 Mutants of the highly promiscuous lantipeptide synthetase ProcM phosphorylate a wide range of peptides attached to ProcA leader peptides, both in vitro and in E. coli. As such, the ProcM mutants are useful enzymes for the enzymatic preparation of phosphorylated peptides.
Mutants of the highly promiscuous lantipeptide synthetase ProcM phosphorylate a wide range of peptides attached to ProcA leader peptides, both in vitro and in E. coli. As such, the ProcM mutants are useful enzymes for the enzymatic preparation of phosphorylated peptides.
Abstract
Phosphorylation is an abundant post-translational modification involved in a myriad of cell signaling pathways. Herein, we have engineered the class II lantipeptide synthetase ProcM to generate a variety of peptides containing O-phosphoserine (pSer) and O-phosphothreonine (pThr) residues, either in vitro or in vivo.
Protein phosphorylation primarily occurs on serine, threonine, and tyrosine residues. Approximately 1.5–2.5% of the genes in higher organisms encode for kinases, underscoring the importance of this post-translational modification for normal cellular function.1,2 Reversible protein phosphorylation allows cells to regulate the functions of their proteomes and plays a significant role in signal transduction cascades and cell cycle regulation.3 Although many techniques have been developed to study phosphorylation and its effects on protein and cellular function, it remains difficult to generate proteins and peptides with defined phosphorylation patterns for detailed in vitro studies or antibody generation.
The site-selective incorporation of phosphate groups into peptides has been achieved mainly through solid phase peptide synthesis.4–7 These methods have some limitations such as often complex mixtures of crude reaction products.8 In an alternative approach, stop codon suppression and non-natural amino acid incorporation has been used to site-selectively place a pSer residue into the human mitogen-activated ERK activating kinase 1.9 Thus far, the efficiency of this approach for preparation of phosphopeptides has not been investigated.
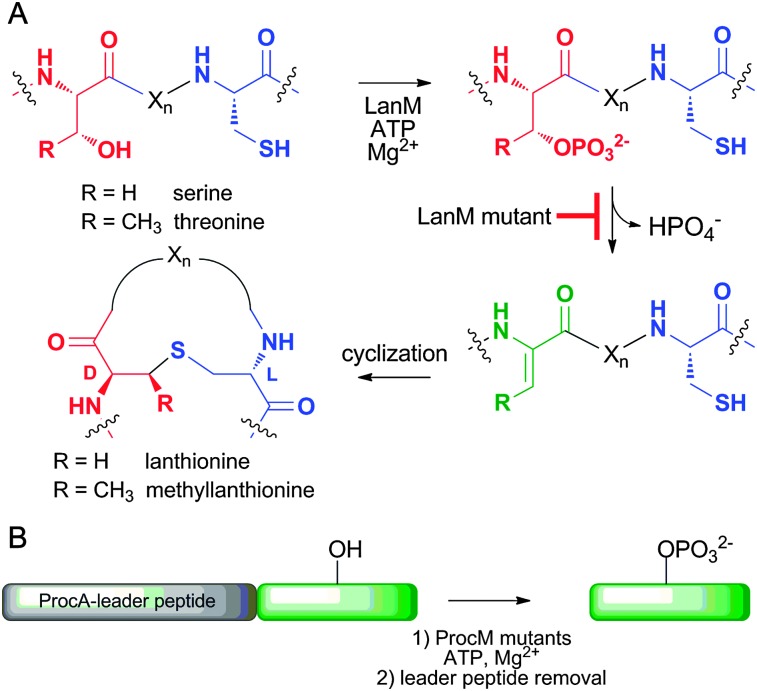
In an effort to develop potentially complementary methods for phosphopeptide synthesis, we have previously exploited the unique catalytic properties of the class II lantipeptide synthetases (termed LanM) for the in vitro synthesis of peptides with homogeneous phosphorylation patterns. LanM enzymes catalyse several steps during the maturation of lantipeptides, including a cryptic ATP-dependent phosphorylation step (Scheme 1). The peptide substrates of LanM enzymes (termed LanA peptides) are composed of an N-terminal leader peptide that plays a role in LanM binding and activation,10 and a C-terminal core peptide that harbors the post-translational modification sites (Scheme 1B). While investigating the mechanism of LctM involved in the biosynthesis of lacticin 481, two mutants (R399M and T405A) were identified that were deficient in the phosphate elimination step.11 These mutants were shown to catalyse in vitro phosphorylation of a variety of peptide substrates attached to the LctA leader peptide.12 Despite our initial success using LctM as a general Ser/Thr kinase, the LctM system has several drawbacks, which include low solubility of the LctA leader peptide, limited tolerance of variations in the sequence context of the Ser/Thr to be phosphorylated, and difficult scalability of an in vitro process that requires purified protein and substrate.
