Abstract
Transcription-coupled repair employs components of the transcription machinery to identify DNA lesions and initiate their repair. These repair pathways are complex so their mechanistic features remain poorly understood. Bacterial transcription-coupled repair is initiated when RNA polymerase stalled at a DNA lesion is removed by Mfd, an ATP-dependent DNA translocase [1–3]. Here we use single-molecule DNA nanomanipulation to observe the dynamic interactions of E. coli Mfd with RNA polymerase elongation complexes stalled by a cyclopyrimidine dimer or by nucleotide starvation. We show that Mfd acts by catalyzing two irreversible, ATP-dependent steps with different structural, kinetic, and mechanistic features. Mfd remains bound to the DNA in a long-lived complex that could serve as a marker for sites of DNA damage, directing assembly of subsequent DNA repair factors. These results provide a framework for considering the kinetics of transcription-coupled repair in vivo, and open the way to reconstruction of complete DNA repair pathways at single-molecule resolution.
In bacteria the Mfd protein couples transcription and nucleotide excision repair (NER). Mfd is a superfamily 2 ATP-dependent DNA translocase [1, 2] (Supplementary Figure S1) with two distinct functions: first, it recognizes a stalled RNAP and uses the energy of ATP hydrolysis to dissociate the RNAP-DNA elongation complex (RDe). Second, Mfd recruits UvrA, a component of the Uvr(A)BC excinuclease machinery. As a consequence of Mfd action, bulky lesions that stall RNAP are repaired more efficiently than similar lesions repaired by the transcription-independent global NER pathway [3]. Transcription-coupled repair in eukaryotes is more complex, but the functional homolog of Mfd in humans (CSB) is also a superfamily 2 class of DNA translocase [4].
Although the role of Mfd in the recognition and dissociation of a stalled RDe has been well-studied genetically [2, 5, 6], biochemically [1, 7–10] and structurally [11–14], the mechanism whereby Mfd promotes RDe dissociation has not been determined. Because Mfd can rescue backtracked RDe it is thought that it acts by pushing RNAP downstream, in what is termed the forward-translocation mechanism [7]. Furthermore, identification of a specific interaction between Mfd and RNAP that is required for Mfd function led to the proposal that Mfd simultaneously interacts with RNAP (via its RNAP interacting domain; RID) and with DNA (via its ATPase domains) [7, 11], allowing its translocase activity to generate positive torque on the DNA, wrenching shut the transcription bubble and thus destabilizing RDe [15, 16].
To investigate the mechanism of action of Mfd on stalled RDe, we developed a single-molecule assay allowing us to monitor Mfd interactions with single stalled E. coli RNAP molecules in real time (Figure 1A and Supplementary Fig. S2) [17]. Here the mechanical properties of a nanomanipulated DNA are used to detect a single RNAP initiating transcription, progressing to form RDe, and dissociating upon reaching a terminator [17] (Fig. 1B). By omitting CTP, we can stall RNAP at position +20 of the template for an indefinite amount of time (Fig. 1C and Supplementary Fig. S3) [7, 20].
Figure 1. Experimental approach and real-time dissociation of stalled RDe by Mfd.
(A) A linear, 2kb dsDNA containing a promoter, a transcribed region and a terminator is extended and supercoiled by anchoring it to a glass surface at one end and a 1-μm magnetic bead at the other, and manipulating the bead using a magnetic field. RNAP-DNA interactions are observed in real-time using videomicroscopy-based bead tracking to measure changes in DNA conformation (see Supplementary Fig. S2 and [17]). (B) Single-molecule time-trace showing two complete transcription events. Green points, raw DNA extension data (31 Hz); red points, data averaged over ~1 s. Black lines indicate the DNA extension for the initial state, the RNAP/promoter initially transcribing complex (RPitc) during which DNA scrunching occurs, and the ternary elongation complex (RDe). The increase in extension from RPitc to RDe corresponds to promoter escape, and the increase in extension from RDe back to the initial state corresponds to termination. (C) Representative time-trace obtained when CTP is omitted. RNAP stalls ~5 bp (~0.5 s [17]) after promoter escape. Termination is not observed. (D) Representative time-trace obtained as in the prior panel, but in the presence of 100 nM Mfd. Return to the initial state takes place via two distinct increases in DNA extension which occur, respectively, t1 and t1+t2 seconds after promoter escape. In the intervening time a stable, long-lived intermediate is formed which we label I.
