Abstract
Vertebrate Tob/BTG proteins inhibit cell proliferation when overexpressed in tissue culture cells, and they can function as tumor suppressors in mice. The single Caenorhabditis elegans Tob/BTG ortholog, FOG-3, by contrast, was identified from its loss-of-function phenotype as a regulator of sperm fate specification. Here we report that FOG-3 also regulates proliferation in the germline tissue. We first demonstrate that FOG-3 is a positive regulator of germline proliferation. Thus, fog-3 null mutants possess fewer germ cells than normal, a modest but reproducible decrease observed for each of two distinct fog-3 null alleles. A similar decrease also occurred in fog-3/+ heterozygotes, again for both fog-3 alleles, revealing a haplo-insufficient effect on proliferation. Therefore, FOG-3 normally promotes proliferation, and two copies of the fog-3 gene are required for this function. We next overexpressed FOG-3 by removal of FBF, the collective term for FBF-1 and FBF-2, two nearly identical PUF RNA-binding proteins. We find that overexpressed FOG-3 blocks proliferation in fbf-1 fbf-2 mutants: whereas germ cells stop dividing and instead differentiate in fbf-1 fbf-2 double mutants, they continue to proliferate in fog-3; fbf-1 fbf-2 triple mutants. Therefore, like its vertebrate Tob/BTG cousins, overexpressed FOG-3 is “antiproliferative”. Indeed, some fog-3; fbf-1 fbf-2 mutants possess small tumors, suggesting that FOG-3 can act as a tumor suppressor. Finally, we show that FOG-3 and FBF work together to promote tumor formation in animals carrying oncogenic Notch mutations. A similar effect was not observed when germline tumors were induced by manipulation of other regulators; therefore this FOG-3 tumor-promoting effect is context-dependent. We conclude that FOG-3 can either promote or inhibit proliferation in a manner that is sensitive to both genetic context and gene dosage. The discovery of these FOG-3 effects on proliferation has implications for our understanding of vertebrate Tob/BTG proteins and their influence on normal development and tumorigenesis.
Keywords: FOG-3, Tob/BTG, proliferation, tumor formation, germ cells, C. elegans
Introduction
Vertebrate Tob/BTG (for transducer of ERBB2/B-cell translocation gene) proteins share two defining features — an N-terminal Tob/BTG domain and “antiproliferative activity” when overexpressed in tissue-culture cells (1–3). Tob/BTG proteins can also be tumor suppressors (reviewed by 3). For example, murine mutants lacking either Tob1 or BTG3 have an increased frequency of spontaneous tumors (4, 5), and ectopically expressed Tob1 or BTG2 can inhibit tumor formation in mouse models (6, 7). Consistent with that tumor suppressor activity, Tob/BTG abundance is dramatically lowered in several cancers (4, 5, 8–14). Tob/BTG proteins are therefore emerging as important proliferation regulators and tumor suppressors.
Analysis of Tob/BTG proteins in vertebrate organisms has been constrained by the complexity of vertebrate tissues and the existence of multiple homologs (e.g. Tob1, Tob2, BTG1, BTG2, BTG3 and BTG4 in humans). Here we take advantage of the genetic power and relative simplicity of the nematode Caenorhabditis elegans to investigate the role of its single Tob/BTG ortholog in the regulation of proliferation. This ortholog (Figure 1a) was identified originally for its role in sperm fate specification and named FOG-3 for its null mutant phenotype (feminization of the germline) (15, 16). The only defect observed in fog-3 mutants was sexual transformation of the germline; in the absence of FOG-3, germ cells that normally differentiate as sperm instead become oocytes. Most importantly, no conspicuous defect in proliferation was observed in fog-3 mutants.
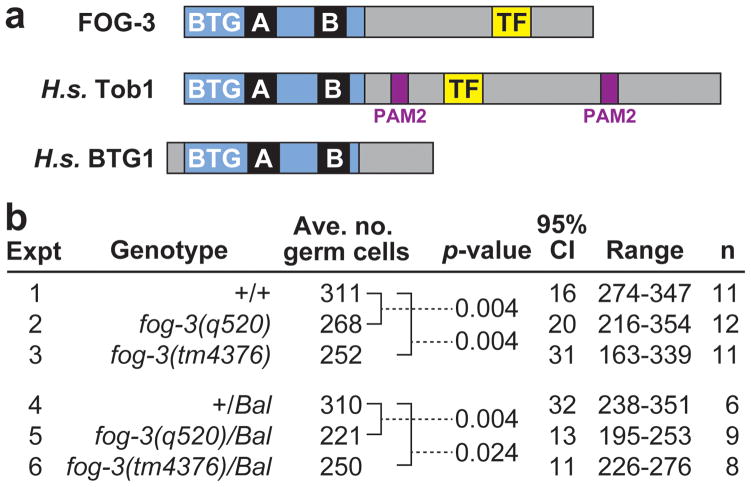
Figure 1.
FOG-3 promotes robust germline proliferation. (a) FOG-3 is a member of the Tob/BTG protein family. The family signature is the BTG domain (blue), which contains two highly conserved regions, Box A and Box B (39). The C-terminal amino acid sequence is not conserved except for the PAM2 motif among vertebrate Tob proteins (40) and the TF motif among vertebrate and C. elegans proteins (16). (b) Average number of germ cells per gonadal arm in mid-L4 larvae of genotypes as designated. Bal, balancer chromosome used to maintain heterozygotes. The p-values are given for comparisons of germ cell counts marked by brackets; these p-values were generated using a Mann-Whitney test; similar numbers were also obtained using a two-tailed t-test. CI, confidence interval in number of germ cells from average. n, number of gonadal arms counted for each genotype.
Given the designation of the Tob/BTG family as “antiproliferative”, we expected FOG-3 might inhibit proliferation. However, our first experiments showed that FOG-3 is normally a positive regulator of proliferation: fog-3 homozygotes and fog-3/+ heterozygotes both have fewer germ cells than normal. This decrease was modest but reproducible and observed for two separate fog-3 alleles. Therefore, FOG-3 promotes proliferation in a dose-sensitive manner. Subsequent experiments revealed that FOG-3 can also inhibit proliferation or promote tumor formation, depending on context. Overexpressed FOG-3 inhibits proliferation and suppresses tumor formation, and in oncogenic Notch strains, FOG-3 promotes tumor formation. We conclude that FOG-3 can either promote or inhibit proliferation and discuss the implications of these findings for vertebrate Tob/BTG proteins.
