Abstract
Sarcoidosis is a chronic granulomatous disease with a wide spectrum of symptoms. Genome-wide association studies in European populations have reported significant associations between sarcoidosis and single nucleotide polymorphisms (SNPs) located in the inter-genic region between the C10ORF67 and OTUD1 genes on chromosome 10p12, and the ANXA11 gene (chromosome 10q22). We carried out fine-mapping at 10p12 and 10q22 to assess associations of genetic variants in those regions with sarcoidosis risk in African American women, based on 486 sarcoidosis cases and 943 age- and geography-matched controls in a nested case-control study within the Black Women’s Health Study. There were no significant associations with variants of the ANXA11 gene (P=0.17). Haplotypic analyses of the C10ORF67-OTUD1 inter-genic region revealed a strong inverse association of the variants rs1398024 and rs11013452 with sarcoidosis (OR=0.52; P=0.01). Both SNPs are located inside a ~300 kb low recombination region of chromosome 10p12, suggesting that both SNPs are tagging the same causal variant. Our top SNP (rs11013452) is located inside a smaller LD block in HapMap YRI, further narrowing the position of the causal SNP to a region of ~ 8kb on chromosome 10p12. The present findings confirm the potential importance of the 10p12 locus in the etiology of sarcoidosis.
Keywords: granuloma, lung disease, polymorphism, single nucleotide, African continental ancestry, ancestry informative markers
INTRODUCTION
Sarcoidosis is a chronic, granulomatous disease that causes a wide spectrum of symptoms and illnesses.1 It occurs worldwide and affects men and women of all ages and races.1–3 In the United States, incidence and mortality from sarcoidosis are highest in black women.1, 3, 4
A 100k genome-wide association study (GWAS) by Franke et al.,5 of German subjects with either sarcoidosis or Crohn’s disease reported a significant association with the rs1398024 SNP (chromosome 10p12), which is located in the inter-genic region between the C10ORF67 and OTUD1 genes.5 A second GWAS, in the same population but with >440k SNPs,6 identified several SNPs in strong linkage disequilibrium (LD) in the annexin (ANXA11) gene (chromosome 10q22): the leading rs2789679 SNP located in the 3′ UTR and a common nonsynonymous SNP (rs1049550, C>T, Arg230Cys) were associated with sarcoidosis risk.6 More recently, two case-control studies conducted in a different German population7 and in a Czech population8 found an association of ANXA11 variants rs1049550 and rs2573346 with sarcoidosis.
To our knowledge, neither C10ORF67 nor ANXA11 variants have been assessed in relation to sarcoidosis in African-Americans. We carried out fine-mapping at 10p12 and 10q22 to assess whether genetic variants on those regions are associated with sarcoidosis risk in the Black Women’s Health Study (BWHS), a follow-up study of U.S. black women.
RESULTS
The present analysis included 486 sarcoidosis cases and 972 controls. The mean age of cases and controls was 43. Twenty-seven percent lived in the Northeast, 31% in the South, 26% in the Midwest, and 15% in the West. The median % African ancestry was 82% (range: 33% – 97%). Mean % African ancestry was 81% in cases and 80% in controls.
