Abstract
Background. The mechanisms underlying smallpox vaccine-induced variations in immune responses are not well understood, but are of considerable interest to a deeper understanding of poxvirus immunity and correlates of protection.
Methods. We assessed transcriptional messenger RNA expression changes in 197 recipients of primary smallpox vaccination representing the extremes of humoral and cellular immune responses.
Results. The 20 most significant differentially expressed genes include a tumor necrosis factor–receptor superfamily member, an interferon (IFN) gene, a chemokine gene, zinc finger protein genes, nuclear factors, and histones (P ≤ 1.06E−20, q ≤ 2.64E−17). A pathway analysis identified 4 enriched pathways with cytokine production by the T-helper 17 subset of CD4+ T cells being the most significant pathway (P = 3.42E−05). Two pathways (antiviral actions of IFNs, P = 8.95E−05; and IFN-α/β signaling pathway, P = 2.92E−04), integral to innate immunity, were enriched when comparing high with low antibody responders (false discovery rate, < 0.05). Genes related to immune function and transcription (TLR8, P = .0002; DAPP1, P = .0003; LAMP3, P = 9.96E−05; NR4A2, P ≤ .0002; EGR3, P = 4.52E−05), and other genes with a possible impact on immunity (LNPEP, P = 3.72E−05; CAPRIN1, P = .0001; XRN1, P = .0001), were found to be expressed differentially in high versus low antibody responders.
Conclusion. We identified novel and known immunity-related genes and pathways that may account for differences in immune response to smallpox vaccination.
Despite the availability of effective smallpox vaccines and the eradication of smallpox in 1980, orthopoxviruses still remain legitimate public health threats, given the ongoing concerns of emerging orthopoxvirus diseases and the possible use of poxviruses as bioweapons [1–3]. There is a continued need for poxvirus research in virus biology, pathogenesis, and host response, including vaccine-induced immunity and correlates of protection [1, 3–8].
The duration and magnitude of protective immunity following smallpox immunization is still a topic of debate ranging from either lifelong protective immunity to only several years of persistence [9]. Even in populations with a documented vaccine vesicular “take,” or development of a pustule at the vaccination site, a small proportion (up to 2%) fail to mount a strong neutralizing antibody response [10]. The mechanisms underlying protection and the observed wide range of humoral and cellular immune host responses following smallpox vaccination and/or infection are still not understood, although they are of considerable interest to a deeper and more comprehensive understanding of poxvirus immunity and correlates of protection, as well as for the development of safer and more effective vaccines and antiviral therapies.
In this study, we assessed changes in messenger RNA (mRNA) expression using high-throughput microarray transcriptional profiling of 197 recipients of primary smallpox Dryvax vaccination with humoral and cellular immune responses at the extremes of the biological spectrum (out of 1076 healthy vaccinated subjects). We identified specific transcriptome signatures and pathways that might account for differences in immune response to smallpox vaccine.
MATERIALS AND METHODS
Study Subjects
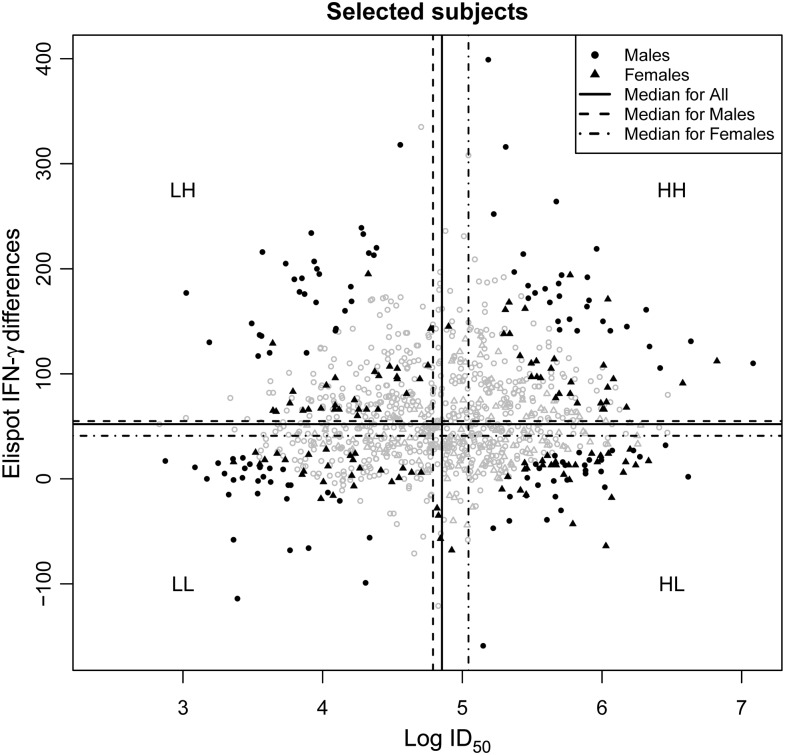
For gene expression profiling, we selected 200 subjects from the extremes (high and low) of the humoral (neutralizing antibody) and cellular (interferon γ [IFN-γ] Elispot) responses to primary smallpox vaccination out of 1076 healthy, eligible individuals (ages 18–40 years) whose demographic and immune variables were previously described [10]. As previously reported, these subjects were recruited as participants in a smallpox immunization program at the Naval Health Research Center (NHRC) in San Diego, CA, and the Department of Health and Human Services civilian healthcare worker smallpox immunization program at Mayo Clinic (Rochester, MN) [10, 11]. All study subjects were immunized with a single dose of Dryvax smallpox vaccine (Wyeth Laboratories, Marietta, PA) within 4 years prior to recruitment and had a documented vaccine vesicular “take.” Specifically, 4 sex-specific quadrants were defined on the basis of sex-specific medians to define high (above the median) and low (below the median) responses as demonstrated in Figure 1. For each of the humoral and cellular responses, the squared difference from the median was calculated per subject and then scaled to range from 0 to 1 for all 1076 subjects. For each subject, the product of the humoral and cellular scaled, squared deviations was then computed. Twenty-five males and 25 females having the largest of these values in each quadrant were chosen for study. The institutional review boards of the Mayo Clinic and NHRC granted approval to the study, and written informed consent was obtained from each study participant.
