Abstract
Background. We conducted a case-control study to identify risk factors for efflux overexpression, an important mechanism of fluoroquinolone resistance, among patients with fluoroquinolone-resistant Escherichia coli (FQREC) gastrointestinal tract colonization.
Methods. Three annual fecal surveillance surveys were performed hospital-wide, and all patients colonized with FQREC (levofloxacin minimum inhibitory concentration, ≥8 μg/mL) were included in the study. Cases and controls were defined on the basis of overexpression of the AcrAB efflux pump, as measured by the organic solvent tolerance (OST) assay. A multivariable logistic regression model was developed to identify risk factors for OST positivity among patients with FQREC colonization.
Results. Eighty-nine patients were colonized with FQREC: 44 (49.4%) and 45 (50.6%) patients had isolates that were OST-positive and OST-negative, respectively. On multivariable analyses, location on the surgical service was significantly associated with recovery of an OST-positive isolate (odds ratio, 7.36; 95% confidence interval, 1.82–29.7; P = .005). Furthermore, patients who had received a first-generation cephalosporin in the 30 days prior to sampling were less likely to have an OST-positive isolate (odds ratio, 0.20; 95% confidence interval, .04–.94; P = .04).
Conclusions. Among phenotypically identical FQREC isolates, different factors may drive the emergence of different resistance mechanisms. Further studies are needed to elucidate the relationship between antimicrobial use and specific resistance mechanisms.
Fluoroquinolones (FQs) are among the most commonly prescribed antimicrobial agents in both inpatient and outpatient healthcare settings in the United States [1, 2]. The increasing prevalence of FQ resistance is of significant public health concern given the association between FQ resistance and poor clinical outcomes, including increased mortality [3, 4]. FQ resistance is of particular concern for Escherichia coli [1, 5], given it is the most common gram-negative pathogen in both hospital and community settings [6, 7]. In addition, FQ-resistant organisms are often resistant to other antimicrobial classes, thereby further limiting treatment options for associated infections [1, 8].
In the laboratory, resistance to FQs in E. coli has typically been demonstrated to arise in a stepwise fashion [9, 10]. In the clinical setting, this stepwise selection of resistance likely occurs at the level of gastrointestinal tract colonization (eg, as a result of antimicrobial selection pressure) [11, 12]. The primary mechanism of FQ resistance in E. coli is chromosomal mutation in one or more of the genes encoding the target bacterial enzymes DNA gyrase and topoisomerase IV [10]. These mutations occur predominantly in the gyrA and parC genes in the quinolone resistance-determining region (QRDR) [10]. Efflux-mediated antimicrobial resistance is also an important mechanism leading to FQ resistance in gram-negative bacteria. For example, overproduction of the AcrAB-TolC pump in E. coli has been shown to contribute to clinical FQ resistance through increased drug efflux [13]. A recent study demonstrated that of a total of 149 E. coli isolates with reduced FQ susceptibility (levofloxacin minimum inhibitory concentration [MIC], >0.125 μg/mL) identified via hospital-wide fecal surveillance, 59 (39.6%) isolates demonstrated efflux overexpression by organic solvent tolerance. Furthermore, the isolates that exhibited efflux overexpression were significantly more likely to demonstrate resistance to other antimicrobial agents [14]. Other studies have also confirmed an association between efflux overexpression in E. coli and increased rates of resistance to other antimicrobial agents [15, 16].
These findings have clear implications for the clinical setting given the large proportion of isolates that exhibit efflux overexpression. In addition, FQ-resistant E. coli (FQREC) isolates with efflux overexpression may present a significant therapeutic challenge given limitations in available antimicrobial treatment options. Along these lines, improved understanding of risk factors for efflux overexpression among FQREC would help elucidate the environmental pressures, including use of antimicrobial agents that can inhibit or activate expression of efflux pumps, which contribute to selection of this particular resistance mechanism. However, to our knowledge, only 2 studies have explored risk factors for efflux overexpression among FQREC [17, 18]. Furthermore, these studies demonstrated conflicting results and were characterized by small sample sizes [17], failure to conduct multivariable analyses [18], and a focus on only clinical urinary isolates [18]. Therefore, we conducted this study to determine risk factors for efflux overexpression as a mechanism of resistance among inpatients with FQREC gastrointestinal tract colonization.