C10ORF67-OTUD1 intergenic region
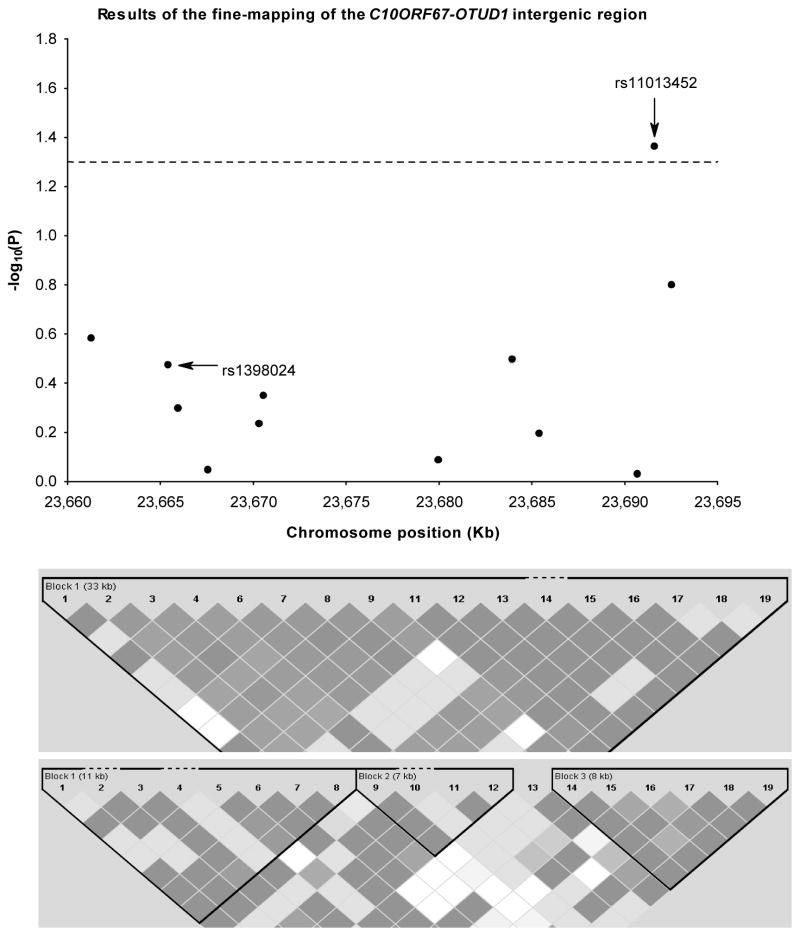
Fine-mapping revealed an 8% nonsignficiant reduction in sarcoidosis risk associated with the A-allele of the index rs1398024 SNP and sarcoidosis, as compared with the 19% reduction observed by Franke et al. 5 (Table 1). We identified rs11013452 as the top SNP in the BWHS associated with sarcoidosis at the nominal α = 0.05 level of significance. The rs11013452 SNP is located in a small 8 kb LD block in the HapMap YRI9 population (Figure 1). Each copy of the A-allele of the rs11013452 SNP was associated with a 20% reduction of sarcoidosis risk (p=0.04). There was no appreciable variation by % African ancestry: the odds ratios (OR) and 95% confidence intervals (CI) were 0.85 (0.62–1.16) among women with % African ancestry below the median (<82%) and 0.76 (0.57–1.01) among women with % African ancestry at or above the median (≥82%). Haplotypic analysis revealed a stronger inverse association based on the two SNPS: the haplotype carrying the A-allele of the index rs1398024 SNP and the A-allele of the rs11013452 SNP was associated with a 48% reduction in sarcoidosis risk (permutated p value=0.01). Separate analyses of incident and prevalent sarcoidosis cases yielded almost identical odds ratios.
Table 1.
Fine-mapping and haplotype analysis of the C10ORF67/10p12 locus in 486 sarcoidosis cases and 943 controls in the Black Women’s Health Study.
| C10ORF67/10p12 | Risk allele |
|
|||||
|---|---|---|---|---|---|---|---|
| Allele frequency (%) | OR2 (95%CI) | P for trend | P permutated3 | ||||
| Cases (n) | Controls (n) | ||||||
| rs1398024 (index SNP)1 | A | 45.5 (485) | 47.3 (941) | 0.92 (0.78–1.07) | 0.28 | - | |
| rs11013452 (best SNP in BWHS) | A | 15.5 (486) | 18.6 (942) | 0.80 (0.65–0.99) | 0.04 | - | |
|
|
|||||||
| Haplotype Analysis | |||||||
| rs1398024 | rs11013452 | Haplotype frequency (%) | |||||
| C | G | 45.0 | 43.0 | 1.00 (reference) | |||
| C | A | 9.8 | 10.0 | 1.13 (0.81–1.57) | 0.89 | 0.99 | |
| A | G | 40.0 | 39.0 | 1.04 (0.87–1.25) | 0.61 | 0.92 | |
| A | A | 5.7 | 8.6 | 0.52 (0.35–0.77) | 0.006 | 0.01 | |
OR=0.81 (95%CI: 0.69–0.96) Franke et al., Gastroenterology 2008.
Adjusted for % of African ancestry, age, and region of residence.