Figure 1.
Scatterplot of subjects selected for microarray studies, based on their vaccinia virus–specific humoral and cellular immune responses. Subjects selected for microarray studies from the overall cohort of 1076 subjects were dichotomized based on the magnitude of their cellular (differences in interferon γ [IFN-γ] Elispot counts in stimulated vs unstimulated cells; vertical axis) and humoral (log neutralizing antibody titer that inhibits 50% of virus activity [ID50; horizontal axis) responses. Fifty subjects (25 male and 25 female) were selected from each of 4 quadrants. The quadrants are defined as HH (high humoral and cellular responses), HL (high humoral and low cellular response), and LH (low humoral and high cellular response), and LL (low responses for both) according to assay medians. Vertical and horizontal dashed lines represent the median for males, overall, and for females, respectively, from left-to-right for vertical lines and from top-to-bottom for horizontal lines. Subjects with the extremes of the humoral and cellular response distributions were chosen for microarray analysis as described in the Study Subjects subsection of Materials and Methods. The dark colored filled symbols represent patients used in the microarray study (vs open symbols), while the circles and triangles represent males and females, respectively.
Viral Stocks and Immune Assays
For Elispot and cytokine secretion assays, we used vaccinia virus grown from Dryvax vaccine (a multiclonal vaccine containing a mixture of closely related vaccinia virus strains), and for measuring vaccinia-specific neutralizing antibody titers, we used the recombinant vaccinia virus vSC56, expressing β-galactosidase (a gift from B. Moss, National Institute of Allergy and Infectious Diseases). We propagated both viruses in HeLa S3 cells (ATCC) and purified, titered, and inactivated (when needed) the stocks as described previously [10].
Vaccinia-Specific Neutralizing Antibody Assay
We quantified neutralizing antibodies to vaccinia virus using a neutralization assay as previously described [10, 12]. Assay results were defined as the serum dilution that inhibits 50% of virus activity (ID50), as previously described [10]. The mean coefficient of variation (CV), based on 3 measurements, was 6.9% in our laboratory [10, 12].
IFN-γ Elispot Assay and Cytokine Measurements
We measured the frequencies of IFN-γ–producing cells in peripheral blood mononuclear cell (PBMC) cultures using total and CD8+ human IFN-γ Elispot kits (R & D Systems, Minneapolis, MN) as previously described and following the manufacturer's protocol [13]. The intraclass correlation coefficients comparing multiple observations per subject were 0.94 for stimulated values and 0.85 for unstimulated values, indicating good measurement reproducibility. We quantified vaccinia virus–specific secreted cytokines in PBMC cultures by enzyme-linked immunosorbent assay, as previously described [13].
Cell Culture, RNA Extraction, and Microarray Experiments
To improve cell viability, we rested PBMCs overnight with 50 IU/mL of interleukin 2, as described previously [13]. For the experiments, we stimulated half of the cells (the other half were left unstimulated as controls) with inactivated vaccinia virus at a multiplicity of infection of 0.5 plaque-forming units per cell for 18 hours, as described in our Taqman gene expression optimization study [14]. We stabilized the cells using RNAprotect cell reagent (Qiagen, Valencia, CA) and extracted total RNA using the RNeasy Plus mini kit (Qiagen, Valencia, CA). We assessed RNA quantity and quality by Nanodrop spectrophotometry (Thermo Fisher Scientific, Wilmington, DE) and Agilent 2100 Bioanalyzer chip kit analysis (Agilent, Palo Alto, CA). All microarray experiments were performed at the Mayo Advanced Genomics Technology Center Microarray Shared Resource core facility using hybridization to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays (Affymetrix, Santa Clara, CA) following a standardized operating procedure and the manufacturer's specifications.
Statistical Methods and Analysis
Sample run order was randomized to ensure the sex, ethnicity, and level of humoral and cellular responses were balanced across assay batches. Stimulated and unstimulated samples for a given subject were run in the same batch for all subjects. Analyses were performed on the perfect match data on the log2 scale. Data were assessed for quality using Affymetrix quality control metrics as well as graphical methods for assessing the existence of and functional form of bias and the success of normalization [15]. Neither stimulation status, sex, ethnicity, nor immune response status were expected to result in a global mRNA abundance shift in the cells. This assumption was evaluated by examining the distributions of percentage of Affymetrix present calls and scaling factors between stimulated and unstimulated samples, between sexes, and between immune status groups (quadrants). As these distributions did not differ between each of these groups, data from all arrays were normalized together. Data were normalized via the fast loess algorithm [16], a model-based, intensity-dependent algorithm similar to cyclic loess [15]; both methods use a loess curve (ie, a moving average as a function of expression level) to empirically estimate the specimen-specific intensity-dependent bias, but fast loess can be computed in a fraction of the time.
Linear mixed effects models [17–19] were used to assess significance of the effect of stimulation for each gene while accounting for the correlation between paired observations on the same subject. The fixed model terms were stimulation status (0 = unstimulated, 1 = stimulated), sex (male/female), and immune response (three 0/1 indicator variables for high humoral/high cellular, high humoral/low cellular, and low humoral/high cellular, with the low humoral/low cellular response group as the reference), and all 2-way interactions between these terms. Subject was included as a random effect in order to account for the correlation between paired specimens. Contrast statements were used to test the null hypothesis of no effect of stimulation overall and within sex or immune response quadrant. P values and false discovery rates (FDRs) [20, 21] were used to rank genes in order of significance. Genes and pathways with FDR < 0.05 were considered statistically significant. Statistical analyses were performed using the R software package (WU, Vienna, Austria). Pathway analysis was performed using the MetaCoreTM software from GeneGo (St. Joseph, MI). The GeneGo canonical pathway maps are a set of >650 known signaling and metabolic networks related to biological processes in humans.
