Summary
The clinical success of EGFR inhibitors in lung cancer patients is limited by the inevitable development of treatment resistance. Two reports in this issue of Cancer Discovery uncover additional mechanisms by which EGFR mutant lung cancers escape from EGFR kinase inhibitor treatment. These findings pave the way for clinical testing of new rational therapeutic strategies to prevent or overcome resistance to EGFR kinase inhibitors in the clinic.
A new paradigm for the development of systemic cancer therapy has emerged over the last decade with the move away from broadly cytotoxic agents to targeted therapies. This has been enabled by the identification of specific alterations that drive oncogenesis in a wide variety of tumor types and the development of small molecules or antibodies that specifically target these “oncogenic drivers”. Cancer cells driven by an oncogene are dependent on its activity for their growth and survival such that the cells die without it (oncogene dependence). Thus, clinical practice has moved into an era of precision medicine in which many cancer patients are treated with therapies that target a specific driver oncogene in the tumor. Despite the success of oncogene-targeted therapies, single-agent treatment with an inhibitor of an oncogenic driver is not curative in patients (with a few notable exceptions) because of both innate and acquired treatment resistance. This challenge provides strong motivation to discover the molecular mechanisms that tumors use to evade driver oncogene inhibition. The identification of these molecular events pinpoints potential biomarkers of response to oncogene inhibitor treatment and rational therapeutic targets to prevent or overcome resistance to oncogene inhibition in patients.
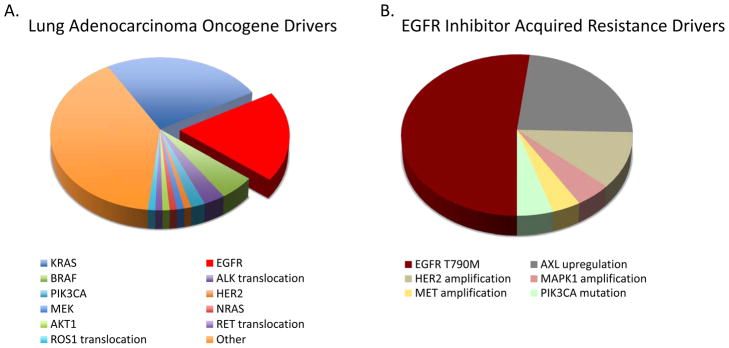
Lung cancers with activating mutations in the kinase domain of EGFR serve as a paradigm for the field of targeted therapeutics and precision cancer medicine. Tumors from patients with advanced non-small cell lung cancer (NSCLC) are routinely screened for the presence of these mutations in EGFR, which most commonly occur in exon 19 or exon 21 in the form of an in-frame deletion or a point mutation (L858R), respectively. These somatic mutations in EGFR occur in approximately 10–30 percent of NSCLC patients (Figure 1A)(1). In EGFR mutant lung cancer patients with advanced disease, treatment with an EGFR kinase inhibitor (erlotinib or gefitinib) is superior to standard cytotoxic chemotherapy and has therefore become first-line therapy (2). While the vast majority of patients initially respond to EGFR TKI treatment, acquired resistance to therapy inevitably develops in patients. Prior work by several groups has uncovered the cause of acquired resistance in many cases. In approximately 50–60 percent of cases, the mechanism of acquired resistance to EGFR TKI therapy is the acquisition of a second site T790M “gate keeper” mutation in the kinase domain of EGFR, in addition to the primary activating kinase domain mutation (3, 4). The second site T790M mutation in EGFR alters the binding of erlotinib and gefitinib to the ATP-binding pocket and therefore these inhibitors are unable to block EGFR signaling. Other mechanisms of acquired resistance to erlotinib and gefitinib include: 1) upregulation of the AXL kinase in approximately 20–25 percent of cases (5), 2) amplification of the MET kinase in approximately 5 percent of cases (3, 4), 3) activating mutations in the PIK3CA gene in approximately 5% of cases(6), and 4) histologic and phenotypic transformation to small cell lung cancer in approximately 5 percent of cases (6). The mechanisms of acquired resistance to first line EGFR TKI treatment are unclear in the remaining 15–20 percent of cases. Moreover, the potential ways in which EGFR mutant lung cancers may evade treatment with next generation EGFR kinase inhibitors developed to overcome EGFR T790M driven resistance and that are entering into the clinic are unknown. Two elegant studies by Ercan and colleagues (7) and by Takezawa and colleagues (8) in the current issue of Cancer Discovery shed new light on the mechanisms of acquired resistance to EGFR kinase inhibitors.
