Abstract
Benign Prostatic Hyperplasia (BPH)is the most common urologic disease in men over age 50. Symptoms include acute urinary retention, urgency to urinate and nocturia. For patients with severe symptoms, surgical treatment is used to remove the affected tissue. Interestingly, the presence of histologic BPH does not always correlate with symptoms. The molecular basis of symptomatic BPH and how it differs from asymptomatic BPH is unknown. Investigation into the molecular players involved in symptomatic BPH will likely give insight into novel therapeutic, and potentially preventative, targets. We determined the expression of genes involved in the innate anti-viral immune response in tissues from patients undergoing surgery to alleviate the symptoms of BPH, and compared the results to prostate tissue with histologic BPH, but from patients with few urinary issues (asymptomatic BPH). We found that expression of CFI, APOBEC3G, OAS2, and IFIT1, four genes whose protein products are involved in the innate anti-viral immune response, were significantly transcriptionally upregulated in symptomatic BPH. Additionally we observe hypomethylation and concomitant expression of ancient retroviral-like sequences, the LINE-1 retrotransposons, in symptomatic BPH when compared to normal prostate tissue. These findings merit further investigation into the anti-viral immune response in symptomatic BPH.
Keywords: benign prostatic hyperplasia, BPH, CFI, APOBEC3G, anti-viral immune response, LINE-1
Introduction
BPH is a condition that affects millions of men in the United States every year and costs the US $4 billion annually.1 The prevalence of BPH increases with age with 50% of men in their 50’s exhibiting symptoms of BPH, and 70% of men 70 years old having symptomatic BPH.2
BPH is a condition in which an imbalance between cell proliferation and cell death occurs in stromal and glandular prostatic tissue, leading to enlargement of the prostate. This abnormal growth occurs in the transitional zone of the prostate surrounding the urethra. Thus, enlargement of the prostate due to BPH can constrict the urethra and lead to lower urinary tract symptoms (LUTS) such as urgency to urinate, strain to begin urination, and a weak urine stream.3 Current treatments for this symptomatic BPH include pharmacological as well as surgical options. 5-alpha reductase inhibitors and/or alpha1-adrenoceptor antagonists are frequently used to treat mild to moderate symptomatic BPH. Moderate to severe cases of symptomatic BPH commonly require invasive procedures including transurethral resection of the prostate (TURP), transurethral microwave thermotherapy (TUMT), or transurethral needle ablation (TUNA). Because of the undesirable side effects of transurethral procedures to treat moderate to severe symptomatic BPH, other therapeutic options are necessary.
Remarkably, only some men with histologic BPH present with LUTS. Men who have histologic BPH but do not present with LUTS have what is termed asymptomatic BPH. Typically symptomatic BPH is associated with a larger prostate size than asymptomatic BPH. It is currently unknown what differentiates symptomatic from asymptomatic BPH at the molecular level. Examining the differences between symptomatic and asymptomatic BPH may uncover what is leading to LUTS, potentially providing novel therapeutic targets for the treatment of BPH.
The molecular pathogenesis of BPH is largely unknown. The limited number of molecular studies examining BPH have demonstrated that androgens play a role in this disease, as well as various growth factors such as fibroblast growth factor 2 and insulin growth factor.4–6 Additionally, inflammation is frequently associated with BPH. Several cytokines and inflammatory mediators are upregulated in BPH including interleukin 1 alpha (IL1alpha), IL2, IL8, IL15, IL17, nuclear factor kappa B, chemokine (C-X-C motif) ligand 5 (CXCL5), and CXCL12.7 In a study that examined gene expression differences between cultured normal prostate transitional zone stromal cells compared to cultured stromal cells from BPH affected tissues, genes involved in the innate immune response, including complement factor I (CFI) and 2’5’-oligoadenylate synthetase2 (OAS2), were found to be upregulated in the BPH-affected cells when compared to the normal transitional zone stromal cells.8
The goal of this current study is to determine molecular differences between symptomatic and asymptomatic BPH in terms of the innate anti-viral immune response. To accomplish this, we compare gene expression differences in symptomatic BPH patient tissue samples from TURP and asymptomatic BPH patient tissue samples from patients undergoing prostatectomy for prostate cancer that had accompanying asymptomatic BPH. BPH typically arises in the transitional zone of the prostate, whereas prostate cancer most often occurs in the peripheral zone. Since these two diseases arise in different zones of the prostate, molecular alterations due to prostate cancer will likely not be present in BPH-affected tissue, although this cannot be completely ruled out. Because a previous study had found upregulation of genes involved in the innate immune response in cells isolated from BPH affected tissues and cultured in vitro8, we wanted to determine if these findings were applicable to patient tissue samples. Our results indicate that an innate anti-viral immune response is activated in symptomatic BPH, which distinguishes patients with BPH requiring surgery from those who are either asymptomatic or able to endure the symptoms.
Results
Activation of innate anti-viral immune response genes in symptomatic BPH
Relative complement factor I (CFI) mRNA expression was determined by real time PCR analysis using symptomatic BPH (n = 17)or asymptomatic BPH samples (n = 9). The patients with asymptomatic BPH reported AUA scores of less than or equal to 12. Specifications regarding patient samples can be found in Supplemental Table 1. CFI inactivates C3b and C4b; two key players in the innate immune systems complement pathway activation.9 Analysis of the CFI real time PCR data using the BPH1 cell line as a calibrator and GAPDH as a reference gene reveals that the symptomatic BPH samples exhibit a 5-fold increase in CFI expression compared to the asymptomatic BPH samples (Figure 1a; p = 0.0012, t-test).
Figure 1.