RESULTS
Subjects Demographic Characteristics and Immune Responses
The study subjects were predominantly white (57%) and African American (16%), and 78% were non-Hispanic. The median age at enrollment was 24 years (interquartile range [IQR], 22–27 years), and the median time from vaccination to blood draw was 16.2 months (IQR, 9.0–34.6 months). Table 1 presents vaccine-specific immune responses for the subjects in the study, characterized by a stronger proinflammatory/innate response and a T-helper 1 cell (Th1)–biased cytokine response pattern as previously described for the overall population of 1076 subjects [13].
Table 1.
Vaccinia Virus–Specific Immune Responses in 197 Subjects Selected for Microarray Studies
| Variablea | Low Antibody Group | High Antibody Group | Low IFN-γ Elispot Group | High IFN-γ Elispot Group | All |
|---|---|---|---|---|---|
| Neutralizing antibody, ID50 | 54 (37–76) | 305 (243–408) | 132 (42–305) | 134 (60–302) | 134 (54–302) |
| Elispot CD8+ IFN-γ, spots/5 × 105 cells | 8 (−2 to 30) | 15 (−1 to 45) | 1 (−8 to 8) | 35 (13–59) | 10 (−2 to 39) |
| Elispot total IFN-γ, spots/2 × 105 cells | 44 (6–125) | 66 (13–141) | 9 (−13 to 16) | 138 (97–176) | 60 (9–138) |
| IFN-α, pg/mL | 54 (19–103) | 63 (11–119) | 38 (6–92) | 84 (25–140) | 56 (16–110) |
| IFN-β, pg/mL | 2 (−6 to 6) | 0 (−6 to 5) | 0 (−7 to 5) | 1 (−5 to 6) | 1 (−6 to 5) |
| IFN-γ, pg/mL | 117 (−30 to 1312) | 341 (41–1495) | 90 (−73 to 646) | 598 (78–1811) | 271 (0–1312) |
| IL-10, pg/mL | 3 (−1 to 11) | 3 (0–10) | 3 (0–7) | 3 (−1 to 15) | 3 (−1 to 10) |
| IL-12p40, pg/mL | 61 (25–118) | 62 (30–125) | 43 (21–99) | 76 (31–126) | 61 (27–119) |
| IL-12p70, pg/mL | 2 (1–5) | 4 (1–6) | 2 (1–4) | 4 (1–6) | 3 (1–5) |
| IL-18, pg/mL | 1 (−2 to 4) | 1 (−2 to 4) | 1 (−3 to 3) | 1 (−2 to 4) | 1 (−2 to 4) |
| IL-1β, pg/mL | 54 (23–126) | 46 (23–98) | 41 (23–85) | 58 (25–147) | 50 (23–113) |
| IL-2, pg/mL | 15 (4–35) | 21 (5–42) | 7 (0–27) | 24 (10–43) | 18 (4–39) |
| IL-4, pg/mL | 0 (−4 to 4) | 1 (−3 to 4) | 0 (−4 to 3) | 1 (−1 to 5) | 1 (−3 to 4) |
| IL-6, pg/mL | 1010 (623–1945) | 926 (517–1553) | 805 (442–1285) | 1163 (652–1924) | 977 (580–1836) |
| TNF-α, pg/mL | 169 (97–279) | 134 (70–310) | 132 (63–213) | 197 (101–374) | 149 (89–288) |
aData are median values (interquartile ranges). Elispot response and secreted cytokine response is defined as the subject-specific median vaccinia virus–stimulated response (measured in triplicates) minus the median unstimulated response (also measured in triplicates). Negative values indicate that stimulated values were on average smaller than unstimulated values.
Abbreviations: ID50, titer that inhibits 50% of virus activity; IFN, interferon; IL, interleukin; TNF, tumor necrosis factor.
Microarrays from 3 subjects were excluded because of higher background values, leaving a total of 197 subjects with 394 microarrays for further analysis (1 unstimulated and 1 vaccinia virus–stimulated sample/array per subject).
Overall Gene Expression in Response to Vaccinia Virus Stimulation
We compared overall differences between probe sets in all 197 stimulated versus all 197 unstimulated samples (regardless of immune response status) to assess overall response to stimulation in our study cohort. We identified 2103 statistically significant genes with an FDR of < 0.05 that were up/downregulated upon viral stimulation. The 20 most significant hits/genes differentially expressed in response to vaccinia virus stimulation are presented in Table 2 and include a tumor necrosis factor (TNF)–receptor superfamily member, an IFN gene and a chemokine gene, zinc finger protein genes, nuclear factors, and histones. We observed highly significant P values and FDR q values, although the observed estimates of the fold-change were relatively small but consistent between samples. Table 3 summarizes the immune response genes and families found among the top 200 differentially expressed genes, such as chemokines, cytokine and cytokine receptors, Toll-like receptors (TLRs), IFNs, antiviral proteins, transcription factors, and cytotoxic and other molecules that have documented roles in antiviral immunity. The analysis identified 29 704 out of the 54 613 transcripts as present reference genes for the pathway assessment (transcripts with absent calls in <50% of the 197 samples, either stimulated or unstimulated, were excluded). The 1022 genes/transcripts used as target genes for the pathway analysis were filtered based on a cutoff P value <.05 (5325 genes passed this filter) and a fold-change > 1.1. The pathway analysis, summarized in Table 4, identified 4 enriched pathways upon vaccinia virus stimulation that passed a FDR filter of 0.05 (P ≤ 2.05E−04) with cytokine production by the T-helper 17 (Th17) subset of CD4+ T cells being the most significant pathway involved (P = 3.42E−05).
Table 2.
