Abstract
Background
The genome of P. marneffei, the most important thermal dimorphic fungus causing respiratory, skin and systemic mycosis in China and Southeast Asia, possesses 23 polyketide synthase (PKS) genes and 2 polyketide synthase nonribosomal peptide synthase hybrid (PKS-NRPS) genes, which is of high diversity compared to other thermal dimorphic pathogenic fungi. We hypothesized that the yellow pigment in the mold form of P. marneffei could also be synthesized by one or more PKS genes.
Methodology/Principal Findings
All 23 PKS and 2 PKS-NRPS genes of P. marneffei were systematically knocked down. A loss of the yellow pigment was observed in the mold form of the pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants. Sequence analysis showed that PKS11 and PKS12 are fungal non-reducing PKSs. Ultra high performance liquid chromatography-photodiode array detector/electrospray ionization-quadruple time of flight-mass spectrometry (MS) and MS/MS analysis of the culture filtrates of wild type P. marneffei and the pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants showed that the yellow pigment is composed of mitorubrinic acid and mitorubrinol. The survival of mice challenged with the pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants was significantly better than those challenged with wild type P. marneffei (P<0.05). There was also statistically significant decrease in survival of pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants compared to wild type P. marneffei in both J774 and THP1 macrophages (P<0.05).
Conclusions/Significance
The yellow pigment of the mold form of P. marneffei is composed of mitorubrinol and mitorubrinic acid. This represents the first discovery of PKS genes responsible for mitorubrinol and mitorubrinic acid biosynthesis. pks12 and pks11 are probably responsible for sequential use in the biosynthesis of mitorubrinol and mitorubrinic acid. Mitorubrinol and mitorubrinic acid are virulence factors of P. marneffei by improving its intracellular survival in macrophages.
Author Summary
Penicillium marneffei is the most important thermal dimorphic fungus causing respiratory, skin and systemic mycosis in China and Southeast Asia. Its genome possesses a large number of polyketide synthase (PKS) genes, which should be responsible for synthesis of secondary metabolites such as pigments, antibiotics and mycotoxins. Using state-of-the-art gene knockdown and ultra high performance liquid chromatography-photodiode array detector/electrospray ionization-quadruple time of flight-mass spectrometry technologies, we discovered that the yellow pigment of P. marneffei was composed of mitorubrinol and mitorubrinic acid and was synthesized by two PKS genes, named pks12 and pks11. This represents the first discovery of PKS genes responsible for mitorubrinol and mitorubrinic acid biosynthesis, in which pks12 and pks11 are probably responsible for sequential use in the biosynthesis of mitorubrinol and mitorubrinic acid. Using a mouse model and human and mouse macrophage cell line models for P. marneffei infection, we also discovered that mitorubrinol and mitorubrinic acid are virulence factors of P. marneffei by improving its intracellular survival in macrophages.
Introduction
Penicillium marneffei is the most important thermal dimorphic fungus causing respiratory, skin and systemic mycosis in China and Southeast Asia [1], [2], [3], [4]. After the discovery of P. marneffei in 1956, only 18 cases of human diseases were reported until 1985 [5]. The appearance of the HIV pandemic, especially in China and Southeast Asian countries, saw the emergence of the infection as an important opportunistic mycosis in HIV positive patients. About 8% of AIDS patients in Hong Kong are infected with P. marneffei [6]. In northern Thailand, penicilliosis is the third most common indicator disease of AIDS, following tuberculosis and cryptococcosis [2]. Besides HIV positive patients, P. marneffei infections have been reported in other immunocompromised patients, such as transplant recipients, patients with systemic lupus erythematosus and those on corticosteroid therapy [7], [8], [9], [10].
Polyketides are a diverse group of secondary metabolites produced by microorganisms. Some of the best known secondary metabolites include pigments, antibiotics and mycotoxins. Polyketides are synthesized by complex enzymatic systems called polyketide synthases (PKS). In the neighborhood of the PKS genes also include additional genes that encode modifying enzymes forming biosynthetic clusters. The availability of more and more fungal genome sequences have enabled us to identify an unprecedented number of fungal PKS genes. However, relatively few fungal PKS genes have been definitely linked to the biosynthesis of specific polyketide secondary metabolites.
In 2002, the complete genome sequencing project of P. marneffei was started. At the moment, a 6× coverage of the genome has been completed [11]. Based on the genome sequence, we have assembled the complete mitochondrial genome sequence and analyzed the phylogeny, predicted the presence of a potential sexual cycle and developed a highly discriminative multilocus sequence typing scheme for P. marneffei [12], [13], [14]. Recently, we also reported a high diversity of PKS in the P. marneffei genome and characterized its melanin biosynthesis gene cluster and confirmed it as a virulence factor in P. marneffei [15]. Since the P. marneffei genome possesses 23 PKS and 2 PKS nonribosomal peptide synthase hybrid (PKS-NRPS) genes, which is of high diversity compared to other thermal dimorphic pathogenic fungi such as Histoplasma capsulatum (with only one PKS gene) and Coccidioides immitis (with 10 PKS genes), we hypothesized that the yellow pigment in the mold form of P. marneffei could also be synthesized by one or more PKS genes. To test this hypothesis, we systematically knocked down all the 23 PKS and 2 PKS-NRPS genes of P. marneffei and observed for the loss of yellow pigment in the knockdown mutants. The culture extracts of the wild type strain and the mutant strains with loss of yellow pigment were characterized using ultra high performance liquid chromatography-photodiode array detector/electrospray ionization-quadruple time of flight-mass spectrometry (UHPLC-DAD/ESI-Q-TOF-MS) to determine its chemical nature. The possible role and mechanism of the yellow pigment in virulence was also examined in a mouse model and macrophage cell line models respectively.
Methods
Ethics statement
The experimental protocols were approved by the Committee on the Use of Live Animals in Teaching and Research, The University of Hong Kong, in accordance with the Guidelines laid down by the NIH in the USA regarding the care and use of animals for experimental procedures.
Strain and culture conditions
P. marneffei strain PM1 was obtained from an already-existing collection from the clinical microbiology laboratory in Queen Mary Hospital and the strain was anonymized. The yeast form of PM1 was used for knockdown of the PKS genes. A single colony of the fungus grown on Sabouraud dextrose agar at 37°C was inoculated into yeast peptone broth and incubated in a shaker at 37°C for 10 days.
Knockdown of PKS genes
DNA extraction was performed using the DNeasy Plant Mini Kit according to manufacturer's instructions (Qiagen, Hilden, Germany). The extracted DNA was eluted in 50 µl of AE buffer and the resultant mixture was diluted 10× and 1 µl of the diluted extract was used for PCR.