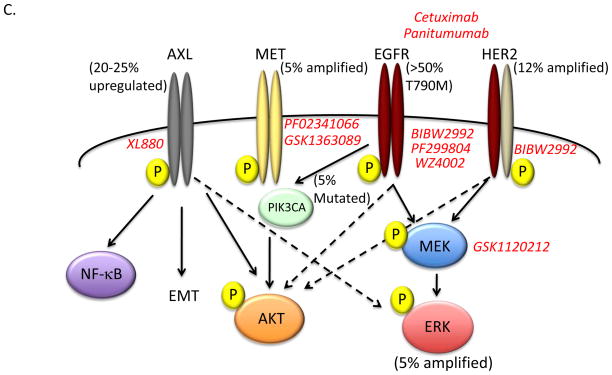
Figure 1. Mechanisms of acquired resistance to EGFR inhibitors and emerging pharmacologic approaches to overcome resistance.
(A) The relative frequency of specific oncogenic driver mutations in lung adenocarcinomas. Red wedge indicates the frequency of somatic activating mutations in EGFR (L858R or in frame exon 19 deletion). (B) The spectrum and frequency of known drivers of acquired resistance to EGFR inhibitor therapy in lung cancer. Two new drivers of acquired resistance are described in this issue of Cancer Discovery: MAPK1 amplification was seen in ~ 5% of patients (Ercan et al.) and HER2 amplification in ~ 12% of patients (Takezawa et al). Transformation to small cell lung cancer and epithelial to mesenchymal transition (EMT) have also been described as resistance mechanisms, however the frequency and degree to which these events drive EGFR TKI acquired resistance and the molecular pathways underlying these events have not been fully defined. (C) Schematic of pathways to EGFR inhibitor acquired resistance and pharmacologic approaches in development to overcome them. EGFR T790M mutation is the dominant driver of EGFR inhibitor resistance (50–60%). Second generation EGFR TKI inhibitors BIBW2992 (afatinib), PF299804 (dacomitinib), and WZ4002 covalently bind to EGFR and have shown promise as EGFRT790M inhibitors in preclinical studies. HER2 amplification may promote acquired resistance through heterodimerization with EGFR and activation of downstream signaling events (i.e. ERK and AKT). The combination of BIBW2992 together with the EGFR monoclonal antibody cetuximab or panitumumab can inhibit both EGFR and HER2 activity in cellular and murine models of EGFR-mutant driven lung cancer. MAPK1 amplification leads to increased ERK expression and elevated phospho-ERK levels. This may promote acquired resistance by promoting EGFR internalization. Inhibition of MEK activity by GSK-1120212 decreases ERK phosphorylation and overcomes acquired resistance driven by ERK overexpression in cellular and murine models. Upregulation AXL kinase activity occurs in 20–25% of patients with acquired resistance to EGFR inhibitors and may act through downstream effectors including NF-kappaB and AKT or by promoting EMT. XL-880 is an AXL inhibitor in clinical development that can overcome AXL-mediated resistance to erlotinib in models of EGFR-mutant lung cancer. MET amplification and PIK3CA mutations have has been described in ~ 5% of patients with acquired resistance. Several MET and PI3K inhibitors are in clinical development and testing.
Ercan et al focus on the clinical problem of EGFR T790M mediated resistance. In prior work, these authors developed a novel class of EGFR kinase inhibitors based on a pyrimidine scaffold that covalently bind and irreversibly inhibit mutant EGFR, including EGFR T790M, but not wild type EGFR (9). These inhibitors, which include a lead candidate WZ4002, are thus mutant selective and were designed to circumvent the limitations of other irreversible EGFR inhibitors including BIBW2992 (afatinib) (10) and PF299804 (dacomitinib) (11). In the current report, Ercan and colleagues used several established human cell line models of EGFR mutant lung cancer to determine the molecular events that could lead to resistance to WZ4002 treatment in EGFR mutant lung cancers. The group used a previously established isogenic model of acquired resistance to gefitinib that contains an EGFR exon 19 deletion/T790M compound mutant and exposed the cells to prolonged WZ4002 treatment to establish individual clones resistant to WZ4002 (WZR cells). Treatment of the WZR cells with WZ4002 resulted in suppression of EGFR phosphorylation, however, the authors noted persistently elevated levels of both phosphorylated and total ERK2. Through genome wide copy number analysis, the authors found that the WZR cells harbored amplification of the region of chromosome 22 harboring the MAPK1 gene that encodes ERK2. ERK2 was required for resistance in this system because genetic or pharmacologic inhibition of ERK2 restored sensitivity to WZ4002 treatment. Moreover, downregulation of several negative regulators of MAPK signaling, including the dual specificity phosphatase 6 (DUSP6), in the absence of MAPK1 amplification, was found as a potential alternative mechanism of acquired resistance to EGFR TKI treatment. Treatment with an allosteric MEK inhibitor restored EGFR TKI sensitivity in cellular and murine models of acquired resistance to EGFR TKI treatment that had increased MAPK signaling. Notably, combination therapy with WZ4002 and a MEK inhibitor prevented the emergence of resistance in EGFR mutant lung cancer cellular models in vitro. Mechanistic studies revealed that MAPK1 amplification could promote EGFR TKI resistance, at least in part, by enhancing internalization of EGFR. To clinically validate the preclinical findings, the authors investigated whether MAPK1 amplification occurred in clinical lung cancer specimens. Indeed, MAPK1 amplification was found in approximately 5 percent (1/21) of clinical specimens from patients with acquired resistance to EGFR TKI treatment and in which there was no evidence of EGFR T790M or MET amplification. Together the data indicate that hyperactivation of MAPK signaling can promote acquired resistance to EGFR TKI treatment. Additional studies are warranted to determine the full extent to which MAPK1 activation, either through MAPK1 amplification or downregulation of DUSP6 or other negative regulators of MAPK signaling, occurs in EGFR mutant lung cancer patients with acquired EGFR TKI resistance. Collectively, the data provide rationale for clinical testing of a MEK inhibitor in combination with WZ4002 or other EGFR TKIs to prevent or overcome EGFR TKI resistance in EGFR mutant lung cancer patients.