Permutation 10,000 times.
Figure 1.
Scatterplot and LD map of the genotyped tagging SNPs along the 33 kb LD block in the C10ORF67/OTUD1 intergenic region. The upper panel shows the association in the logarithmic scale. The dotted line indicates the threshold of nominal significance α = 0.05. Position of the index SNP (rs1398024) is indicated as well as the newly identified SNP (rs11013452). The lower panels show the D′ pair-wise values in both CEU and YRI HapMap samples. The newly identified SNP is located in a smaller 8 kb LD block in YRI HapMap sample.
ANXA11 gene
The T-allele of the index rs2789679 SNP showed a non-significant reduction of 14% in sarcoidosis risk as compared to the 40% reduction in the GWAS conducted by Hofmann et al 6 (Table 2). Fine-mapping in the BWHS revealed rs2819941 to be the top SNP, associated with a non-significant reduction (p=0.06) in sarcoidosis risk (17% reduction per each copy of the C-allele). Estimates did not vary according to % African ancestry: the OR (95%CI) for women with % African ancestry below the median (<82%) was 0.81 (0.61–1.08) while that for those with African ancestry at or above the median (≥82%) was 0.84 (0.64–1.11). Haplotypes carrying the C-allele of the rs2819941 SNP were less frequent in cases than controls resulting in a 19% reduction of sarcoidosis risk (permutated p value=0.17). Results were similar when prevalent and incident sarcoidosis cases were assessed separately.
Table 2.
Fine-mapping and haplotype analysis of the ANXA11/10q22.3 locus in 486 sarcoidosis cases and 943 controls in the Black Women’s Health Study.
| ANXA11/10q22.3 | Risk allele |
|
|||||
|---|---|---|---|---|---|---|---|
| Allele frequency (%) | OR2 (95% CI) | P for trend | P permutated3 | ||||
| Cases (n) | Controls (n) | ||||||
| rs2789679 (index SNP)1 | T | 17.0 (486) | 19.4 (943) | 0.86 (0.70–1.05) | 0.14 | - | |
| rs2819941 (best SNP in BWHS) | C | 16.7 (486) | 19.7 (943) | 0.83 (0.68–1.01) | 0.06 | - | |
|
|
|||||||
| Haplotype Analysis | |||||||
| rs2789679 | rs2819941 | Haplotype frequency (%) | |||||
| A | T | 80.0 | 77.0 | 1.00 (reference) | |||
| T | C | 13.4 | 16.0 | 0.81 (0.65–1.01) | 0.06 | 0.17 | |
| A | C | 3.3 | 3.7 | 0.86 (0.56–1.34) | 0.59 | 0.94 | |
| T | T | 3.6 | 3.3 | 1.01 (0.66–1.56) | 0.74 | 0.99 | |
OR=0.60 (95%CI: 0.52–0.69) Hofmann et al., Nat Genet 2008.
Adjusted for % of African ancestry, age, and region of residence.
Permutation 10,000 times.
Analyses according to measures of disease severity
We assessed whether the C10ORF67-OTUD1 or ANXA11 SNPs were associated with disease severity as determined by self-reported symptoms (Table 3). The association of the C10ORF67-OTUD1 rs11013452 SNP with sarcoidosis risk did not vary consistentely from the overall risk in the various categories considered. The ORs were similar to the overall estimate among women who reported ≥4 symptoms at diagnosis, current fair/poor health, or severe fatigue, while there was a significant 35% reduction among women reporting a high level of pain (≥5) and a nonsignificant 50% reduction among women who reported little or no ability to perform daily physical activities. For ANXA11 rs 2819941, the associations among women with more severe disease were generally stronger than those in the overall sample. Specifically, the C-allele of the ANXA11 rs2819941 SNP was associated with a significant 32% reduction in sarcoidosis risk among women who reported 4 or more symptoms at diagnosis (p<0.05), a non-significant 34% reduction of sarcoidosis risk among women who reported fair/poor general health, a nonsiginificant 24% reduction among those who reported little or no ability to carry out everyday physical activity, and a nonsignificant 47% reduction among women reporting severe fatigue.
Table 3.
