]
- 22.Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. Myc and cancer metabolism. Clin Cancer Res. 2012;18:X. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le A, Rajeshkumar NV, Maitra A, Dang CV. Conceptual framework for cutting the pancreatic cancer fuel supply. Clin Cancer Res. 2012;18:4285–90. doi: 10.1158/1078-0432.CCR-12-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metabol. 2012;15:110–21. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu S, Balakrishnan A, Bok RA, Anderton B, Larson PEZ, Nelson SJ, et al. 13C-pyrivate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metabolism. 2011;14:131–42. doi: 10.1016/j.cmet.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward PS, Thompson CB. Signaling in control of cell growth and metabolism. Cold Spring Harb Perspect Biol. 2012 doi: 10.1101/cshperspect.a006783. doi: 10.1101/cshperspect.a006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhatia B, Potts CR, Guldal C, Choi SP, Korshunov A, Pfister S, Kenney AM, Nahle ZA. Hedgehog-mediated regulation of PPARγ controls metabolic patterns in neural precursors and shh-driven medulloblastoma. Acta Neuropathol. 2012;123:587–600. doi: 10.1007/s00401-012-0968-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamanaka RB, Chandel NS. Targeting glucose metabolism for cancer therapy. J Exp Med. 2012;209:211–5. doi: 10.1084/jem.20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Formentini L, Martinez-Reyes I, Cuezva JM. The mitochondrial bioenergetics capacity of carcinomas. IUBMB Life. 2010;62:554–60. doi: 10.1002/iub.352. [DOI] [PubMed] [Google Scholar]
- 32.Formentini L, Sanchez-Arago M, Sanchez-Cenizo L, Cuezva JM. The mitochondrial ATPase inhibitory factor 1 triggers a ROS-mediated retrograde prosurvival and proliferative response. Molec Cell. 2012;45:731–42. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Kroemer G. Mitochondrion in cancer. Oncogene. 2006;25:4630–32. doi: 10.1038/sj.onc.1209589. [DOI] [PubMed] [Google Scholar]
- 34.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al. Reductive carboxylation supports growth in tumor cells with defective mitochondria. Nature. 2011;481:385–8. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gottlieb E, Tomlinson IPM. Mitochondrial tumor suppressors: a genetic and biochemical update. Nature Rev Cancer. 2005;5:857–66. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 36.Costantini P, Jacotot E, Decaudin D, Kroemer G. Mitochondrion as a novel target of anticancer. J Natl Cancer Inst. 2000;92:1042–53. doi: 10.1093/jnci/92.13.1042. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Reyes I, Sanchez-Arago M, Cuezva JM. AMPK and GCN2-ATF4 signal the repression of mitochondria in colon cancer cells. Biochem J. 2012;444:249–59. doi: 10.1042/BJ20111829. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez-Angulo AM, Meric-Bernstam F. Metformin: a therapeutic opportunity in breast cancer. Clin Cancer Res. 2010;16:1695–700. doi: 10.1158/1078-0432.CCR-09-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan DA, Sutphin PD, Nguyen P, Turcotte S, Lai EW, Banh A, et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Trans Med. 2011;3:94ra70. doi: 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meijer TWH, Schurbiers OCJ, Kaanders JHAM, Looijen-Salamon MG, de Geus-Oei LF, Verhagen AFTM, et al. Differences in metabolism between adeno- and squamous cell non-small cell lung carcinomas: spatial distribution and prognostic value of GLUT1 and MCT4. Lung Cancer. 2012;76:316–23. doi: 10.1016/j.lungcan.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Tamada M, Suematsu M, Saya H. Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clin Cancer Res. 2012;18:X. doi: 10.1158/1078-0432.CCR-12-0859. [DOI] [PubMed] [Google Scholar]