METHODS
Study Design and Setting
This case-control study was conducted at 2 hospitals in the University of Pennsylvania Health System in Philadelphia: the Hospital of the University of Pennsylvania (HUP), a 725-bed academic tertiary care medical center, and Penn Presbyterian Medical Center, a 344-bed urban community hospital. As described elsewhere [14, 19], 3 annual fecal surveillance surveys were performed hospital-wide at the 2 hospitals during the study years 2002, 2003, and 2004. For the present study, a subject could only be included once, with inclusion of only the first sample for each subject. The study was approved by the institutional review board of the University of Pennsylvania.
Detection of FQREC from fecal samples, as well as evaluation for specific mechanisms of FQ resistance, was performed as described elsewhere [14, 19]. For the present study, E. coli isolates with a levofloxacin MIC of ≥8 μg/mL were classified as FQ resistant. Only FQREC isolates were included in this study. As previously validated [14], overexpression of AcrAB was measured indirectly by the organic solvent tolerance assay [20, 21]. Cases and controls were defined solely on the basis of organic solvent tolerance (OST), with case patients designated as those OST-positive and control patients as those OST-negative. Finally, the genetic relatedness of E. coli isolates was determined by molecular typing using pulsed-field gel electrophoresis (PFGE) [14], with all results analyzed using the Fingerprinting II Informatix software (version 3.0; Bio-Rad Laboratories) and interpreted according to established criteria [22].
Data Collection
Data were abstracted from the Pennsylvania Integrated Clinical and Administrative Research Database [23], which includes demographic, laboratory, pharmacy, and billing information. Information for all patients was collected on the following: baseline demographics, year of culture, location within the hospital and service (ie, medical vs surgical) at the time of sampling, and number of hospital and intensive care unit days prior to sampling. The presence of the following comorbid conditions was documented at the time of the sampling date: diabetes mellitus, malignancy, renal insufficiency (creatinine level of ≥2.0 mg/dL or the requirement of dialysis), human immunodeficiency virus (HIV) infection, and solid organ or hematopoietic stem cell transplant. In addition, the Charlson comorbidity index was calculated for each subject [24]. Furthermore, data on antimicrobial therapy, chemotherapy, and steroids or other immunosuppressive agents administered during the 30 days prior to fecal sampling were ascertained as described elsewhere [19]. Antimicrobial agents were categorized by specific agents (if only one agent of a given class was used) and by class, including aminoglycosides (ie amikacin, gentamicin, and tobramycin), extended-spectrum cephalosporins (ie, ceftriaxone, ceftazidime, and cefepime), first-generation cephalosporins (ie, cefazolin and cephalexin), extended-spectrum penicillins (ie, piperacillin-tazobactam), other penicillins (ie, ampicillin, amoxicillin, ampicillin-sulbactam, and amoxicillin-clavulanate), macrolides (ie, azithromycin, clarithromycin, and erythromycin), and carbapenems (ie, meropenem and imipenem). Receipt of proton-pump inhibitors (eg, omeprazole and lansoprazole) and salicylates (ie, aspirin) in the 30 days prior to fecal sampling was also ascertained (eg, commonly prescribed efflux pump inhibitors and inducers, respectively) [13, 25].
Statistical Analysis
Cases and controls were characterized by potential risk factors, including demographic variables, comorbid conditions, prior antimicrobial use, and receipt of proton-pump inhibitors or aspirin in the 30 days prior to sampling. Continuous variables were compared using the Student t test or Wilcoxon rank-sum test, and categorical variables were compared using the χ2 test or Fisher exact test. Bivariable analyses were then performed to determine the association between potential risk factors and OST positivity. An odds ratio (OR) and 95% confidence interval (CI) were calculated to evaluate the strength of any association.
Multivariable analyses were subsequently performed using multiple logistic regression [26], with calculation of adjusted ORs with 95% CIs. As there was no a priori hypothesized risk factor for isolation of OST-positive vs OST-negative FQREC (eg, to minimize the potential for exclusion of risk factors in analyses), all variables with P < .20 on bivariable analyses were maintained in the final explanatory model [27]. For all calculations, a 2-tailed P value of <.05 was considered to be significant. All statistical calculations were performed using commercially available software (STATA version 11.0; StataCorp).