Response to Vaccinia Virus Stimulation in Peripheral Blood Mononuclear Cells From 197 Vaccinees After Primary Smallpox Vaccinationa
| Genea | Gene Descriptiona | Log2 FCb | Pc | qc |
|---|---|---|---|---|
| TNFRSF10D | Tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain | 0.220 | 3.10E−48 | 8.46E−44 |
| IKZF1 | IKAROS family zinc finger 1 (Ikaros) | 0.397 | 8.63E−42 | 1.18E−37 |
| IKZF2 | IKAROS family zinc finger 2 (Helios) | 0.304 | 1.30E−40 | 1.42E−36 |
| Unknown | Unknown | 0.113 | 5.46E−34 | 4.98E−30 |
| Unknown | Unknown | 0.119 | 4.65E−31 | 3.63E−27 |
| NFE2L3 | Nuclear factor (erythroid-derived 2)–like 3 | 0.152 | 4.07E−30 | 2.78E−26 |
| POLR2C | Polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | 0.140 | 1.72E−28 | 1.04E−24 |
| HIST1H4E | Histone cluster 1, H4e | 0.106 | 1.02E−26 | 5.56E−23 |
| FAM71B | Family with sequence similarity 71, member B | 0.107 | 2.74E−25 | 1.36E−21 |
| RSRC1 | Arginine/serine-rich coiled-coil 1 | 0.096 | 3.46E−25 | 1.58E−21 |
| HERC5 | Hect domain and RLD 5 | 0.259 | 9.65E−23 | 4.06E−19 |
| IFNA1 | Interferon, alpha 1 | 0.223 | 2.38E−22 | 9.30E−19 |
| SOBP | Sine oculis binding protein homolog (Drosophila) | 0.191 | 1.18E−21 | 4.30E−18 |
| NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 0.132 | 2.79E−21 | 9.55E−18 |
| CD160 | CD160 molecule | 0.120 | 5.25E−21 | 1.69E−17 |
| XCL1 | Chemokine (C motif) ligand 1 | 0.131 | 5.56E−21 | 1.69E−17 |
| KIAA1967 | KIAA1967 | 0.122 | 6.66E−21 | 1.92E−17 |
| NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 0.131 | 7.10E−21 | 1.94E−17 |
| NEXN | Nexilin (F actin binding protein) | 0.244 | 9.36E−21 | 2.44E−17 |
| TNK2 | Tyrosine kinase, nonreceptor, 2 | 0.099 | 1.06E−20 | 2.64E−17 |
The analysis compares overall differences between probe sets in 197 stimulated and 197 unstimulated samples (regardless of immune response status) to assess overall response to stimulation in the study subjects within 4 years after primary Dryvax vaccination.
a Provided for gene identification/annotation.
b Defined as the log2 of the estimate of the fold-change for stimulated relative to unstimulated values.
c Measures of statistical significance, with q calculated to correct for the false discovery rate.
Table 3.
Immune Response Genes Identified in Comparison of Stimulated and Unstimulated Peripheral Blood Mononuclear Cells (PBMCs) from 197 Smallpox Vaccine Recipients
| Immune Response Genes, Differentially Expressed in Response to Vaccinia Stimulation | ||
|---|---|---|
| Gene Family | Gene(s)a | P Range |
| Chemokines | CCL20, CCL4, CXCL2, XCL1, XCL2 | 1.15E−19–2.4E−12 |
| Toll-like receptors | TLR3, TLR7 | 6.21E−15–2.55E−13 |
| Cytokines and cytokine receptors | IL-6, 1L12B, 1L23A, TNF, TNFSF10, TNFRSF10D | 3.10E−48–4.91E−12 |
| Antiviral proteins | EIF2AK2, IFI44, IFIT1, IFIT2, IFIT3, IFIT5, ZC3HAV1, ISG15, ISG20, MX1, MX2, OAS1, OAS2, OAS3, OASL, RSAD2, SP110, USP18, DDX58, IFIH1 | 4.11E−19–5.67E−11 |
| Interferons | IFNA1, IFNB1 | 2.38E−22–3.59E−11 |
| Transcription factors | IRF7 | 8.00E−16 |
| Other | FASLG, GZMB, CD36, CD160 | 5.25E−21–7.82E−11 |
The table summarizes the genes with known immune function among the top 200 most significant genes, identified in comparison of stimulated and unstimulated PBMCs from 197 subjects after primary smallpox vaccination.
a All presented genes passed the false discovery rate threshold.
Table 4.
Major Enriched Immune Response Pathways Expressed in 197 Subjects After Documented Primary Smallpox Vaccination
| Pathway | Pa | Network Objects |
|---|---|---|
| Enriched pathways upon vaccinia virus stimulation, overall analysis | ||
| 1. Cytokine production by T-helper 17 cellsa | 3.42E−05 | 11/26 |
| 2. Immune response: Antiviral actions of interferonsa | 5.95E−05 | 10/23 |
| 3. Immune response: Innate immune response to RNA viral infectiona | 1.02E−04 | 9/20 |
| 4. Immune response: PGE2 signaling in immune responsea | 2.05E−04 | 10/26 |
| Enriched pathways in high vs low antibody responders | ||
| 1. Immune response: Antiviral actions of interferonsa | 8.95E−05 | 6/25 |
| 2. Immune response: Interferon α/β signaling pathwaya | 2.92E−04 | 5/20 |
a All presented pathways passed the false discovery rate of < 0.05.
Response to Viral Stimulation in High Versus Low Antibody Responders to Smallpox Vaccination
We characterized and compared change in mRNA expression (from unstimulated to stimulated) between high and low antibody responders to primary smallpox vaccination and identified 3376 genes with a P value < .05. The top 20 most significant findings (P ≤ .0003) are presented in Table 5. These identified genes include immune function–related genes and transcription factors/transcriptional regulators (TLR8, P = .0002; DAPP1, P = .0003; LAMP3, P = 9.96E−05; NR4A2, P ≤ .0002; EGR3, P = 4.52E−05), as well as other genes with a possible impact on immune responses (LNPEP, P = 3.72E−05; CAPRIN1, P = .0001; XRN1, P = .0001), although none of the identified genes passed the threshold for FDR of 0.05. Genes that were differentially expressed in high versus low antibody responders to smallpox vaccination were further analyzed for pathway interactions using the MetaCore software, with 454 filtered genes (P < .05 and |FC| > 1.1) as target genes for the generation of results. Two pathways presented in Table 4 (antiviral actions of IFNs, P = 8.95E−05; and IFN-α/β signaling pathway, P = 2.92E−04) integral to innate immunity passed the threshold for FDR (P < .05) and were considered to be significant pathways in high versus low antibody responders to smallpox vaccine.
