Abstract
A small proportion (1%–1.5%) of 2009 pandemic influenza A/H1N1 virus strains (A[H1N1]pdm09) are oseltamivir resistant, almost exclusively because of a H275Y mutation in the neuraminidase protein. However, many individuals infected with resistant strains had not received antivirals. Whether drug-resistant viruses are initially present as minor variants in untreated individuals before they emerge as the dominant strain in a virus population is of great importance for predicting the speed at which resistance will arise. To address this issue, we used ultra-deep sequencing of viral populations from serial nasopharyngeal specimens from an immunocompromised child and from 2 individuals in a household outbreak. We observed that the Y275 mutation was present as a minor variant in infected hosts before the onset of therapy. We also found evidence for the transmission of this drug-resistant variant with drug-susceptible viruses. These observations provide important information on the relative fitness of the Y275 mutation in the absence of oseltamivir treatment.
The 2009 pandemic influenza A/H1N1 virus (A[H1N1]pdm09) emerged after reassortment between 2 swine viruses circulating in North America and Eurasia [1]. From 1% through 1.5% of influenza A(H1N1)pdm09 strains analyzed to date have been found to be resistant to oseltamivir, a neuraminidase (NA) inhibitor that constitutes the current standard of care [2]. Virtually all oseltamivir-resistant influenza A(H1N1)pdm09 viruses contain an H275Y amino acid substitution in the viral NA gene [3]. Among the drug-resistant strains recovered from immunocompetent patients, approximately one-third have been recovered from untreated individuals [4]. Whether drug-resistant variants are initially present as minor variants in untreated individuals because of transmission from a host harboring a minority drug-resistant population or whether they emerge after de novo replication is of great importance for predicting the speed at which resistance will arise; the selection of resistant mutations will occur more rapidly if they are already present in hosts as pre-existing minor variants [5]. In addition, the presence (or not) of the H275Y mutation in pretreatment samples provides important information on the relative fitness of drug resistance mutations in the absence of oseltamivir treatment.
To determine whether the H275Y mutation is present as a minor variant in hosts infected with influenza A virus, we performed ultra-deep sequencing of viral populations from nasopharyngeal specimens from 2 sets of individuals infected with influenza A(H1N1)pdm09 virus. First, we examined longitudinal samples collected from an immunocompromised child who remained infected for >6 weeks, during which time a drug-resistant strain came to dominate the virus population. Second, we analyzed the emergence of oseltamivir-resistant virus in a household outbreak of influenza A(H1N1)pdm09 infection in which the contact patient developed influenza symptoms 24 hours after starting post-exposure oseltamivir prophylaxis [6].
MATERIALS AND METHODS
Study 1: Immunocompromised Child
A 31-month-old boy weighing 13.4 kg who received a diagnosis of medulloblastoma 3 months earlier was admitted on 5 January 2011 for consolidation chemotherapy in preparation for the first of 3 consecutive autologous bone marrow transplants (ABMT). At admission, the child presented with rhinorrhea and mild cough but was afebrile. Members of his immediate family, including his older sister and his father, had cold-like symptoms 1–2 weeks before his admission; none of the family members, including the patient, had received the 2010–2011 influenza vaccine, the monovalent influenza A(H1N1)pdm09 vaccine, or any antiviral drug. A nasopharyngeal aspirate (NPA) specimen collected at admission was positive for the influenza A(H1N1)pdm09 virus by real-time reverse-transcription polymerase chain reaction (RT-PCR) [7] and by viral culture on A549 and Mink lung cells. Treatment with oseltamivir (30 mg, twice daily) was started on January 6. The following day, the patient developed fever (max. 39.2°C), coincident with decreasing neutrophil counts. The child received his first ABMT on January 10. NPA specimens collected throughout hospitalization remained positive for influenza A(H1N1)pdm09 virus by RT-PCR (Table 1). Oseltamivir therapy was continued during the hospitalization and after discharge on January 22. The patient was readmitted from 27 January through 14 February 2011 for his second ABMT. An NPA specimen collected on January 28 was positive for influenza A(H1N1)pdm09 by RT-PCR. Because of persistent viral excretion, oseltamivir was replaced by zanamivir (25 mg inhaled 4 times daily) on 1 February and continued until negative RT-PCR results on 17 February. The patient received a third ABMT on 18 February, and he recovered from his influenza virus infection without complications.