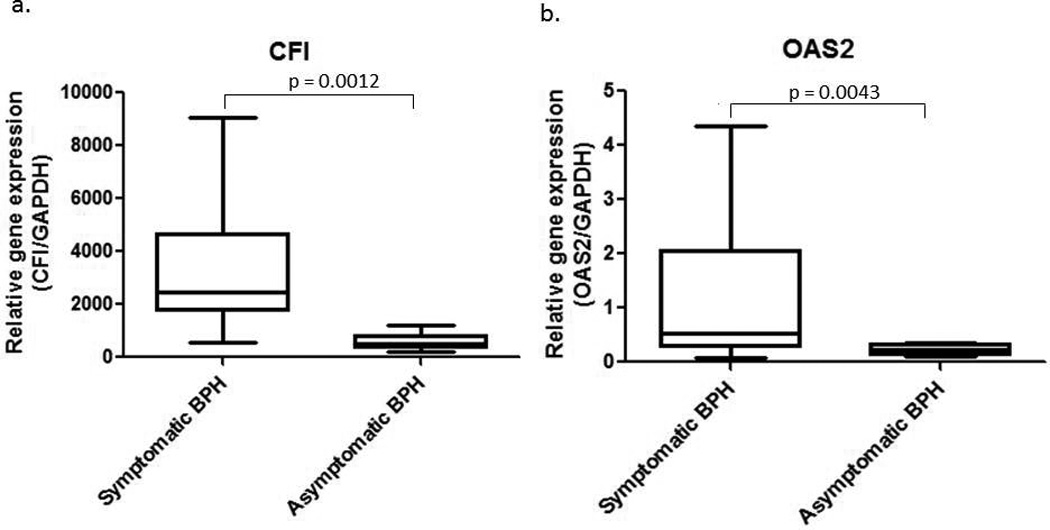
a. CFI real time PCR results for symptomatic BPH compared to asymptomatic BPH. The student’s t test was utilized in the statistical analysis and resulted in a statistically significant p value of 0.0012. b. Real time PCR for OAS2 gene expression in symptomatic and asymptomatic BPH. Statistical analysis was performed using the Mann-Whitney test, which resulted in a statistically significant difference between the two groups, p = 0.0043.
The expression profile of OAS2 was then examined. OAS2 recognizes virally produced double-stranded RNA and produces 2’-5’ oligoadenylates to initiate RNA destabilization through activation of RNaseL, thus contributing to the anti-viral response. Quantitative PCR for OAS2revealsa greater than 2-fold increase in OAS2 gene expression in symptomatic BPH (n = 16) when compared to the asymptomatic BPH (n = 9)(Figure 1b; p = 0.0043, Mann-Whitney).
Long interspersed nuclear element 1 (LINE-1) retroelements are methylated in normal prostate tissue but frequently become demethylated in cancer.10 We discovered that marked demethylation of LINE-1 also occurs in BPH tissue from TURPs when we used BPH affected tissue as a “normal” prostate control for our studies ofLINE-1 methylation in prostate cancer (Figure 2a, compare symptomatic BPH lanes 3–5 with donors lanes 9–11). Methylation is measured by combined bisulfite restriction analysis involving isolation of genomic DNA, bisulfite conversion of the DNA which converts unmethylated cytosines to uracil and conserves methylated cytosines, and LINE-1 PCR followed by restriction digestion of the PCR product by an enzyme that preferentially cuts based on the presence of a cytosine that was conserved during bisulfite conversion due to methylation. Therefore digested product indicates the presence of methylation. A statistically significant difference exists between symptomatic BPH samples (n = 11) and histologically normal prostate tissue from organ donors (n = 9) (Figure 2b; p = 0.040, Mann-Whitney). Next we examined LINE-1 methylation status in asymptomatic BPH tissues. LINE-1 is demethylated and expressed in prostate cancer and a field defect exists in prostate cancer in which normal prostate tissue adjacent to the tumor also demonstrates demethylation of LINE-1 (unpublished data). Since the asymptomatic BPH samples come from patients undergoing prostatectomy due to prostate cancer, any demethylation of LINE-1 observed in the asymptomatic BPH samples may be due to the field defect from the prostate cancer. Although the average relative methylation of the asymptomatic BPH samples was higher than symptomatic BPH samples, it did not reach statistical significance (Figure 2b).
Figure 2.
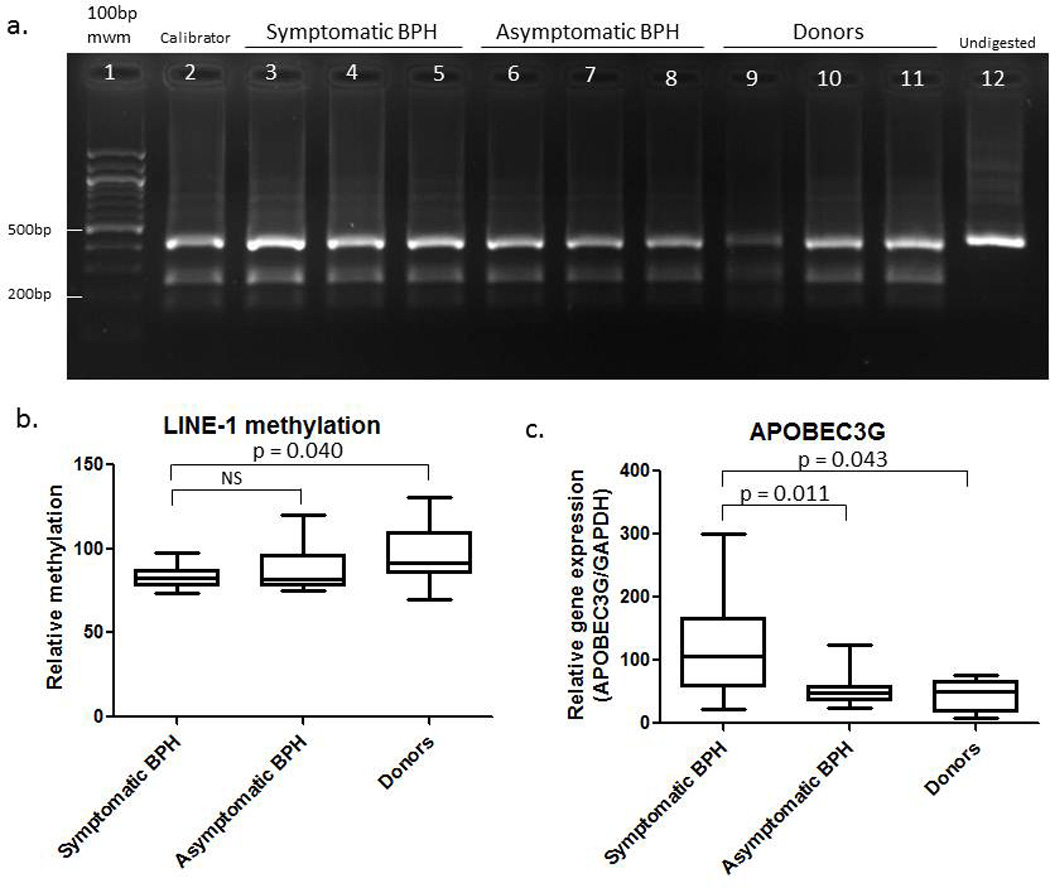
a. Combined bisulfite restriction analysis of LINE-1 in symptomatic BPH TURP samples, asymptomatic BPH, and donors. Lane 1 is a 100 base pair (bp) molecular weight marker (mwm), lane 2 is the calibrator, lanes 3–5 are symptomatic BPH TURP samples, lanes 6–8 are asymptomatic BPH, lanes 9–11 are donor and lane 12 is undigested PCR product. DNA samples were bisulfite treated, subjected to Line1 PCR and subsequent digestion with HinfI. The undigested top band at 450bp represents unmethylated DNA whereas the digestion products at 275bp and 180bp indicate methylated DNA. b. Quantification of the percent methylation compared to the calibrator of 11 symptomatic BPH from TURP, 10 asymptomatic BPH and 9 donors. Data is represented as a box and whisker plot. A statistically significant difference exists between symptomatic BPH and donors groups (p = 0.040, Mann Whitney), whereas there is no statistically significant difference between symptomatic BPH and asymptomatic BPH. c. APOBEC3G real time PCR results for symptomatic BPH, asymptomatic BPH, and donor samples. The Mann-Whitney rank-sum test was used to determine the statistically significant p value of 0.011 between symptomatic BPH and donors and the t-test was used to determine the statistically significant difference between symptomatic BPH and donors (p = 0.043).