- 42.Goldberg MS, Sharp PA. Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. J Exp Med. 2012;209:217–24. doi: 10.1084/jem.20111487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–83. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herzig S, Raemy E, Montessuit S, Veuthey JL, Zamboni N, Westermann B, et al. Identification and functional expression of the mitochondrial pyruvate carrier. Sciencexpress. 2012 doi: 10.1126/science.1218530. 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- 45.Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, et al. A mitochondrial pyrivate carrier required for pyruvate uptake in yeast, drosophila and humans. Sciencexpress. 2012 doi: 10.1126/science.1218099. 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diers AR, Broniowska KA, Chang CF, Hogg N. Pyruvate fuels mitochondrial respiration and proliferation of breast cancer cells: effect of monocarboxylate transporter inhibition. Biochem J. 2012;444:561–71. doi: 10.1042/BJ20120294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, Thompson CB. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci. 2012;109:6904–9. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–6. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 49.Tamada M, Nagano O, Tateyama S, Ohmura M, Yae T, Ishimoto T, et al. Modulation of glucose metabolism by CD44 contributes to antioxidant status and drug resistance in cancer cells. Cancer Res. 2012;72:1438–48. doi: 10.1158/0008-5472.CAN-11-3024. [DOI] [PubMed] [Google Scholar]
- 50.Oermann EK, Wu J, Guan KL, Xiong Y. Alterations of metabolic genes and metabolites in cancer. Semin Cell Develop Biol. 2012 doi: 10.1016/j.semcdb.2012.01.013. doi: 10.1016/j.semcdb.2012.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bourgeron T, Rustin P, Chretien D, Birch-Machin M, Bourgeois M, Viegas-Pequignot E, et al. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nature Genetics. 1995;11:144–9. doi: 10.1038/ng1095-144. [DOI] [PubMed] [Google Scholar]
- 52.King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–82. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- 53.Miettinen M, Killian JK, Wang ZF, Lasota J, Lau C, Jones L, Walker R, Pineda M, Zhu YJ, Kim SY, Helman L, Meltzer P. Immunohistochemical loss of succinate dehydrogenase subunit A (SDHA) in gastrointestinal stromal tumors (GISTs) signals SDHA germline mutation. Amer J Surg Pathol. 2012 doi: 10.1097/PAS.0b013e3182671178. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janeway KA, Kim SY, Lodish M, Nose V, Rustin P, Gaal J, et al. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc Natl Acad Sci. 2011;108:314–8. doi: 10.1073/pnas.1009199108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasini B, Stratakis CA. SDH mutations in tumorigenesis and inherited endocrine tumors: lesson from the phaeochromocytoma-paraganglioma syndromes. J Intern Med. 2009;266:19–42. doi: 10.1111/j.1365-2796.2009.02111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Develop. 2012;26:1326–38. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frezza C, Zheng L, Tennant DA, Papkovsky DB, Hedley BA, Kalna G, Watson DG, Gottlieb E. Metabolic profiling of hypoxic cells revealed a catabolic signature required for cell survival. PlosOne. 2011;6:e24411. doi: 10.1371/journal.pone.0024411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frezza C, Zheng L, Folger O, Rajagopalan KN, MacKenzie ED, Jerby L, et al. Haem oxygenase is synthetically lethal with the tumor suppressor fumarate hydratase. Nature. 2011;477:225–8. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- 59.Tong WH, Sourbier C, Kovtunovych G, Jeong SY, Vira M, Ghosh M, et al. The glycolytic shift in fumarate-hydrtase0deficient kidney cancer lowers AMPK levels, increases anabolic propensities and lowers cellular iron levels. Cancer Cell. 2011;20:315–27. doi: 10.1016/j.ccr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costa B, Dettori D, Lorenzato A, Bardella C, Coltella N, Martino C, et al. Fumarase tumor suppressor gene and MET oncogene cooperate in upholding transformation and tumorigenesis. FASEB J. 2010;24:2680–88. doi: 10.1096/fj.09-146928. [DOI] [PubMed] [Google Scholar]