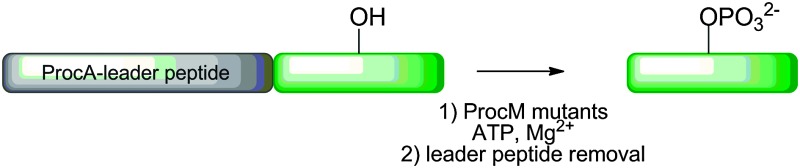
Scheme 1. (A) LanM-catalysed introduction of lanthionines and methyllanthionines into the core peptide of their LanA substrates via a cryptic phosphorylation step. Several mutations in these enzymes generate proteins that still phosphorylate but no longer eliminate. (B) General presentation of the strategy used herein. A peptide of interest was expressed fused to the C-terminus of ProcA leader peptides, phosphorylated with a ProcM mutant, and the leader peptide was removed by proteolysis.
In an attempt to phosphorylate peptides of interest with a wider substrate scope, in this study we chose to utilise the recently characterized class II lantipeptide synthetase ProcM.
This enzyme is involved in the biosynthesis of 29 different prochlorosins in Prochlorococcus MIT9313.13 The 29 ProcA substrates have strongly conserved leader peptides, but their core peptides are highly diverse. Collectively the 29 core peptides contain serines and threonines at every position from the 1st residue to the 24th residue, and nearly all of these residues are dehydrated. 13 Thus, ProcM appears to have no sequence or positional specificity with respect to dehydration (and hence phosphorylation, Scheme 1A) of Ser/Thr residues in the core peptide. Thus, we anticipated that ProcM would be able to modify peptides with a wide range of sequences, provided that a ProcA-like leader peptide is conjugated to the N-terminus of the target peptide.
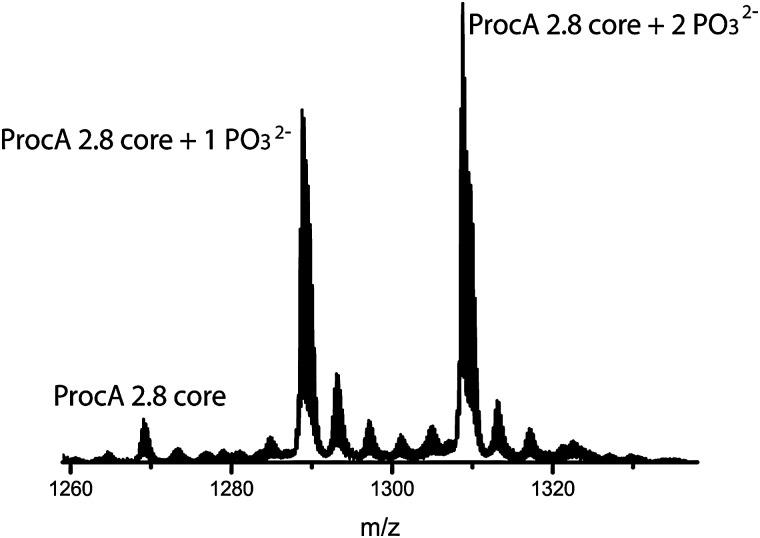
To obtain a ProcM-derived kinase, we performed an amino acid sequence alignment of LctM and ProcM and identified Arg510 and Thr516 of ProcM as analogous to Arg399 and Thr405 of LctM. These residues of ProcM were mutated to methionine and alanine, respectively. The mutant proteins were expressed in Escherichia coli as N-terminally hexa-histidine tagged fusion proteins for use in in vitro phosphorylation assays. Unfortunately, the R510M variant could not be purified because of solubility problems, and therefore all in vitro phosphorylation experiments were performed with the T516A mutant. Initially, the catalytic competence of T516A was tested with the natural ProcM substrates ProcA 1.1 and ProcA 2.8 (Fig. 1 and Fig. S1, ESI†). Reaction products were analysed by liquid chromatography electrospray ionization mass spectrometry (LC-ESI MS) following purification and proteolytic digestion of the peptide products to remove the leader peptide (Scheme 1B). In each assay, both mono- and diphosphorylated products were observed.