Table 1.
Virological Testing of Nasopharyngeal Aspirate Samples Obtained from a Young Boy Undergoing Autologous Bone Marrow Transplantation and Infected with Influenza A(H1N1)pdm09 Virus
| Sample | Antiviral therapy (43 d) | Multiplex real time diagnostic PCR (pH1N1) | Discriminatory real-time RT-PCR |
Deep Sequencing |
|||||
|---|---|---|---|---|---|---|---|---|---|
| H275 copies/mL ± stdev (%) | Y275 copies/mL ± stdev (%) | H275% | Y275% | C.I.± | Coverage #reads | Phenotypic drug susceptibility | |||
| 05–01–2011 - S1 | None | Positive | 1.78 × 107 ± 0.71 × 107 (99.91) | 1.44 × 104 ± 0.29 × 104 (0.08) | 95.7 | 3.7 | 1.7 | 488 | n.e. |
| 05–01–2011 - CM2 | None | Positive | 3.15 × 1010 ± 2.05 × 1010 (99.99) | 3.16 × 105 ± 3.41 × 105 (0.001) | 96.9 | 2.7 | .7 | 1914 | Susceptible to oseltamivir, zanamivir, and peramivir |
| 10–01–2011 - S2 | Oseltamivir | Positive | 5.99 × 108 ± 3.15 × 108 (99.75) | 1.53 × 106 ± 1.26 × 106 (0.25) | 94.6 | 4.4 | 1.0 | 1552 | n.e. |
| 17–01–2011 - S3 | Oseltamivir | Positive | 4.21 × 105 ± 4.09 × 105 (3.13) | 1.30 × 107 ± 6.57 × 106 (96.87) | 2.3 | 97 | .8 | 1341 | n.e. |
| 20–01–2011 - S4 | Oseltamivir | Positive | 2.24 × 106 ± 1.56 × 106 (4.08) | 5.26 × 107 ± 3.67 × 107 (95.92) | 3.3 | 96 | 1.0 | 1170 | n.e. |
| 20–01–2011 - CM1 | Oseltamivir | Positive | 6.25 × 105 ± 5.73 × 105 (4.62) | 1.29 × 107 ± 0.83 × 107 (95.38) | 3.3 | 96.5 | .8 | 1838 | n.e. |
| 20–01–2011 - CM2 | Oseltamivir | n.e. | 0 | 9.40 × 109 ± 3.24 × 109 (100.00) | n.e. | n.e. | n.e. | n.e. | Resistant to peramivir and oseltamivir, susceptible to zanamivir |
| 28–01–2011 - S5 | Zanamivir | Positive | 2.85 × 104 ± 1.15 × 104 (16.54) | 1.44 × 105 ± 0.53 × 105 (83.46) | 9.9 | 90 | 2.4 | 131 | n.e. |
| 08–02–2011 - S6 | Zanamivir | Positive | 8.23 × 105 ± 4.98 × 105 (11.53) | 6.32 × 106 ± 3.18 × 106 (88.47) | 6.1 | 93.9 | 6.1a | 66 | n.e. |
| 17–02–2011 - S7 | Zanamivir | Negative | 0 | 0 | n.e. | n.e. | n.e. | n.e. | n.e. |
Note: Real-time reverse-transcription polymerase chain reaction (RT-PCR) assays were performed in triplicate. Abbreviations: CI, confidence interval; CM1 and CM2, culture passages 1 and 2; n.e., not evaluated; S, primary specimen (nasopharyngeal aspirate specimens).
a Not significant with 95% confidence.