Since we observed a statistically significant difference in LINE-1 methylation between symptomatic BPH and donors, we were interested in pursuing a potential cause for this demethylation. It is not known what mechanism(s) lead to demethylation of DNA, however recent evidence suggests that apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like proteins (APOBECs) may target cellular genomic DNA leading to the demethylation of cytosines.11 APOBEC3G is a cytidine deaminase that can mutate nucleic acids by deaminating cytidines and converting them into uridines. APOBEC3G typically performs this function on the viral genomes of viruses that have infected the cell.12 Since APOBEC3G might be involved in both DNA demethylation and the innate antiviral response, we examined its expression in our BPH tissue samples. Real time PCR for APOBEC3Gwas performed on symptomatic BPH (n = 16), asymptomatic (n = 13) BPH, and donor (n = 6) samples with the BPH1 cell line used as the calibrator. There is over a 2-fold increase in APOBEC3G expression in symptomatic BPH when compared to asymptomatic BPH (Figure 2c; p = 0.011, Mann-Whitney) and donors (p = 0.043, t-test).
In an attempt to distinguish activation of these genes in response to virus versus other pathogens, we examined the expression profile of interferon-induced tetratricopeptide 1 (IFIT1). IFIT1 expression is activated in response to viruses as well as IFNalpha/beta, which are induced in response to virus. The function of IFIT1 is to inhibit cellular and viral processes such as translation, migration, proliferation, signaling and viral replication.13 Real time PCR for IFIT1revealed no statistically significant difference between the symptomatic (n = 16) and asymptomatic BPH (n = 13) groups (Figure 3a). However, we observed that the asymptomatic BPH samples with the largest mass had the highest fold change in IFIT1. Large prostate size is oftentimes associated with LUTS. When we remove the 4 samples that came from asymptomatic BPH patients with a prostate mass greater than 60g, there is a statistically significant 2-fold increase in IFIT1 expression in symptomatic BPH samples (n = 17) when compared to asymptomatic BPH (n = 9) (Figure 3b; p = 0.012, Mann-Whitney).
Figure 3.
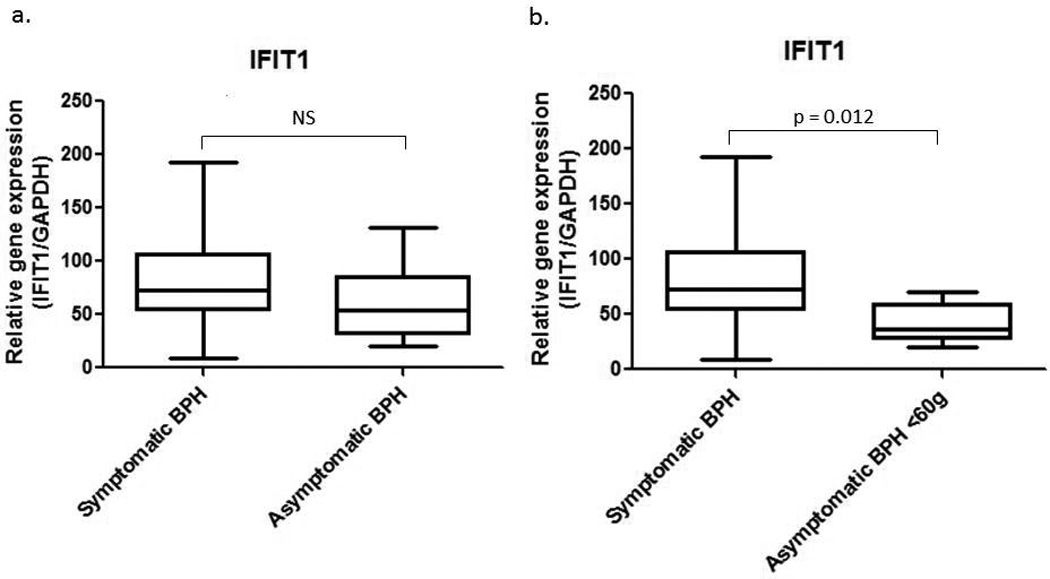
a. IFIT1 real time PCR with symptomatic and asymptomatic BPH samples. The Mann-Whitney statistical test was utilized for statistical analysis and determined that there was no statistically significant difference between the two groups. b. Real time PCR for IFIT1 in symptomatic BPH and asymptomatic BPH samples in which the prostate mass was less than 60g. Statistical analysis using the Mann-Whitney rank-sum test demonstrates a statistically significant difference between the two groups, p = 0.0118.
Correlations observed in gene expression data and patient parameters
When analyzing our IFIT1 data we observed that the asymptomatic BPH samples with large masses had the highest fold changes in IFIT1 expression. Therefore we wanted to determine if there was a significant correlation between IFIT1 fold change and the mass in grams of the asymptomatic BPH prostates. We found that there is a strong positive correlation between IFIT1 expression and the mass of the asymptomatic BPH prostates (Pearson’s r = 0.65), and this was found to be statistically significant (Figure 4a; p = 0.017).
Figure 4.