- 61.Bratslavsky G, Sudarshan S, Neckers L, Linehan WM. Pseudohypoxic pathways in renal cell carcinoma. Clin Cancer Res. 2007;13:4667–71. doi: 10.1158/1078-0432.CCR-06-2510. [DOI] [PubMed] [Google Scholar]
- 62.Yang Y, Valera VA, Padilla-Nash HM, Sourbier C, Vocke CD, Vira MA, et al. UOK 262 cell line, fumarate hydratase deficient (FH-/FH-) hereditary leiomyomatosis renal cell carcinoma: in vitro and in vivo model of an aberrant energy metabolic pathway in human cancer. Cancer Genetics Cytogenetics. 2010;196:45–55. doi: 10.1016/j.cancergencyto.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sudarshan S, Sourbier C, Kong HS, Block K, Romero VAV, Yang Y, et al. Fumarate hydratase deficiency in renal cancer induces glycolytic addiction and hypoxia-inducible transcription factor 1a stabilization by glucose-dependent generation of reactive oxygen species. Molec Cell Biol. 2009;29:4080–90. doi: 10.1128/MCB.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Y, Valera V, Sourbier C, Vocke CD, Wei M, Pike L, et al. A novel fumarate hydratase-deficient HLRCC kidney cancer cell line, UOK268: a model of the Warburg effect in cancer. Cancer Genetics. 2012;205:377–90. doi: 10.1016/j.cancergen.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ashrafian H, O'Flaherty L, Adam J, Steeples V, Chung YL, East P, et al. Expression profiling in progressive stages of fumarate-hydratase deficiency: the contribution of metabolic changes to tumorigenesis. Cancer Res. 2010;70:9153–65. doi: 10.1158/0008-5472.CAN-10-1949. [DOI] [PubMed] [Google Scholar]
- 66.Yang H, Ye D, Guan K, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18:X. doi: 10.1158/1078-0432.CCR-12-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prensner JR, Chinnaiyan AM. Metabolism unhinged: IDH mutations in cancer. Nature Med. 2011;17:291–3. doi: 10.1038/nm0311-291. [DOI] [PubMed] [Google Scholar]
- 68.Kranendijk M, Struys EA, van Schaftingen E, Gibson KM, Kanhai WA, van der Knaap MS, et al. IDH2 mutations in patients with D-2-hydroxyglutaric aciduria. Sciencexpress. 2010 doi: 10.1126/science.1192632. 10.1126/science.1192632. [DOI] [PubMed] [Google Scholar]
- 69.Labussiere M, Sanson M, Idbaih A, Delattre JY. IDH1 gene mutations: a new paradigm in glioma prognosis and therapy? The Oncologist. 2010;15:196–9. doi: 10.1634/theoncologist.2009-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ichimura K, Pearson DM, Kocialkowski S, Backlund LM, Chan R, Jones DTW, Collins VP. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro-Oncol. 2009;11:341–7. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Metellus P, Colin C, Taieb D, Guedj E, Nanni-Metellus I, de Paula AM, et al. IDH mutation status impact on in vivo hypoxia biomarkers expression: new insights from a clinical, nuclear imaging and immunohistochemical study in 33 glioma patients. J Neurooncol. 2011;105:591–600. doi: 10.1007/s11060-011-0625-2. [DOI] [PubMed] [Google Scholar]
- 72.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gorovets D, Kannan K, Shen R, Kastenhuber ER, Islamdoust N, Campos C, et al. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower-grade diffuse astrocytic glioma. Clin Cancer Res. 2012;18:2490–501. doi: 10.1158/1078-0432.CCR-11-2977. [DOI] [PubMed] [Google Scholar]
- 74.Sahm F, Capper D, Jeibman A, Habel A, Paulus W, Troost D, van Deimling A. Addressing diffuse glioma as a systemic brain disease with single-cell analysis. Arch Neurol. 2011 doi: 10.1001/archneurol.2011.2910. doi:101001/archneurol.2011.2910. [DOI] [PubMed] [Google Scholar]