Study 2: Transmission in Household
A detailed description of the familial cluster of infections with influenza A(H1N1)pdm09 virus has been reported elsewhere [6]. In brief, a 13-year-old asthmatic male developed infection with influenza A(H1N1)pdm09 virus, confirmed by RT-PCR testing of an NPA specimen. The child initiated oseltamivir treatment (60 mg twice daily for 5 days) and was discharged home on the same day. Simultaneously to treatment of the index patient, postexposure oseltamivir prophylaxis (75 mg once daily for 10 days) was prescribed to the 59-year-old father with chronic obstructive pulmonary disease. Approximately 24 hours after beginning oseltamivir prophylaxis, the father developed influenza-like symptoms. On day 8 of oseltamivir prophylaxis, he consulted his general practitioner for persistent cough. An NPA specimen collected at that time was positive by RT-PCR and by culture for influenza A(H1N1)pdm09. The father had an uneventful clinical course, and an NPA sample obtained at the end of his illness was negative for the virus. The son's influenza A(H1N1)pdm09 isolate collected before oseltamivir therapy was susceptible to oseltamivir (50% inhibitory concentration [IC50], 0.27 nM), whereas the father's influenza A(H1N1)pdm09 isolate was highly resistant to oseltamivir (IC50, >400 nM). The complete (consensus sequence) virus genomes of the father (GenBank accession number FN434454) differed by 1 amino substitution (H275Y) in the NA protein from the virus present in the son (GenBank accession number FN434445).
Informed Consent
Written consent was obtained for report of the case described in study 1. Samples used in study 2 were obtained as part of an investigation by the Public Health Department of the Ministry of Health, Quebec, Canada.
Clinical Specimens and Viral Culture
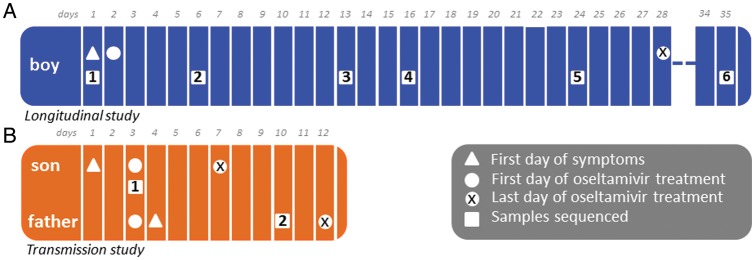
In study 1 (immunocompromised child), 7 NPA specimens were collected from 5 January through 17 February 2011 for RT-PCR testing (Table 1 and Figure 1). Viral isolates were also obtained by culture from NPA specimens obtained on 5 January and 20 January. In study 2 (household transmission), the NPA specimen from the index patient (son) was collected before oseltamivir treatment, whereas the NPA specimen from his father was obtained on day 8 of oseltamivir prophylaxis (Figure 1).
Figure 1.
Outline of studies indicating day of onset, day when oseltamivir treatment was started, and sampling timeline. (A) Study 1: Immunocomprised 31-month-old boy. (B) Study 2: Son-father transmission.
NA Inhibition Assay
The drug resistance phenotype to NA inhibitors was determined by NA inhibition assays [8]. The IC50 values were determined from the dose-response curve. A virus was considered to be resistant to a drug if its IC50 value was 10-fold greater than that of the wild-type (WT) virus [9].
RNA Extraction
Total RNA was extracted from 200 µL of thawed specimen or culture with use of the MagNA Pure instrument and the MagNA Pure LC total nucleic acid isolation kit (Roche Applied Science), according to the manufacturer's instructions, and was stored at −80°C.
Discriminative Real-Time PCR Assay
To discriminate between WT and H275Y oseltamivir-resistant strains of influenza A(H1N1)pdm09, a modified version of a previously-reported real-time RT-PCR method [10] was used to test samples. This technique requires a reverse (panN1-H275-sense 5′–cagtcgaaatgaatgcccctaa-3′) and a forward (panN1-H275-antisense 5′–tgcacacacatgtgatttcactag-3′) primer for both the WT and the H275Y viruses and 2 labelled allele-specific probes: panN1-275H-probe (5′–ttaTCActAtgAggaatga-6-FAM/BHQ-1) and panN1-275Y-probe (5′–ttaTTActAtgAggaatga-HEX/BHQ-1). In the aforementioned probe sequences, locked nucleic acid nucleotides are denoted in upper case, DNA nucleotides are denoted in lower case, and the single nucleotide polymorphism is underlined. The limits of detection for the assay are 50 copies for the H275Y target and 10–50 copies for the WT target. RT-PCR conditions are available upon request. Data acquisition was performed in both FAM and HEX filters during the annealing/extension step. Standard curves were constructed using 10-fold serial dilutions of pJET1.2-NA-Y275 and pJET1.2-NA-H275 plasmids.