Results
FOG-3 appears to promote robust germline proliferation
To investigate fog-3 effects on proliferation, we focused on the hermaphrodite germline tissue for two reasons: FOG-3 acts in this tissue (Supplementary Figure 1) (16; this work) and most studies of germline proliferation have been conducted in the hermaphrodite sex (17).
Prior work failed to find any effect on germline proliferation in fog-3 mutants (15). To investigate the possibility of a subtle effect, we counted germ cell number at the mid-L4 stage, when germline proliferation is extensive but gametes are not yet made. Two deletion mutants, both likely null alleles, were used: fog-3(q520) is a small 4-bp deletion in the Tob/BTG domain that shifts the reading frame (16), and fog-3(tm4376) is a 364 bp deletion that removes the N-terminal half of the Tob domain and also shifts the reading frame (this work; see Methods).
Based on the established antiproliferative activity of vertebrate Tob/BTG proteins (see Introduction), we predicted that germlines in fog-3 null mutants might have more germ cells than normal. However, germ cell number in fog-3 mutants was lower than normal: whereas wild type had 311 germ cells per gonadal arm on average at the mid-L4 stage, the fog-3 mutants had either 268 (fog-3(q520)) or 252 (fog-3(tm4376)) germ cells per gonadal arm on average. The decrease was modest but statistically significant (p=0.004) in both mutants (Figure 1b, lines 1–3).
The simplest explanation of this reduction in germ cell number is that FOG-3 normally functions to promote robust proliferation. One alternate explanation is that germ cells lacking FOG-3 activity begin dividing later than normal. To test this idea, we examined timing of the first germ cell division relative to somatic neighbors. In wild-type L1 larvae, germ cells initiate divisions before somatic gonadal cells. Similarly in 7/7 fog-3(q520) L1s found at the diagnostic developmental stage, germ cells divided before somatic gonadal cells, indicating that the onset of proliferation was not delayed. Moreover, germ cell size and number appeared normal throughout the L1 stage. A second alternate explanation is that germline sex affects proliferation: wild-type L4 germlines are spermatogenic but fog-3 mutant L4 germlines are oogenic. We were unable to address this possibility in homozygotes, because no condition has yet been found that allows fog-3 null mutants to be spermatogenic (15, 16; Kimble lab, unpublished). However, we were able do exclude this explanation in heterozygotes (see below).
A single fog-3 gene copy is not sufficient for normal germline proliferation
We next found that fog-3/+ mid-L4 animals also generate fewer germ cells than wild-type: fog-3(q520)/+ had 221 and fog-3(tm4376)/+ had 250 germ cells per gonadal arm on average, numbers significantly lower than the 310 germ cells in controls scored in parallel (Figure 1b, lines 4–6). Indeed the proliferation defect in fog-3/+ heterozygotes was remarkably similar to that observed in fog-3 homozygotes (compare Figure 1b, lines 2,3 to lines 5,6). We also examined when germ cells start dividing in fog-3(q520)/+ heterozygotes to ask whether their reduced numbers in L4s might be explained by a delayed start in germ cell divisions. As in wild-type, the fog-3(q520)/+ germ cell progenitor cells always divided before the somatic gonadal progenitor cells (n=7), and germ cell number and size appeared normal throughout the L1 stage. Therefore, the reduced proliferation observed in L4s is unlikely to result from a delay in the start of germ cell divisions.
We also note that this experiment with fog-3/+ heterozygotes does not suffer from the germline-sex caveat raised above for fog-3 homozygotes. The fog-3/+ hermaphrodites not only make sperm, but their brood sizes do not differ significantly from wild type, indicating that they make the same number of sperm as wild-type hermaphrodites (Supplementary Table 1) (15). Therefore germline sex determination is equivalent in wild-type and fog-3/+ hermaphrodites. We conclude that the wild-type fog-3 gene promotes proliferation and that its wild-type gene dosage is crucial for that control.
Undetectable FOG-3 in the mitotic zone depends on FBF
We next overexpressed FOG-3 to analyze its effect on proliferation. Attempts to generate animals that harbored transgenes designed to overexpress FOG-3 were not successful. As an alternative method, we tested the idea that removal of the FBF repressor might result in overexpressed FOG-3. FBF is the collective term for FBF-1 and FBF-2, two nearly identical PUF RNA-binding proteins that are largely redundant (18). Three lines of evidence had previously suggested that FBF might repress fog-3: an FBF binding element was found in the fog-3 3′UTR (19); fog-3 mRNA immunoprecipitates with FBF-1 (20); and the FBF and FOG-3 proteins reside in largely non-overlapping regions of the germline (21–23). Yet no assay had been performed to show that FBF removal would lead to overexpressed FOG-3, because no reagent had been available to examine FOG-3 protein.
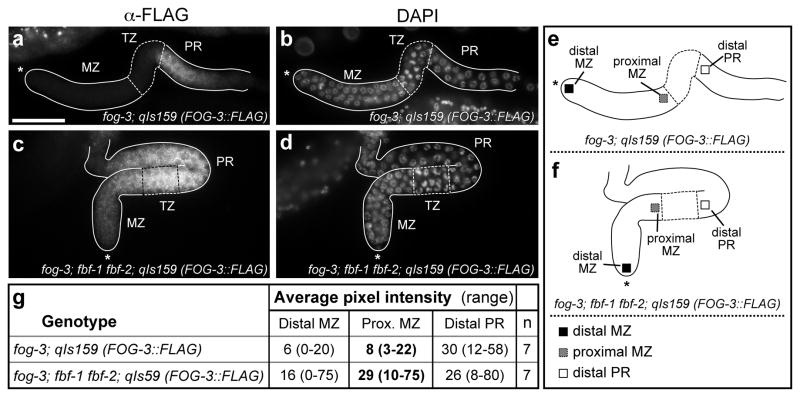
We visualized FOG-3 using an epitope-tagged transgenic FOG-3::FLAG protein that rescues fog-3 null mutants to fertility (23). This FOG-3::FLAG protein was assayed in a strain that is null for endogenous FOG-3 to ensure that FOG-3::FLAG maintains its rescuing activity. To ask if FBF affects fog-3 expression in the mitotic zone, we compared the FOG-3::FLAG expression pattern in a strain wild-type for FBF (fog-3; qIs159[FOG-3::FLAG]) to the expression pattern in a strain without FBF (fbf-1 fbf-2; fog-3; qIs159[FOG-3::FLAG]). This comparison was made in early L4 larvae, when both strains possessed actively proliferating germ cells in their distal gonad and spermatogenic germ cells in their proximal gonad.