Association of the C10ORF67-OTUD1 (rs11013452) and ANXA11 (rs2819941) single nucleotide polymorphisms with self-reported measures of sarcoidosis severity.
| Cases(n) | Per Allele OR (95% CI)
|
||
|---|---|---|---|
| C10ORF67-OTUD1 (rs11013452) | ANXA11 (rs2819941) | ||
| ≥4 symptoms at diagnosis1 | 133 | 0.83 (0.58–1.17) | 0.68 (0.47–0.97) |
| Fair or poor health2 | 80 | 0.89 (0.58–1.36) | 0.66 (0.42–1.04) |
| Little or no ability to carry out everyday physical activities2 | 35 | 0.50 (0.23–1.09) | 0.76 (0.40–1.46) |
| Severe or very severe fatigue2 | 44 | 0.82 (0.46–1.47) | 0.53 (0.27–1.04) |
| ≥5 on the pain scale2 | 107 | 0.65 (0.43–0.98) | 0.90 (0.63–1.30) |
Data obtained from BWHS Sarcoidosis Supplemental Survey.
Data obtained from BWHS 2011 questionnaire.
DISCUSSION
The non-caseating granuloma is a pathophysiologic hallmark shared by sarcoidosis and Crohn’s disease,10,11 a chronic inflammatory disorder of the gastrointestinal tract.12 There is considerable clinical overlap between the two diseases10, 13 including manifestation of both disorders in a single patient.14 Franke et al. conducted a combined GWAS of 393 Crohn’s disease and 400 sarcoidosis patients and 399 controls in a German population, hypothesizing that any common susceptibility loci, if present, would demonstrate a stronger association peak.5 Their analysis identified a common SNP, rs1398024, located in the inter-genic region between the C10ORF67 and OTUD1 genes on chromosome 10p12 that was associated with both sarcoidosis and Crohn’s disease. They estimated a 19% reduction in disease risk in association with the A allele of rs1398024. We failed to replicate this association in our data, but did observe a non-significant 20% risk reduction associated with each A-allele of the top BWHS SNP: rs11013452. Further, haplotypic analyses of the rs1398024 and rs11013452 SNPs in the C10ORF67-OTUD1 inter-genic region on chromosome 10p12 revealed a strong inverse association with sarcoidosis. It is unclear which genes, if any, are directly responsible for the observed relationship with sarcoidosis. Besides the C10ORF67 and OTUD1 genes, the genomic neighborhood around the rs1398024 and rs11013452 SNPs contains several genes such as PTF1A, MSRB2, ARMC3, and C10ORF115 with no obvious functional link to sarcoidosis biology.5 Both our top SNP and the index rs1398024 SNP are located inside a ~300 kb low recombination region reported by Franke and colleagues5 suggesting that both SNPs are tagging the same causal variant. Because our top SNP is located inside a smaller LD block in HapMap YRI, our results further narrow the position of the causal SNP to a region of ~ 8kb.
The annexin gene family has been associated with the etiology of several chronic and autoimmune disorders including rheumatoid arthritis, systemic lupus erythematosus, and Sjogren’s syndrome.15, 16 It has been hypothesized that dysfunction of ANXA11 may affect the apoptosis pathway in individuals with sarcoidosis, thereby affecting the balance between cell death and survival of activated inflammatory cells. A GWAS by Hofmann and colleagues6 of 499 sarcoidosis cases and 490 controls identified two SNPs in the ANXA11 gene, rs2789679 and rs1049550, associated with risk of sarcoidosis. This association has been replicated in two recent studies involving a separate German population (325 sarcoidosis cases and 364 healthy controls)7 and a Czech population (245 sarcoidosis cases and 254 healthy controls).8 Those two SNPs are in complete LD in the HapMap YRI population and are also in perfect LD in the HapMap population of African Americans from the South Western United States (ASW).9 Therefore, we genotyped only rs2789679 which will likely have tagged most of the common variation in African Americans. We observed a non-significant 14% reduction in sarcoidosis risk associated with the T-allele of rs2789679, a much smaller reduction than the 40% reduction observed by Hofmann et al.6 Thus, our findings are in the same direction as the inverse association observed by Hofmann,6 but did not achieve statistical significance.