RESULTS
Over the 3-year study period, a total of 774 unique subjects were included, as described elsewhere [19]. Of these patients, 89 (11.5%) were colonized with FQREC. A total of 44 (49.4%) and 45 (50.6%) of these patients had isolates that were OST-positive and OST-negative, respectively. Of the OST-negative isolates, 39 isolates had mutations in gyrA, and 25 isolates had mutations in parC. In addition, 18 isolates from control patients had mutations in both gyrA and parC. Furthermore, the proportion of FQREC isolates with OST varied across study years (31.8% in 2002, 52.3% in 2003, and 15.9% in 2004; P = .06).
On bivariable analyses (Table 1), there was a borderline significant association between OST positivity and location on a surgical service (OR, 2.42; 95% CI, .88–6.87; P = .06). The hospital length of stay prior to the sampling date was not significantly different between case and control patients, with a mean of 14.2 days (standard deviation [SD], 23.6 days) and 12.9 days (SD, 19.1 days), respectively (P = .86). There was no significant association between OST positivity and use of FQs (OR, 1.16; 95% CI, .40–3.36; P = .76) or any antimicrobial therapy (OR 1.15; 95% CI, .46–2.89; P = .74) in the 30 days prior to the culture date. In addition, OST status was not associated with previous receipt of either a proton pump inhibitor (OR, 1.51; 95% CI, .37–6.58; P = .55) or aspirin (OR, 0.70; 95% CI, .16–2.82; P = .76). On subsequent multivariable analyses (Table 2), location on the surgical service was an independent risk factor for recovery of OST-positive isolates. Receipt of a first-generation cephalosporin in the 30 days prior to sampling was significantly associated with a reduced risk for OST positivity.
Table 1.
Bivariable Analyses of Risk Factors for Recovery of Isolates With Organic Solvent Tolerance Among Patients Colonized With Fluoroquinolone-Resistant Escherichia coli
| No. (%) of patients |
||||
|---|---|---|---|---|
| Variable | OST-positive isolates (n = 44) | OST-negative isolates (n = 45) | OR (95% CI) | Pa |
| Year of culture | ||||
| 2002 | 14 (31.8) | 12 (26.7) | .06 | |
| 2003 | 23 (52.3) | 16 (35.6) | ||
| 2004 | 7 (15.9) | 17 (37.8) | ||
| Nonwhite race | 28 (63.6) | 23 (51.1) | 1.67 (.66–4.27) | .23 |
| Surgical service | 18 (40.9) | 10 (22.2) | 2.42 (.88–6.87) | .06 |
| Malignancy | 10 (22.7) | 17 (37.8) | 0.48 (.17–1.34) | .12 |
| Receipt of chemotherapy ≤30 d prior to sampling | 2 (4.6) | 6 (13.3) | 0.31 (.03–1.89) | .27 |
| Receipt of first-generation cephalosporin ≤30 d prior to samplingb | 7 (15.9) | 13 (28.9) | 0.47 (.14–1.45) | .20 |
Abbreviations: CI, confidence interval; OR, odds ratio; OST, organic solvent tolerance.
a Only variables with P < .30 are shown.
b Cefazolin or cephalexin.
Table 2.
Multivariable Model of Risk Factors for Recovery of Isolates With Organic Solvent Tolerance Among Patients Colonized With Fluoroquinolone-Resistant Escherichia coli
| Variable | OR (95% CI) | P |
|---|---|---|
| Year of culture | 0.53 (.27–1.02) | .06 |
| Surgical service | 7.36 (1.82–29.7) | .005 |
| Malignancy | 0.66 (.24–1.82) | .42 |
| Receipt of first-generation cephalosporin ≤30 d prior to samplinga | 0.20 (.04–.94) | .04 |
Abbreviations: CI, confidence interval; OR, odds ratio; OST, organic solvent tolerance.