Normally FBF is most abundant in mitotically-dividing germ cells at the distal end of the gonad (21, 24), and FOG-3::FLAG is most abundant more proximally in germ cells that have left the mitotic zone and entered meiotic prophase (23). We therefore focused on the mitotic zone, the region of actively dividing germ cells, to ask if FBF affects FOG-3::FLAG expression. Confirming previous results (23), FOG-3::FLAG was not detected in the mitotic zone of a germline harboring wild-type FBF (Figures 2a and b). By contrast, in an fbf-1 fbf-2 double mutant germline with no FBF, FOG-3::FLAG became easily detectable in the proximal mitotic zone (Figures 2c and d). Quantitation with ImageJ confirmed this FOG-3::FLAG increase (Figures 2e and f). We do not know if this FBF effect is direct or indirect. Nonetheless, FBF clearly affects FOG-3::FLAG expression in the mitotic zone.
Figure 2.
The undetectable level of FOG-3::FLAG in the mitotic zone is dependent upon FBF. (a–d) Dissected gonads from early L4 hermaphrodites harboring a FLAG-tagged FOG-3 transgene to rescue their lack of endogenous fog-3; (e) schematic of gonad in (a,b); (f) schematic of gonad in (c,d). Gonads are outlined and germline regions marked: mitotic zone (MZ), transition zone (TZ) and pachytene region (PR). Most MZ germ cells are in the mitotic cell cycle; all TZ and PR germ cells have entered the meiotic cell cycle with TZ cells in early meiotic prophase and PR germ cells progressing through the pachytene stage of meiotic prophase. See Kimble and Crittenden (17) for details. Asterisk indicates gonadal distal end. (a,c) Staining with anti-FLAG antibody to visualize transgenic FOG-3::FLAG rescuing protein; (b,d) DAPI staining to visualize all nuclei and visualize boundaries of germline regions. Widefield images were taken at same magnification with the same settings; scale bar in (a) represents 50 μM and serves for all images. (a,b) FOG-3::FLAG is not detectable in the mitotic zone (MZ) in a genetic strain with wild-type FBF. (c,d) FOG-3::FLAG becomes easily detectable in the proximal part of the MZ in a genetic strain lacking FBF. The images shown are representative for this dramatic strain difference in the MZ. We note that this image also shows a staining difference in the proximal part of the pachytene region, which was not quantitated and is beyond the scope of this study. (e,f) Positions at which FOG-3::FLAG staining was quantitated using ImageJ: black square, distal MZ; grey square, proximal MZ; and open square, distal PR; (g) ImageJ quantitation of FOG-3::FLAG staining, including both average pixel intensity and range.
One interpretation of this experiment might have been that the overexpressed FOG-3::FLAG was the result of a difference in germline sex. Although both germlines were spermatogenic at the early L4 stage when the staining was performed, the germline with wild-type FBF was destined to make oocytes during adulthood, while the germline lacking FBF was destined to cease proliferation and never make oocytes. One might postulate that the germline with wild-type FBF had a feminized mitotic zone in preparation for its later switch into oogenesis. To control for this possibility, we compared FOG-3::FLAG levels in the early L4 mitotic zones of two strains that both express wild-type FBF but that differ in the type of gamete generated in adults. The first strain, fog-3; FOG-3::FLAG, is spermatogenic at the L4 stage and oogenic in adulthood. The second strain, fem-3(gf); fog-3; FOG-3::FLAG, is spermatogenic during both the L4 stage and adulthood and never makes oocytes. The abundance of FOG-3::FLAG was equivalent in the early L4 mitotic zones of these two strains (Supplementary Figure 2). Therefore, the FOG-3::FLAG overexpression in fbf-1 fbf-2 mitotic zones results from loss of FBF, not from loss of the switch to oogenesis. We conclude that FBF is required to lower fog-3 expression in the mitotic zone.
FOG-3 inhibits proliferation when overexpressed in fbf-1 fbf-2 mutants
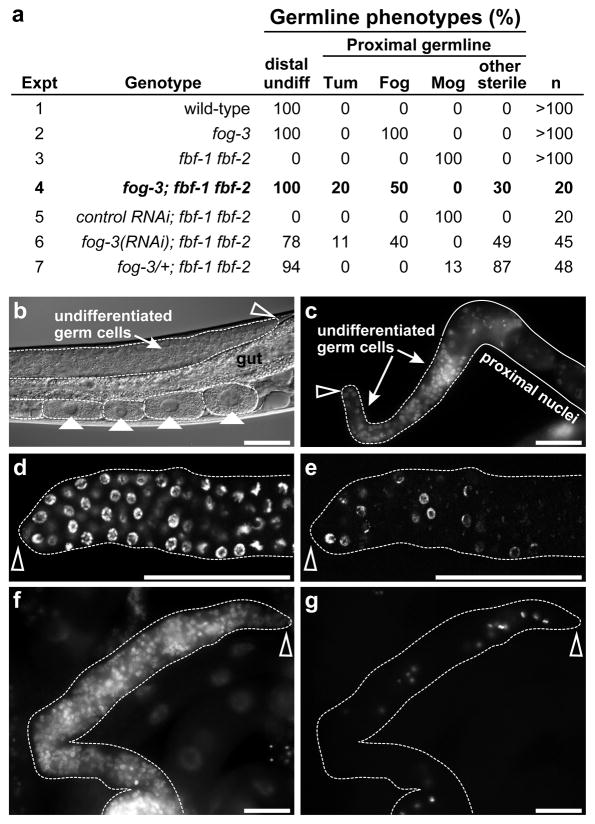
To assess the effect of overexpressed FOG-3 on germline proliferation, we compared fbf-1 fbf-2 null mutant germlines with and without FOG-3. For these studies, we focused on adults, 24 hours past the L4 stage, and first asked whether germ cells were undifferentiated or differentiated in the distal gonad (Figure 3a, Distal undiff). Prior work showed that the distal gonads of wild-type animals and fog-3 null mutants maintain undifferentiated germ cells throughout adulthood (15, 25, 26), but that the distal gonads of late L4 fbf-1 fbf-2 larvae contain no undifferentiated germ cells because all their germ cells differentiate into sperm (21). We first confirmed these conclusions for the distal germlines of wild-type, fog-3 and fbf-1 fbf-2 late L4 larvae (Figure 3a, lines 1–3). We then examined the effect of removing FOG-3 in fbf-1 fbf-2 mutants. In fog-3; fbf-1 fbf-2 triple mutant adults, the distal germ cells all appeared undifferentiated (Figure 3a, line 4; Figures 3b and c). A similar effect was seen when FOG-3 was depleted using RNAi (Figure 3a, line 6) or reduced by one gene copy (Figure 3a, line 7). To ask if these distal undifferentiated cells in fog-3; fbf-1 fbf-2 germlines were cycling, we treated animals with the thymidine analog EdU, an S-phase marker, and found that nuclei among the undifferentiated germ cells were labeled (Figures 3d and e). Therefore, these undifferentiated nuclei are progressing through the cell cycle.