Plasmid construction was performed according to our previous publication. For knockdown of pks1, plasmid pSilent-1 [16], obtained from the Fungal Genetics Stock Center, was used to construct the pPW1459 plasmid. First, the internal pks1 fragment (sense) was amplified using primers LPW11663 5′-CCGCTCGAGTTTCAACGACCTATCGCCCACTCAA-3′ and LPW11664 5′-CCCAAGCTTGGGACAACAGCACCAAGCAGTGTGGACA-3′ (Invitrogen, USA) (Table 1). The PCR mixture (25 µl) contained P. marneffei DNA, PCR buffer (10 mM Tris-HCl pH 8.3, 50 mM KCl, 2 mM MgCl2 and 0.01% gelatin), 200 µM of each deoxynucleoside triphosphates and 1.0 U Taq polymerase (Applied Biosystem, USA). The mixtures were amplified in 32 cycles of 95°C for 30 s, 56°C for 30 s and 72°C for 40 s, and a final extension at 72°C for 10 min in an automated thermal cycler (Applied Biosystem, Foster City, CA, USA). The PCR product was purified using the QIAquick Gel Extraction kit (QIAgen, Germany), digested with XhoI and HindIII, and cloned into the XhoI-HindIII site of the pSilent-1 plasmid, resulting in pPW1459-1. Second, the internal pks1 fragment (antisense) was amplified with primers LPW11665 5′-GGGGTACCTTTCAACGACCTATCGCCCACTCAA-3′ and LPW11666 5′-GAAGATCTACAACAGCACCAAGCAGTGTGGACA-3′ (Invitrogen, USA), using the PCR conditions described above. This amplified fragment was purified as described above, digested with BglII and KpnI, and cloned into the BglII-KpnI site of the pPW1459-1, resulting in pPW1459. The wild type P. marneffei strain PM1 was transformed with linearized pPW1459, using 200 µg/ml hygromycin for selection. For the other PKS genes, they were knocked down using the same protocol described above with primers listed in Table 1.
Table 1. Primers and plasmids used to construct polyketide knockdown mutants in present study.
| Gene | Primer | RE site | F/R | Sequence (5′-3′) | Plasmid |
| pks1 | LPW 11663 | XhoI | F | CCGCTCGAGTTTCAACGACCTATCGCCCACTCAA | pPW1459 |
| LPW 11664 | HindIII | R | CCCAAGCTTGGGACAACAGCACCAAGCAGTGTGGACA | ||
| LPW 11665 | KpnI | F | GGGGTACCTTTCAACGACCTATCGCCCACTCAA | ||
| LPW 11666 | BglII | R | GAAGATCTACAACAGCACCAAGCAGTGTGGACA | ||
| pks2 | LPW 11381 | XhoI | F | CCGCTCGAGATAAGCTGGTTTTGGTCGAATCGGC | pPW1320 |
| LPW 11382 | HindIII | R | CCCAAGCTTGGGGGATTGAATGGTTGTTGGTCCCGAT | ||
| LPW 11383 | KpnI | F | GGGGTACCATAAGCTGGTTTTGGTCGAATCGGC | ||
| LPW 11384 | BglII | R | GAAGATCTGGATTGAATGGTTGTTGGTCCCGAT | ||
| pks3 | LPW 9873 | XhoI | F | CCGCTCGAGCCTTCTCTTTCGGATCTCTTC | pPW1294 |
| LPW 9874 | KpnI | F | GGGGTACCCCTTCTCTTTCGGATCTCTTC | ||
| LPW 9875 | BglII | R | GAAGATCTGCCTAATGTCAAGCTTTTCG | ||
| pks4 | LPW 9506 | XhoI | F | CCGCTCGAGCCAAACCACTCAGAGTAGCC | pPW1302 |
| LPW 9507 | HindIII | R | CCCAAGCTTGGGACCCTGGTAGAGGAGATTCC | ||
| LPW 9508 | KpnI | F | GGGGTACCCCAAACCACTCAGAGTAGCC | ||
| LPW 9509 | BglII | R | GAAGATCTACCCTGGTAGAGGAGATTCC | ||
| pks5 | LPW 11385 | XhoI | F | CCGCTCGAGTCGACAACTCATCCAACAGATGCCA | pPW1321 |
| LPW 11386 | KpnI | F | GGGGTACCTCGACAACTCATCCAACAGATGCCA | ||
| LPW 11387 | BglII | R | GAAGATCTTTCAACCTGAAGCTTCCGGGAGAAT | ||
| pks6 | LPW 11667 | XhoI | F | CCGCTCGAGTGACCACTCAACAAAGTTCTGGGCC | pPW1460 |
| LPW 11668 | KpnI | F | GGGGTACCTGACCACTCAACAAAGTTCTGGGCC | ||
| LPW 11669 | BglII | R | GAAGATCTCGCATTCATCGGATATGTGCAAGCT | ||
| pks7 | LPW 11670 | XhoI | F | CCGCTCGAGTGCAAGTGTCACCGTATCTGGCGA | pPW1461 |
| LPW 11671 | HindIII | R | CCGCTCGAGTGCAAGTGTCACCGTATCTGGCGA | ||
| LPW 11672 | KpnI | F | GGGGTACCTGCAAGTGTCACCGTATCTGGCGA | ||
| LPW 11673 | BglII | R | GAAGATCTAGCCCCTGTCCGTGGAAAGTTGATA | ||
| pks8 | LPW 13722 | XhoI | F | CCGCTCGAGGCCGCATGTGGACAACATAT | pPW1322 |
| LPW 13723 | HindIII | R | CCCAAGCTTGGGTTTTGGCCCTGCTGAGCT | ||
| LPW 13724 | KpnI | F | GGGGTACCGCCGCATGTGGACAACATAT | ||
| LPW 13725 | BglII | R | GAAGATCTTTTTGGCCCTGCTGAGCT | ||
| pks9 | LPW 11674 | XhoI | F | CCGCTCGAGTGCAGGAAAGCAACTTCGGCCCTA | pPW1515 |
| LPW 11675 | HindIII | R | CCCAAGCTTGGGGCATTATCACCTCGCGCAGCTCATA | ||
| LPW 11676 | KpnI | F | GGGGTACCTGCAGGAAAGCAACTTCGGCCCTA | ||
| LPW 11677 | BglII | R | GAAGATCTGCATTATCACCTCGCGCAGCTCATA | ||
| pks10 | LPW 11678 | XhoI | F | CCGCTCGAGAGAATGGCATCGACTGCCACAGGA | pPW1516 |
| LPW 11679 | HindIII | R | CCCAAGCTTGGGGCCAAACTGGAAGAGCATGCGGTAT | ||
| LPW 11680 | KpnI | F | GGGGTACCAGAATGGCATCGACTGCCACAGGA | ||
| LPW 11681 | BglII | R | GAAGATCTGCCAAACTGGAAGAGCATGCGGTAT | ||
| pks11 | LPW 9278 | XhoI | F | CCGCTCGAGAAGAACCTAAGGGATTATGGAG | pPW1303 |