A single-molecule time-trace obtained when Mfd is present during the RNAP stall indicates that Mfd forms a remarkably long-lived intermediate, denoted I (Fig. 1D). Analysis of changes in DNA extension during formation and resolution of I provides information on its structural properties (Supplementary Fig. S4 and [18,19]), revealing that, in the intermediate, at least 2/3 of the transcription bubble is rewound and the DNA is bent by approximately 90°. As permanganate footprinting suggests that the transcription bubble is rapidly and fully rewound by Mfd action (Supplementary Fig. S5) the DNA deformation observed in the intermediate is likely due to bending/wrapping interactions between proteins and DNA rather than residual unwinding in a partially collapsed transcription bubble.
Pulse-chase experiments show that both formation and resolution of the intermediate require ATP binding (Fig. 2A and B) and hydrolysis (Supplementary Figs. S6 and S7). Moreover when we trap the intermediate with a wash step which removes both ATP and free Mfd from solution, we find that adding back only ATP is sufficient to allow resolution of the trapped intermediate (Fig. 2B). Thus, the same molecule of Mfd is responsible for formation and resolution of the intermediate. Finally, we occasionally observe that following formation of the intermediate, a second RNAP can bind to the promoter, initiate transcription, stall, and become a target for a second molecule of Mfd (Figure 2C). No such reloading events are observed when RNAP is stalled in the absence of Mfd (Supplementary Fig. S3). Thus as Mfd modifies the structure of RDe it also clears the transcription start site for a new RNAP to initiate [7, 15].
Figure 2. Mfd ATP usage and displacement of stalled RNAP.
Arrows indicate wash steps for trapping and release of the intermediate. For clarity we present 200-s snapshots of the states thus obtained. (A) ATP is required for formation of the intermediate. Stalled RDe is formed, then trapped by washing out (blue arrow) free RNAP and NTPs. Upon addition of Mfd (200 nM), DNA extension is unchanged. When supplemented with ATP (2 mM) (orange arrow), the intermediate forms. (B) ATP is required for release of the intermediate. Stalled RDe is formed and trapped as above. Upon addition of both Mfd and ATP (first orange arrow) the intermediate forms. Upon washing out free Mfd and ATP (second blue arrow) the intermediate is stable for thousands of seconds. Upon adding back ATP (second orange arrow) the intermediate is resolved. (C) Upon intermediate formation stalled RNAP is displaced from its promoter-proximal stall site. Black arrows indicate transcription initiation events; red arrows indicate intermediate formation. After the first initiation event (first black arrow) is followed by intermediate formation (first red arrow), a second RNAP can initiate transcription (second black arrow) and become stalled, forming a new substrate for another Mfd to displace (second red arrow). No reloading by a second RNAP occurs in the absence of Mfd (Supplementary Fig. S3).
Surprisingly, single molecule experiments with Mfd alone show that in the nucleotide-bound state (ie in the presence of ATP-γ-S but not in the presence of ATP) the protein distorts the DNA in a pattern characteristic of unwinding or wrapping (Supplementary Figs. S6 and S8). In the presence of ATP, Mfd distorts the DNA provided that core RNAP is present at high concentration in solution (Supplementary Fig. S8). Since permanganate attack of DNA in the presence of Mfd and ATP-γ-S detects no unwinding (Supplementary Fig. S6) this distortion is most likely due to partial, left-handed wrapping of DNA about Mfd. However, in contrast to the remarkably stable nature of the Mfd-containing TCR intermediate in all nucleotide conditions tested, the Mfd/DNA complexes in Supplementary Figs. S6 and S8 are unstable as they do not persist after free Mfd is washed out.
Real-time monitoring of the formation and resolution of the repair intermediate allows for its precise kinetic description. Although the mean lifetime <t1> of the stalled RDe decreases as Mfd concentration increases (Fig. 3A), the mean lifetime <t2> of the intermediate complex is unchanged (Supplementary Table), confirming that the same molecule of Mfd is present throughout the reaction (Fig. 2B). Thus formation of the intermediate is expected to obey Michaelian kinetics according to
Figure 3. Kinetic characterization of the action of Mfd on RDe.
(A) Mean of RDe lifetime <t1> vs. Mfd concentration, and fit to Michaelis-Menten model (error bars = S.E.M). (B) Full distributions of t1 for [Mfd] = 50 nM (n = 273 events) and [Mfd] = 750 nM (n=281 events). Red curves: global fits with a difference-of-two-exponentials according to the single-molecule Michaelis-Menten model (see Supplementary Materials and Supplementary Fig. S9). (C) Distribution of reaction intermediate lifetime t2 (n = 146 events). Red curve, fit with a normal distribution model with a mean of 335 ± 3 s and a standard deviation of 181 seconds. Experiments were performed on DNA supercoiled by +4 turns.
| (Eq. 1) |
while disassembly of the intermediate is expected to depend on simple rate constants (see below). Fitting the mean lifetime data for <t1> (Fig. 3A) yields KM = 760 ± 350 nM and 1/k3= 19±7 s. Thus formation of the intermediate is characterized by weak binding of Mfd to RDe followed by a remarkably slow catalytic step.