Figure 3.
Overexpressed FOG-3 blocks proliferation in fbf-1 fbf-2 mutants. (a) Germlines were scored for presence of undifferentiated cells in the distal gonad (Distal undiff) and for gamete formation in adults (Proximal germline); Mog (masculinization of germline), sperm only; Fog, oocytes only; Tum, undifferentiated throughout germline; other sterile. (b–g) Adult fog-3; fbf-1 fbf-2 gonads, all 1 day past mid-L4. Scale bars represent 50 μM. Gonads are outlined. Empty arrowhead marks distal end. (b) Nomarski micrograph of Fog germline with oocytes marked by solid arrowheads, shown in intact animal. The distal germline has many undifferentiated cells. Germlines of this appearance could be fertile and make viable progeny when crossed to wild-type males, showing that FBF is not required for oogenesis or embryogenesis. (c) Hoechst-stained germline from the “other sterile” group. The germline has distal undifferentiated germ cells and proximal germ cells that had deteriorated (outlined by solid line). (d,e) Distal region of same Fog gonad contains undifferentiated cells with nuclear morphology characteristic of mitotic cell cycle. (d) DAPI-stained. (e) EdU incorporation highlights actively cycling cells. (f,g) Same gonad with a tumorous germline. (f) DAPI-stained. (g) Stained with anti-phosphorylated histone H3 to mark nuclei of cells in late G2 and M-phase (41).
We next examined the proximal germline. Whereas all fog-3; fbf-1 fbf-2 germlines had undifferentiated cells distally (Figure 3a, line 4), their proximal regions were variable (Figure 3a; Proximal germline). In most, the proximal germ cells had left the mitotic cell cycle, which in this system signals differentiation. Some made oocytes (Fog), while others were disorganized and difficult to classify (collectively termed “other sterile”); none made any sperm. More strikingly, 20% appeared tumorous (Tum). We use the term “tumorous” in this context to mean a tissue abnormally transformed to a state that is composed entirely of dividing undifferentiated cells, a classic hallmark of tumors. These Tum germlines did not possess differentiated gametes or disorganized germ cells in their proximal region, but instead all their germ cells appeared proliferative. All their nuclei had a chromosomal morphology typical of the mitotic cell cycle, and none had a morphology typical of entry into the meiotic cell cycle, as assayed using Hoechst or DAPI staining (Figure 3f). Moreover, cells positive for phosphorylated histone H3 were scattered from distal to proximal (Figures 3f and g), and they incorporated EdU randomly from distal to proximal, indicating that their cells were progressing through the cell cycle. These fog-3; fbf-1 fbf-2 Tum germlines were relatively small, however, and therefore not typical of aggressive tumors.
To confirm that germ cells were continuing to proliferate into adulthood in fog-3; fbf-1 fbf-2 animals, we compared germ cell number between L4 larvae and adults. In adults, we focused on Fog and Tum germlines, because germ cells in the “other sterile” class were often nebulous and difficult to count. Germ cell number per gonadal arm increased from an average of 86 in fog-3; fbf-1 fbf-2 L4s (range, 45–144; n=7) to an average of 130 in fog-3; fbf-1 fbf-2 Fog adults (n=5; range, 65–195) and to an average of 590 in fog-3; fbf-1 fbf-2 Tum adults (n=6; range, 310–823). We conclude that FOG-3 removal relieves the block to proliferation typical of fbf-1 fbf-2 mutants, although proliferation is not restored to a normal level. We also conclude that FOG-3 removal can lead to formation of small non-aggressive tumors in at least one genetic context.
We note that an earlier study reported that fog-3; fbf-1 fbf-2 mutants made fewer germ cells than fbf-1 fbf-2 mutants (19). Our more rigorous analysis of the same triple mutant strain suggests that the previous report was not correct. It is likely that an incorrectly marked fog-1; fbf-1 fbf-2 strain was analyzed in the earlier study, not the reported fog-3; fbf-1 fbf-2 strain. No other discrepancies were found between the two analyses. For example, we confirmed that fog-1; fbf-1 fbf-2 generates only a few germ cells that all differentiate as oocyte-like cells (Supplementary Table 2). Therefore, FOG-1 and FOG-3 have opposite effects on proliferation in an fbf-1 fbf-2 background, the first indication that these two proteins do not control germ cells identically.
FOG-3 can promote tumor formation
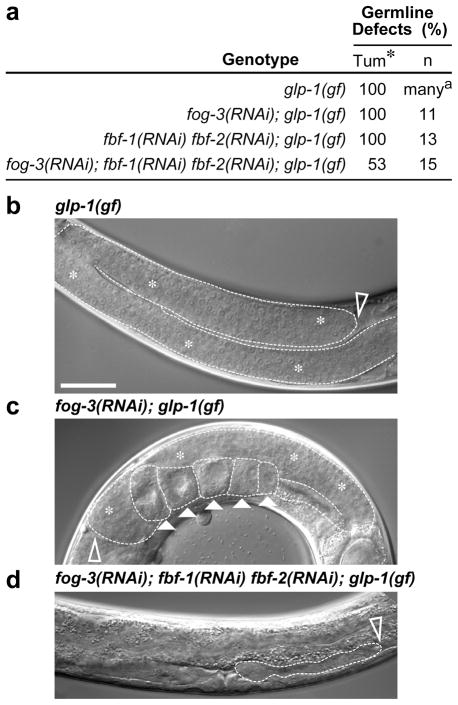
We explored the idea that FOG-3 might influence tumor formation in mutants that normally generate very large germline tumors. We first used animals that harbor an oncogenic gain-of-function (gf) mutation in the gene encoding the GLP-1 Notch receptor (27). All glp-1(gf) homozygotes make germline tumors and are sterile, but they can be propagated from glp-1(gf)/+ heterozygotes, which are often fertile as young adults (27). We used RNAi to deplete fog-3 in glp-1(gf)/+ L4 larvae and examined their glp-1(gf) homozygous progeny in the next generation. The fog-3(RNAi); glp-1(gf) animals formed large germline tumors, similar to those in glp-1(gf) controls (Figures 4a-c, white asterisks). However, unlike glp-1(gf) germlines, the fog-3(RNAi); glp-1(gf) germline tumors sometimes made one or a few oocytes, confirming RNAi efficacy (Figure 4c, arrowheads). Therefore, FOG-3 depletion had no major effect on glp-1(gf) tumor formation. We then removed both FOG-3 and FBF with a much more dramatic result. Whereas RNAi knockdown of fbf-1 and fbf-2 had no effect (Figure 4a), as previously shown (19), the simultaneous depletion of both FOG-3 and FBF could abolish germline tumors: essentially no germ cells remained in ~50% of the fog-3(RNAi); fbf-1(RNAi); fbf-2(RNAi); glp-1(gf) homozygotes (Figure 4a, 4d). This ~50% penetrance likely reflects the well-known character of RNAi to reduce rather than eliminate gene expression. The loss of glp-1(gf) tumor formation might result from either a decrease in proliferation or increase in cell death. Regardless, FBF and FOG-3 work together to promote glp-1(gf) tumor formation—an activity consistent with a role in proliferation, but that may involve other developmental mechanisms.