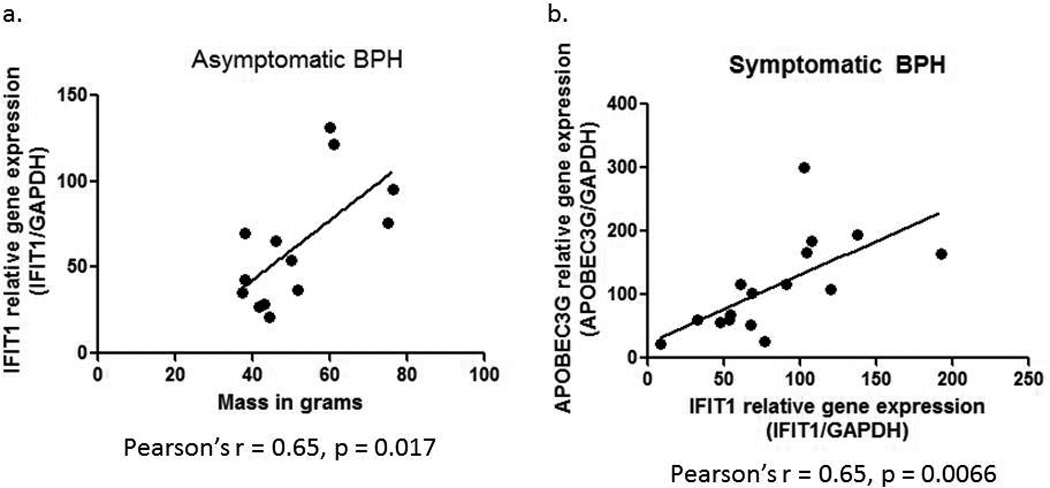
a. Correlation between IFIT1 real time PCR results (IFIT1 relative gene expression – IFIT1/GAPDH) and mass in grams of the asymptomatic BPH samples. Statistical analysis was carried out using a Pearson’s correlation, with the Pearson’s r = 0.65 and p = 0.0172. b. Correlation between APOBEC3G and IFIT1 relative gene expression (normalized to GAPDH) in the symptomatic BPH samples. Pearson’s correlation statistical analysis resulted in a Pearson’s r = 0.65 and p = 0.0066.
Additionally, we observed that the symptomatic BPH samples demonstrating a strong induction of IFIT1 expression also showed strong induction of APOBEC3G. Therefore we examined whether there was a strong correlation between expression of these two genes in the symptomatic BPH samples. Indeed we found that there is a strong correlation between IFIT1 fold change and APOBEC3G fold change in the symptomatic BPH samples (Figure 4b; Pearson’s r = 0.65, p = 0.0066). We were unable to correlate IFIT1 expression with prostate size for symptomatic BPH samples, as only the affected tissue and not the entire prostate is removed.
LINE1 retrotransposon expression is not elevated in symptomatic BPH samples
After observing the activation of an innate antiviral immune response in symptomatic BPH, we wanted to determine the mechanism potentially causing this response. Since we know that LINE-1 is demethylated in symptomatic BPH samples, we hypothesized that LINE-1 retroelements were being expressed and activating the anti-viral immune response in symptomatic BPH. Examination of LINE1ORF2 (LINE-1 open reading frame 2) expression in symptomatic BPH samples compared to donors demonstrates that there is a statistically significant difference in LINE-1 expression between symptomatic BPH (n = 19) and donor tissue (n = 8) (Figure 5; p = 0.015, Mann-Whitney). Examination of asymptomatic BPH samples (n = 14) indicates that the mean was lower than the symptomatic BPH mean but did not reach statistical significance (Figure 5). Again, LINE-1 expression in asymptomatic BPH may be due to the field defect observed in prostate cancer.
Figure 5.
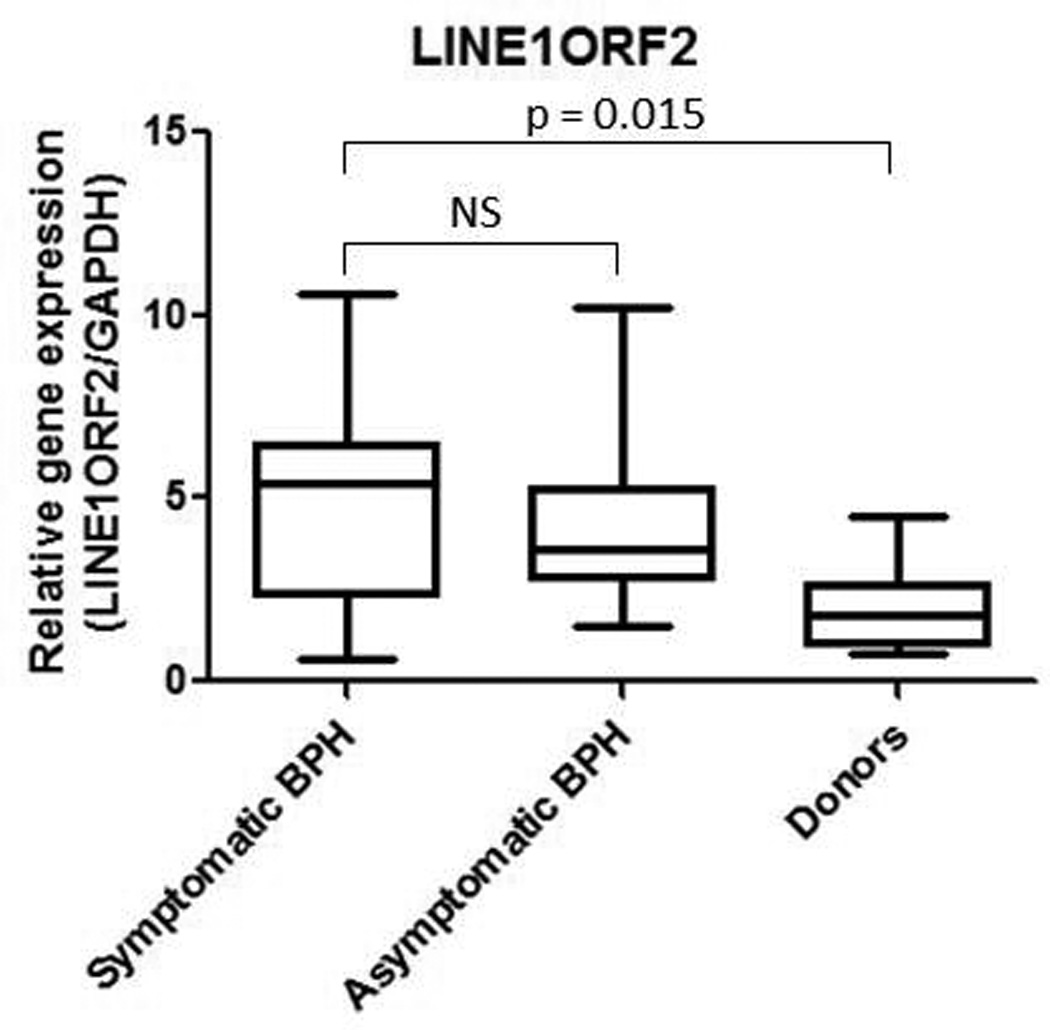
a. LINE1ORF2 real time PCR for symptomatic BPH, asymptomatic BPH and normal prostate tissue from organ donors. The Mann-Whitney statistical test was performed, determining that a statistically significant difference exists between symptomatic BPH and donors (p = 0.015), however there is no statistically significant difference between symptomatic and asymptomatic BPH.