The identification of cases and controls from the same nationally drawn base population of African American women is a strength of the present study. Furthermore, we controlled for geographic region of residence, nativity, and African admixture, mitigating concern about confounding due to population stratification. In addition, follow-up of the study population has been satisfactory and participants who provided a DNA sample were similar to those who did not with regard to numerous characteristics, lessening concerns about selective participation. Sarcoidosis cases were identified by self-report since it is infeasible in large observational studies to examine all participants for disease. Our validation effort in a subset of women showed a high degree of accuracy of self-report. Misdiagnosis of sarcoidosis or incorrect reporting of the disease by participants was unlikely to have been related to genetic status because the physicians and women were unaware of their status with regard to the genetic variants under study. Random misclassification would have resulted in the dilution of effects (i.e., ORs closer to the null).
A limitation of the present study was the lack of clinical phenotype data available to investigate the association with disease severity, course, or organ involvement. However, we were able to assess the associations within strata of self-reported symptoms as well as with self-reported measures of general health, ability to perform everyday activities, fatigue, and pain using a validated instrument.17 Our findings of an inverse association of of ANXA11 with sarcoidosis risk among women with more symptoms at diagnosis or current poor health are consistent with those of Mrazek et al., who reported that the T-allele of the rs1049550 SNP was less frequent in Czech patients with stage II-IV chest radiographs.8
Our study population was limited to females and we therefore can not assess whether our results are generalizable to men. However, while some studies suggest that the incidence of sarcoidosis is greater among women than men,18–20 large population-based incidence studies in the U.S. and Scandinavia have found similar rates in men and women,21–23 which is consistent with genetic factors not being linked to gender.
Our study is the first to assess these genetic loci in relation to sarcoidosis risk in an African ancestry population. The present findings from the BWHS confirm the potential importance of the 10p12 locus in the etiology of sarcoidosis.
MATERIALS AND METHODS
Study population
The human subjects’ protocol for this study was approved by the Boston University Medical Center Institutional Review Board. We conducted a nested case-control study within the Black Women’s Health Study cohort,24, which was established in 1995 when 59,000 women aged 21–69 years enrolled through postal health questionnaires. Cohort participants self-identified as “black”. Diagnosis of sarcoidosis was ascertained by self-report. However, validation in a subset of women showed a high level of agreement between self-report and physician report/records.24
Assessment of disease severity
A supplemental sarcoidosis survey sent to both incident and prevalent cases asked if they had experienced any of a list of 9 symptoms at the time of their diagnosis (e.g., shortness of breath, cough, fatigue).24 The 2011 BWHS follow-up questionnaire included questions assessing current general health and physical function, and levels of fatigue and pain within the past 7 days, based on a previously developed and validated instrument.17 Subjects were asked “In general, would you say your health is …” (Excellent, very good, good, fair, poor); “To what extent are you able to carry out your everyday physical activities such as walking, climbing stairs, carrying groceries, or moving a chair?” (completely, mostly, moderately, a little, not at all); “In the past 7 days, how would you rate your fatigue on average?” (none, mild, moderate, severe, very severe); and, “In the past 7 days, how would you rate your pain on average?” (0=no pain through 10=worst imaginable pain).
Collection of DNA Samples
Mouthwash-saliva samples, from which DNA was extracted, were obtained from BWHS participants by the mouthwash-swish method,25 with all samples stored freezers at −80°C. Approximately 50% of participants, 26,800 women, provided a mouthwash-saliva sample. Women who provided samples were slightly older than women who did not, but the two groups were similar with regard to educational level, geographic region of residence, and body mass index.
Cases and Controls for the Present Study
There were 500 women diagnosed with sarcoidosis through the end of the 2009 follow-up cycle who provided a DNA sample; 486 were successfully genotyped (see below), of which 291 were prevalent cases at baseline in 1995 and 195 had occurred during follow-up. We selected up to two matched controls per case among BWHS participants who had provided a DNA sample and who were free of sarcoidosis at the end of the 2009 follow-up period, for a total of 972 controls. Controls were matched to cases on year of birth (± 1 year) and geographical region of residence (Northeast, South, Midwest, and West). As described below, 943 controls were successfully genotyped.