Figure 4.
FOG-3 and FBF can promote tumor formation. (a) Effects of fog-3, fbf-1 and fbf-2 depletion on tumor formation in animals homozygous for an oncogenic Notch mutation, glp-1(gf). Tum, tumorous; *, some fog-3(RNAi) tumorous germlines made a few oocytes; n, number of germlines scored; a Berry et al. (27). (b–d) Young adult gonads in intact animals, 1 day past L4. Dotted line, outline of gonadal tissue. Scale bar, 50 μM. Empty arrowheads, distal end. (b) Tumorous germline with no apparent differentiation. (c) Tumorous germline with oocytes (solid arrowheads) after fog-3 depletion. (d) Germline tumor abolished after fog-3 and fbf depletion.
We next assayed tumor formation in double mutants lacking gld-1 and gld-2, two genes that act downstream of GLP-1/Notch to promote differentiation (28). For these experiments, we used mutants rather than RNAi to remove FOG-3 and FBF. All gld-1 gld-2 double mutants made germline tumors (Supplementary Table 2, line 1), as expected (28), and all gld-1 gld-2 fog-3 triple mutants and gld-1 gld-2 fog-3; fbf-1 fbf-2 quintuple mutants also made germline tumors (Supplementary Table 2, lines 2 and 3). Moreover, germline tumors also formed when fog-3; fbf-1 fbf-2 triple mutants were depleted for gld-1 and gld-2 using RNAi (Supplementary Table 2, lines 4 and 5). We conclude that FOG-3 and FBF are not required to promote formation of all germline tumors.
Discussion
This work demonstrates that FOG-3, the single C. elegans Tob/BTG ortholog, can either promote or block germline proliferation, and that this effect is exquisitely sensitive to both gene dosage and genetic context. We also provide evidence that FOG-3 can suppress, enhance or have no effect on germline tumor formation, again depending on genetic context. Previous work showed that FOG-3 is an essential regulator of sperm cell fate specification, including both initiation and maintenance of that fate (15, 23). Together these FOG-3 studies provide a model for how a Tob/BTG protein may normally function within a developing tissue (Figure 5). Our discussion focuses on these antagonistic FOG-3 roles and their implications for function of vertebrate Tob/BTG proteins in development and cancer.
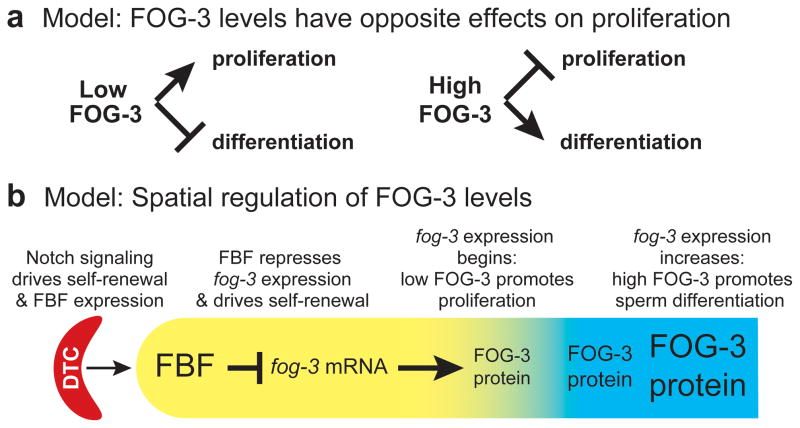
Figure 5.
Models for FOG-3 controls of proliferation. (a) Low FOG-3 promotes proliferation (left) but high FOG-3 promotes differentiation (right). (b) Diagram of C. elegans distal gonad to show spatial control of FOG-3 abundance. Somatic distal tip cell (DTC) provides stem cell niche (red); germline region containing stem cells and transit-amplifying cells (yellow); region with cells in meiotic cell cycle and differentiating (blue). GLP-1/Notch signaling from the DTC promotes FBF expression and FBF represses fog-3 expression; FOG-3 protein is first expressed at a low level when germ cell differentiation begins and accumulates to a high level as differentiation continues.
The C. elegans Tob/BTG protein FOG-3 can promote proliferation
We have found that FOG-3 is a positive regulator of germline proliferation in normal cells and a tumor-promoter in one genetic context. Thus, FOG-3 promotes robust proliferation during normal development, and FOG-3 and FBF together promote tumor formation in the context of oncogenic GLP-1/Notch signaling. The mechanism used by FOG-3 to promote proliferation and tumor formation is not understood. Vertebrate Tob/BTG proteins shuttle between the nucleus and cytoplasm and have been implicated in both transcriptional and translational regulation (1–3, 29, 30), but FOG-3 has not yet been subjected to similar analyses. One clue is that FOG-3 is cytoplasmic, consistent with a post-transcriptional role (23). Based on its positive effect on germline proliferation, a simple prediction is that the FOG-3 protein should be expressed in proliferating germ cells. However, abundant FOG-3 protein is only found in germ cells that have begun to differentiate (23; this work). Two distinct models could explain this paradox. One possibility is that FOG-3 promotes proliferation when expressed at a level that is not easily detectable. Consistent with this idea, very faint FOG-3::FLAG protein was observed in mitotically-dividing germ cells just prior to their entry into the meiotic cell cycle (23), and its role in promoting proliferation is dose-dependent (this work). An alternative idea is that FOG-3 controls proliferation from a distance (e.g. by generating a mitogenic factor). To our knowledge, no experimental evidence yet supports this latter model, but our understanding of FOG-3 and its downstream effecters remains in its infancy. Regardless of the mechanism, the finding that FOG-3 promotes proliferation in a dose-dependent manner was unexpected and has heuristic value for thinking about the action of Tob/BTG proteins in mammals.