Table 5.
Differential Gene Expression in High Versus Low Antibody Responders to Primary Smallpox Vaccination
| Genea | Gene Descriptiona | Log2 FCb | Pc | qc |
|---|---|---|---|---|
| LNPEP | Leucyl/cystinyl aminopeptidase | 0.228 | 3.72E−05 | 0.586 |
| EGR3 | Early growth response 3 | 0.188 | 4.52E−05 | 0.586 |
| TRIB1 | Tribbles homolog 1 (Drosophila) | 0.168 | 6.14E−05 | 0.586 |
| NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 0.197 | 9.58E−05 | 0.586 |
| LAMP3 | Lysosomal-associated membrane protein 3 | 0.247 | 9.96E−05 | 0.586 |
| RABGAP1L | RAB GTPase activating protein 1-like | 0.183 | .0001 | 0.586 |
| CAPRIN1 | Cell cycle–associated protein 1 | −0.115 | .0001 | 0.586 |
| XRN1 | 5′-3′ exoribonuclease 1 | 0.231 | .0001 | 0.586 |
| SOBP | Sine oculis binding protein homolog (Drosophila) | 0.273 | .0001 | 0.586 |
| NCOA7 | Nuclear receptor coactivator 7 | 0.256 | .0002 | 0.586 |
| GPBP1 | GC-rich promoter binding protein 1 | 0.188 | .0002 | 0.586 |
| NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 0.188 | .0002 | 0.586 |
| GCNT2 | Glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) | 0.183 | .0002 | 0.586 |
| KCNS2 | Potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 | −0.085 | .0002 | 0.586 |
| SAMD9 | Sterile alpha motif domain containing 9 | 0.258 | .0002 | 0.586 |
| TLR8 | Toll-like receptor 8 | 0.253 | .0002 | 0.586 |
| WIPF1 | WAS/WASL interacting protein family, member 1 | 0.178 | .0002 | 0.586 |
| EMP1 | Epithelial membrane protein 1 | 0.221 | .0002 | 0.586 |
| ETV6 | Ets variant gene 6 (TEL oncogene) | 0.191 | .0002 | 0.586 |
| DAPP1 | Dual adaptor of phosphotyrosine and 3-phosphoinositides | 0.197 | .0003 | 0.586 |
The analysis compares change in messenger RNA expression (from unstimulated to stimulated) between high and low antibody responders within 4 years after primary Dryvax vaccination.
a The gene symbol and gene description are provided for gene identification/annotation.
b Defined as the log2 of the estimate of the fold-change for high antibody stimulated/high antibody unstimulated relative to low antibody stimulated/low antibody unstimulated.
c Measures of statistical significance, with q calculated to correct for the false discovery rate.
We also compared mRNA expression changes between the extremes of the cellular immune responses (CMI, as measured by the frequencies of specific IFN-γ–producing cells in the IFN-γ Elispot assay), but none of the identified genes and/or pathways passed the FDR threshold (Supplementary Table 1).
DISCUSSION
We and others have highlighted the important role of host genetic factors and host gene expression in the heterogeneity of immune response and adverse events following smallpox immunization [10, 11, 22–27]. However, the complex causes underlying interindividual differences in host response to vaccinia (and, presumably, to smallpox or other orthopoxviruses), including immunity and/or infection progression and outcome, still remain largely unknown, and research is warranted in view of recently emerging human and animal orthopoxvirus infections and bioterrorism threats [1, 4–8].
In this microarray-based study, we assessed and compared mRNA expression patterns in 197 high and low responders to smallpox vaccine (selected as “immune extremes” from a cohort of 1076 subjects) as determined by their humoral (neutralizing antibody) and cellular (IFN-γ Elispot) immune responses a median of 16 months (IQR, 9–34.6 months) after primary immunization. To the best of our knowledge, there are no smallpox/vaccinia-specific gene expression studies with such a large sample size that evaluate immune phenotype at the extremes of the biological spectrum.
We evaluated overall gene expression in response to viral stimulation (regardless of immune response status) and observed numerous differentially expressed genes (5325 genes with P values < .05 and 1843 genes with q values < 0.05), including genes with highly significant P values and FDR q values as shown for the top 200 differentially expressed genes with P values ≤ 8.04E−11 and q values ≤ 2.20E−8. These genes included classical immune genes (summarized in Table 3), such as chemokines, cytokines and cytokine receptors, TLRs, IFNs, antiviral proteins, transcription factors, and cytotoxic and other immune molecules. We also observed novel genes (with unknown relation to immune function), such as nuclear factors, nuclear receptors, histones, zinc finger proteins, and actin binding proteins, which may account for observed phenotypic changes upon vaccinia virus encounter.
A number of studies have analyzed the global host transcriptional responses (although with a very limited number of samples) after immunization, following in vitro infection of human cells with vaccinia virus and MVA, or during a disease in a nonhuman primate model with variola virus [28–32]. Consistent with our results, most of these studies provide evidence for the differential expression of many type I IFN-activated genes, viral sensors (TLRs, cytosolic pattern recognition receptors), effectors with antiviral activity, type I IFN signaling pathway genes and transcription factors, as well as apoptosis and signal transduction–related genes in different cell types under different conditions.
The top gene in our study, TNFRSF10D (TRAIL4; P = 3.10E−48, q = 8.46E−44; Table 2), encodes a protein of the TNF-receptor superfamily with a truncated cytoplasmic death domain and an extracellular TNF-related apoptosis-inducing ligand (TRAIL)–binding domain and has been demonstrated to reduce TRAIL-induced apoptosis [33]. Similar to our results, a study by Yang et al [34] identified 2 important genes, TNFRSF10D and EMP1 (epithelial membrane protein EMP1 gene), which are related to apoptosis, as upregulated 2 hours following vaccinia virus infection, thus strengthening our observations. Inhibition and/or modulation of host-programmed cell death can enhance viral replication and was recently shown to augment innate immunity and inflammatory reaction to vaccinia virus by the changing cytokine/chemokine milieu and supporting the recruitment of immune effector γδ T cells, which may potentially alter vaccinia virus–induced adaptive immune responses [35].