Sequencing and Analysis
RNA isolated from 2 cultured isolates and 7 primary specimens collected for study 1 (Figure 1A) and 2 primary specimens for study 2 (Figure 1B) was subjected to a multisegment RT-PCR (M-RT-PCR) step [11] and random priming with barcoding using the sequence-independent single-primer amplification (SISPA) protocol [12]. For each RNA sample, we performed 2 M-RT-PCRs using the One-Step Superscript III RT kit (Invitrogen). Reactions were purified independently using the Qiagen MinElute kit and were quantitated on a Nanodrop spectrophotometer; 100–200 ng of each purified M-RT-PCR was used in 2 separate SISPA reactions with 2 different barcode tags for a total of 4 tagged reactions per original RNA sample. Products were then separated on a 1% agarose gel, and fragments from 200–400 bp were purified using the Qiagen MinElute kit. Pooled samples were sent for paired-end library preparation and 100 base sequencing on the Illumina Hi-Seq2000 platform.
The barcoded amplification products were sequenced on 1 lane of the sequence run. Analyses were performed to reduce the distortion caused by SISPA amplification, account for both PCR and sequencing errors, and provide a clean comparison between the mapped reads of the experimental samples. The trimmed reads were mapped to A/Quebec/144147/2009(H1N1) (GenBank accession FN434457-FN434464) with use of the bowtie short-read aligner [13].
The frequency of each codon observed in the set of mapped reads from each amplification replicate was tabulated across each of the 10 influenza genes. To account for sequence-specific errors [14, 15], the variant counts for the forward and reverse direction reads were calculated separately, and only those variants for which counts were within 50% of each other in both directions were retained. For these summaries, the unique reads from all amplification replicates were pooled, and total coverage is reported for each codon site. The proportion of codons expected to differ from the consensus because of background mutation and technical error was estimated from a separate cell culture of the PR8 strain that was otherwise processed in exactly the same manner as the specimens in this study. This proportion, found to be 0.00392, lies well outside of the 95% confidence interval for any variant codon in our study that is (1) represented by >4 sequence reads and (2) found in at least 2% of all sequence reads mapped to that position. The lower limit of the 95% confidence interval determined by computing the inverse of the appropriate cumulative β distribution is 0.00813.
RESULTS
Presence of Drug-Resistant Viruses before Drug Treatment in an Immunocompromised Child (Study 1)
The results of the NA gene H275Y discriminatory real-time RT-PCR assay performed on the 7 primary specimens and the 2 viral isolates (5 and 20 January) are presented in Table 1. In the first NPA specimen collected on 5 January (day 1), before antiviral therapy (initiated on 6 January), 99.9% of the viral population was WT at NA position 275 by our discriminatory assay. Nevertheless, a very small subpopulation of H275Y mutant was also detectable (0.08%). The corresponding viral isolate (05–01–2011 – CM2 in Table 1) contained 99.9% of WT virus and was susceptible to oseltamivir (IC50 = 0.77 nM ± 0.02), zanamivir (IC50 = 0.15 nM ± 0.02), and peramivir (IC50 = 0.05 nM ± 0.01). Of note, the H275Y mutation could not be detected by conventional RT-PCR and Sanger sequencing in the original sample. A second NPA specimen collected on 10 January (day 6) also demonstrated a predominance of the WT population (99.8%). However, the proportion of the H275Y mutant detected in NPA specimens collected on 17, 20, and 28 January increased to 96.9%, 95.9%, and 83.5%, respectively, during continuous oseltamivir treatment. Furthermore, the second passage on Madin Darby canine kidney cells of the 20 January viral isolate (20–01–2011-CM2 in Table 1) resulted in 100% H275Y mutant population, compared with 95.4% from the primary culture recovered from A549 and Mink lung cells. This viral isolate exhibited an IC50 of 556.75 nM ± 61.32 for oseltamivir, 0.22 nM ± 0.01 for zanamivir, and 34.81 nM ± 5.77 for peramivir, which indicates a resistance phenotype to oseltamivir and peramivir. Antiviral therapy was changed to zanamivir on 1 February. The 8 February sample contained a predominance of 88.5% of H275Y mutant virus, whereas the last NPA specimen collected on 17 February was negative for influenza A(H1N1)pdm09 by RT-PCR.