Discussion
In the current study examining molecular differences between symptomatic BPH and asymptomatic BPH, we found four genes involved in the innate anti-viral immune response to be upregulated in symptomatic BPH: CFI, OAS2, APOBEC3G, and IFIT1. CFI is a major regulator of the complement system in the innate immune response. Activation of the complement pathway leads to various signaling cascades, all of which converge at the point of C3b and C4b production. CFI is a protease that cleaves C3b and C4b yielding C3 and C4 fragments resulting in inhibition of complement signaling.9 Interestingly, some viruses such as vaccinia, mumps and Epstein-Barr virus utilize CFI during infection to prevent complement activation and allow for successful infection.14–16 Additionally, some cancer cells such as glioma cells and Non-small cell lung cancer cells express increased levels of CFI.17, 18 Expression of CFI by cancer cells may be contributing to protection of these altered cells from complement attack. In the current study, we observe a statistically significant increase in CFI expression in symptomatic BPH samples when compared to asymptomatic BPH. In fact the average fold change of the symptomatic BPH samples is five times higher than the average fold change of the asymptomatic BPH samples.
It is interesting that a potent inhibitor of one arm of the immune system, the complement cascade, is activated in symptomatic BPH since BPH is a disease currently characterized by a strong immune/inflammatory response. Importantly, factors involved in inflammation, namely IL6 and interferon gamma (IFNgamma), are known activators of CFI.19, 20 It is possible that CFI is playing a similar role in symptomatic BPH as it is in glioma and non-small cell lung cancer cells: CFI may be contributing to the protection of altered symptomatic BPH cells from complement attack. Protection from the complement system in symptomatic BPH may allow for tissue remodeling and growth, thus leading to LUTS. This finding warrants further investigation into the role CFI is playing in symptomatic BPH and provides insight into a potential pathway to target in symptomatic BPH.
OAS2, APOBEC3G and IFIT1 are all induced by Type I interferons and all are involved in the innate anti-viral response. OAS2 is an antiviral enzyme that recognizes virally produced dsRNA and initiates RNA degradation through activation of RNaseL within infected cells. APOBEC3G is critical in the innate anti-viral response, because it recognizes and subsequently mutates the viral genome, and with the exception of HIV, usually renders the virus non-functional.12 Finally, IFIT1 is strongly induced by Type I interferons and viral infection and can lead to inhibition of cellular and viral protein synthesis and replication.13 Recently IFIT1 has also been shown to be involved in binding viral 5’ triphosphate-containing RNA.21 In the present study, we demonstrate a statistically significant upregulation of APOBEC3G, OAS2 and IFIT1 in symptomatic BPH when compared to asymptomatic BPH. Additionally, IFIT1 expression correlates with the size of the prostate in the asymptomatic BPH group, with the largest prostates resulting in the biggest induction of IFIT1. Larger prostates are more likely to be associated with LUTS, therefore it is consistent with our results that IFIT1 expression would correlate with prostate size. Another correlation that we observe is between IFIT1 and APOBEC3G. It is not surprising to observe a correlation in the expression of two genes involved in response to the same/similar stimuli.
It is interesting that an anti-viral response is observed in symptomatic BPH, since this condition has not been linked to viral infection of the prostate. Studies have, however, demonstrated a link between sexually transmitted viral infections and LUTS.22, 23 The significance of this correlation is currently unknown. The anatomical location of the prostate in the urogenital tract makes it possible that sexually transmitted viral infection may contribute to the innate anti-viral immune response in symptomatic BPH. In addition, it is possible that inflammation associated with BPH results in the production of endogenous ligands (“Sterile Inflammation”) which activate the innate immune receptors to induce Type I IFN and/or these anti-viral innate immunity associated genes.
Activation of the innate anti-viral immune response in symptomatic BPH could also be due to activation of endogenous retroelements. Methylation of LINE-1 suppresses expression of these elements. There was a statistically significant demethylation of LINE-1 in symptomatic BPH samples when compared to histologically normal prostate tissue from organ donors (Figure 2a). We also found that there was a significant upregulation of LINE1 expression in the symptomatic BPH samples when compared to donors (Figure 5), indicating that demethylation of LINE-1 may be playing an important role in symptomatic BPH. Asymptomatic BPH samples did show a higher level of LINE-1 methylation and lower LINE-1 expression than symptomatic BPH; however these did not reach statistical significance. This may be due to the field defect observed in prostate cancer in regards to methylation of LINE-1. Future studies examining the role of LINE-1 in symptomatic BPH could utilize samples from patients with prostate cancer and accompanying symptomatic BPH versus asymptomatic BPH. It will be important to tease out the role of LINE-1 expression in symptomatic BPH, as expression of LINE-1retroelements may be triggering activation of the innate anti-viral immune response.
Two examples of diseases in which LINE-1 is demethylated, expressed and believed to be responsible for an innate anti-viral immune response are the autoimmune disorders Sjorgens syndrome (SS) and systemic lupuserythematosus (SLE).24 Inflammation is present in SS and SLE, as well as in BPH. The anti-viral immune response in SS and SLE is likely responsible for the inflammation present in these diseases, therefore the anti-viral immune response may be responsible for the inflammation observed in BPH.24 Previously it has been suggested that BPH does have characteristics of an autoimmune disease, because significantly more IgC antibodies to prostate specific antigen were present in BPH samples when compared to controls.25 As we see in BPH, part of the anti-viral immune response in SS is the expression of APOBEC3 family members. APOBECs may be playing multiple roles in regard to LINE-1 hypomethylation and expression. Some APOBEC family members are able to target RNA of cellular retroelements, providing a line of defense against aberrantly expressed retrotransposons.12 Recently, however, it has been suggested that some APOBECs may be able to demethylate cellular DNA.11 Upregulation of APOBECs could then be contributing to demethylation of LINE-1. Determination of the effect that APOBECs have on LINE-1 demethylation and expression in BPH, SS and SLE is critical. It will be important to answer the question of whether APOBECs are being upregulated and demethylating LINE-1 or is LINE-1 being demethylated by some other cause, which is leading to the upregulation of APOBECs, allowing for subsequent targeting of LINE-1 RNAs.