- 75.Narasimhaiah D, Miquel C, Verhamme E, Desclee P, Cosnard G, Godfraind C. IDH1 mutation, a genetic alteration associated with adult gliomatosis cerebri. Neuropathol. 2012;32:30–7. doi: 10.1111/j.1440-1789.2011.01216.x. [DOI] [PubMed] [Google Scholar]
- 76.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not other mesenchymal tumors. J Pathol. 2011;224:334–43. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 77.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsai HC, Van Neste L, Cai Y, Robert C, Rassool FV, Shin JJ, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–46. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci. 2011;108:3270–5. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of a-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:1–14. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Damm F, Thol F, Hollink I, Zimmermann M, Reinhardt K, van den Heuvel-Eibrink MM, et al. Prevalence and prognostic value of IDH1 and IDH2 mutations in childhood AML: a study of the AML-BFM and DCOG study groups. Leukemia. 2011;25:1704–10. doi: 10.1038/leu.2011.142. [DOI] [PubMed] [Google Scholar]
- 83.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Borodovsky A, Seltzer MJ, Riggins GJ. Altered cancer cell metabolism in gliomas with mutant IDH1 or IDH2. Curr Opin Oncol. 2012;24:83–9. doi: 10.1097/CCO.0b013e32834d816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. G Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pansuriya TC, van Eijk R, d'Adamo P, van Ruler MAJH, Kuijjer ML, Oosting J, et al. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nature Genetics. 2011;43:1256–61. doi: 10.1038/ng.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Amary MF, Damato S, Halai D, Eskandapour M, Berisha F, Bonar F, et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nature Genetics. 2011;43:1262–5. doi: 10.1038/ng.994. [DOI] [PubMed] [Google Scholar]
- 88.Bar-Even A, Flamholz A, Noor E, Milo R. Rethinking glycolysis: on the biochemical logic of metabolic pathways. Nature Chem Biol. 2012;8:509–17. doi: 10.1038/nchembio.971. [DOI] [PubMed] [Google Scholar]
- 89.Jerby L, Ruppin E. Predicting drug-targets and biomarkers of cancer via genome-scale metabolic modeling. Clin Cancer Res. 2012;18:X. doi: 10.1158/1078-0432.CCR-12-1856. [DOI] [PubMed] [Google Scholar]
- 90.Folger O, Jerby L, Frezza C, Gottlieb E, Ruppin E, Shlomi T. Predicting selective drug targets in cancer through metabolic networks. Molec Systems Biol. 2011;7:501. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15:431–40. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meijer TW, Kaanders JH, Span PN, Bussink J. Targeting hypoxia, HIF-1 and tumor glucose metabolism to improve radiotherapy efficacy. Clin Cancer Res. 2012;18:X. doi: 10.1158/1078-0432.CCR-12-0858. [DOI] [PubMed] [Google Scholar]
- 93.Vriens D, Disselhorst JA, Oyen WJG, de Geus-Oei LF, Visser EP. Quantitative assessment of heterogeneity in tumor metabolism using FDG-PET. Int J Rad Oncol Biol Phys. 2012;82:e725–31. doi: 10.1016/j.ijrobp.2011.11.039. [DOI] [PubMed] [Google Scholar]
- 94.Andersen EKF, Hole KH, Lund KV, Sundfor K, Kristensen GB, Lyng H, Malinen E. Dynamic contrast-enhanced MRI of cervical cancers: temporal percentile screening of contrast enhancement identifies parameters for prediction of chemoresistance. Int J Rad Oncol Biol Phys. 2012;82:e485–92. doi: 10.1016/j.ijrobp.2011.05.050. [DOI] [PubMed] [Google Scholar]
- 95.Shaw RJ, Cantley LC. Decoding key nodes in the metabolism of cancer cells: sugar & spice and all things nice. F1000 Biol Rep. 2012;4:2. doi: 10.3410/B4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]