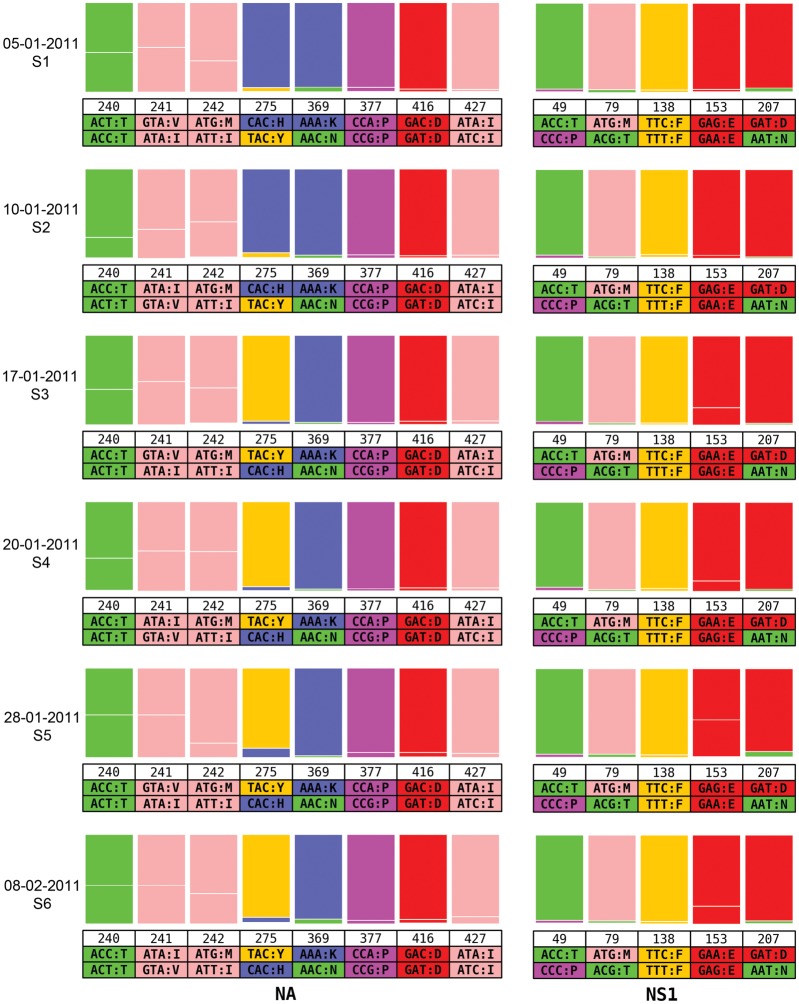
A number of the primary specimens (from 5, 10, 17, 20, and 28 January and 8 February, corresponding to samples 1–6 in Figure 1A) for which an M-RT-PCR product could be generated, as well as the viral isolates, were subjected to deep sequencing to better evaluate the genetic diversity of the viral population, including the presence of drug-resistant mutants. On the basis of the mean depth of coverage across each of the virus segments, we highlighted codons represented by at least 2% of the sequence reads covering each position (Supplementary Table 1). This percentage is conservative enough that, even in low-coverage areas, it excludes potential sequence and PCR errors.
The positions on the NA and NS1 proteins that display evidence for the presence of minor variants at a frequency of ≥2% in >1 sample are shown in Figure 2. Similar patterns are observed for all other proteins (Supplementary Table 1). Over time, the ratios of the minor variants to the dominant codon remain relatively stable, except for NA position 275, where a shift of H to Y was apparent on 17 January 2011. The ratios are similar to the ones observed in the real-time discriminatory RT-PCR assays for each of the samples tested (Table 1), although values across both assays are not identical. No other position on the NA protein appears to covary with the 275Y variant. The same pattern is observed in the culture isolates (05–01–2011-CM2 and 20–01–2011-CM1 in Supplementary Table 2). However, position 153 in NS1 displays a similar switch, although involving a synonymous mutation (from codon GAG to GAA, for E [glutamic acid]). Therefore, the sample from the original infection contained a drug-associated minor variant before the onset of treatment, and this minor variant differed from the dominant strain by only 2 nucleotide positions. Because of drug-associated selection pressure, this minor variant eventually became dominant in the host. The variant codons observed at the other positions are also possibly representative of other minor variants in the original virus population; however, because they remained minor members of the viral population, they are unlikely to have a selective advantage.
Figure 2.