Fine-mapping of the C10ORF67 and ANXA11 genes
We performed fine-mapping of the intergenic region between C10ORF67 and OTUD1 genes(10p12) and the ANXA11 gene (10q22). For the C10ORF67-OTUD1 intergenic region we first identified a 33 kb LD block in HapMap CEU samples that contains the index rs1398024 SNP and then downloaded SNPs covering the LD block from HapMap YRI database.9 For the ANXA11 gene we downloaded SNPs covering the entire gene from the HapMap Yoruba (YRI) database.9 We used the Tagger software26 implemented in Haploview version 4.127, 28 (http://www.broadinstitute.org/haploview/haploview) to select all tagging SNPs with a minor allele frequency (MAF) ≥ 10% and r2 ≥ 0.9. The index SNPs (rs2789679 and rs1049550) in the ANXA11 gene are in complete LD (r2=1.0) in HapMap YRI. We therefore included only rs2789679 in the set of SNPs to be genotyped. In addition, we included the index SNP rs1398024 of the C10ORF67-OTUD1 intergenic region, 16 tagging SNPs in the ANXA11 gene, and 12 tagging SNPs in the C10ORF67-OTUD1 intergenic region.
Genotyping and Quality Control
DNA was isolated from the mouthwash-saliva samples by use of the QIAAMP DNA Mini Kit (Qiagen, Valencia, CA, USA, www.quiagen.com). Whole genome amplification was performed with the Qiagen RePLI-g Kits using the method of multiple displacement amplification. Amplified samples underwent purification and Pico Green quantification at the Broad Institute Center for Genotyping and Analysis (Cambridge, MA) before being plated for genotyping.
Genotyping was carried out at the Broad Institute Center for Genotyping and Analysis using the Sequenom MassArray iPLEX technology. There were 1,472 DNA samples (500 cases and 972 controls); thirty-two blinded duplicate samples were included to assess reproducibility of the genotypes. An average reproducibility of 98.5% was obtained among the blinded duplicates. All SNPs with calling rate < 90% or a deviation from Hardy-Weinberg equilibrium in the control samples at P < 0.001 were excluded. We also excluded samples with calling rates < 80% (14 cases and 29 controls), leaving 486 cases and 943 controls. The final analysis included 12 tagging SNPs in the C10ORF67-OTUD1 intergenic region and 15 tagging SNPs in the ANXA11 gene in 1,429 samples. Mean call rate in the final data set for both SNPs and samples was 99.8%.
African admixture proportions
We also selected 30 ancestral informative markers (AIMs) to estimate the percent African ancestry among the participants in order to control for population stratification due to African admixture. 29 These 30 AIMs are the top SNPs of the >1500 “phase 3” admixture panel.30 We have previously shown that ancestry estimates derived from the set of 30 AIMs are highly correlated with estimates derived from the entire “phase 3” admixture panel (r=0.89, p<0.0001).31
Data analysis
We tested each SNP for association with sarcoidosis risk using the Cochran-Armitage trend test of an additive genetic model. We adjusted for multiple testing by using 10,000 permutations as implemented in the PLINK software.28 We used logistic regression analysis (PROC LOGISTIC, SAS statistical software version 9.1.3, SAS Institute Inc., Cary, NC, USA) to estimate per-allele odds ratios (ORs), odds ratios for heterozygosity and homozygosity of the risk alleles, and 95% confidence intervals. We controlled for age, geographical region of residence (Northeast, South, Midwest, West), nativity (US, foreign country), and African admixture proportion. Haplotype ORs were estimated using an exception substitution approach32, 33 which estimates the probabilities of all possible haplotype configurations of each individual in the sample, conditional on their genotype and case-control status. Haplotypes with an estimated frequency of <5% were pooled in a single group and the most common haplotype was used as the reference haplotype.
We estimated individual admixture proportions using a Bayesian approach as implemented in open-source ADMIXMAP software.34, 35 Prior allele frequencies of the African and European populations were taken from the International HapMap project.9
In subanalyses, we assessed the association of the C10ORF67-OTUD1 and ANXA11 SNPs and sarcoidosis risk according to the number of symptoms reported at diagnosis (≥4), general health (fair or poor), ability to carry out daily physical activities (little or no ability), fatigue (severe or very severe), and pain (≥5 on the pain scale).