Overexpressed FOG-3 can inhibit proliferation in fbf-1 fbf-2 mutants
We also found that overexpressed FOG-3 can block germline proliferation. Although we could not find a way to overexpress FOG-3 in the proliferating germ cells of a wild-type animal, when the FBF RNA-binding protein was removed in fbf-1 fbf-2 mutants, FOG-3 became overexpressed in the germline mitotic zone. These fbf-1 fbf-2 mutants typically stop germ cell divisions at the L4 stage, but when FOG-3 was removed, the fbf-1 fbf-2 germ cells continued proliferation into adulthood. Indeed, small germline tumors were sometimes seen in fog-3; fbf-1 fbf-2 triple mutants. Therefore, FOG-3 protein is critical for blocking proliferation in the fbf-1 fbf-2 mutant context.
FOG-3 overexpression in fbf-1 fbf-2 mutants is likely accompanied by overexpression of many other FBF targets (20, 31). Nonetheless, removal of a different well-established FBF target, FOG-1, did not have the same effect as FOG-3 removal: fog-1; fbf-1 fbf-2 germlines did not continue to proliferate and did not form tumors (19; this work). FOG-1 is appropriate for this comparison, because it has the same biological function as FOG-3 with respect to sperm fate specification, but belongs to a different protein family (17). Therefore, the proliferation block is specific to FOG-3.
The idea that FOG-3 can have opposite effects on proliferation, depending on context, is underscored by results with fog-3/+ heterozygotes: fog-3/+ animals have fewer germ cells than wild type controls (Figure 1), but fog-3/+; fbf-1 fbf-2 animals have more germ cells than fbf-1 fbf-2 controls (Supplementary Table 2). The FOG-3 dose-dependence is therefore context dependent. We do not understand the difference but suggest one simple possibility based on the idea that low FOG-3 promotes proliferation. In a wild-type background, halving the FOG-3 dose may drive its already undetectable level in the mitotic zone too low for proliferation; in the fbf-1 fbf-2 background, halving its dose may reduce its overexpressed level in the mitotic zone to a lower level that is appropriate for proliferation.
The discovery that FOG-3 can have opposite effects on proliferation — promoting proliferation in one context and inhibiting it in another — was not predicted from studies of Tob/BTG proteins in other organisms. We do not understand how FOG-3 exerts its dual effects but suggest that FOG-3 likely acts within multiple complexes. Since FOG-3 dosage is critical, one can imagine formation of one complex when FOG-3 abundance is low (perhaps nucleated by a high-affinity binder), but formation of a different complex when FOG-3 abundance is high (perhaps dependent on a low-affinity binder). Testing this idea will depend on the identification of FOG-3 complexes and their functions within the proliferating germline.
Implications for vertebrate Tob/BTG proteins
The effects of nematode FOG-3 on proliferation have intriguing implications for vertebrate Tob/BTG proteins. In particular, FOG-3 can have either positive or negative effects on proliferation, and its effects are dose-sensitive and context dependent. Expression of Tob1, BTG2 and BTG3 mRNA is reduced in several human cancers (4, 5, 8, 11–13), and a decrease in BTG4 expression has been found in colon cancer (14). By analogy with FOG-3, we surmise that lowered Tob/BTG expression in cancers may in some cases be able to promote proliferation and stimulate tumor formation — an idea with potentially important consequences for therapeutic manipulation of human Tob/BTG proteins. Moreover, Tob/BTG haplo-insufficiency may have little effect on its own, but depending on context, may enhance or suppress tumor formation when other genes have gone awry.
The defining function of vertebrate Tob/BTG proteins is their “antiproliferative” activity when overexpressed in tissue culture cells (1). We have found that FOG-3 can also be “antiproliferative” when assayed in a genetic context that leads to its overexpression in cells that normally would be proliferative. A clear understanding of how Tob/BTG proteins exert antiproliferative activity remains elusive. FOG-3 has a major role in initiation and maintenance of sperm fate specification (15, 23). Similarly, some vertebrate Tob/BTG members drive differentiation (32–34). A simple and unifying explanation is that overexpression of a Tob/BTG protein releases its pro-differentiation activity.
Materials and Methods
Nematode strains and methods
We used wild-type strain N2 and the following mutations: LGI: fog-3(q520 or tm4376) (15; this work), LGII: fbf-1(ok91) fbf-2(q704) (21), LGIII: unc-32(e189) glp-1(oz112 gf) (27), LGIV: fem-3(q20 ts, gf) (35). FOG-3::FLAG was carried on the qIs159 transgene (23). Strains were maintained at 20 C as described (36), unless noted. Animals of genotype fog-3(q520); fbf-1 fbf-2 and fog-3(q520)/+; fbf-1 fbf-2 were distinguished from each other with marked balancers and a restriction site difference between fog-3(+) and fog-3(q520) (details available upon request).
The fog-3(tm4376) mutant has a fully penetrant Fog phenotype; its molecular lesion is a two base pair insertion and 364 base pair deletion, starting before exon 1 and ending in exon 3 (positions 10,016,746 to 10,017,107). Brood sizes for fog-3(q520)/+ and fog-3(tm4376)/+ heterozygotes were determined by standard methods (e.g. 15).
Germ Cell Counts
Germ cell counts in Figure 1b and Supplementary Table 2 were done at mid-L4 when vulvas displayed a “Christmas tree” shape (37). Extruded gonads were stained with Hoechst or DAPI to visualize nuclei; germ cells were counted on all focal planes by marking individual nuclei with the point measurement tool in Openlab 5.5.2 software (PerkinElmer). For fog-3; fbf-1 fbf-2 and fog-3/+; fbf-1 fbf-2 comparisons, germ cells were counted and subsequently genotyped for fog-3 by PCR of DNA prepared from the individual scored animal followed by Sac I digestion. Confidence intervals and p-values from a two-tailed t-test were generated in Numbers ’09 (Apple). In addition, p-values were generated from a Mann-Whitney test with VassarStats (http://faculty.vassar.edu/lowry/VassarStats.html).
Real-Time Quantitative PCR
Real-time qPCR was performed as previously described (23, 25).