Longitudinal study of variant codon prevalence across multiple times in an infected immunocompromised child. Ratios of major and minor codons are represented at each position where the variant codons appear in >2% of the deep sequence data reads in at least 2 of the times. Data collection dates are represented on the left side, and each group corresponds to the positions where variant residues are observed in the NA and NS1. Codons and single-letter amino acid codes are indicated below the position number.
Evidence for Transmission of Drug-Resistant Virus in a Household (Study 2)
In a separate study, we observed a similar phenomenon in which oseltamivir resistance emerged quickly in the household contact (father) of an index patient (son). Both family members initiated oseltamivir treatment on the same day (ie, twice daily treatment for the son and once daily prophylaxis for the father) (Figure 1B). The latter developed influenza-like symptoms 24 hours after treatment was begun. Such a rapid clinical presentation suggests that he was already infected at the time that prophylaxis was initiated and that drug-resistant virus was most likely to have already been present.
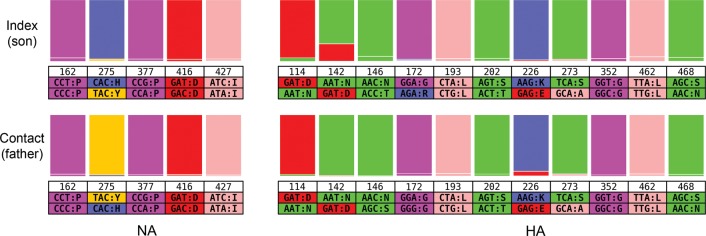
We characterized the genetic diversity of the virus populations in both individuals by deep sequencing. An example for the HA and NA genes in which most of the variants seen in the son are also observed in the father is shown in Figure 3. Although the dominant viruses were drug susceptible in the son and drug resistant in the father, apparent by the switch from H275 to 275Y, it is striking that a minor population of viruses in the son already carried the drug resistance mutation; minor drug resistant variant residue 275Y is present in >2.4% of the reads in the son (which was not detected by conventional RT-PCR and Sanger sequencing). Therefore, it is likely that viruses carrying this mutation were transmitted to the father along with drug-susceptible viruses and became dominant in that individual after selection associated with a subtherapeutic (prophylactic) dose of oseltamivir.
Figure 3.
Transmission study of variant codon prevalence compared between son and father specimens. Ratios of major and minor codons are represented at each position of the neuraminidase (NA) and hemagglutinin (HA) where the variant codons appear in >2% of the deep sequence data reads in at least 1 of the samples. Codons and single letter amino acid codes are indicated below the position number.
Of note, the same minor variants were found in both the father and the son at 60 residue positions across all 10 viral proteins (Supplementary Table 3). We estimate that there were 8 days of replication in the father from the time that he was possibly infected by the son (assuming that infection occurred 24 hours before any symptoms) to the time that the specimen was collected. Over that time, variant representation could have fluctuated, such that the set of 60 variants seen in both samples is likely to underestimate the true number. Although the number of conserved variants indicates possible transmission and the probability that the same variants could appear in both the son and the father by chance alone is extremely low, we do not have other potential contacts or index cases to test to confirm this observation.
DISCUSSION
The most striking observation from both of these studies is that the mutation most commonly associated with resistance to oseltamivir (H275Y) is present in the viral population of some individuals before the onset of treatment. In addition, this minor drug-resistant population could not be revealed by conventional methods, such as phenotypic resistance tests and Sanger sequencing. This observation is important for a number of reasons. First, the prior existence of Y275 means that the selection for drug resistance will proceed much more rapidly after the onset of drug selection pressure than if only WT viruses are present in the population, because there is no waiting time for the correct mutation to appear [5]. Furthermore, the presence of the Y275 mutation in untreated hosts indicates that this mutation is not strongly deleterious in the absence of oseltamivir treatment and likely does not need compensatory mutations to enable its fixation [16–18]. Indeed, in both cases studied here, we observed no amino acid changes that were fixed concordant with Y275 and only a single synonymous mutation (in NS1), in the case of the immunocomprised child. In these circumstances, the pre-existence of Y275 means that oseltamivir resistance will likely spread rapidly as soon as there is drug selection pressure, especially in immunocompromised individuals and when suboptimal antiviral dosage is used.