| LPW 9279 | KpnI | F | GGGGTACCAAGAACCTAAGGGATTATGGAG | ||
| LPW 9280 | BglII | R | GAAGATCTGATTCAGTTCCTTTGCCAAC | ||
| pks12 | LPW 11824 | XhoI | F | CCGCTCGAGTGGAATTTCACGGTTCGCAGCA | pPW1517 |
| LPW 11825 | HindIII | R | CCCAAGCTTGGGTCGCCAGCAACATGTGATTCGCT | ||
| LPW 11826 | KpnI | F | GGGGTACCTGGAATTTCACGGTTCGCAGCA | ||
| LPW 11827 | BglII | R | GAAGATCTTCGCCAGCAACATGTGATTCGCT | ||
| pks13 | LPW 11682 | XhoI | F | CCGCTCGAGTAACGCTTTCGACAGGGTTGGCTTC | pPW1462 |
| LPW 11683 | HindIII | R | CCCAAGCTTGGGGCAGCTCCACGATTGCAGCAATAGA | ||
| LPW 11684 | KpnI | F | GGGGTACCTAACGCTTTCGACAGGGTTGGCTTC | ||
| LPW 11685 | BglII | R | GAAGATCTGCAGCTCCACGATTGCAGCAATAGA | ||
| pks14 | LPW 11686 | XhoI | F | CCGCTCGAGTTATCAAATGCTCGCAGTACGGGCC | pPW1477 |
| LPW 11687 | KpnI | F | GGGGTACCTTATCAAATGCTCGCAGTACGGGCC | ||
| LPW 11688 | BglII | R | GAAGATCTGCGTTGTGCAAAAGAGCCAAGCTT | ||
| pks15 | LPW 9879 | XhoI | F | CCGCTCGAGTTGACGTGAACAATACTTCC | pPW1295 |
| LPW 9880 | KpnI | F | GGGGTACCTTGACGTGAACAATACTTCC | ||
| LPW 9881 | BglII | R | GAAGATCTTGTGTCGAGACTCAAGCT | ||
| pks16 | LPW 9882 | XhoI | F | CCGCTCGAGCTTAGGAGAGGCGAATAAGAAG | pPW1296 |
| LPW 9883 | KpnI | F | GGGGTACCCTTAGGAGAGGCGAATAAGAAG | ||
| LPW 9884 | BglII | R | GAAGATCTTGGCTATCTGCACAAGCT | ||
| pks17 | LPW 9885 | XhoI | F | CCGCTCGAGGCGGGATATCACAATGCA | pPW1518 |
| LPW 9886 | KpnI | F | GGGGTACCGCGGGATATCACAATGCA | ||
| LPW 9887 | BglII | R | GAAGATCTTTCGTGAACCAAGAAGCC | ||
| LPW 9888 | HindIII | R | CCCAAGCTTGGGTTCGTGAACCAAGAAGCC | ||
| pks18 | LPW 11689 | XhoI | F | CCGCTCGAGCAAATCGTCTTATCAGAGGGACTGC | pPW1463 |
| LPW 11690 | KpnI | F | GGGGTACCCAAATCGTCTTATCAGAGGGACTGC | ||
| LPW 11691 | BglII | R | GAAGATCTGGAAGACCACCGATTGTGCAAGCT | ||
| pks19 | LPW 11692 | XhoI | F | CCGCTCGAGTTTCATGGACCAAATCTCTCGCCG | pPW1478 |
| LPW 11693 | HindIII | R | CCCAAGCTTGGGCCGTTACGGTTGATGCTCTCCATGA | ||
| LPW 11694 | KpnI | F | GGGGTACCTTTCATGGACCAAATCTCTCGCCG | ||
| LPW 11695 | BglII | R | GAAGATCTCCGTTACGGTTGATGCTCTCCATGA | ||
| pks20 | LPW 11696 | XhoI | F | CCGCTCGAGTTGAGGCGTATTATGACCCTTCGGG | pPW1323 |
| LPW 11697 | KpnI | F | GGGGTACCTTGAGGCGTATTATGACCCTTCGGG | ||
| LPW 11698 | BglII | R | GAAGATCTAACGTCTTGCATCCTCCTGTTGGG | ||
| pks21 | LPW 12317 | XhoI | F | CCGCTCGAGAAGACAGAACTGTGCCGGGTTGATG | pPW1519 |
| LPW 12318 | HindIII | R | CCCAAGCTTGGGTGAATGATTTCGCAGACTGCTGTCG | ||
| LPW 12319 | KpnI | F | GGGGTACCAAGACAGAACTGTGCCGGGTTGATG | ||
| LPW 12320 | BglII | R | GAAGATCTTGAATGATTTCGCAGACTGCTGTCG | ||
| pks22 | LPW 11703 | XhoI | F | CCGCTCGAGATCGCAAGCTCATCGCCAAACG | pPW1479 |
| LPW 11704 | HindIII | R | CCCAAGCTTGGGTGGCCATTACGCCACAACTGGACT | ||
| LPW 11705 | KpnI | F | GGGGTACCATCGCAAGCTCATCGCCAAACG | ||
| LPW 11706 | BglII | R | GAAGATCTTGGCCATTACGCCACAACTGGACT | ||
| pks23 | LPW 11828 | XhoI | F | CCGCTCGAGGTCGGATTGAACTTTCGTGACGTCG | pPW1562 |
| LPW 11829 | HindIII | R | CCCAAGCTTGGGATCTGCGTAATGCTTCCCCAGCCA | ||
| LPW 11830 | KpnI | F | GGGGTACCGTCGGATTGAACTTTCGTGACGTCG | ||
| LPW 11831 | BglII | R | GAAGATCTATCTGCGTAATGCTTCCCCAGCCA | ||
| pks24 | LPW 11991 | XhoI | F | CCGCTCGAGATAACGCTTGGACAGAGCACTG | pPW1563 |
| LPW 11992 | HindIII | R | CCCAAGCTTGGGCCTTTCCTGGTTGGGTCTCA | ||
| LPW 11993 | KpnI | F | GGGGTACCATAACGCTTGGACAGAGCACTG | ||
| LPW 11994 | BglII | R | GAAGATCTCCTTTCCTGGTTGGGTCTCA | ||
| pks25 | LPW 12638 | XhoI | F | CCGCTCGAGAGATGAAGACCTTGGCGGCTT | pPW1564 |
| LPW 12639 | HindIII | R | CCCAAGCTTGGGTCACCACGAGCATAGCCATTGG | ||
| LPW 12640 | KpnI | F | GGGGTACCAGATGAAGACCTTGGCGGCTT | ||
| LPW 12641 | BglII | R | GAAGATCTTCACCACGAGCATAGCCATTGG |
To construct the pks11pks12 double knockdown mutant, the pks11 fragment (sense) was amplified using primers LPW20378 5′-CCGCTCGAGGTATCAACACGGAAACCGACAA-3′ and LPW20379 5′-TGGCATGTGTGGTTGGTCTGCCATTCTGCGTTATCGGTAGAA-3′. The pks12 fragment (sense) was amplified using primers LPW20380 5′- CAGACCAACCACACATGCCA-3′ and LPW20381 5′- CCCAAGCTTGGGCAGGACAAGTCTCACTGCTATTGA-3′. The two PCR products were used as template for fusion PCR to construct the pks11pks12 fragment (sense) by using primers LPW20378 and LPW20381. The pks11pks12 fragment (antisense) was also amplified by the same method by replacing LPW20378 with LPW20382 5′- GGGGTACCGTATCAACACGGAAACCGACAA-3′ and LPW20381 with LPW20383 5′-GAAGATCTCAGGACAAGTCTCACTGCTATTGA-3′. The sense and antisense pks11pks12 fragments were cloned into pSilent-1 as described above.