Other differentially expressed gene candidates that are directly related to adaptive immune response and/or viral immunity are CD160 (P = 5.25E−21, q = 1.69E−17), a molecule expressed on cells with cytolytic effector activity (natural killer cells and CD8+ T lymphocytes) that binds classic and nonclassic MHC class I molecules and provides a proliferative signal for activated T cells, and XCL1/lymphotactin (P = 5.56E−21, q = 1.69E−17), which is a member of the C-chemokine family and is specifically chemotactic for T cells.
Other highly significant genes include the Ikaros family, zinc finger DNA binding proteins (IKZF1 and IKZF2; P ≤ 1.30E−40, q ≤ 1.42E−36) that are hematopoietic-specific transcription factors associated with chromatin remodeling and regulation of lymphocyte differentiation [36] with plausible but unknown relation to infection and vaccine response.
We identified several enriched gene pathways after viral stimulation, which were mostly related to immunity (including innate immunity to viral infection). The most significant pathway identified in our study, cytokine production by the Th17 subset of CD4+ T cells (P = 3.42E−05; Table 4), was directly linked to the production of interleukin 17 (IL-17), a cytokine with an important role in the progression of smallpox vaccine–induced lesion that was demonstrated to specifically alter vaccinia virus–specific humoral and cellular immune responses [37]. The fourth-ranked pathway discovered in our study, the prostaglandin E2 (PGE2) signaling in immune response pathway, is also directly related to Th17, since PGE2-EP4 signaling was previously found to promote inflammation through expansion of the Th17 subset and through Th1 differentiation [38].
To further dissect smallpox vaccine–induced immunity, we characterized and contrasted the transcriptome patterns observed in high vaccine antibody responders compared to low vaccine antibody responders. Although the identified single genes did not pass the threshold for FDR (which might be a type II error due to limited sample size), analyses in high versus low antibody responders suggested specific transcriptome signatures of known and novel immunity-related genes (Table 5) that might account for differences in immune response to smallpox vaccine, such as Toll-like receptor 8/TLR8, DAPP1 (dual adaptor of phosphotyrosine and 3-phosphoinositides), NR4A2 (essential transcription factor, triggering the production of inflammatory cytokines and involved in Th-17 production), LAMP3 (DC-lysosome-associated membrane glycoprotein), LNPEP (zinc-dependent aminopeptidase involved in cleaving of several peptide hormones), XRN1 (5′-3′ exoribonuclease, involved in mRNA metabolism), EGR3 (transcriptional regulator), CAPRIN1 (cell cycle–associated protein) and EMP1 (epithelial membrane protein 1). Little is known about the importance of these genes in immunity and/or viral immunity. TLR8 was demonstrated to play a key role in controlling immune responses through regulation of Treg cells, while DAPP1 functions as a B lymphocyte adaptor molecule critical for B-cell receptor/BCR downstream signaling, that is regulating BCR internalization and linking BCR to ERK and JNK activation in B cells [39, 40]. The lysosome-associated membrane glycoprotein 3 (LAMP3) gene was previously described as a virus-inducible gene, which directly modulates influenza A virus replication in human cells and therefore can potentially alter viral antigen load and virus-induced immunity [41]. Similar to the study by Yang et al [34], the apoptosis-related EMP1 gene was found to be upregulated upon viral stimulation in our study, with significant differences in gene expression between high antibody responders and low antibody responders (P = .0002; Table 5). Although the single gene analysis comparing immune phenotype extremes did not identify single genes that passed the FDR threshold, we identified 2 classical pathways central to innate immunity and host primary antiviral defense mechanisms that were found significant in high versus low antibody responders. These significant pathways—antiviral actions of IFNs and IFN-α/β signaling pathway (P ≤ 2.92E−04, FDR q value <0.05, Table 4)—were represented by multiple IFN-α/β signaling and/or antiviral pathway–related genes, such as IFNA1, IDO1, MX1, OAS1, STAT1, STAT2, WARS, IFIT2, PML, and USP18. IFNs and IFN-induced antiviral effectors are key in antiviral immunity [42, 43]. Type I IFNs bind to the IFN-α/β receptor and initiate signal transduction through associated Janus kinases (JAK1 and Tyk2) and signal transducers and activators of transcription (STAT1 and STAT2), leading to the induction and expression of various ISREs (IFN-stimulated response elements)–containing genes with direct antiviral and immunoregulatory activities. It was demonstrated that vaccinia virus specifically activates a TLR-independent pathway for the production of type I IFNs, which are central for effective vaccinia-specific innate and adaptive antiviral immune responses in vivo and therefore might account for the observed differences in immune response phenotype after vaccination [44].
We have also previously shown that polymorphisms in some IFN-induced antiviral genes (OAS1, RNASEL, MX1, DDX58, ADAR, TRIM5) are associated with immune response variations in measles and rubella vaccine recipients [45–48], which is consistent with the pathway gene expression differences observed for the “immune extremes” phenotypes in our smallpox vaccine study.
The strengths of our study include a comprehensive evaluation of gene expression in a relatively large cohort of subjects after primary smallpox vaccination, with “immune extreme” phenotypes. The study design included optimized experimental conditions for time and multiplicity of infection [14] and comprehensive statistical design and analysis addressing potential confounding variables. We also used primary human cells (PBMCs) reflecting the antiviral response in diverse cell populations, which is likely to reproduce what happens in vivo. Our results are in agreement and confirm findings from previous related gene expression studies, although the unique study design (examining immune extreme phenotypes in primary cell populations) and experimental conditions may explain some observed differences. In addition, some of the subjects' humoral responses in our study are near the middle of the humoral immunity distribution, which may dampen the observed effects. Our results provide a deeper insight into vaccine response to a live viral orthopoxvirus vaccine by explaining and characterizing interindividual differences in gene expression of single genes and pathways. Our study's primary limitation is functional validation of results, which is currently being planned. Such studies will aid in the identification, characterization, discrimination, and/or prediction of immune response gene profiles, as well as signatures and immune phenotypes related to host immune response variations after immunization and/or infection, thus promoting the directed and rational development of next-generation vaccines for prevention of orthopoxvirus and other viral diseases [49].