Acknowledgments
This work was supported by grant K01HL088709 from the National Heart, Lung, and Blood Institute and grant CA058420 from the Division of Cancer Control and Population Science, National Cancer Institute (http://www.cancercontrol.cancer.gov).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest
Slone Epidemiology Center at Boston University, Boston, Massachusetts (Yvette C. Cozier, Edward A Ruiz-Narvaez, Julie R. Palmer, Lynn A. Rosenberg, Julie R. Palmer); The Pulmonary Center, Boston University School of Medicine, Boston, Massachusetts (Jeffrey S. Berman); and the Boston University Graduate School of Arts and Sciences, Graduate Research Assistant Scholarship Program (GRASP) (Craig J McKinnon).
The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health.
References
- 1.Iannuzzi MC, Rybicki BA, Teirstein AS. Sarcoidosis. N Engl J Med. 2007;357(21):2153–65. doi: 10.1056/NEJMra071714. [DOI] [PubMed] [Google Scholar]
- 2.Newman LS, Rose CS, Maier LA. Sarcoidosis. N Engl J Med. 1997;336(17):1224–34. doi: 10.1056/NEJM199704243361706. [DOI] [PubMed] [Google Scholar]
- 3.Rybicki BA, Maliarik MJ, Major M, et al. Epidemiology, demographics, and genetics of sarcoidosis. Semin Respir Infect. 1998;13(3):166–73. [PubMed] [Google Scholar]
- 4.Swigris JJ, Olson AL, Huie TJ, Fernandez-Perez ER, Solomon J, Sprunger D, et al. Sarcoidosis-related mortality in the United States from 1988 to 2007. Am J Respir Crit Care Med. 2011;183(11):1524–30. doi: 10.1164/rccm.201010-1679OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franke A, Fischer A, Nothnagel M, Becker C, Grabe N, Till A, et al. Genome-wide association analysis in sarcoidosis and Crohn’s disease unravels a common susceptibility locus on 10p12. 2. Gastroenterology. 2008;135(4):1207–15. doi: 10.1053/j.gastro.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann S, Franke A, Fischer A, Jacobs G, Nothnagel M, Gaede KI, et al. Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat Genet. 2008;40(9):1103–6. doi: 10.1038/ng.198. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Pabst S, Kubisch C, Grohe C, Wollnik B. First independent replication study confirms the strong genetic association of ANXA11 with sarcoidosis. Thorax. 2010;65(10):939–40. doi: 10.1136/thx.2010.138743. [DOI] [PubMed] [Google Scholar]
- 8.Mrazek F, Stahelova A, Kriegova E, Fillerova R, Zurkova M, Kolek V, et al. Functional variant ANXA11 R230C: true marker of protection and candidate disease modifier in sarcoidosis. Genes Immun. 2011;12(6):490–4. doi: 10.1038/gene.2011.27. [DOI] [PubMed] [Google Scholar]
- 9.The International HapMap Project. Nature. 2003;426(6968):789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 10.Levine JB, Lukawski-Trubish D. Extraintestinal considerations in inflammatory bowel disease. Gastroenterol Clin North Am. 1995;24(3):633–46. [PubMed] [Google Scholar]
- 11.Zumla A, James DG. Granulomatous infections: etiology and classification. Clin Infect Dis. 1996;23 (1):146–58. doi: 10.1093/clinids/23.1.146. [DOI] [PubMed] [Google Scholar]
- 12.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361(21):2066–78. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fellermann K, Stahl M, Dahlhoff K, Amthor M, Ludwig D, Stange EF. Crohn’s disease and sarcoidosis: systemic granulomatosis? Eur J Gastroenterol Hepatol. 1997;9(11):1121–4. doi: 10.1097/00042737-199711000-00018. [DOI] [PubMed] [Google Scholar]
- 14.Oakley JR, Lawrence DA, Fiddian RV. Sarcoidosis associated with Crohn’s disease of ileum, mouth and oesophagus. J R Soc Med. 1983;76(12):1068–71. doi: 10.1177/014107688307601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes MJ, Longbottom RE, Evans MA, Moss SE. Annexinopathies. Subcell Biochem. 2007;45:1–28. doi: 10.1007/978-1-4020-6191-2_1. [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen CS, Levantino G, Houen G, Jacobsen S, Halberg P, Ullman S, et al. Determination of autoantibodies to annexin XI in systemic autoimmune diseases. Lupus. 2000;9(7):515–20. doi: 10.1177/096120330000900707. [DOI] [PubMed] [Google Scholar]