Real-time quantitative RT-PCR
Total RNA was extracted using RiboPure-Yeast (Ambion, USA). The RNA was eluted in 70 µl of RNase-free water and was used as the template for real-time RT-PCR. Reverse transcription was performed using the SuperScript III kit (Invitrogen, USA). Real-time RT-PCR assays was performed as described previously [17], for pks1 fragment with primers LPW11663 and LPW11664 (Table 1), using actin with primers LPW8614 5′-CAYACYTTCTACAAYGARCTCC-3′ and LPW8615 5′-KGCVARRATRGAACCACC-3′ for normalization. cDNA was amplified in a LightCycler 2.0 (Roche, Switzerland) with 20 µl reaction mixtures containing FastStart DNA Master SYBR Green I Mix reagent kit (Roche, Switzerland), 2 µl cDNA, 2 mM MgCl2 and 0.5 mM primers at 95°C for 10 min followed by 50 cycles of 95°C for 10 s, 57°C (55°C for actin gene) for 5 s and 72°C for 23 s (36 s for actin gene). For the other PKS genes, real-time RT-PCR was performed using the protocol described above with primers listed in Table 1.
Sequence and phylogenetic analysis of PKS11 and PKS12
Introns were predicted by performing pairwise alignment with the annotated Talaromyces stipitatus (teleomorph of Penicillium emmonsii) complete genome sequence. Domains of PKS11 and PKS12 were predicted using the Conserved Domains Database of NCBI and PFAM (http://pfam.sanger.ac.uk/search?tab=searchSequenceBlock) and manual inspection of multiple alignments of PKS11 and PKS12 and their homologous sequences.
Extraction of yellow pigment
Conidia of seven-day-old cultures of wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutant strains of P. marneffei were collected gently and immersed into 5 µl Milli-Q water. Milli-Q water without conidia was used as negative control. The mixtures were vortexed and filtered with 0.22 µm filters. Metabolic activities in the filtrates were quenched by incubating the filtrates in liquid nitrogen for 10 min. The filtrates were used for UV-Vis spectroscopic examination and UHPLC-DAD/ESI-Q-TOF-MS analysis respectively.
UV-Vis spectroscopic analysis
The maximum absorbance of the extracts was examined by a UV-Vis spectrometer (NanoDrop 1000 spectrophotometer, Thermo scientific, USA). The absorbance was measured by using the 0.2 mm path.
UHPLC-DAD/ESI-Q-TOF-MS analysis
For liquid chromatography, the separation was performed by a 1290 Agilent UHPLC with an Infinity DAD (Agilent Technologies, USA) and a C18 RRHD (Agilent Zorbax Eclipse Plus 2.1 mm×100 mm, 1.8 µm) column. The column temperature was maintained at 40°C. 15 µl of reconstituted sample was injected into the UHPLC instruments. The mobile buffer A consisted of 0.05% acetic acid and 5 mM ammonium acetate in water and mobile buffer B is methanol. Gradient started from 2% buffer B from 0 to 1 min, 2% to 40% buffer B from 1 to 10 min, 40% to 95% buffer B from 10 to 17 min, maintained at 95% buffer B from 17 to 20 min and equilibrated from 20 to 22 min. The flow rate was 0.35 ml/min. Separation was coupled to a 6540 Agilent Q-TOF mass spectrometer (Agilent Technologies, USA) with a jet stream ESI mode and infinity DAD. UV spectrum was collected from 200 to 640 nm with a 4-nm silt.
For MS, mass spectrometer acquisition was operated in the positive ESI mode with accurate mass ranged from 110 to 1700 m/z. Fragmentor was at 135 V and skimmer at 50 V. Drying gas flow rate was kept at 10 l/min at 300°C, the sheath gas flow was 10 l/min at 325°C, capillary voltage was 3500 V, and nebulizer was 45 psi. MS/MS acquisition was operated in the same parameter in MS acquisition. Collision energy was 5 eV for fragmentation of the targeted compounds. Mass spectrometry data were acquired at extended dynamic range with 4 spectra/s and 8 spectra/s in MS/MS mode. Mass accuracy was enhanced by automated calibrant system with two internal reference masses (121.0509 and 922.0098).
Animal experiments
Balb/c (H-2d) mice (6 to 8 weeks old, 18 to 22 g) were obtained from the Laboratory Animal Unit, The University of Hong Kong [18]. The experimental protocols were approved by the Committee on the Use of Live Animals in Teaching and Research, The University of Hong Kong, in accordance with the Guidelines laid down by the NIH in the USA regarding the care and use of animals for experimental procedures. The number of animals used was kept at the minimum that still ensured statistical significance of survival differences between the experimental groups. Mice were housed in cages, under standard conditions with regulated day length, temperature and humidity, and were given pelleted food and tap water ad libitum. Ten mice were challenged intravenously with 8×106 spores of wild type P. marneffei and another 10 mice each with pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants respectively. Survival of the mice was recorded daily for 60 days and analyzed by Kaplan-Meier method and Log-rank test. P<0.05 was regarded as statistically significant. The experiment was performed in duplicate.
Intracellular survival assays in J774 and THP1 macrophages
J774 macrophages (Sigma-Aldrich, USA) were grown in DMEM (Gibco, USA) supplemented with 10% fetal bovine serum and THP1 monocytes (THP1, American type culture collection, ATCC) were grown in suspension in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum at 37°C and 5% CO2.