Fig. 1. ESI mass spectrum illustrating the phosphorylation of ProcA 2.8 by ProcM-T516A. Expected and experimental m/z values are listed in Table S1, ESI.† .
We next assessed whether the engineered ProcM kinase could accept non-native peptides as substrates (Table 1). Most of the early constructs contained the ProcA 1.7 leader sequence, but we later switched to the ProcA 2.8 leader sequence because of superior solubility (for amino acid sequences, see Table S2, ESI†). Fused to the C-terminus of the leader peptide was the peptide of interest. All substrates were expressed in E. coli as N-terminally hexa-histidine tagged peptides and purified by Ni2+-affinity chromatography and HPLC. The substrates were incubated with the T516A mutant as described in the ESI methods section. Following the reactions, the leader peptide was removed proteolytically using endoproteinases Lys-C or Glu-C (Table 1 and Table S2, ESI†), demonstrating that the phosphorylations took place in the core peptide by either LC ESI-MS or matrix assisted laser desorption ionization time of flight (MALDI-ToF) MS.
Table 1. Amino acid sequences attached to the leader peptides.
| Name | Protease a | ProcA-leader-peptide sequence b |
| In vitro | ||
| ProcA 1.1 | LysC | 1.1-FFCVQGTANRFTINVC |
| ProcA 2.8 | LysC | 2.8-AACHNHAPSMPPSYWEGEC |
| S1 | LysC | 2.8-SYNFNLFNNYNNNCY |
| S6 | LysC | 2.8-YHHYNSYNFNLFNNYNNNCY |
| S14 | LysC | 2.8-CYNFNLFNNYNNNSY |
| S19 | LysC | 2.8-YHHYNCYNFNLFNNYNNNSY |
| RW | LysC | 1.7-RWVRSALLI |
| RLT | GluC | 1.7-RLIKTFAYV |
| Proc-AKT | LysC | 1.7-ARKRERTYSFGHHA |
| Proc-CaM | LysC | 2.8-MRHQETVDCLK |
| Proc-PKCμ | LysC | 2.8-AALVRQMSVAFFFK |
| Proc-LR | LysC | 1.7-LRRASVA |
| Proc-PKG | GluC | 1.7-RKRSRAE |
| In vivo | ||
| Proc-CaM | LysC | 2.8-MRHQETVDCLK |
| Proc-PKCμ | LysC | 2.8-AALVRQMSVAFFFK |
| Proc-H1 | GluC | 2.8-GGGPATPKKAKKL |
| Proc-PKC | LysC | 2.8-RRGRTGRGRRGIFR |
aFor specific sites of proteolytic cleavage, see Table S2, ESI.
bPhosphorylated residues are shown in bold underlined font.
The peptide substrates S1, S6, S14, and S19 were used to explore the positional specificity of phosphorylation while keeping the flanking residues the same (Table 1). Peaks corresponding to the phosphorylated (+79.9) core peptides were observed for all four substrates as the major product (Fig. S2a–d, ESI†). This series of peptides demonstrates that the T516A kinase is flexible with respect to the position of phosphorylation sites in non-natural substrates. ProcM-T516A efficiently phosphorylated Ser residues adjacent to the leader sequence (S1) and as far as 19 residues C-terminal to the leader sequence (S19) (Fig. S2, ESI†). The RW and RLT peptides were previously shown to be phosphorylated by LctM-T405A when expressed as fusions to the LctA leader sequence.12 Treatment of RW and RLT sequences fused to the leader peptide of ProcA with the ProcM T516A mutant in vitro also resulted in efficient phosphorylation of the Ser residue (in RW) and of the Thr residue in RLT (Fig. S3, ESI†).
Having demonstrated the ability of T516A to efficiently phosphorylate non-native peptide sequences, we next tested whether this mutant could phosphorylate peptide sequences derived from naturally occurring phosphoproteins. For our study, we chose peptide sequences modified by AKT kinase (AKT), CaM kinase (CaM), protein kinase C (PKCμ), cAMP-dependent protein kinase (PKA and LR – also known as kemptide), and protein kinase G (PKG). As the aberrant activity of these kinases is linked to certain types of cancers, there is currently great interest in identifying phosphopeptide inhibitors that selectively target their activities.14–20 Therefore, these phosphorylated peptides are available commercially (Anaspec), but at considerable cost. The Proc-AKT, Proc-CaM, Proc-PKCμ, Proc-LR, and Proc-PKG peptides were all efficiently phosphorylated by ProcM T516A in vitro (Fig. S4, ESI†). These promising results suggest that the engineered ProcM enzyme could be a useful tool for the synthesis of biomedically relevant, homogeneous phosphopeptides.