Taken together, we observe an innate anti-viral immune response involving the upregulation of CFI, OAS2, APOBEC3G and IFIT1 in symptomatic BPH when compared to asymptomatic BPH. Additionally, LINE-1 retroelements are demethylated and expressed in symptomatic BPH tissue compared to donors. This study gives insight into the molecular changes in symptomatic BPH, a disease in which knowledge at the molecular level is lacking. Further investigation into the cause of this anti-viral immune response and how this response contributes to LUTS is necessary. Targeting a factor involved in this anti-viral immune response may be therapeutically beneficial in the prevention and/or treatment of symptomatic BPH.
Materials and Methods
Tissue acquisition
Symptomatic BPH samples were collected from patients undergoing TURP to alleviate symptoms. These samples were de-identified and supplied to us by the University of Pittsburgh Health Sciences Tissue Bank (HSTB) after Institutional Review Board approval. Asymptomatic BPH samples were collected from patients undergoing prostatectomy due to prostate cancer. These patients also had accompanying histologic BPH. Tissues utilized as asymptomatic BPH were from patients with an American Urological Association (AUA) score of less than or equal to 12. A University of Pittsburgh HSTB genito-urinary pathologist isolated the transitional zone containing histologic BPH from the asymptomatic BPH samples and gave us the de-identified tissue. Organ donor prostate tissue was also provided by the University of Pittsburgh HSTB and a genito-urinary pathologist confirmed that they were histologically normal. The zone of origin of the donor prostate tissue is unknown. All samples were stored at −70°C.
The same symptomatic and asymptomatic BPH samples were used for the CFI, OAS2, APOBEC3G and IFIT1 real time PCR. Additional asymptomatic BPH samples were prepared and used for the APOBEC3G and IFIT1 real time PCR to increase statistical power. A large overlap in samples exist in the LINE-1 real time PCR and LINE-1 methylation assays, however there are some additional samples used because samples for these assays were prepared differently (RNA was DNase treated for the LINE-1 real time PCR).
The AUA symptom score is determined by a questionnaire that patients fill out regarding the severity of their LUTS. Questions range from how many times the patient urinates at night to how often the patient has to strain to urinate. The patients answer each question on a scale of 0–5, with 0 being not at all and 5 representing almost always. Scores for all of the questions are then added up and the total is the patients AUA symptom score, with a score of 35 being the maximum possible. Scores between 0 and 7 indicate mild symptoms; 8–19 are moderate, and 20–35 severe.
Ribonucleic acid (RNA) isolation
RNA was isolated from TURP or asymptomatic BPH tissue using the Trizol method. Briefly, approximately 50mg of tissue was placed in 500µLTrizol(Invitrogen, Grand Island, NY, USA) and was homogenized. The homogenized tissue was allowed to incubate in Trizol for 5 minutes at room temperature. 100µL of chloroform was then added to the tissue/Trizol. Samples were shaken vigorously for 15 seconds and then allowed to incubate at room temperature for 3 minutes. Samples were then centrifuged at 12,000×g for 15 minutes at 4°C. The upper aqueous phase was carefully transferred to a new tube, at which point 2µL of glycogen and 250µL of isopropyl alcohol was added. The samples were gently mixed and allowed to incubate at room temperature for 10 minutes. The samples were then centrifuged at 12,000×g for 10 minutes at 4°C allowing for the precipitation of the RNA. The supernatant was removed and the RNA pellet was washed with 500µL of cold 75% ethanol. This was followed by centrifugation at 7,500×g for 5 minutes at 4°C. The supernatant was again removed and the RNA pellet was allowed to air dry for approximately 30 minutes. The RNA pellet was then resuspended in diethylpyrocarbonate-treated water and the concentration was read on a Nanodrop 2000C spectrophotometer (Thermo Scientific, Asheville, NC, USA). 500ng of RNA was then run on a 0.8% agarose/tris-borate-ethylenediaminetetraacetic acid gel and visualized to ensure intact 28S and 18S ribosomal RNA.
DNase treatment of RNA
RNA utilized to make complementary DNA (cDNA) for real time PCR for CFI, OAS2, APOBEC3G and IFIT1 were not DNase treated whereas RNA used to make cDNA for LINE1ORF2 real time PCR was DNase treated. This is because the primers designed for the LINE1ORF2 real time PCR recognize genomic DNA as well as cDNA, so it is essential to remove all contaminating genomic DNA for this particular assay. All other real time PCR primers were designed to not recognize genomic DNA. 4ug of RNA was treated with 1µL of Ambion/Life Technologies (Grand Island, NY, USA) TURBO DNase at 37°C for 30 minutes. The RNA was extracted from the enzyme via Phenol-Chloroform extraction. RNA concentration was then read using the nanodrop and 500µg of RNA was run on a 0.8% agarose/TBE gel and visualized to confirm RNA integrity.
cDNA production
For the non-DNase treated RNA samples, 1µg of RNA was utilized to make cDNA using random hexamers and the SuperScript III First-Strand Synthesis System for Reverse Transcription-Polymerase Chain Reaction (RT-PCR) (Invitrogen, Grand Island, NY, USA) per the manufacturer’s instructions.
The DNase treated RNA samples produced cDNA starting with 1µg of DNase treated RNA and utilization of random primers and the Promega (Madison, WI, USA) M-MLV RT First-Strand Synthesis of cDNA kit.