J774 macrophages were seeded to 24-well tissue culture plates at 4×105 cells per well. THP1 monocytes were seeded to 24-well tissue culture plates at 1×106 cells per well and differentiated into macrophages by incubating in RPMI 1640 medium supplemented with 100 nM PMA. All cell cultures were incubated at 37°C with 5% CO2 for 24 h before adding fungal strains. Fresh culture media were replaced before addition of fungal strains. Infection was carried out by inoculating the conidia of wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants of P. marneffei at a multiplicity of infection of 1 and incubated for 2 h to allow adhesion and invasion to occur. After 2 h, the monolayers were washed with 240 U/ml of nystatin (Sigma-Aldrich) to kill the extracellular conidia. The monolayers were then washed with warm Hank's buffered salt solution (HBSS) to remove the nystatin. Macrophages were supplemented with fresh media and incubated for 24 h. After 24 h post infection, macrophages were lysed with 1% Triton X-100 (Sigma-Aldrich) for colony forming unit (CFU) count. Cell lysates were diluted and plated on Sabouraud dextrose agar and incubated at 37°C. The CFUs recovered from cell lysates after 2 h of phagocytosis were considered as the initial inocula and were used as the baseline values for intracellular survival analysis. CFUs recovered at 24 h were used to calculate the recovery rate of fungal cells in macrophages. Experiments were repeated in triplicate to calculate the mean of intracellular survival of conidia.
Intracellular survival assays in human neutrophils
Unpooled peripheral blood of three healthy blood donors was obtained from the Hong Kong Red Cross Blood Transfusion Service. Neutrophils were isolated according to published protocols with some modifications [19]. Human blood was diluted in HBSS without calcium and magnesium ions and undergone Ficoll-Paque density gradient centrifugation. The bottom layer which contained red blood cells (RBC) with granulocytes was collected and mixed with 3% dextran in 0.9% sodium chloride for 30 min sedimentation. Neutrophils rich supernatant at upper layer was collected and trace amount of RBC in the neutrophils layer was lysed by using RBC lysis buffer.
Conidia of wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants of P. marneffei were preincubated in YPD broth for 4 h at 37°C and preopsonized by incubation in autologous human serum for 30 min at 37°C [20]. Preopsonized conidia were kept at 4°C for at least 1 h before challenge. Equal number of human neutrophils and conidia were incubated in 1 ml RPMI medium containing 10% autologous serum for 2 h. After 2 h, the cultures were washed with 240 U/ml of nystatin to kill the extracellular conidia. The cultures were then washed with HBSS. Neutrophils were supplemented with fresh media and incubated for 18 h. After 18 h post-infection, Neutrophils were lysed with 1% Igepal-CA-630 for CFU count. Cell lysates were diluted and plated on Sabouraud dextrose agar. The CFUs recovered from cell lysates after 2 h of phagocytosis were considered as the initial inocula and were used as the baseline values for intracellular survival analysis. CFUs recovered at 18 h were used to calculate the recovery rate of fungal cells in neutrophils. Experiments were repeated in triplicate to calculate the mean of intracellular survival of conidia.
Susceptibility to hydrogen peroxide killing
Conidial suspensions of wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants of P. marneffei respectively were adjusted to 4×103 cells/ml in 100 mM potassium phosphate buffer (PBS) (pH 7.0) containing 25 mM hydrogen peroxide [15], [21]. At 5-min intervals, aliquots were taken, diluted in 100 mM PBS, and plated onto Sabouraud dextrose agar plates. The experiment was performed in triplicate.
Susceptibility to ultraviolet light killing
Conidial suspensions of wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants of P. marneffei respectively were adjusted to 4×103 cells/ml. Appropriate dilutions of cells were plated on Sabouraud dextrose agar plates and exposed to UV light (254 nm) generated in a Crosslinker (UVP, CA, USA) at various energy settings. Percentage survival was determined by comparing the number of colonies on irradiated plates to those on non-irradiated plates [15], [21]. The experiment was performed in triplicate.
Killing assay of antifungal peptides
Histatin 5 and PGLa were dissolved at a concentration of 1 mg/ml in 10 mM PBS, pH 7.0 respectively and stored at −20°C. Conidial suspensions of wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants of P. marneffei were adjusted to 2×106 cells/ml respectively in 100 mM PBS with a dilution series of peptides. After 1 h incubation, aliquots were taken, diluted in 100 mM PBS, and plated onto Sabouraud dextrose agar plates to determine viabilities. The experiment was performed in triplicate.
Results
pks11 and pks12 are responsible for yellow pigment production in P. marneffei
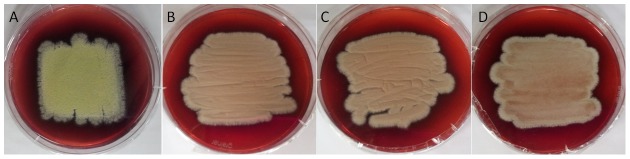
All 23 PKS and 2 PKS-NRPS genes of P. marneffei were systematically knocked down. The median transcription levels of the 25 knockdown mutants were 12.6% (range 0.1% to 40%) of that in wild type. A loss of the yellow pigment was observed exclusively in the mold form of the pks11 and pks12 knockdown mutants, which have pks11 and pks12 transcription levels 5.4% and 10.0% respectively of that in wild type (Fig. 1). A pks11pks12 double knockdown mutant was also constructed. A loss of the yellow pigment was also observed (Fig. 1). The transcription levels of pks11 and pks12 were 26.5% and 9.0% respectively of that in wild type.
Figure 1. Yellow pigment production of wild type, pks11, pks12 and pks11pks12 knockdown mutants of P. marneffei.
(A) Wild type, (B) pks11, (C) pks12 and (D) pks11pks12 knockdown mutants of P. marneffei were grown on Sabouraud dextrose agar after 7 days incubation at 25°C.
Sequence and phylogenetic analysis of PKS11 and PKS12
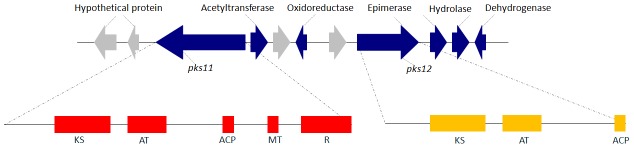
The pks11 gene is 7780 bp in length. It has one intron of 52 bp (from 607 to 658 bp). The resultant putative mRNA encodes 2575 amino acid residues with a predicted molecular mass of 282.6 kDa. The pks12 gene is 5485 bp in length. It has one intron of 63 bp (from 439 to 501 bp). The resultant putative mRNA encodes 1806 amino acid residues with a predicted molecular mass of 197.9 kDa. PKS11 has one ketosynthase, one acyltransferase, one acyl carrier protein, one methyltransferase and one thioester reductase domains whereas PKS12 only has one ketosynthase, one acyltransferase and one acyl carrier protein domain (Fig. 2). The domain organization of PKS11 and PKS12 showed that both are fungal non-reducing PKSs.
Figure 2. Yellow pigment biosynthesis gene cluster and domain structures of pks11 and pks12 in P. marneffei.
Each arrow indicates the direction of transcription and relative sizes of the ORFs. Putative function is indicated for each gene. ACP, acyl carrier protein; AT, acyltransferase; KS, ketosynthase; MT, methyltransferase; R, thioester reductase.