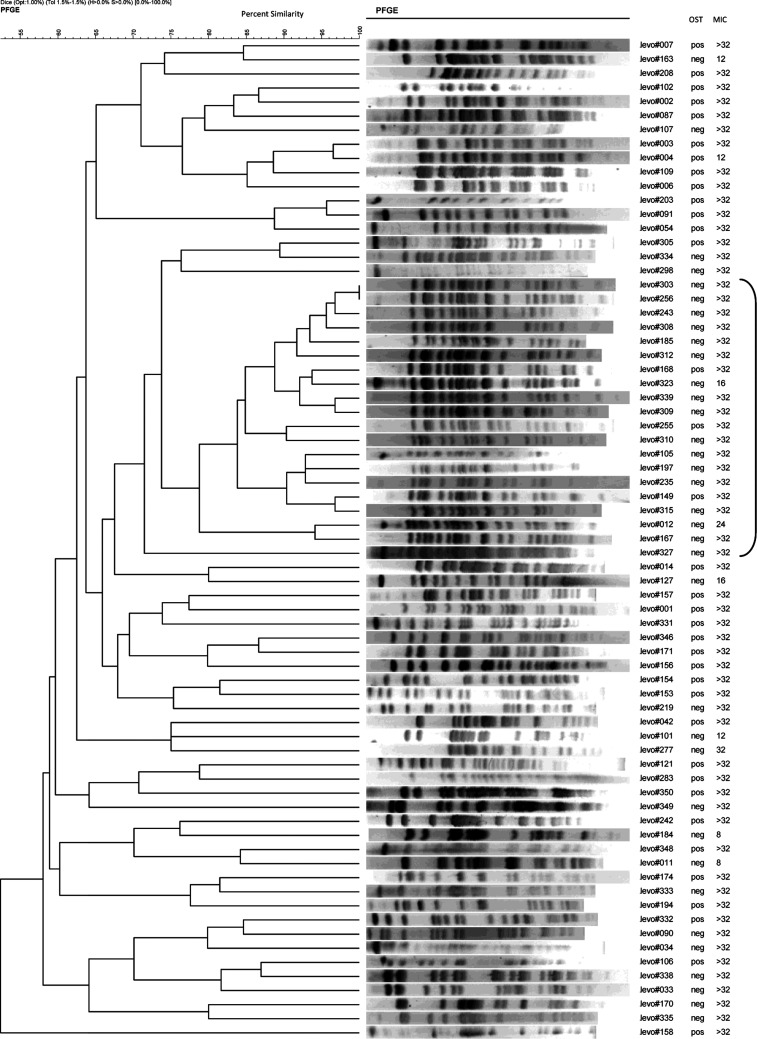
Of the 89 unique FQREC E. coli isolates, 71 were successfully characterized by PFGE (Figure 1). Nineteen isolates demonstrated clonal relatedness. Among these, OST-negative isolates were more likely to be clonally related compared with OST-positive isolates (16 and 3 isolates, respectively; P = .001). Ten (52.6%) and 9 (47.4%) patients were hospitalized on medical and surgical services, respectively. Seven patients from HUP had durations of hospitalization that demonstrated temporal overlap (ie, from 31 December 2003 through 18 March 2004). Of these patients, 3 were on medical services and 4 were on surgical services; however, there was no geographic overlap (ie, they were hospitalized on different floors of different buildings). Notably, all of the isolates from these 7 patients were OST-negative.
Figure 1.
Pulsed-field gel electrophoresis (PFGE) patterns of fluoroquinolone-resistant Escherichia coli isolates. Abbreviations: MIC, minimum inhibitory concentration; neg, negative; OST, organic solvent tolerance; pos, positive.
DISCUSSION
In this 3-year study, we found that 49.4% of a total of 89 FQREC isolates were OST-positive. Furthermore, the proportion of OST-positive isolates changed over time, such that the proportion of FQREC isolates that were OST-positive was notably lowest in the last year of the study (ie, 2004). In the final multivariable model, OST positivity was significantly associated with location on the surgical service and receipt of a first-generation cephalosporin in the 30 days prior to sampling. Finally, 19 of 71 isolates that were characterized by PFGE demonstrated clonal relatedness, the majority of which were OST-negative.