- 17.Cella D, Riley W, Stone A, Rothrock N, Reeve B, Yount S, et al. The Patient-Reported Outcomes Measurement Information System (PROMIS) developed and tested its first wave of adult self-reported health outcome item banks: 2005–2008. J Clin Epidemiol. 2010;63(11):1179–94. doi: 10.1016/j.jclinepi.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bresnitz EA, Strom BL. Epidemiology of sarcoidosis. Epidemiol Rev. 1983;5:124–56. doi: 10.1093/oxfordjournals.epirev.a036255. [DOI] [PubMed] [Google Scholar]
- 19.Sutherland I, Mitchell DN, Hart PD. Incidence of Intrathoracic Sarcoidosis among Young Adults Participating in a Trial of Tuberculosis Vaccines. Br Med J. 1965;5460:497–503. doi: 10.1136/bmj.2.5460.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terris M, Chaves AD. An epidemiologic study of sarcoidosis. Am Rev Respir Dis. 1966;94(1):50–5. doi: 10.1164/arrd.1966.94.1.50. [DOI] [PubMed] [Google Scholar]
- 21.Henke CE, Henke G, Elveback LR, et al. The epidemiology of sarcoidosis in Rochester, Minnesota: a population-based study of incidence and survival. Am J Epidemiol. 1986;123(5):840–5. doi: 10.1093/oxfordjournals.aje.a114313. [DOI] [PubMed] [Google Scholar]
- 22.Hillerdal G, Nou E, Osterman K, Schmekel B. Sarcoidosis: epidemiology and prognosis. A 15-year European study. Am Rev Respir Dis. 1984;130(1):29–32. doi: 10.1164/arrd.1984.130.1.29. [DOI] [PubMed] [Google Scholar]
- 23.Byg KE, Milman N, Hansen S. Sarcoidosis in Denmark 1980–1994. A registry-based incidence study comprising 5536 patients. Sarcoidosis Vasc Diffuse Lung Dis. 2003;20(1):46–52. [PubMed] [Google Scholar]
- 24.Cozier YC, Berman JS, Palmer JR, Boggs DA, Serlin DM, Rosenberg L. Sarcoidosis in black women in the United States: data from the Black Women’s Health Study. Chest. 2011;139(1):144–50. doi: 10.1378/chest.10-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cozier YC, Palmer JR, Rosenberg L. Comparison of methods for collection of DNA samples by mail in the Black Women’s Health Study. Ann Epidemiol. 2004;14(2):117–22. doi: 10.1016/S1047-2797(03)00132-7. [DOI] [PubMed] [Google Scholar]
- 26.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37(11):1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, et al. A high-density admixture map for disease gene discovery in african americans. Am J Hum Genet. 2004;74(5):1001–13. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005;37(10):1113–8. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz-Narvaez EA, Rosenberg L, Wise LA, Reich D, Palmer JR. Validation of a small set of ancestral informative markers for control of population admixture in African Americans. Am J Epidemiol. 2011;173(5):587–92. doi: 10.1093/aje/kwq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered. 2002;53(2):79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
- 33.Stram DO, Leigh Pearce C, Bretsky P, Freedman M, Hirschhorn JN, Altshuler D, et al. Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case-control study of unrelated individuals. Hum Hered. 2003;55(4):179–90. doi: 10.1159/000073202. [DOI] [PubMed] [Google Scholar]
- 34.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, et al. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72(6):1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKeigue PM, Carpenter JR, Parra EJ, Shriver MD. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann Hum Genet. 2000;64(Pt 2):171–86. doi: 10.1017/S0003480000008022. [DOI] [PubMed] [Google Scholar]