UV-Vis spectroscopic analysis
By UV-Vis spectroscopic analysis, an absorption maximum at 360 nm was recognized in the filtrate of wild type P. marneffei. This absorption maximum was not observed in the culture filtrates of the pks11 knockdown, pks12 knockdown or pks11pks12 double knockdown mutants.
UHPLC-DAD-MS analysis
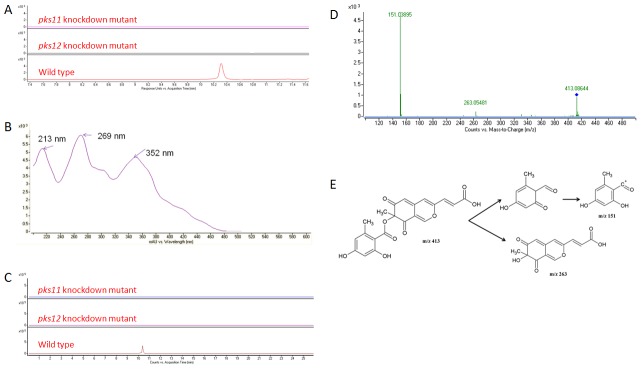
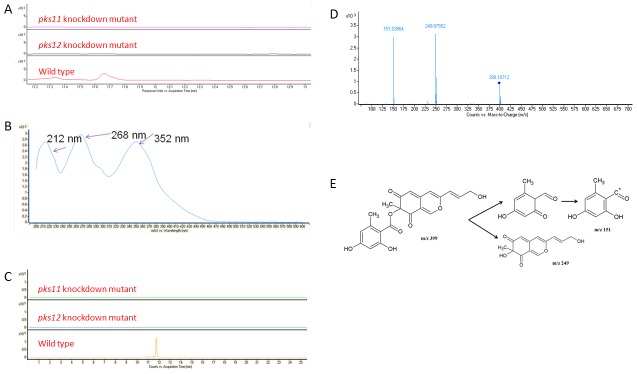
The filtrates from wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants of P. marneffei were monitored by UV-Vis spectroscopy from 200 to 640 nm and positive ion electrospray MS. To detect the presence of yellow pigment in wild type P. marneffei but not in the pks11 knockdown, pks12 knockdown or pks11pks12 double knockdown mutants, the UHPLC profiles were monitored by DAD using UV-visible absorption at 360 nm. Peaks at 10.2 min and 11.7 min were present in wild type P. marneffei but not the pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants (Fig. 3A and Fig. 4A). The two peaks were subjected to UV absorption, MS and MS/MS analyses. For the peak at 10.2 min, UV absorption analysis showed that the isolated molecule has three absorbance maxima at 213 nm, 269 nm and 352 nm respectively (Fig. 3B). MS analysis showed that this molecule has m/z of 413.0876 [M+H]+, which matched for C21H16O9 (Fig. 3C). This chemical formula was compatible with mitorubrinic acid (Fig. 3E). The fragmentation pattern of the MS/MS analysis showed a peak at m/z 151.03895, which corresponded to C8H6O3 and another at m/z 263.05481, which corresponded to C13H10O6, further confirming the isolated molecule is mitorubrinic acid (Fig. 3D and 3E). For the peak at 11.7 min, UV absorption analysis showed that the isolated molecule has three absorbance maxima at 212 nm, 268 nm and 352 nm respectively (Fig. 4B). MS analysis showed that this molecule has m/z of 399.10834 [M+H]+ matched for C21H18O8 (Fig. 4C). This chemical formula was compatible with mitorubrinol (Fig. 4E). The fragmentation pattern of the MS/MS analysis showed a peak at m/z 151.0386, which corresponded to C8H6O3 and another at m/z 249.0755, which corresponded to C13H12O5, further confirming the isolated molecule is mitorubrinol (Fig. 4D and 4E).
Figure 3. Detection of mitorubrinic acid by UHPLC-DAD/ESI-Q-TOF-MS and MS/MS analysis.
(A) HPLC profiles monitored by photodiode array detector and illustrated at 360 nm, (B) UV absorption spectrum, (C) extracted ion chromatograms (m/z 413.0876), (D) MS/MS fragmentation pattern and (E) MS/MS fragmentation pathway showing the presence of mitorubrinic acid detected and identified in wild type of P. marneffei but not in pks11 or pks12 knockdown mutants.
Figure 4. Detection of mitorubrinol by UHPLC-DAD/ESI-Q-TOF-MS and MS/MS analysis.
(A) HPLC profiles monitored by photodiode array detector and illustrated at 360 nm, (B) UV absorption spectrum, (C) extracted ion chromatograms (m/z 399.10834), (D) MS/MS fragmentation pattern and (E) MS/MS fragmentation pathway showing the presence of mitorubrinol detected and identified in wild type of P. marneffei but not in pks11 or pks12 knockdown mutants.
Animal experiments
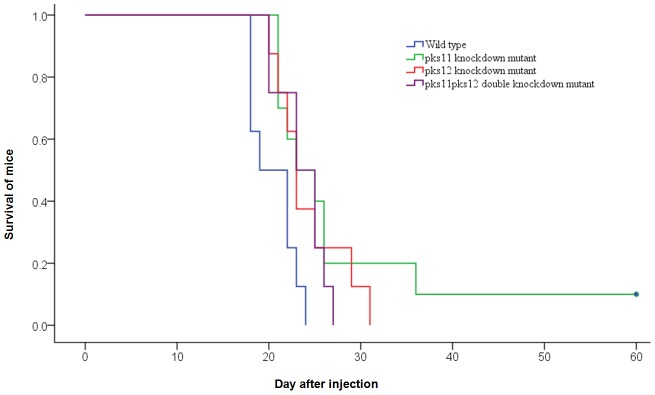
The survival of mice after intravenous challenge with wild type P. marneffei or the pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants on day 60 was summarized in Fig. 5. The survival of mice challenged with the pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants were significantly better than those challenged with wild type P. marneffei (P<0.05).
Figure 5. Survival of mice challenged with wild type P. marneffei or pks11/pks12/pks11pks12 knockdown mutants.
Groups of 10 BALB/c mice were challenged intravenously with 8×106 spores. Survival was recorded daily for 60 days.
Intracellular survival assays in J774 and THP1 macrophages
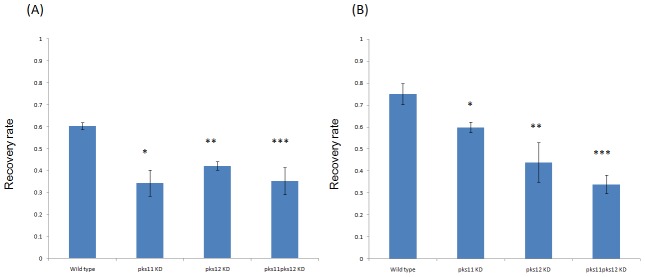
The survival of wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants of P. marneffei in J774 and THP1 macrophages is shown in Fig. 6. There was statistically significant decrease in survival of pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants compared to wild type P. marneffei in both J774 and THP1 macrophages (P<0.05).