Previous studies have demonstrated conflicting results in regard to risk factors for efflux pump overexpression in FQREC. A study evaluating 153 clinical E. coli isolates (approximately 80% of which were FQREC) from a tertiary care hospital in the United States [18] found no association between efflux pump overexpression and patient age, sex, hospital location, or culture source. However, this study was limited to bivariable analyses and did not evaluate prior use of antimicrobial agents as potentially selecting for specific resistance mechanisms. Furthermore, selection bias was a potential concern as the study was largely limited to evaluation of clinical urinary cultures (ie, approximately 80% of isolates). Another study performed in Japan [17] evaluated urinary FQREC isolates from 38 patients with urinary tract infections; results of the study implicated female sex and treatment in the urologic department on multivariable analysis as risk factors for overexpression of the marA gene, a transcriptional activator associated with increased expression of the AcrAB efflux pump. However, this study was limited by a small sample size as well as potential selection bias given evaluation solely of clinical urinary isolates.
In contrast, our study evaluated risk factors for efflux pump overexpression in a large sample of patients with gastrointestinal tract colonization with FQREC as assessed via fecal cultures performed hospital-wide. FQ resistance likely arises at the level of gastrointestinal tract colonization, particularly in response to selection pressure from use of specific antimicrobial agents (eg, FQs) [11, 12]. We found a significant association between efflux pump overexpression and recent use of a first-generation cephalosporin in our study, with patients who received cephalexin or cefazolin less likely to have subsequent recovery of FQREC isolates that were OST-positive. To our knowledge, this is the first study to demonstrate such an association. Interestingly, in an intact cell model of E. coli containing the AcrAB-TolC complex [28], Nagano and Nikaido were able to determine the kinetic behavior for several cephalosporins and thereby evaluate the degree of drug efflux. In this model, the investigators demonstrated that although extensive efflux occurred with many cephalosporins, cefazolin specifically (a cephalosporin with 2 hydrophilic substituents) showed very little evidence for active efflux [28]. Of the patients who received a first-generation cephalosporin in our study, the majority (>90%) were prescribed cefazolin. It is possible that given the lack of active efflux of cefazolin in E. coli, the use of this antibiotic results in a selective disadvantage for FQREC with efflux pump overexpression as the predominant mechanism of FQ resistance. However, further studies are needed to elucidate this relationship, including the kinetic behavior of other commonly used antimicrobials and selection for specific mechanisms of FQ resistance. Nevertheless, given the association between efflux overexpression and resistance to other antimicrobials [14–16, 29], consideration of this finding in institutions with high rates of cefazolin use will be important.
Proton-pump inhibitors and aspirin are commonly prescribed agents in both the outpatient and inpatient healthcare settings [30]. As such, we postulated that the use of these drugs, which act as efflux pump inhibitors and activators, respectively, may be associated with efflux pump overexpression. However, our study did not demonstrate such an association, although this may have been due to small numbers of patients who were prescribed proton-pump inhibitors and aspirin (12 in each group).
The results of our study also demonstrated an increased risk of efflux pump overexpression in FQREC isolates recovered from patients who were on a surgical service at either study hospital. It is unclear why there was a significant association between location on a surgical service and OST positivity. Although 19 isolates were found to be clonally related by PFGE analysis, the patients from whom these isolates were recovered were approximately evenly distributed between surgical and medical services, therefore arguing against an outbreak of a specific, OST-positive E. coli strain among a particular service. It is possible that efflux pump overexpression as a mechanism of resistance may decrease transmissibility. However, further studies are needed to evaluate differences in transmissibility among specific resistance mechanisms in FQREC.
Finally, of the 19 isolates that were genetically related by PFGE analysis, 7 demonstrated temporal overlap in dates of hospitalization of the associated patients (ie, from 31 December 2003 through 18 March 2004). Although all of the patients from whom the 7 isolates were recovered were hospitalized at HUP, building and floor locations differed among the group at the time of fecal sampling. Nevertheless, given the close temporal overlap among these isolates, as well as their OST-negative status, it is possible that acquisition from other patients or the environment may have played a role in these findings.