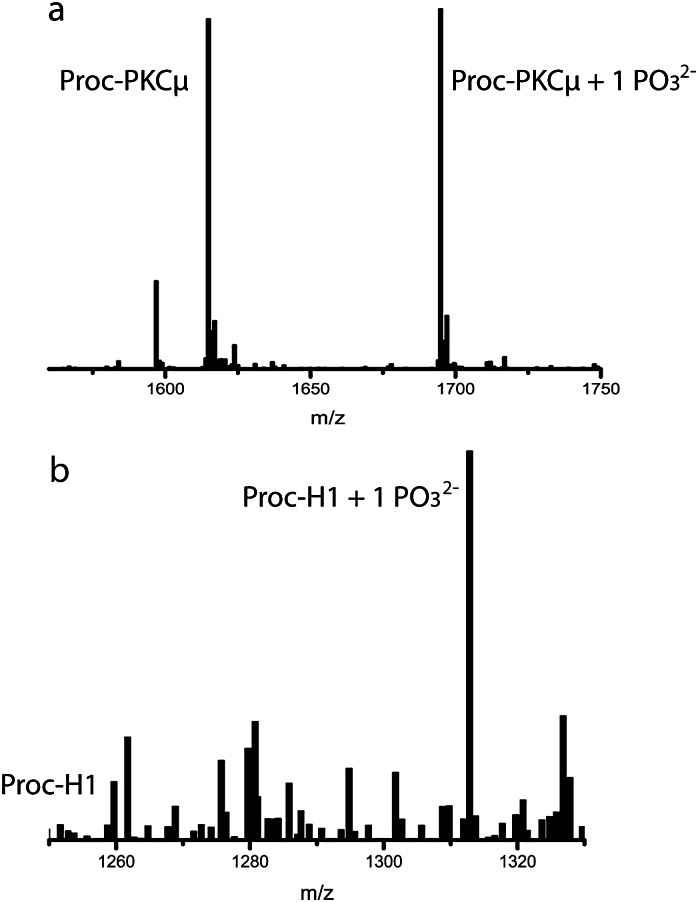
The methodology to prepare phosphorylated peptides in vitro is useful but difficult to scale up. Production of phosphopeptides by fermentation could potentially provide a solution. Prochlorosins have been successfully produced in E. coli by co-expression of ProcM and ProcA.21 Thus, the T516A and R510M ProcM mutants were co-expressed with ProcA2.8 in E. coli to evaluate their ability to produce phosphopeptides. Surprisingly, in these assays the ProcM-R510M mutant, which could not be purified in soluble form during the in vitro studies, clearly performed better than ProcM-T516A. Co-expression of the R510M mutant with the peptides Proc-PKCμ, Proc-H1 (histone H1 tail), Proc-CaM, and Proc-PKC (Table 1) resulted in phosphorylation of these peptides (Fig. 2 and Fig. S5 and S6a, ESI†). We then devoted some effort to larger scale production of a phosphorylated peptide. The crude mixture of phosphorylated and unphosphorylated His6-tagged Proc-PKC produced in E. coli was first purified by Ni-NTA affinity chromatography. The peptide was precipitated out of the imidazole-containing buffer and the ProcA 2.8 leader sequence was removed by proteolytic digestion with LysC. The digested peptide product was applied to a C18 HPLC column and the elution conditions were optimized to achieve base line resolution between phosphorylated and unphosphorylated PKC substrate (see ESI† and Fig. S6, ESI†). Thus, we were able to obtain highly pure, phosphorylated peptide from an in vivo biosynthesis reaction in a simple, two-step purification process. Comparable observations were made for two other peptides that were analyzed in a similar fashion (Proc-LR and Proc-PKCμ; Fig. S6, ESI†).
Fig. 2. Mass spectra for (a) Proc-PKCμ and (b) Proc-H1 after removal of the leader peptide. Calculated and observed m/z values are shown in Table S1, ESI.† .