The statistical distributions of dwell-times preceding formation of the intermediate (Fig. 3B) allow resolution of the Michaelis constant KM into the diffusion-controlled on-rate and the thermodynamically-controlled off-rate of Mfd from RDe. The shape of the distribution – an exponential rise followed by an exponential decay – is characteristic of a reaction with at least two intermediates, namely diffusion/docking of Mfd to RDe and a forward-catalytic event (see Supplementary Materials and [21]). Taking k3 = 0.05 s−1 from our prior analysis of <t1>, we obtain k1 ~ 1.9·105 M−1·s−1 and k2 ~ 0.11 s−1. By performing a global fit to the distributions obtained at four different Mfd concentrations (Supplementary Fig. S9) the constraint on k3 can be relaxed, yielding k1 = 1.6±0.6·105 M−1·s−1, k2 = 0.08 ± 0.04 s−1, and k3 = 0.059 ± 0.01 s−1 with a reduced χ2 = 0.88, supporting the internal consistency of the data and the validity of the kinetic model.
Surprisingly, the statistical distribution of intermediate lifetimes, t2 (corresponding to resolution of the intermediate) is non-exponential (Fig. 3C). It tends instead to a normal distribution characteristic of a reaction consisting of multiple irreversible steps with equivalent rate constants. The data thus give <t2> = 335 ± 3 s and a standard deviation of 181 seconds. Variance analysis of this distribution suggests that a minimum of three irreversible events takes place during this time, each thus having a rate constant k4 = 3/<t2>. Pulse-chase analysis of ATP usage in this reaction indicates that at least one of these steps requires ATP hydrolysis.
The data analysed here were collected using a substrate bearing five successive C residues on the non-template strand at the stall site, but the process leading to disassembly of the intermediate is not sequence-dependent as similar results were obtained when only one C residue was present (Supplementary Figs. S10 and S11). Similarly, disassembly of the intermediate is only weakly torque- dependent, as a long-lived complex was observed using negatively supercoiled DNA: <t2> = 180 +/− 40 s (n = 16; data collected as described in Supplementary Fig. S4). Finally, we analyzed the interactions between Mfd and RNAP stalled on a cyclopyrimidine dimer located on the template strand (Supplementary Fig. S12). The kinetics of the entire pathway are essentially identical to those observed when RNAP is stalled by withholding a nucleotide. Indeed, a crystal structure of RNAP II stalled on a cyclopyrimidine dimer revealed no conformational changes compared to elongation complexes stalled by nucleotide deprivation [22].
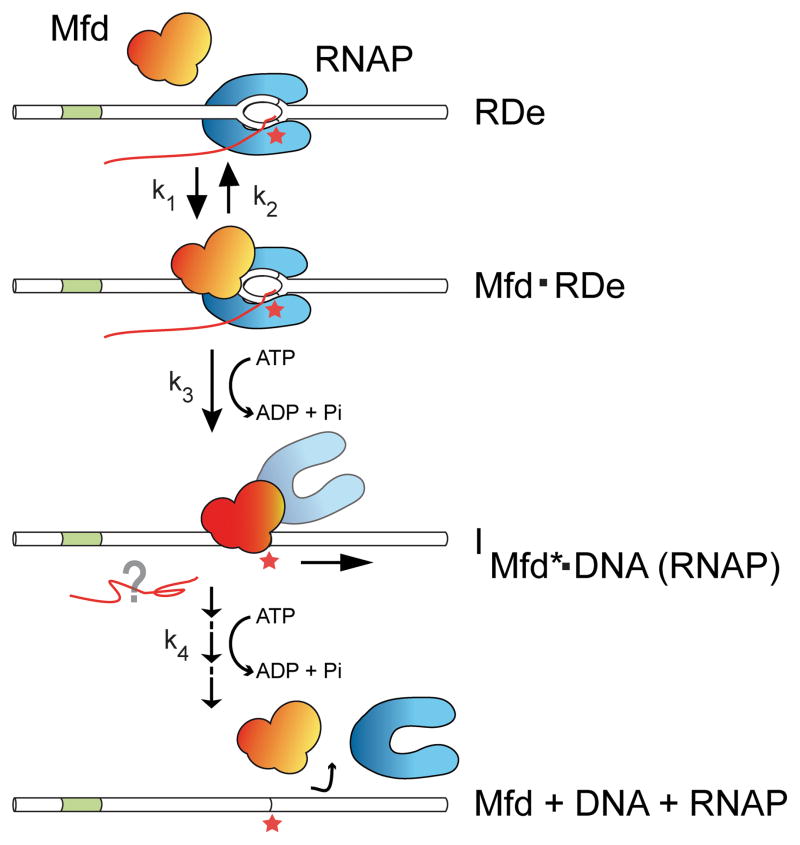
Based on these data, we propose a model to describe RDe displacement during TCR (Fig. 4). From a structural standpoint, the mechanism by which Mfd initiates displacement of the stalled RNAP involves sufficient collapse of the transcription bubble to destabilize the stalled RDe [23]. Concurrently, promoter-proximal DNA is cleared for another RNAP to initiate (Fig. 2C), indicating either forward translocation of RNAP or full dissociation of RNAP. Mfd remains associated with the DNA long after bubble collapse. The source of the DNA deformation observed in the intermediate cannot be fully ascribed from our current data. Based