Figure 6. Survival of wild type P. marneffei and pks11/pks12/pks11pks12 knockdown mutants in J774 and THP1 macrophages.
Panels A and B represent the recovery rates of wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants of P. marneffei in J774 and THP1 macrophages respectively. Error bars represent as mean ± SEM. Statistical significance between groups is indicated. *: wild type versus pks11 knockdown mutant (p<0.05); **: wild type versus pks12 knockdown mutant (p<0.05), ***: wild type versus pks11pks12 double knockdown mutant (p<0.05).
Intracellular survival assays in human neutrophils
No difference was observed between the survivals of wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants of P. marneffei in human neutrophils.
Susceptibility to hydrogen peroxide killing
The relative survival of P. marneffei conidia capable of forming visible colonies was calculated and plotted as a function of time of incubation in 25 mM hydrogen peroxide. No difference was observed between the survivals of wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants of P. marneffei.
Susceptibility to ultraviolet light killing
No difference was observed between the survivals of wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants of P. marneffei exposed to different doses of ultraviolet light.
Killing assay of antifungal peptides
No difference was observed between the survivals of wild type, pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants of P. marneffei exposed to different concentrations of histatin 5 and PGLa peptides.
Discussion
We report the first discovery of PKS genes responsible for mitorubrinol and mitorubrinic acid biosynthesis. Mitorubrinol and mitorubrinic acid are mitorubrin derivatives, a unique subclass of azaphilones isolated from a variety of fungal species, including Penicillium species such as P. rubrum and P. funiculosum and Talaromyces (teleomorph of Penicilllium) species such as T. emodensis, T. hachijoensis, T. wortmannii var. sublevisporus, T. austrocalifornicus and T. convolutus [22], [23], [24]. Although it has been known that mitorubrinol and mitorubrinic acid are polyketides for decades, no PKS genes have been identified for their synthesis [22]. In this study, we systematically knocked down the 23 PKS and 2 PKS-NRPS genes in the P. marneffei genome and observed for loss of yellow pigment in its mold form. Two PKS genes in the same PKS gene cluster were confirmed to be responsible for biosynthesis of yellow pigment in P. marneffei. UV absorption, MS and MS/MS analyses all unambiguously confirmed that this yellow pigment consisted of mitorubrinol and mitorubrinic acid.
pks12 and pks11 are probably responsible for sequential use in the biosynthesis of mitorubrinol and mitorubrinic acid. It is well known that some polyketides, such as lovastatin and zearalenone, were synthesized by two PKSs. The first PKS serves to synthesize a starter unit for the second PKS, which possesses a starter unit ACP transacylase (SAT) domain for utilizing the advanced starter unit [25]. For zearalenone, the starter unit is a highly reduced hexaketide which encoded by a highly reducing PKS gene (PKS13) [26]. The starter unit is utilized by a second PKS gene (PKS4) which possesses SAT domain for further extension. As for mitorubrinic acid and mitorubrinol, we speculate that the first part of the biosynthesis was by PKS12, which synthesized orsellinic acid. This tetraketide is then used as a starter unit for PKS11, which possessed a putative SAT domain, in the second part of the biosynthesis. PKS11, which possessed a methyltransferase domain, also served to methylate the products, using a methyl group from S-adenosylmethionine. Interestingly, PKS12 also possessed putative SAT domain, in line with the fact that most non-reducing PKS also possess potential SAT domains irrespective of whether they require an acetate starter unit or not. Feeding experiments using isotopically or 19F labeled precursors will confirm the use of advanced starter unit for the biosynthesis of mitorubrinic acid and mitorubrinol. Notably, polyketides that are synthesized by two PKS genes commonly require a highly reducing PKS at the early stage of biosynthesis and a non-reducing PKS at the later stage, resulting in a final product consisted of a highly reducing chain and non-reducing rings. Examples include the biosynthesis of zearalenone in Gibberella zeae and asperfuranone in Aspergillus nidulans [26], [27]. However, it is rare to see a polyketide, similar to mitorubrinol and mitorubrinic acid, that is synthesized by two non-reducing PKS in a sequential manner resulting in a non-reduced polyketide product.
Mitorubrinol and mitorubrinic acid are virulence factors of P. marneffei by improving its intracellular survival in macrophages. Although P. marneffei is infecting about 8% of AIDS patients in China and Southeast Asia, the pathogenetic mechanisms of this fungus remained under studied. So far, several molecules, including superoxide dismutase and melanin, have been implicated to be associated with virulence in P. marneffei [15], [28]. Superoxide dismutase converts superoxide radicals into hydrogen peroxide and oxygen, whereas melanin contributed to virulence through decreased susceptibility to killing by hydrogen peroxide [15], [28]. In this study, these two PKS genes in the yellow pigment biosynthesis gene cluster responsible for mitorubrinol and mitorubrinic acid production were shown to be associated with virulence in a mouse model. However, unlike melanin, the mechanism of virulence is not related to increasing resistance to hydrogen peroxide killing and the decrease in virulence in the pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants were also not due to decrease in survival within neutrophils, reduced resistance to antifungal peptides, decrease in growth rates or change in electron microscopic appearances of the mutants (data not shown). On the other hand, the survival of wild type P. marneffei was significantly better than pks11 knockdown, pks12 knockdown and pks11pks12 double knockdown mutants in both murine and human macrophages. Notably, it has been shown that injection of mitorubrin to mice did not result in death of the mice, although livestock fed with feedstuff contaminated with P. rubrum could be poisoned, indicating that mitorubrin itself is not a toxic metabolite [22]. This is in line with the results of the present study, which suggested that the mechanism of virulence was mediated through enhancement of intracellular survival in macrophages. It is also noteworthy that the pks11pks12 double knockdown mutant did not survive better than the pks11 or pks12 knockdown mutants. This is because knocking down either pks11 or pks12 is sufficient to abolish the pathway for mitorubrinol and mitorubrinic acid synthesis.