If the Y275 mutation is present in individual hosts before the onset of treatment, then it is also likely to have been transmitted between individuals as a minor variant. This in turn suggests that there may not often be a severe population bottleneck during the inter-host transmission of influenza virus. Indeed, mixed infection of multiple variants of influenza virus have been observed in both natural human infection [19–21] and experimental animal infection [22, 23] and, thus, may be commonplace. Coinfection with major and minor variants, captured by deep sequencing, has also been observed during the course of human rhinovirus infection [24], indicating that this phenomenon is not unique to influenza. In contrast, sequencing studies of HIV suggest that a small number of viral particles initiate infection, such that most variants are produced after replication in the newly infected host [25].
Such transmission of multiple variants is most clearly documented in the son-father case, in which perhaps 60 mutational variants were passed between these 2 individuals, 1 of which confers oseltamivir resistance. However, the availability of only short sequence reads makes it impossible to determine the exact number of distinct viral haplotypes to which these correspond. In addition, our sampling protocol in the son-father transmission case dictates that we cannot exclude that there was rapid selection of oseltamivir resistance in the son after we sampled his virus population, such that a majority Y275 population was in fact transmitted to the father. However, this would entail extremely rapid selection for resistance and does not change the central observation that multiple variants are transmitted between hosts, because both H275 and Y275 were found in the father.
Presence of the Y275 mutation in the son before oseltamivir treatment and soon after symptom onset suggests that this resistance mutation was also present in the viral population initially transmitted to the son. Similarly, the presence of Y275 in the immunocomprised child suggests that this mutation may have been transmitted to the child in a mixed infection containing both drug-susceptible and -resistant mutations, although it cannot be excluded that the variant appeared de novo. If Y275 is indeed present in the founding population in both individuals, it is possible that this mutation is present as a low-frequency variant in many individuals infected with influenza A(H1N1)pdm09 virus and that its presence reflects the combined action of past selection for drug resistance in patients receiving oseltamivir treatment, incomplete reversion to the WT H275 mutation in patients who are not receiving the drug, and a lack of strongly deleterious fitness effects in the absence of treatment. The large-scale ultra-deep sequencing of additional samples from patients with influenza A(H1N1)pdm09 who have not received oseltamivir treatment will clearly be central to answering this question.
Next-generation ultra-deep sequencing of intra-host viral populations, such as that undertaken here, promises to transform our understanding of the evolution of drug resistance in acute viral infections, allowing the dissection of the mutational spectrum at an unprecedented level of precision. Indeed, it is striking that, in the 2 cases, conventional RT-PCR failed to detect the presence of oseltamivir resistance, even though Y275 was present in the viral population. However, despite its undoubted potential, ultra-deep sequencing also has a number of inherent analytical difficulties. First, because the sequencing protocol leads to the generation of short sequence reads, nucleotide positions cannot be linked either in or among individual genes except if they are close enough to appear on the same sequence read or if they have the same pattern of prevalence. More fundamentally, it is critical to ensure that minor genetic variants are not the result of PCR and/or sequencing artefacts. Amplification leads to the well-known problem of PCR duplicates, sometimes resulting in severe distortion to the observed proportions of true variant subpopulations and the possible creation of false variant sequences through PCR errors. To address these problems, each specimen from our study was amplified in 4 independent reactions using different barcodes, allowing us to track amplification products and their respective sequence reads. Future work will use a simpler and more cost-effective approach using modified primers that include unique tags for each template [26].
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://cid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported in part by National Institute of Allergy and Infectious Diseases at the National Institutes of Health (U54 GM088491), National Institute of General Medical Sciences at the National Institutes of Health (2R01 GM080533-06 to E. C. H.), and the Canadian Institutes of Health Research (to G. B.).