There are potential limitations of our study. Selection bias is a potential concern; although only approximately 60% of eligible subjects were enrolled as detailed in the original study, participants and nonparticipants had similar demographic characteristics apart from median age [14]. Similarly, misclassification bias is a concern in case-control studies. However, testing for specific mechanisms of FQ resistance, including efflux pump overexpression, was performed prior to data collection, and cases and controls were classified solely based on OST. Finally, the present study was conducted in a single healthcare system, and these results may not be generalizable to other institutions.
In conclusion, the results of our study demonstrate a significant association between efflux pump overexpression as the mechanism of resistance in FQREC colonization and prior use of first-generation cephalosporins and location on a surgical service. It is clear that the clinical and molecular epidemiology of FQ resistance is complex, and future studies will need to focus on elucidating the relationship between specific antimicrobial use and resulting selection pressure for various resistance mechanisms. Furthermore, it will be important for future studies to evaluate whether differences in risk factors exist depending on the underlying mechanism of resistance among other antimicrobial-resistant bacteria.
Notes
Financial support. This work was supported by the Centers for Disease Control and Prevention (Public Health Service grant RS1/CCR320627-01 and Prevention Epicenters Program grant U54-CK000163 to E. L.); the National Institutes of Health (grant K24 AI080942 to E. L.); and the Pennsylvania State Department of Health (Commonwealth Universal Research Enhancement Program grant to E. L.).
Role of the funding agency. The funding agencies had no role in the design and conduct of the study; the collection, management, analysis, and interpretation of the data; or the preparation, review, or approval of the manuscript.
Potential conflicts of interest. E. L. has received research grant support from AstraZeneca, Cubist, and 3M. All other authors: no conflicts relevant to this study. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Neuhauser MM, Weinstein RA, Rydman R, Danziger LH, Karam G, Quinn JP. Antibiotic resistance among gram-negative bacilli in US intensive care units: implications for fluoroquinolone use. JAMA. 2003;289:885–8. doi: 10.1001/jama.289.7.885. [DOI] [PubMed] [Google Scholar]
- 2.Linder JA, Huang ES, Steinman MA, Gonzales R, Stafford RS. Fluoroquinolone prescribing in the United States: 1995 to 2002. Am J Med. 2005;118:259–68. doi: 10.1016/j.amjmed.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Camins BC, Marschall J, DeVader SR, Maker DE, Hoffman MW, Fraser VJ. The clinical impact of fluoroquinolone resistance in patients with E coli bacteremia. J Hosp Med. 2011;6:344–9. doi: 10.1002/jhm.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lautenbach E, Metlay JP, Bilker WB, Edelstein PH, Fishman NO. Association between fluoroquinolone resistance and mortality in Escherichia coli and Klebsiella pneumoniae infections: the role of inadequate empirical antimicrobial therapy. Clin Infect Dis. 2005;41:923–9. doi: 10.1086/432940. [DOI] [PubMed] [Google Scholar]
- 5.Lautenbach E, Strom BL, Nachamkin I, et al. Longitudinal trends in fluoroquinolone resistance among Enterobacteriaceae isolates from inpatients and outpatients, 1989–2000: differences in the emergence and epidemiology of resistance across organisms. Clin Infect Dis. 2004;38:655–62. doi: 10.1086/381549. [DOI] [PubMed] [Google Scholar]
- 6.Gaynes R, Edwards JR. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41:848–54. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 7.Sahm DF, Thornsberry C, Mayfield DC, Jones ME, Karlowsky JA. Multidrug-resistant urinary tract isolates of Escherichia coli: prevalence and patient demographics in the United States in 2000. Antimicrob Agents Chemother. 2001;45:1402–6. doi: 10.1128/AAC.45.5.1402-1406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lautenbach E, Strom BL, Bilker WB, Patel JB, Edelstein PH, Fishman NO. Epidemiological investigation of fluoroquinolone resistance in infections due to extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Clin Infect Dis. 2001;33:1288–94. doi: 10.1086/322667. [DOI] [PubMed] [Google Scholar]