In summary, we demonstrate that rationally engineered ProcM mutants can be used to phosphorylate serine and threonine residues located in a variety of peptide sequence contexts both in vitro and in vivo. These mutant enzymes exhibit relaxed substrate specificity in terms of both the position of the phosphorylation site (Ser or Thr) relative to the leader peptide and in the amino acid residues that flank the modification sites. Collectively, in the substrates shown in Table 1, Ser/Thr residues were phosphorylated in positions 1, 5–9, 12–14, and 19 after the end of the leader peptide. Furthermore, Ser residues adjacent to Pro, Gly, hydrophobic (AIVFM), charged (EKR), and polar residues were all phosphorylated. As such, ProcM mutants are more general as leader peptide dependent kinases than LctM mutants reported previously, and they may find utility as enzymatic tools for phosphopeptide synthesis. The ability to perform the phosphorylation in E. coli provides an additional improvement over the previous methodology and suggests potential applications in library synthesis and display techniques.
This work was supported by the National Institutes of Health (GM58822 to WAV). The authors thank Dr. Christopher Thibodeaux for help in editing the manuscript.
Footnotes
References
- Ozlu N., Akten B., Timm W., Haseley N., Steen H., Steen J. A. Wiley Interdiscip. Rev.: Syst. Biol. Med. 2010;2:255. doi: 10.1002/wsbm.41. [DOI] [PubMed] [Google Scholar]
- Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. Science. 2002;298:1912. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Hunter T. Cell. 2000;100:113. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- Vorherr T., Bannwarth W. Bioorg. Med. Chem. Lett. 1995;5:2661. [Google Scholar]
- Singer D., Hoffmann R. Methods Mol. Biol. 2008;494:209. doi: 10.1007/978-1-59745-419-3_12. [DOI] [PubMed] [Google Scholar]
- Kitas E. A., Perich J. W., Wade J. D., Johns R. B., Tregear G. W. Tetrahedron Lett. 1989;30:6229. [Google Scholar]
- Handa B. K., Hobbs C. J. J. Pept. Sci. 1998;4:138. doi: 10.1002/(sici)1099-1387(199804)4:2<138::aid-psc150>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Malik L., Tofteng A. P., Pedersen S. L., Sorensen K. K., Jensen K. J. J. Pept. Sci. 2010;16:506. doi: 10.1002/psc.1269. [DOI] [PubMed] [Google Scholar]
- Park H. S., Hohn M. J., Umehara T., Guo L. T., Osborne E. M., Benner J., Noren C. J., Rinehart J., Soll D. Science. 2011;333:1151. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton G. C., Paul M., Cooper L. E., Chatterjee C., van der Donk W. A. Biochemistry. 2008;47:7342. doi: 10.1021/bi800277d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y. O., van der Donk W. A. Biochemistry. 2007;46:5991. doi: 10.1021/bi602663x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y. O., Levengood M. R., Ihnken L. A., Knowlton A. K., van der Donk W. A. ACS Chem. Biol. 2009;4:379. doi: 10.1021/cb800309v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Sher D., Kelly L., Shi Y., Huang K., Knerr P. J., Joewono I., Rusch D., Chisholm S. W., van der Donk W. A. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10430. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsha H. C., Pandey A. Mol. Oncol. 2010;4:482. doi: 10.1016/j.molonc.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandlin I., Eiseler T., Salowsky R., Johannes F. J. J. Biol. Chem. 2002;277:45451. doi: 10.1074/jbc.M205299200. [DOI] [PubMed] [Google Scholar]
- Pomerantz A. H., Allfrey V. G., Merrifield R. B., Johnson E. M. Proc. Natl. Acad. Sci. U. S. A. 1977;74:4261. doi: 10.1073/pnas.74.10.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T., Yaffe M. B., Leparc G. G., Piro E. T., Maegawa H., Kashiwagi A., Kikkawa R., Cantley L. C. J. Biol. Chem. 2000;275:36108. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Toker A., Johannes F. J., Songyang Z., Cantley L. C. J. Biol. Chem. 1997;272:952. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- Zelada A., Passeron S., Lopes Gomes S., Cantore M. L. Eur. J. Biochem. 1998;252:245. doi: 10.1046/j.1432-1327.1998.2520245.x. [DOI] [PubMed] [Google Scholar]
- Bosco R., Melloni E., Celeghini C., Rimondi E., Vaccarezza M., Zauli G. Mini-Rev. Med. Chem. 2011;11:185. doi: 10.2174/138955711795049899. [DOI] [PubMed] [Google Scholar]
- Shi Y., Yang X., Garg N., van der Donk W. A. J. Am. Chem. Soc. 2011;133:2338. doi: 10.1021/ja109044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.