The companion study by Takezawa et al complements the work of Ercan and colleagues and moves the field forward in parallel by uncovering a potential role for HER2 in acquired resistance to EGFR inhibitor treatment in EGFR mutant lung cancers that do not harbor the EGFR T790M resistance mutation. The authors set out to determine a molecular basis for the apparent synergistic effects of treatment with the EGFR TKI afatinib (BIBW2992) in combination with the anti-EGFR monoclonal antibody cetuximab in EGFR T790M mediated resistance in lung cancer. Using an approach similar to Ercan and colleagues, Takezawa and colleagues studied the basis of EGFR TKI resistance in isogenic cellular models of acquired resistance to afatinib that also harbor the EGFR T790M mutation. Through a comprehensive and integrated series of in vitro and in vivo studies, the group found that HER2 levels were increased in the setting of resistance to multiple EGFR TKIs and that knockdown of HER2 enhanced sensitivity to EGFR TKI treatment in these models. This effect of HER2 suppression occurred even in EGFR mutant cells that were already relatively sensitive to EGFR TKI treatment, offering additional rationale for combined HER2/EGFR blockade. Conversely, the authors demonstrated that forced expression of HER2 promoted resistance to erlotinib but not to afatinib in EGFR mutant lung cancers that are otherwise relatively sensitive to erlotinib treatment. Furthermore, treatment with panitumumab, an alternative EGFR-directed antibody that is fully humanized and approved for clinical use (12), phenocopied the effects of cetuximab treatment in combination with afatinib and provided further support for a role for HER2 in this setting. Using an established in vivo model of EGFR L858R lung cancer in which acquired erlotinib resistance occurred after prolonged treatment, the authors validated their in vitro findings by demonstrating that 37 percent (7/19) of erlotinib resistant tumors had elevated levels of HER2 mRNA by quantitative RT-PCR. The authors validated their preclinical findings using a cohort of EGFR mutant human lung cancer specimens from patients that developed acquired resistance to erlotinib or gefitinib. HER2 amplification was observed by fluorescent in situ hybridization (FISH) in 12 percent (3/26) of the specimens from patients with EGFR TKI resistance in the absence of other known resistance mechanisms, including EGFR T790M. Together, the data suggest that HER2 upregulation is associated with acquired EGFR TKI resistance in EGFR mutant lung cancers. Whether increased levels and activation of HER2 can occur through mechanisms other than genomic amplification, such as epigenetic regulation or activating somatic mutations, in some EGFR TKI resistant lung cancers remains to be determined.
These two important studies in this issue of Cancer Discovery bring us closer to understanding the full spectrum of alterations that lead to clinical relapses in EGFR mutant lung cancer patients treated with an EGFR TKI (Figure 1B and 1C). The findings reported also prompt several new questions and areas for future investigations. First, it is critical to validate MAPK1 and HER2 hyperactivation, through genomic amplification or other mechanisms, as molecular drivers of resistance to EGFR TKI treatment in additional clinical cohorts that contain matched, paired specimens from EGFR TKI treated patients. Unfortunately, these specimens are rare. This is clearly an obstacle to rapid clinical translation of exciting laboratory discoveries, such as those reported by Ercan et al and Takezawa et al. This challenge is a call-to-arms to the oncology community to institute routine serial collection of clinical samples from patients prior to and during the course of therapy. This will undoubtedly accelerate progress in understanding the key determinants of treatment response in patients and enable the rational design of more effective therapeutic strategies to enhance clinical responses. By extension, the analysis of similar cohorts of clinical specimens from patients with other EGFR driven cancers who are treated with EGFR inhibitors could address whether MAPK1 or HER2 amplification may lead to resistance in additional clinical contexts.