- 9.Hooper DC. Emerging mechanisms of fluoroquinolone resistance. Emerg Infect Dis. 2001;7:337–41. doi: 10.3201/eid0702.010239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;41(suppl 2):S120–6. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 11.Donskey CJ. Antibiotic regimens and intestinal colonization with antibiotic-resistant gram-negative bacilli. Clin Infect Dis. 2006;43(suppl 2):S62–9. doi: 10.1086/504481. [DOI] [PubMed] [Google Scholar]
- 12.Richard P, Delangle MH, Raffi F, Espaze E, Richet H. Impact of fluoroquinolone administration on the emergence of fluoroquinolone-resistant gram-negative bacilli from gastrointestinal flora. Clin Infect Dis. 2001;32:162–6. doi: 10.1086/317551. [DOI] [PubMed] [Google Scholar]
- 13.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009;69:1555–623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lautenbach E, Fishman NO, Metlay JP, et al. Phenotypic and genotypic characterization of fecal Escherichia coli isolates with decreased susceptibility to fluoroquinolones: results from a large hospital-based surveillance initiative. J Infect Dis. 2006;194:79–85. doi: 10.1086/503046. [DOI] [PubMed] [Google Scholar]
- 15.Swick MC, Morgan-Linnell SK, Carlson KM, Zechiedrich L. Expression of multidrug efflux pump genes acrAB-tolC, mdfA, and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrob Agents Chemother. 2011;55:921–4. doi: 10.1128/AAC.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keeney D, Ruzin A, McAleese F, Murphy E, Bradford PA. MarA-mediated overexpression of the AcrAB efflux pump results in decreased susceptibility to tigecycline in Escherichia coli. J Antimicrob Chemother. 2008;61:46–53. doi: 10.1093/jac/dkm397. [DOI] [PubMed] [Google Scholar]
- 17.Yasufuku T, Shigemura K, Shirakawa T, et al. Correlation of overexpression of efflux pump genes with antibiotic resistance in Escherichia coli strains clinically isolated from urinary tract infection patients. J Clin Microbiol. 2011;49:189–94. doi: 10.1128/JCM.00827-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan-Linnell SK, Becnel Boyd L, Steffen D, Zechiedrich L. Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob Agents Chemother. 2009;53:235–41. doi: 10.1128/AAC.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lautenbach E, Metlay JP, Weiner MG, et al. Gastrointestinal tract colonization with fluoroquinolone-resistant Escherichia coli in hospitalized patients: changes over time in risk factors for resistance. Infect Control Hosp Epidemiol. 2009;30:18–24. doi: 10.1086/592703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White DG, Goldman JD, Demple B, Levy SB. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Dzink-Fox JL, Chen M, Levy SB. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother. 2001;45:1515–21. doi: 10.1128/AAC.45.5.1515-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goering RV, Tenover FC. Epidemiological interpretation of chromosomal macro-restriction fragment patterns analyzed by pulsed-field gel electrophoresis. J Clin Microbiol. 1997;35:2432–3. doi: 10.1128/jcm.35.9.2432-2433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee I, Fishman NO, Zaoutis TE, et al. Risk factors for fluconazole-resistant Candida glabrata bloodstream infections. Arch Intern Med. 2009;169:379–83. doi: 10.1001/archinte.169.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quan H, Sundararajan V, Halfon P, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care. 2005;43:1130–9. doi: 10.1097/01.mlr.0000182534.19832.83. [DOI] [PubMed] [Google Scholar]
- 25.Cohen SP, Levy SB, Foulds J, Rosner JL. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–62. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosmer D, Lemeshow SL. Applied logistic regression. New York: Wiley and Sons; 1989. [Google Scholar]
- 27.Mickey RM, Greenland S. The impact of confounder selection criteria on effect estimation. Am J Epidemiol. 1989;129:125–37. doi: 10.1093/oxfordjournals.aje.a115101. [DOI] [PubMed] [Google Scholar]
- 28.Nagano K, Nikaido H. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci U S A. 2009;106:5854–8. doi: 10.1073/pnas.0901695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole K. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob Agents Chemother. 2000;44:2233–41. doi: 10.1128/aac.44.9.2233-2241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qato DM, Alexander GC, Conti RM, Johnson M, Schumm P, Lindau ST. Use of prescription and over-the-counter medications and dietary supplements among older adults in the United States. JAMA. 2008;300:2867–78. doi: 10.1001/jama.2008.892. [DOI] [PMC free article] [PubMed] [Google Scholar]