In summary, we identified transcriptome signatures and pathways, including novel and known genes integral to immune function, that characterize the response to vaccinia virus stimulation and may explain the observed differences in immune responses after primary smallpox vaccination. Our data demonstrate the power of high-throughput transcriptional profiling to analyze virus/host interactions and delineate which genes and pathways have the largest impact on variations in immunity to a live smallpox vaccine.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We gratefully acknowledge the volunteers who participated in the study; the Mayo Clinic Vaccine Research Group nurses; Kevin L. Russell, MD, and Megan Ryan, MD; and the NHRC team for subject recruitment. We thank the Mayo Clinic Vaccine Research Group staff for technical help. We also thank V. Shane Pankratz, Robert A. Vierkant, Megan O'Byrne, David A. Watson, and Ying Li for their contribution to statistical analyses.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Financial support. The project described was supported by the National Institutes of Health (grant AI 40065).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Nalca A, Rimoin AW, Bavari S, Whitehouse CA. Reemergence of monkeypox: prevalence, diagnostics, and countermeasures. Clin Infect Dis. 2005;41:1765–71. doi: 10.1086/498155. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy RB, Ovsyannikova IG, Jacobson RM, Poland GA. The immunology of smallpox vaccines. Curr Opin Immunol. 2009;21:314–20. doi: 10.1016/j.coi.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lane JM. Immunity to smallpox and vaccinia: the future of smallpox vaccines. Expert Rev Clin Immunol. 2006;2:325–7. doi: 10.1586/1744666X.2.3.325. [DOI] [PubMed] [Google Scholar]
- 4.Formenty P, Muntasir MO, Damon I, et al. Human monkeypox outbreak caused by novel virus belonging to Congo Basin clade, Sudan, 2005. Emerg Infect Dis. 2010;16:1539–45. doi: 10.3201/eid1610.100713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schatzmayr HG, Costa RV, Goncalves MC, D'Andrea PS, Barth OM. Human and animal infections by vaccinia-like viruses in the state of Rio de Janeiro: a novel expanding zoonosis. Vaccine. 2011;29 Suppl 4:D65–9. doi: 10.1016/j.vaccine.2011.09.105. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds MG, Damon IK. Outbreaks of human monkeypox after cessation of smallpox vaccination. Trends Microbiol. 2012;20:80–7. doi: 10.1016/j.tim.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Moussatche N, Damaso CR, McFadden G. When good vaccines go wild: Feral Orthopoxvirus in developing countries and beyond. J Infect Dev Ctries. 2008;2:156–73. doi: 10.3855/jidc.258. [DOI] [PubMed] [Google Scholar]
- 8.Reynolds MG, Davidson WB, Curns AT, et al. Spectrum of infection and risk factors for human monkeypox, United States, 2003. Emerg Infect Dis. 2007;13:1332–9. doi: 10.3201/eid1309.070175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammarlund E, Lewis MW, Hanifin JM, Mori M, Koudelka CW, Slifka MK. Antiviral immunity following smallpox virus infection: a case-control study. J Virol. 2010;84:12754–60. doi: 10.1128/JVI.01763-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennedy R, Ovsyannikova IG, Pankratz VS, et al. Gender effects on humoral immune response to smallpox vaccine. Vaccine. 2008;27:3319–23. doi: 10.1016/j.vaccine.2009.01.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haralambieva IH, Ovsyannikova IG, Dhiman N, et al. Common SNPs/haplotypes in IL18R1 and IL18 genes are associated with variations in humoral immunity to smallpox vaccination in Caucasians and African Americans. J Infect Dis. 2011;204:433–41. doi: 10.1093/infdis/jir268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy R, Pankratz VS, Swanson E, Watson D, Golding H, Poland GA. Statistical approach to estimate vaccinia-specific neutralizing antibody titers using a high-throughput assay. Clin Vaccine Immunol. 2009;16:1105–12. doi: 10.1128/CVI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Umlauf BJ, Ovsyannikova IG, Haralambieva IH, et al. Correlations between vaccinia-specific immune responses within a cohort of armed forces members. Viral Immunol. 2011;24:415–20. doi: 10.1089/vim.2011.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oberg AL, Dhiman N, Grill DE, Ryan JE, Kennedy RB, Poland GA. Optimizing high dimensional gene expression studies for immune response following smallpox vaccination using Taqman(R) Low density immune arrays. J Immunol Methods. 2011;366:69–78. doi: 10.1016/j.jim.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudoit S, Yang YH, Callow MJ, Speed TP. Statistical methods for identifying genes with differential expression in replicated cDNA microarray experiments. Statistica Sinica. 2002;12:111–39. [Google Scholar]
- 16.Ballman KV, Grill DE, Oberg AL, Therneau TM. Faster cyclic loess: normalizing RNA arrays via linear models. Bioinformatics. 2004;20:2778–86. doi: 10.1093/bioinformatics/bth327. [DOI] [PubMed] [Google Scholar]
- 17.Chu TM, Weir B, Wolfinger R. A systematic statistical linear modeling approach to oligonucleotide array experiments. Math Biosci. 2002;176:35–51. doi: 10.1016/s0025-5564(01)00107-9. [DOI] [PubMed] [Google Scholar]
- 18.Littell RC, Milliken GA, Stroup WW, Wolfinger RD. SAS system for mixed models. Cary, NC: SAS Institute; 2002. [Google Scholar]
- 19.Pinheiro JC, Bates DM. Mixed-Effects Models in S and S-PLUS. New York: Springer; 2004. [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- 21.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanley SL, Jr, Frey SE, Taillon-Miller P, et al. The immunogenetics of smallpox vaccination. J Infect Dis. 2007;196:212–9. doi: 10.1086/518794. [DOI] [PubMed] [Google Scholar]