Second, the clonal dynamics of the emergence or appearance of MAPK1 or HER2 upregulation remain to be clarified. Do these molecular events pre-exist in minor frequencies in tumors prior to therapy and emerge during the selective pressure of EGFR TKI treatment, as suggested for the presence of the EGFR T790M resistance allele (3, 4)? Or does EGFR TKI treatment directly or indirectly induce the de novo appearance of MAPK1 or HER2 amplification or activation in the tumors? Novel applications of next generation sequencing methodologies and single cell analysis of tumor cells within a population may provide answers to these unresolved questions.
Third, do we now have a complete picture of the ways in which EGFR mutant lung cancers can escape from EGFR TKI treatment based on the findings reported here and several prior studies (3, 4)? A more comprehensive analysis of clinical specimens from EGFR TKI treated patients should offer deeper insight into the degree to which the known mechanisms of resistance occur exclusively and concomitantly to promote clinical resistance. This is a key issue to resolve to clinically implement the identification of resistance mechanisms because we will need to determine whether to target individual or multiple drivers of resistance with targeted therapies in patients classified according to the molecular alterations present in the tumor. Such efforts should also clarify whether there are high frequency alterations in EGFR TKI resistant lung cancers that have not yet been identified or whether only lower frequency or private molecular events that contribute to resistance in individual patients remain to be uncovered.
Each of these unresolved issues relates directly to how the deeper understanding of the events that lead to EGFR inhibitor resistance could be leveraged to improve outcomes for lung cancer patients. Clinical trials that test whether inhibition of MAPK signaling, HER2 or other drivers of resistance improves clinical outcomes in EGFR TKI treated lung cancer patients will need to incorporate validated molecular tests performed on clinical samples to identify the secondary resistance alterations. This will enable molecular stratification of patients to the therapy(ies) targeted against the resistance driver(s). The challenge is to develop molecular testing for resistance drivers that is comprehensive, standardized and also practical to facilitate widespread clinical testing of biomarker driven therapeutic strategies in patients. Ultimately, the best way to overcome acquired resistance to any therapy is to prevent it from occurring in the first place. Thus, biomarker driven clinical studies testing appropriate companion therapies in the upfront setting are also warranted, as suggested by Ercan and colleagues. Mechanisms of resistance to targeted therapy are coming into full view in lung and other cancers. The challenge and hope is that we can capitalize on the knowledge gained and, through mechanism-based clinical trials, divide and conquer the molecular basis of resiliency that allows oncogene driven tumors to evade our initial precision attack.
References
- 1.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–80. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 2.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–42. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 3.Workman P, Clarke PA. Resisting targeted therapy: fifty ways to leave your EGFR. Cancer Cell. 2011;19:437–40. doi: 10.1016/j.ccr.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 4.Soria JC, Mok TS, Cappuzzo F, Janne PA. EGFR-mutated oncogene-addicted non-small cell lung cancer: current trends and future prospects. Cancer Treat Rev. 2012;38:416–30. doi: 10.1016/j.ctrv.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, Lee JC, Lin L, Olivas V, Au V, Laframboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–60. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ercan D. Reactivation of ERK Signaling causes resistance to EGFR kinase inhibitors. Cancer Discov. 2012 doi: 10.1158/2159-8290.CD-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takezawa K. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR mutant lung cancers that lack the second-site EGFR T790M mutation. Cancer Discov. 2012 doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, et al. Novel mutantselective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–4. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller VA, Hirsh V, Cadranel J, Chen YM, Park K, Kim SW, et al. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): a phase 2b/3 randomised trial. Lancet Oncol. 2012;13:528–38. doi: 10.1016/S1470-2045(12)70087-6. [DOI] [PubMed] [Google Scholar]
- 11.Ramalingam SS, Blackhall F, Krzakowski M, Barrios CH, Park K, Bover I, et al. Randomized Phase II Study of Dacomitinib (PF-00299804), an Irreversible Pan-Human Epidermal Growth Factor Receptor Inhibitor, Versus Erlotinib in Patients With Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. 2012 doi: 10.1200/JCO.2011.40.9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohenuram M, Saif MW. Panitumumab the first fully human monoclonal antibody: from the bench to the clinic. Anticancer Drugs. 2007;18:7–15. doi: 10.1097/CAD.0b013e32800feecb. [DOI] [PubMed] [Google Scholar]