Real time PCR
Quantitative real time PCR for APOBEC3G, IFIT1, LINE1ORF2 and glyceraldehyde 2-phosphate dehydrogenase (GAPDH) was performed on a Bio-Rad (Hercules, CA, USA) iQ5 Multicolor Real Time PCR detection system real time PCR machine whereas real time PCR for CFI and OAS2 was performed on the Applied Biosystems Step One Plus real time PCR system/machine (Carlsbad, CA, USA). The APOBEC3G primer/probe set from Solaris/Thermo Scientific (Catalog number AX-013072-00-0100; Lafayette, CO, USA) was used at a 1× concentration along with a 1× concentration of the Solaris QPCR Master Mix (Catalog number AB-4350) in a total volume of 20µL. The real time PCR was performed using the manufacturer-recommended thermocycling conditions, which yielded a satisfactory efficiency (103.7%). SYBR green real time PCR was performed for IFIT1. 0.625µM of each primer (F1 – 5’-acagcaaccatgagtacaaatgg-3’ and R1 - 5’-catcgtcatcaatggataactccc-3’) was used with a 1× concentration of Bio-Rad’s iQ SYBR Green Supermix in a 20µL total volume. The following thermocycling conditions resulted in a satisfactory efficiency (94.3%): 94°C for 1 minute and 45 cycles x[94°C for 30 seconds, 60°C for 45 seconds, 76.5°C for 15 seconds). SYBR green real time PCR was also performed for LINE1ORF2 using 0.625µM final concentration of each primer (F1 – 5’-ccagctaacatcataatgacagg-3’ and R1 – 5’-tatccaacttgccagtctgtgtc-3’) and a 1× final concentration of the Bio-Rad iQ SYBR Green Supermix in a total volume of 20µL. Thermocycling conditions consisted of 94°C for 30 seconds and 45 cycles x[94°C for 15 seconds, 52°C for 60 seconds], resulting in an acceptable efficiency (92%). A primer/probe set designed by Sigma-Aldrich (St. Louis, MO, USA) was utilized for the GAPDH real time PCR. 0.25µM of each primer (F2 – 5’-ccatgacaactttggtatc-3’ and R2 – 5’-ggatgatgttctggagag-3’) and 0.1µM of the probe (5’-FAM-aaggactcatgaccacagtccatc-black hole quencher 1) were used along with a final 1× concentration of the iQSupermix (170–8860) from Bio-Rad in a total volume of 20µL. 94°C for 30 seconds and 45 cycles x[94°C for 15 seconds, 52°C for 60 seconds] was the thermocycling conditions allowing for an acceptable efficiency (102%). CFI and OAS2 real time PCR were performed using primer/probe sets from Applied Biosystems (Catalog numbers Hs00989715_m1 and Hs00942643_m1, respectively). The ABI Master Mix was used at a final 1× concentration and manufacturer-recommended thermocycling conditions were used. These conditions resulted in a satisfactory efficiency for CFI (93.6%), however the OAS2 efficiency was only 84.2%.
Real time PCR analysis
The deltadelta Ct method was used to analyze the APOBEC3G, IFIT1, LINE1ORF2, and CFI real time PCR results using BPH1 or LNCaP cell lines as the calibrator. GAPDH was used as a reference gene. The Pfaffl method was used to analyze the OAS2 real time PCR results to account for the reduced efficiency. The BPH1 cell line was used as a calibrator and GAPDH as the reference gene. Statistical analysis included the Student’s t-test which was used for data that was normally distributed, or the Mann-Whitney rank-sum test that was utilized in the case of a non-normal distribution. Pearson’s correlation coefficient was calculated for the correlations performed. Statistical tests were performed using Sigmastat version 3.5.
Global DNA methylation combined bisulfite restriction analysis (COBRA)
LINE-1 elements are found so frequently in the human genome that its methylation status is used as a surrogate marker for global DNA methylation. Genomic DNA was prepared from TURP tissue utilizing the phenol chloroform extraction method. Briefly, samples digested with proteinase K were mixed in equal volume with Phenol/chloroform/isoamyl alcohol reagent at a 25:24:1 dilution. Samples were then shaken vigorously for 15 seconds and centrifuged at maximum speed for 10 minutes at 4°C. The aqueous (top) phase was then put in a new tube. Half of the original volume of 7.5M ammonium acetate and 2 volumes of 100% ethanol were added. The samples were then centrifuged at maximum speed for 20 minutes at 4°C. The supernatant was removed and the pellet washed with 70% ethanol followed by another centrifugation at maximum speed for 10 minutes at 4°C. The wash step and centrifugation was repeated, the pellet air dried and then resuspended in water. Genomic DNA and controls were then bisulfite converted using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA). Bisulfite treatment of DNA converts non-methylated cytosines into uracil via deamination, which is replicated as a thymidine during PCR. 5-methylcytosines are protected and identified as cytosines in the PCR product. Samples were then subjected to a LINE-1 PCR using 1µL bisulfite converted DNA, 0.2µLTitanium Taq Polymerase (Clonetech Mountain View, CA, USA), Titanium Buffer at a 1× concentration, 0.5µM of each primer, and 40µMdNTPs. The LINE-1 primers and this method have been previously reported.26 The LINE-1 PCR products were then subjected to restriction enzyme digestion by HinFI (New England Biolabs, Ipswich, MA, USA). Digestion products were then run on a 1.8% agarose/TBE (Tris base, EDTA) gel and stained with ethidium bromide. Digestion with HinfI indicates methylation of LINE-1, as only methylated and thus conserved(methyl)cytosine during bisulfite conversion creates a HinfI digestion site in the LINE-1 PCR product. The ratio of the top digestion product to the uncut band was determined using Image J and this was utilized to calculate the percent methylation. This was then calculated relative to the calibrator.
Supplementary Material
Acknowledgements
Thank you to Dr. Saumendra Sarkar for helpful scientific discussion. Special thanks to Jennifer Gregg and Katherine Semmler for insightful suggestions. Thank you to the University of Pittsburgh HSTB for the symptomatic and asymptomatic BPH specimens. AAA and this work were funded by the T32 post-doctoral fellowship NIHT32DK007774. DOK, DJB and JLC are funded by NIHR01CA138444 and KMS by DOD Award PC101949. The U.S. Army Medical Research Acquisition Activity, 820 Chandler Street, Fort Detrick MD 21702-5014 is the awarding and administering acquisition office. The content of this publication does not necessarily reflect the position or policy of the government, and no official endorsement should be inferred.
This work was funded by the University of Pittsburgh Urologic Training Program for Scientists T32 5T32DK007774-12, Cancer Center Support Grant P30CA047904, and RO1CA138444.
Footnotes
Supplementary information is available at Genes and Immunity’s website.
Conflict of Interest: none
Contributor Information
Allison A. Madigan, Email: atwoodaa@upmc.edu.
Kathryn M. Sobek, Email: gonyeak@upmc.edu.