Funding Statement
This work was partly supported by the Research Fund for the Control of Infectious Diseases (commissioned study) of the Health, Welfare and Food Bureau of the Hong Kong SAR Government, Research Grant Council Grant, University Development Fund, Committee for Conference and Research Grant, Providence Foundation Limited in memory of the late Dr. Lui Hac Minh, HKU Award for CAE Membership, HKU Medical Faculty Award for CAE Membership, Dr. Hector T.G. Ma. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hsueh PR, Teng LJ, Hung CC, Hsu JH, Yang PC, et al. (2000) Molecular evidence for strain dissemination of Penicillium marneffei: an emerging pathogen in Taiwan. J Infect Dis 181: 1706–1712. [DOI] [PubMed] [Google Scholar]
- 2. Supparatpinyo K, Khamwan C, Baosoung V, Nelson KE, Sirisanthana T (1994) Disseminated Penicillium marneffei infection in southeast Asia. Lancet 344: 110–113. [DOI] [PubMed] [Google Scholar]
- 3. Wong SS, Siau H, Yuen KY (1999) Penicilliosis marneffei–West meets East. J Med Microbiol 48: 973–975. [DOI] [PubMed] [Google Scholar]
- 4. Yuen KY, Wong SS, Tsang DN, Chau PY (1994) Serodiagnosis of Penicillium marneffei infection. Lancet 344: 444–445. [DOI] [PubMed] [Google Scholar]
- 5. Deng ZL, Connor DH (1985) Progressive disseminated penicilliosis caused by Penicillium marneffei. Report of eight cases and differentiation of the causative organism from Histoplasma capsulatum. Am J Clin Pathol 84: 323–327. [DOI] [PubMed] [Google Scholar]
- 6. Low K, Lee SS (2002) The pattern of AIDS Reporting and the implications on HIV surveillance. Public Health Epidemiol Bull 11: 41–49. [Google Scholar]
- 7. Lo CY, Chan DT, Yuen KY, Li FK, Cheng KP (1995) Penicillium marneffei infection in a patient with SLE. Lupus 4: 229–231. [DOI] [PubMed] [Google Scholar]
- 8. Wang JL, Hung CC, Chang SC, Chueh SC, La MK (2003) Disseminated Penicillium marneffei infection in a renal-transplant recipient successfully treated with liposomal amphotericin B. Transplantation 76: 1136–1137. [DOI] [PubMed] [Google Scholar]
- 9. Wong SS, Woo PC, Yuen KY (2001) Candida tropicalis and Penicillium marneffei mixed fungaemia in a patient with Waldenstrom's macroglobulinaemia. Eur J Clin Microbiol Infect Dis 20: 132–135. [DOI] [PubMed] [Google Scholar]
- 10. Woo PC, Lau SK, Lau CC, Chong KT, Hui WT, et al. (2005) Penicillium marneffei fungaemia in an allogeneic bone marrow transplant recipient. Bone Marrow Transplant 35: 831–833. [DOI] [PubMed] [Google Scholar]
- 11. Woo PC, Lau SK, Liu B, Cai JJ, Chong KT, et al. (2011) Draft genome sequence of Penicillium marneffei strain PM1. Eukaryot Cell 10: 1740–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woo PC, Zhen H, Cai JJ, Yu J, Lau SK, et al. (2003) The mitochondrial genome of the thermal dimorphic fungus Penicillium marneffei is more closely related to those of molds than yeasts. FEBS Lett 555: 469–477. [DOI] [PubMed] [Google Scholar]
- 13. Woo PC, Chong KT, Tse H, Cai JJ, Lau CC, et al. (2006) Genomic and experimental evidence for a potential sexual cycle in the pathogenic thermal dimorphic fungus Penicillium marneffei. FEBS Lett 580: 3409–3416. [DOI] [PubMed] [Google Scholar]
- 14. Woo PC, Lau CC, Chong KT, Tse H, Tsang DN, et al. (2007) MP1 homologue-based multilocus sequence system for typing the pathogenic fungus Penicillium marneffei: a novel approach using lineage-specific genes. J Clin Microbiol 45: 3647–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woo PC, Tam EW, Chong KT, Cai JJ, Tung ET, et al. (2010) High diversity of polyketide synthase genes and the melanin biosynthesis gene cluster in Penicillium marneffei. FEBS J 277: 3750–3758. [DOI] [PubMed] [Google Scholar]
- 16. Nakayashiki H, Hanada S, Nguyen BQ, Kadotani N, Tosa Y, et al. (2005) RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol 42: 275–283. [DOI] [PubMed] [Google Scholar]
- 17. Woo PC, Tung ET, Chan KH, Lau CC, Lau SK, et al. (2009) Cytokine Profiles Induced by the Novel Swine-Origin Influenza A/H1N1 Virus: Implications for Treatment Strategies. J Infect Dis [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wong LP, Woo PC, Wu AY, Yuen KY (2002) DNA immunization using a secreted cell wall antigen Mp1p is protective against Penicillium marneffei infection. Vaccine 20: 2878–2886. [DOI] [PubMed] [Google Scholar]
- 19. Maqbool M, Vidyadaran S, George E, Ramasamy R (2011) Optimisation of laboratory procedures for isolating human peripheral blood derived neutrophils. Med J Malaysia 66: 296–299. [PubMed] [Google Scholar]
- 20. Levitz SM, Diamond RD (1985) Mechanisms of resistance of Aspergillus fumigatus Conidia to killing by neutrophils in vitro. J Infect Dis 152: 33–42. [DOI] [PubMed] [Google Scholar]
- 21. Romero-Martinez R, Wheeler M, Guerrero-Plata A, Rico G, Torres-Guerrero H (2000) Biosynthesis and functions of melanin in Sporothrix schenckii. Infect Immun 68: 3696–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buechi G, White JD, Wogan GN (1965) The Structures of Mitorubrin and Mitorubrinol. J Am Chem Soc 87: 3484–3489. [DOI] [PubMed] [Google Scholar]
- 23. Natsume M, Takahashi Y, Marumo S (1985) (-)-Mitorubrinic Acid, a Morphogenic Substance Inducing Chlamydospore-Like Cells, and Its Related New Metabolite, (+)-Mitorubrinic Acid-B, Isolated from Penicillium-Funiculosum. Agricultural and Biological Chemistry 49: 2517–2519. [Google Scholar]
- 24. Suzuki S, Hosoe T, Nozawa K, Yaguchi T, Udagawa S, et al. (1999) Mitorubrin derivatives on ascomata of some talaromyces species of ascomycetous fungi. J Nat Prod 62: 1328–1329. [DOI] [PubMed] [Google Scholar]
- 25. Cox RJ (2007) Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem 5: 2010–2026. [DOI] [PubMed] [Google Scholar]
- 26. Kim YT, Lee YR, Jin J, Han KH, Kim H, et al. (2005) Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol Microbiol 58: 1102–1113. [DOI] [PubMed] [Google Scholar]
- 27. Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, et al. (2009) A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J Am Chem Soc 131: 2965–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thirach S, Cooper CR Jr, Vanittanakom P, Vanittanakom N (2007) The copper, zinc superoxide dismutase gene of Penicillium marneffei: cloning, characterization, and differential expression during phase transition and macrophage infection. Med Mycol 45: 409–417. [DOI] [PubMed] [Google Scholar]