on the observation that free Mfd is capable of distorting DNA, we favor a model in which the transcription bubble is fully collapsed upon formation of the intermediate, and the residual DNA deformation observed in the long-lived intermediate is caused by Mfd (Fig. 4). Since Mfd alone does not form long-lived complexes on DNA, we propose that the intermediate is stabilised by a combination of Mfd:DNA interactions, Mfd:RNAP interactions (mediated by the RID) and, potentially, interaction of the Mfd-tethered core RNAP with DNA (Fig. 4). Binding of Mfd to RNAP derepresses the DNA translocation domains of the helicase, and so the nature of Mfd:DNA interactions in this activated ternary complex likely differs from those made by the isolated protein [8, 13, 24].
Figure 4. Model of RNAP displacement by Mfd during TCR.
Mfd kinetically discriminates the stalled RDe, and upon ATP hydrolysis disrupts RDe by collapsing its transcription bubble and clearing the RNAP from the stall site. Subsequently, additional ATP hydrolysis events are required for activated Mfd to dissociate from DNA. RNA presumably dissociates upon transcription bubble collapse, but the moment of RNAP disassembly from DNA and Mfd is unclear.
From a kinetic standpoint, the rate constants k1, k2 and k3 indicate that Mfd attempts binding to RDe several times before engaging productively, and the rate-limiting step itself is slow. This suggests that Mfd kinetically discriminates stalled RDe from paused RDe [25], as only stalled RDe is sufficiently long-lived to serve as an Mfd target. This model is supported by our observation that the kinetics of displacement are the same for RNAP stalled at a CPD lesion or by nucleotide starvation. The slow catalytic event could be related to derepression of Mfd, which likely involves large conformational changes [8, 24]. The intermediate I is formed in an ATP hydrolysis-dependent step, and Mfd then performs at least three irreversible steps before releasing the DNA. As ATP hydrolysis is required to proceed out of this intermediate state, we propose that Mfd hydrolyzes a second ATP molecule, and possibly more, before dissociating.
The lifetime of the intermediate is surprisingly long for an organism that repairs its DNA in much shorter times and which can divide every 20 minutes; in vivo, downstream repair proteins may capture and process the intermediate, reducing this period. Nevertheless, the fact that the intermediate is reliably long-lived could serve to ensure that repair components have ample opportunity to assemble.
The recruitment of UvrA by Mfd to the damage site is not well understood. The interaction between UvrA and Mfd is inhibited by inter-domain contacts within free Mfd, and the interaction between Mfd and RNAP likely overcomes this inhibition by domain repositioning (Supplementary Fig. S1 and [8, 11, 24, 26–28]). As Mfd plays a central role in the recruitment of the NER machinery, it will be interesting to see how addition of subsequent players (such as UvrA, UvrB and UvrC) affects the temporal properties of the system, and from there to pursue bottom-up reconstruction of the full DNA repair pathway at single-molecule resolution.
Methods Summary
See legend to Figure 1 for methods summary; full Methods can be found in the Supplementary Information.