Potential conflict of interest. G. B. has received a research grant from GlaxoSmithKline. All other authors: No conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Garten RJ, Davis CT, Russell CA, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pizzorno A, Abed Y, Boivin G. Influenza drug resistance. Seminars in Respiratory and Critical Care Medicine. 2011;32:409–22. doi: 10.1055/s-0031-1283281. [DOI] [PubMed] [Google Scholar]
- 3.Pizzorno A, Bouhy X, Abed Y, Boivin G. Generation and characterization of recombinant pandemic influenza A(H1N1) viruses resistant to neuraminidase inhibitors. J Infect Dis. 2011;203:25–31. doi: 10.1093/infdis/jiq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. Weekly update on oseltamivir resistance in influenza A(H1N1)2009 viruses. 2011. 7 September 2011 ed http://www.who.int/influenza/surveillance_monitoring/updates/2011_09_09_weekly_web_update_oseltamivir_resistance.pdf. [Google Scholar]
- 5.Bonhoeffer S, Nowak MA. Pre-existence and emergence of drug resistance in HIV-1 infection. Proc Biol Sci. 1997;264:631–7. doi: 10.1098/rspb.1997.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baz M, Abed Y, Papenburg J, Bouhy X, Hamelin ME, Boivin G. Emergence of oseltamivir-resistant pandemic H1N1 virus during prophylaxis. N Engl J Med. 2009;361:2296–7. doi: 10.1056/NEJMc0910060. [DOI] [PubMed] [Google Scholar]
- 7.Semret M, Fenn S, Charest H, McDonald J, Frenette C, V L. A real-time RT-PCR assay for detection of influenza H1N1 (swine-type) and other respiratory viruses. 2009 26th International Congress of Chemotherapy and Infection. Toronto, ON. [Google Scholar]
- 8.Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Analytical biochemistry. 1979;94:287–96. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 9.Mishin VP, Hayden FG, Gubareva LV. Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob Agents Chemother. 2005;49:4515–20. doi: 10.1128/AAC.49.11.4515-4520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Vries E, Jonges M, Herfst S, et al. Evaluation of a rapid molecular algorithm for detection of pandemic influenza A (H1N1) 2009 virus and screening for a key oseltamivir resistance (H275Y) substitution in neuraminidase. J Clin Virol. 2010;47:34–7. doi: 10.1016/j.jcv.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou B, Donnelly ME, Scholes DT, et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol. 2009;83:10309–13. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djikeng A, Halpin R, Kuzmickas R, et al. Viral genome sequencing by random priming methods. BMC Genomics. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minoche AE, Dohm JC, Himmelbauer H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and Genome Analyzer systems. Genome Biol. 2011;12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura K, Oshima T, Morimoto T, et al. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39:e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamelin ME, Baz M, Abed Y, et al. Oseltamivir-resistant pandemic A/H1N1 virus is as virulent as its wild-type counterpart in mice and ferrets. PLoS Pathog. 2010;6:e1001015. doi: 10.1371/journal.ppat.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Memoli MJ, Hrabal RJ, Hassantoufighi A, et al. Rapid selection of a transmissible multidrug-resistant influenza A/H3N2 virus in an immunocompromised host. J Infect Dis. 2010;201:1397–403. doi: 10.1086/651610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seibert CW, Kaminski M, Philipp J, et al. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J Virol. 2010;84:11219–26. doi: 10.1128/JVI.01424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghedin E, Fitch A, Boyne A, et al. Mixed infection and the genesis of influenza virus diversity. J Virol. 2009;83:8832–41. doi: 10.1128/JVI.00773-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghedin E, Laplante J, DePasse J, et al. Deep sequencing reveals mixed infection with 2009 pandemic influenza A (H1N1) virus strains and the emergence of oseltamivir resistance. J Infect Dis. 2011;203:168–74. doi: 10.1093/infdis/jiq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pajak B, Stefanska I, Lepek K, et al. Rapid differentiation of mixed influenza A/H1N1 virus infections with seasonal and pandemic variants by multitemperature single-stranded conformational polymorphism analysis. J Clin Microbiol. 2011;49:2216–21. doi: 10.1128/JCM.02567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murcia PR, Baillie GJ, Daly J, et al. Intra- and interhost evolutionary dynamics of equine influenza virus. J Virol. 2010;84:6943–54. doi: 10.1128/JVI.00112-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murcia PR, Hughes J, Battista P, et al. Evolution of an Eurasian avian-like influenza virus in naive and vaccinated pigs. PLoS Pathogens. 2012;8:e1002730. doi: 10.1371/journal.ppat.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cordey S, Junier T, Gerlach D, et al. Rhinovirus genome evolution during experimental human infection. PLoS ONE. 2010;5:e10588. doi: 10.1371/journal.pone.0010588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc Natl Acad Sci USA. 2011;108:20166–71. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.