Microscopy
Nomarski and widefield images were obtained on a Zeiss Axioskop with a Hamamatsu Orca camera using Openlab 5.5.2 software (PerkinElmer). Confocal images were acquired on a Zeiss LSM510 confocal microscope. For FOG-3::FLAG quantitation, samples were processed concurrently using the same reagents, and images were taken with the same settings. Gain was set for little or no saturation. Pixel intensity was measured in a 6x6 micron square centered within each of three germline regions, as depicted in the diagrams accompanying the images. All three regions, the distal mitotic zone (MZ), the proximal MZ and the distal pachytene region, were quantified in the same germlines. ImageJ was used for quantitation, and the data were copied into Numbers ’09 (Apple) and averaged. Publication images were processed in Adobe Photoshop. (Images used for comparisons were processed identically.) Immunohistochemistry and EdU labeling were done as previously described (23, 25).
Mosaic analysis
Genetic mosaics were scored in a fog-3 null mutant strain carrying an extrachromosomal array that harbored wild-type fog-3 and an ubiquitously expressed GFP marker (38). Loss of the array from the germline progenitor was confirmed by crossing mosaic females with wild-type males and observing that no cross progeny carried the array.
Supplementary Material
FOG-3 functions in the germline. The newly fertilized zygote follows an invariant embryonic cell lineage to generate somatic founder cells (AB, MS, E, C, D) and a germline founder cell (P4). Tissues made by each founder cell are shown as are results of the genetic mosaics. Animals were fertile if FOG-3 was able to specify sperm; animals were sterile if FOG-3 was not able to specify sperm and only oocytes were made. For example, two animals were found that had lost the array in AB, a somatic founder that generates mostly hypodermis and neurons; because both AB mosaics were fertile, FOG-3 is not required in AB descendants to specify the sperm fate. No mosaics were found that lost the array in D, which is a minor somatic founder that generates 20 body wall muscle cells.
Control to show that FOG-3::FLAG is expressed similarly in the mitotic zones of early L4 germlines destined for production of different gametes in adults. (a-d) Gonads dissected from early L4 hermaphrodites. Widefield images were taken at same magnification with the same settings; scale bar in (a) represents 50 μM. In each panel, gonad is outlined, empty arrowhead marks distal end and germline regions are indicated: mitotic zone (MZ), transition zone (TZ) and pachytene region (PR). (a,b) Early L4 fog-3; qIs159(FOG-3::FLAG) germlines are spermatogenic but these germlines will switch to oogenesis in adults. (c,d) Early L4 fog-3; fem-3(gf); qIs159(FOG-3::FLAG) germlines are spermatogenic and will continue spermatogenesis in adults and never switch to oogenesis. (e) Diagrams of gonads in a–b (above) or c–d (below) with squares depicting quantitated regions. (f) ImageJ quantitation of FOG-3::FLAG staining.
Brood sizes of fog-3 heterozygotes do not differ from those of wild type. p-values were generated using a two-tailed t-test.
Removal of FOG-1 and FOG-3 have opposite effects on germline proliferation in fbf-1 fbf-2 double mutants. p-values were generated using a Mann-Whitney test.
Removal of FOG-3 and FBF does not abolish germline tumors in gld-1 gld-2 double mutants. Therefore the ability of FOG-3 to promote tumor formation is context dependent.
Acknowledgments
We thank the Japanese consortium for generation of fog-3(tm4376) and the CGC for strains. We also thank members of the Kimble lab for helpful discussions, Laura Vanderploeg for help with figures and Anne Helsley for help preparing the manuscript. J.J.S. was supported by NIH Training Grant 5T32GM007215 in Molecular Biosciences. J.K. is supported by NIH R01GM069454. J.V. is supported by NIH 5F32GM095036. J.K. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Jia S, Meng A. Tob genes in development and homeostasis. Dev Dyn. 2007;236:913–921. doi: 10.1002/dvdy.21092. [DOI] [PubMed] [Google Scholar]
- 2.Mauxion F, Chen CY, Seraphin B, Shyu AB. BTG/TOB factors impact deadenylases. Trends Biochem Sci. 2009;34:640–647. doi: 10.1016/j.tibs.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winkler GS. The mammalian anti-proliferative BTG/Tob protein family. J Cell Physiol. 2010;222:66–72. doi: 10.1002/jcp.21919. [DOI] [PubMed] [Google Scholar]
- 4.Yoneda M, Suzuki T, Nakamura T, Ajima R, Yoshida Y, Kakuta S, et al. Deficiency of antiproliferative family protein Ana correlates with development of lung adenocarcinoma. Cancer science. 2008;100:225–232. doi: 10.1111/j.1349-7006.2008.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida Y, Nakamura T, Komoda M, Satoh H, Suzuki T, Tsuzuku JK, et al. Mice lacking a transcriptional corepressor Tob are predisposed to cancer. Genes Dev. 2003;17:1201–1206. doi: 10.1101/gad.1088003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farioli-Vecchioli S, Tanori M, Micheli L, Mancuso M, Leonardi L, Saran A, et al. Inhibition of medulloblastoma tumorigenesis by the antiproliferative and pro-differentiative gene PC3. FASEB J. 2007;21:2215–2225. doi: 10.1096/fj.06-7548com. [DOI] [PubMed] [Google Scholar]
- 7.Yanagie H, Tanabe T, Sumimoto H, Sugiyama H, Matsuda S, Nonaka Y, et al. Tumor growth suppression by adenovirus-mediated introduction of a cell-growth-suppressing gene tob in a pancreatic cancer model. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2009;63:275–286. doi: 10.1016/j.biopha.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Ficazzola MA, Fraiman M, Gitlin J, Woo K, Melamed J, Rubin MA, et al. Antiproliferative B cell translocation gene 2 protein is down-regulated post-transcriptionally as an early event in prostate carcinogenesis. Carcinogenesis. 2001;22:1271–1279. doi: 10.1093/carcin/22.8.1271. [DOI] [PubMed] [Google Scholar]
- 9.Ito Y, Suzuki T, Yoshida H, Tomoda C, Uruno T, Takamura Y, et al. Phosphorylation and inactivation of Tob contributes to the progression of papillary carcinoma of the thyroid. Cancer letters. 2005;220:237–242. doi: 10.1016/j.canlet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Iwanaga K, Sueoka N, Sato A, Sakuragi T, Sakao Y, Tominaga M, et al. Alteration of expression or phosphorylation status of tob, a novel tumor suppressor gene product, is an early event in lung cancer. Cancer letters. 2003;202:71–79. doi: 10.1016/j.canlet.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Kawakubo H, Brachtel E, Hayashida T, Yeo G, Kish J, Muzikansky A, et al. Loss of B-cell translocation gene-2 in estrogen receptor-positive breast carcinoma is associated with tumor grade and overexpression of cyclin d1 protein. Cancer Res. 2006;66:7075–7082. doi: 10.1158/0008-5472.CAN-06-0379. [DOI] [PubMed] [Google Scholar]