Supplementary Material
Acknowledgments
We thank K. Neumann for showing us how to perform global fitting with the Igor software package. KH was supported by a PhD scholarship from the Frontieres Interdisciplinaires du Vivant Doctoral Program and the Fondation pour la Recherche Médicale. Work in the lab of NS was supported by BBSRC Grant BB/1003142/1, and work by in the lab of SD was supported by NIH grant GM073829. This work was also made possible by a EURYI grant, in addition to CNRS and University of Paris Diderot core funding, to TS.
Footnotes
Contributions
KH, NS and TS devised and carried out experiments; SD, KH, NJ, NS, AS, TS and LW provided unique reagents and AS carried out further control experiments; KH, WG, NS, TS and SZ analyzed data; and SD, NS and TS wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
The PDF file contains full Methods, Supplementary Figures S1–12, and the Supplementary Table.
References
- 1.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 2.Witkin EM. Time, temperature, and protein synthesis: a study of ultraviolet-induced mutation in bacteria. Cold Spring Harb Symp Quant Biol. 1956;21:123–140. doi: 10.1101/sqb.1956.021.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 4.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 5.Selby CP, Sancar A. Gene- and strand-specific repair in vitro: partial purification of a transcription-repair coupling factor. Proc Natl Acad Sci USA. 1991;88:8232–8236. doi: 10.1073/pnas.88.18.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selby CP, Sancar A. Transcription-repair coupling and mutation frequency decline. J Bacteriol. 1993;175:7509–7514. doi: 10.1128/jb.175.23.7509-7514.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JS, Marr MT, Roberts JW. E coli Transcription Repair Coupling Factor (Mfd Protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 8.Smith AJ, Szczelkun MD, Savery NJ. Controlling the motor activity of a transcription-repair coupling factor: autoinhibition and the role of RNA polymerase. Nucleic Acids Res. 2007;35:1802–1811. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith AJ, Savery NJ. Effects of the bacterial transcription-repair coupling factor during transcription of DNA containing non-bulky lesions. DNA Repair (Amst) 2008;7:1670–1679. doi: 10.1016/j.dnarep.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 10.Chambers AL, Smith AJ, Savery NJ. A DNA translocation motif in the bacterial transcription--repair coupling factor, Mfd. Nucleic Acids Res. 2003;31:6409–6418. doi: 10.1093/nar/gkg868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deaconescu AM, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 12.Deaconescu AM, Darst SA. Crystallization and preliminary structure determination of Escherichia coli Mfd, the transcription-repair coupling factor. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:1062–1064. doi: 10.1107/S1744309105035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westblade LF, et al. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 2010;38:8357–8369. doi: 10.1093/nar/gkq692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deaconescu AM, Savery NJ, Darst SA. The bacterial transcription repair coupling factor. Curr Opin Struct Biol. 2007;17:96–102. doi: 10.1016/j.sbi.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savery NJ. The molecular mechanism of transcription-coupled DNA repair. Trends Microbiol. 2007;15:326–333. doi: 10.1016/j.tim.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Revyakin A, Liu CY, Ebright RH, Strick TR. Abortive Initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Revyakin A, Ebright RH, Strick TR. Promoter unwinding and promoter clearance by RNA polymerase: detection by single-molecule DNA nanomanipulation. Proc Natl Acad Sci USA. 2004;101:4776–4780. doi: 10.1073/pnas.0307241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Revyakin A, et al. Single-molecule DNA nanomanipulation: detection of promoter-unwinding events by RNA polymerase. Methods Enzymol. 2003;370:577–598. doi: 10.1016/S0076-6879(03)70049-4. [DOI] [PubMed] [Google Scholar]
- 20.Smith AJ, Savery NJ. RNA polymerase mutants defective in the initiation of transcription-coupled DNA repair. Nucleic Acids Res. 2005;33:755–764. doi: 10.1093/nar/gki225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kou SC, et al. Single-molecule Michaelis-Menten equations. J Phys Chem B. 2005;109:19068–19081. doi: 10.1021/jp051490q. [DOI] [PubMed] [Google Scholar]
- 22.Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 23.Komissarova N, et al. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 24.Srivastava DB, Darst SA. Derepression of bacterial transcription-repair coupling factor is associated with a profound conformational change. J Mol Biol. 2011;406:275–284. doi: 10.1016/j.jmb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 26.Manelyte L, et al. Regulation and rate enhancement during transcription-coupled DNA repair. Mol Cell. 2010;40:714–724. doi: 10.1016/j.molcel.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor I Structural domains and binding properties. J Biol Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 28.Deaconescu AM, Sevostyanova A, Artsimovitch I, Grigorieff N. Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface. Proc Natl Acad Sci USA. 2012;109:3353–3358. doi: 10.1073/pnas.1115105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.