Jessica L. Cummings, Email: cummingsjl@upmc.edu.
William R. Green, Email: w.ross.green@gmail.com.
Dean J. Bacich, Email: bacidj@upmc.edu.
References
- 1.Saigal CS, Joyce G. Economic costs of benign prostatic hyperplasia in the private sector. J Urol. 2005;173(4):1309–1313. doi: 10.1097/01.ju.0000152318.79184.6f. [DOI] [PubMed] [Google Scholar]
- 2.Blanker MH, Bohnen AM, Groeneveld FP, Bernsen RM, Prins A, Ruud Bosch JL. Normal voiding patterns and determinants of increased diurnal and nocturnal voiding frequency in elderly men. J Urol. 2000;164(4):1201–1205. [PubMed] [Google Scholar]
- 3.Ventura S, Oliver V, White CW, Xie JH, Haynes JM, Exintaris B. Novel drug targets for the pharmacotherapy of benign prostatic hyperplasia (BPH) Br J Pharmacol. 2011;163(5):891–907. doi: 10.1111/j.1476-5381.2011.01332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ropiquet F, Giri D, Lamb DJ, Ittmann M. FGF7 and FGF2 are increased in benign prostatic hyperplasia and are associated with increased proliferation. J Urol. 1999;162(2):595–599. [PubMed] [Google Scholar]
- 5.Dong G, Rajah R, Vu T, Hoffman AR, Rosenfeld RG, Roberts CT, et al. Decreased expression of Wilms' tumor gene WT-1 and elevated expression of insulin growth factor-II (IGF-II) and type 1 IGF receptor genes in prostatic stromal cells from patients with benign prostatic hyperplasia. J Clin Endocrinol Metab. 1997;82(7):2198–2203. doi: 10.1210/jcem.82.7.4067. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson TM, Ricke WA. Androgens and estrogens in benign prostatic hyperplasia: past, present and future. Differentiation. 2011;82(4–5):184–199. doi: 10.1016/j.diff.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schauer IG, Rowley DR. The functional role of reactive stroma in benign prostatic hyperplasia. Differentiation. 2011;82(4–5):200–210. doi: 10.1016/j.diff.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao H, Ramos CF, Brooks JD, Peehl DM. Distinctive gene expression of prostatic stromal cells cultured from diseased versus normal tissues. J Cell Physiol. 2007;210(1):111–121. doi: 10.1002/jcp.20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janeway C, Travers P, Walport M, Shlomchik M. Immunobiology, The Immune System in Health and Disease. 5th edn. New York: Garland Science; 2001. [Google Scholar]
- 10.Santourlidis S, Florl A, Ackermann R, Wirtz HC, Schulz WA. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate. 1999;39(3):166–174. doi: 10.1002/(sici)1097-0045(19990515)39:3<166::aid-pros4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 11.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146(6):866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu YL, Greene WC. APOBEC3G: an intracellular centurion. Philos Trans R Soc Lond B Biol Sci. 2009;364(1517):689–703. doi: 10.1098/rstb.2008.0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fensterl V, Sen GC. The ISG56/IFIT1 gene family. J Interferon Cytokine Res. 2011;31(1):71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahu A, Isaacs SN, Soulika AM, Lambris JD. Interaction of vaccinia virus complement control protein with human complement proteins: factor I-mediated degradation of C3b to iC3b1 inactivates the alternative complement pathway. J Immunol. 1998;160(11):5596–5604. [PubMed] [Google Scholar]
- 15.Johnson JB, Grant K, Parks GD. The paramyxoviruses simian virus 5 and mumps virus recruit host cell CD46 to evade complement-mediated neutralization. J Virol. 2009;83(15):7602–7611. doi: 10.1128/JVI.00713-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mold C, Bradt BM, Nemerow GR, Cooper NR. Epstein-Barr virus regulates activation and processing of the third component of complement. J Exp Med. 1988;168(3):949–969. doi: 10.1084/jem.168.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okroj M, Hsu YF, Ajona D, Pio R, Blom AM. Non-small cell lung cancer cells produce a functional set of complement factor I and its soluble cofactors. Mol Immunol. 2008;45(1):169–179. doi: 10.1016/j.molimm.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Gasque P, Julen N, Ischenko AM, Picot C, Mauger C, Chauzy C, et al. Expression of complement components of the alternative pathway by glioma cell lines. J Immunol. 1992;149(4):1381–1387. [PubMed] [Google Scholar]
- 19.Minta JO, Fung M, Paramaswara B. Transcriptional and post-transcriptional regulation of complement factor I (CFI) gene expression in Hep G2 cells by interleukin-6. Biochim Biophys Acta. 1998;1442(2–3):286–295. doi: 10.1016/s0167-4781(98)00189-4. [DOI] [PubMed] [Google Scholar]
- 20.Timár KK, Junnikkala S, Dallos A, Jarva H, Bhuiyan ZA, Meri S, et al. Human keratinocytes produce the complement inhibitor factor I: Synthesis is regulated by interferon-gamma. Mol Immunol. 2007;44(11):2943–2949. doi: 10.1016/j.molimm.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Pichlmair A, Lassnig C, Eberle CA, Górna MW, Baumann CL, Burkard TR, et al. IFIT1 is an antiviral protein that recognizes 5'-triphosphate RNA. Nat Immunol. 2011;12(7):624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 22.Sutcliffe S, Rohrmann S, Giovannucci E, Nelson KE, De Marzo AM, Isaacs WB, et al. Viral infections and lower urinary tract symptoms in the third national health and nutrition examination survey. J Urol. 2007;178(5):2181–2185. doi: 10.1016/j.juro.2007.06.041. [DOI] [PubMed] [Google Scholar]
- 23.Breyer BN, Van den Eeden SK, Horberg MA, Eisenberg ML, Deng DY, Smith JF, et al. HIV status is an independent risk factor for reporting lower urinary tract symptoms. J Urol. 2011;185(5):1710–1715. doi: 10.1016/j.juro.2010.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mavragani CP, Crow MK. Activation of the type I interferon pathway in primary Sjogren's syndrome. J Autoimmun. 2010;35(3):225–231. doi: 10.1016/j.jaut.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 25.Zisman A, Zisman E, Lindner A, Velikanov S, Siegel YI, Mozes E. Autoantibodies to prostate specific antigen in patients with benign prostatic hyperplasia. J Urol. 1995;154(3):1052–1055. [PubMed] [Google Scholar]
- 26.Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32(3):e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