- 23.Reif DM, McKinney BA, Motsinger AA, et al. Genetic basis for adverse events after smallpox vaccination. J Infect Dis. 2008;198:1–7. doi: 10.1086/588670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinney BA, Reif DM, Rock MT, et al. Cytokine expression patterns associated with systemic adverse events following smallpox immunization. J Infect Dis. 2006;194:444–53. doi: 10.1086/505503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reif DM, Motsinger-Reif AA, McKinney BA, Rock MT, Crowe JE, Jr, Moore JH. Integrated analysis of genetic and proteomic data identifies biomarkers associated with adverse events following smallpox vaccination. Genes Immun. 2009;10:112–9. doi: 10.1038/gene.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis NA, Crowe JE, Jr, Pajewski NM, McKinney BA. Surfing a genetic association interaction network to identify modulators of antibody response to smallpox vaccine. Genes Immun. 2010;11:630–6. doi: 10.1038/gene.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA. Human leukocyte antigen genotypes in the genetic control of adaptive immune responses to smallpox vaccine. J Infect Dis. 2011;203:1546–55. doi: 10.1093/infdis/jir167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubins KH, Hensley LE, Jahrling PB, et al. The host response to smallpox: analysis of the gene expression program in peripheral blood cells in a nonhuman primate model. Proc Natl Acad Sci U S A. 2004;101:15190–5. doi: 10.1073/pnas.0405759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harenberg A, Guillaume F, Ryan EJ, Burdin N, Spada F. Gene profiling analysis of ALVAC infected human monocyte derived dendritic cells. Vaccine. 2008;26:5004–13. doi: 10.1016/j.vaccine.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherer CA, Magness CL, Steiger KV, et al. Distinct gene expression profiles in peripheral blood mononuclear cells from patients infected with vaccinia virus, yellow fever 17D virus, or upper respiratory infections. Vaccine. 2007;25:6458–73. doi: 10.1016/j.vaccine.2007.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerra S, Najera JL, Gonzalez JM, et al. Distinct gene expression profiling after infection of immature human monocyte-derived dendritic cells by the attenuated poxvirus vectors MVA and NYVAC. J Virol. 2007;81:8707–21. doi: 10.1128/JVI.00444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guerra S, Lopez-Fernandez LA, Conde R, Pascual-Montano A, Harshman K, Esteban M. Microarray analysis reveals characteristic changes of host cell gene expression in response to attenuated modified vaccinia virus Ankara infection of human HeLa cells. J Virol. 2004;78:5820–34. doi: 10.1128/JVI.78.11.5820-5834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheikh MS, Fornace AJ., Jr Death and decoy receptors and p53-mediated apoptosis. Leukemia. 2000;14:1509–13. doi: 10.1038/sj.leu.2401865. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc Natl Acad Sci U S A. 2010;107:11513–8. doi: 10.1073/pnas.1006594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Challa S, Woelfel M, Guildford M, Moquin D, Chan FK. Viral cell death inhibitor MC159 enhances innate immunity against vaccinia virus infection. J Virol. 2010;84:10467–76. doi: 10.1128/JVI.00983-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georgopoulos K, Winandy S, Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu Rev Immunol. 1997;15:155–76. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- 37.Patera AC, Pesnicak L, Bertin J, Cohen JI. Interleukin 17 modulates the immune response to vaccinia virus infection. Virology. 2002;299:56–63. doi: 10.1006/viro.2002.1400. [DOI] [PubMed] [Google Scholar]
- 38.Yao C, Sakata D, Esaki Y, et al. Prostaglandin E2-EP4 signaling promotes immune inflammation through Th1 cell differentiation and Th17 cell expansion. Nat Med. 2009;15:633–40. doi: 10.1038/nm.1968. [DOI] [PubMed] [Google Scholar]
- 39.Peng G, Guo Z, Kiniwa Y, et al. Toll-like receptor 8-mediated reversal of CD4+ regulatory T cell function. Science. 2005;309:1380–4. doi: 10.1126/science.1113401. [DOI] [PubMed] [Google Scholar]
- 40.Niiro H, Allam A, Stoddart A, Brodsky FM, Marshall AJ, Clark EA. The B lymphocyte adaptor molecule of 32 kilodaltons (Bam32) regulates B cell antigen receptor internalization. J Immunol. 2004;173:5601–9. doi: 10.4049/jimmunol.173.9.5601. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Z, Xue Q, Wan Y, Yang Y, Wang J, Hung T. Lysosome-associated membrane glycoprotein 3 is involved in influenza A virus replication in human lung epithelial (A549) cells. Virol J. 2011;8:384. doi: 10.1186/1743-422X-8-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 43.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–68. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu J, Martinez J, Huang X, Yang Y. Innate immunity against vaccinia virus is mediated by TLR2 and requires TLR-independent production of IFN-beta. Blood. 2007;109:619–25. doi: 10.1182/blood-2006-06-027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ovsyannikova IG, Haralambieva IH, Dhiman N, et al. Polymorphisms in the vitamin A receptor and innate immunity genes influence the antibody response to rubella vaccination. J Infect Dis. 2010;201:207–13. doi: 10.1086/649588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ovsyannikova IG, Dhiman N, Haralambieva IH, et al. Rubella vaccine-induced cellular immunity: evidence of associations with polymorphisms in the Toll-like, vitamin A and D receptors, and innate immune response genes. Hum Genet. 2010;127:207–21. doi: 10.1007/s00439-009-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haralambieva IH, Dhiman N, Ovsyannikova IG, et al. 2′-5′-Oligoadenylate synthetase single-nucleotide polymorphisms and haplotypes are associated with variations in immune responses to rubella vaccine. Hum Immunol. 2010;71:383–91. doi: 10.1016/j.humimm.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haralambieva IH, Ovsyannikova IG, Umlauf BJ, et al. Genetic polymorphisms in host antiviral genes: associations with humoral and cellular immunity to measles vaccine. Vaccine. 2011;29:8988–97. doi: 10.1016/j.vaccine.2011.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haralambieva IH, Poland GA. Vaccinomics, predictive vaccinology and the future of vaccine development. Future Microbiol. 2010;5:1757–60. doi: 10.2217/fmb.10.146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.