- 12.Majid S, Dar AA, Ahmad AE, Hirata H, Kawakami K, Shahryari V, et al. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009;30:662–670. doi: 10.1093/carcin/bgp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Struckmann K, Schraml P, Simon R, Elmenhorst K, Mirlacher M, Kononen J, et al. Impaired expression of the cell cycle regulator BTG2 is common in clear cell renal cell carcinoma. Cancer Res. 2004;64:1632–1638. doi: 10.1158/0008-5472.can-03-1687. [DOI] [PubMed] [Google Scholar]
- 14.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 15.Ellis RE, Kimble J. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics. 1995;139:561–577. doi: 10.1093/genetics/139.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen P-J, Singal A, Kimble J, Ellis RE. A novel member of the Tob family of proteins controls sexual fate in Caenorhabditis elegans germ cells. Dev Biol. 2000;217:77–90. doi: 10.1006/dbio.1999.9521. [DOI] [PubMed] [Google Scholar]
- 17.Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- 18.Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, et al. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 19.Thompson BE, Bernstein DS, Bachorik JL, Petcherski AG, Wickens M, Kimble J. Dose-dependent control of proliferation and sperm specification by FOG-1/CPEB. Development. 2005;132:3471–3481. doi: 10.1242/dev.01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kershner AM, Kimble J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc Natl Acad Sci USA. 2010;107:3936–3941. doi: 10.1073/pnas.1000495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, et al. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 22.Lamont LB, Kimble J. Developmental expression of FOG-1/CPEB protein and its control in the Caenorhabditis elegans hermaphrodite germ line. Dev Dyn. 2007;236:871–879. doi: 10.1002/dvdy.21081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee M-H, Kim KW, Morgan CT, Morgan DE, Kimble J. Phosphorylation state of a Tob/BTG protein, FOG-3, regulates initiation and maintenance of the Caenorhabditis elegans sperm fate program. Proc Natl Acad Sci USA. 2011;108:9125–9130. doi: 10.1073/pnas.1106027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamont LB, Crittenden SL, Bernstein D, Wickens M, Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Crittenden SL, Leonhard KA, Byrd DT, Kimble J. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol Biol Cell. 2006;17:3051–3061. doi: 10.1091/mbc.E06-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cinquin O, Crittenden SL, Morgan DE, Kimble J. Progression from a stem cell-like state to early differentiation in the C. elegans germ line. Proc Natl Acad Sci USA. 2010;107:2048–2053. doi: 10.1073/pnas.0912704107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berry LW, Westlund B, Schedl T. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development. 1997;124:925–936. doi: 10.1242/dev.124.4.925. [DOI] [PubMed] [Google Scholar]
- 28.Kadyk LC, Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- 29.Rodier A, Rochard P, Berthet C, Rouault JP, Casas F, Daury L, et al. Identification of functional domains involved in BTG1 cell localization. Oncogene. 2001;20:2691–2703. doi: 10.1038/sj.onc.1204398. [DOI] [PubMed] [Google Scholar]
- 30.Kawamura-Tsuzuku J, Suzuki T, Yoshida Y, Yamamoto T. Nuclear localization of Tob is important for regulation of its antiproliferative activity. Oncogene. 2004;23:6630–6638. doi: 10.1038/sj.onc.1207890. [DOI] [PubMed] [Google Scholar]
- 31.Byrd DT, Kimble J. Scratching the niche that controls Caenorhabditis elegans germline stem cells. Semin Cell Dev Biol. 2009;20:1107–1113. doi: 10.1016/j.semcdb.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Busson M, Carazo A, Seyer P, Grandemange S, Casas F, Pessemesse L, et al. Coactivation of nuclear receptors and myogenic factors induces the major BTG1 influence on muscle differentiation. Oncogene. 2005;24:1698–1710. doi: 10.1038/sj.onc.1208373. [DOI] [PubMed] [Google Scholar]
- 33.Rodier A, Marchal-Victorion S, Rochard P, Casas F, Cassar-Malek I, Rouault JP, et al. BTG1: a triiodothyronine target involved in the myogenic influence of the hormone. Exp Cell Res. 1999;249:337–348. doi: 10.1006/excr.1999.4486. [DOI] [PubMed] [Google Scholar]
- 34.el-Ghissassi F, Valsesia-Wittmann S, Falette N, Duriez C, Walden PD, Puisieux A. BTG2(TIS21/PC3) induces neuronal differentiation and prevents apoptosis of terminally differentiated PC12 cells. Oncogene. 2002;21:6772–6778. doi: 10.1038/sj.onc.1205888. [DOI] [PubMed] [Google Scholar]
- 35.Barton MK, Schedl TB, Kimble J. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics. 1987;115:107–119. doi: 10.1093/genetics/115.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Podbilewicz B. WormBook. The C. elegans Research Community. WormBook; 2006. Cell fusion. [Google Scholar]
- 38.Thompson BE, Lamont LB, Kimble J. Germ-line induction of the Caenorhabditis elegans vulva. Proc Natl Acad Sci USA. 2006;103:620–625. doi: 10.1073/pnas.0510264103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guéhenneux F, Duret L, Callanan MB, Bouhas R, Hayette S, Berthet C, et al. Cloning of the mouse BTG3 gene and definition of a new gene family (the BTG family) involved in the negative control of the cell cycle. Leukemia. 1997;11:370–375. doi: 10.1038/sj.leu.2400599. [DOI] [PubMed] [Google Scholar]
- 40.Albrecht M, Lengauer T. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 2004;569:18–26. doi: 10.1016/j.febslet.2004.03.126. [DOI] [PubMed] [Google Scholar]
- 41.Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FOG-3 functions in the germline. The newly fertilized zygote follows an invariant embryonic cell lineage to generate somatic founder cells (AB, MS, E, C, D) and a germline founder cell (P4). Tissues made by each founder cell are shown as are results of the genetic mosaics. Animals were fertile if FOG-3 was able to specify sperm; animals were sterile if FOG-3 was not able to specify sperm and only oocytes were made. For example, two animals were found that had lost the array in AB, a somatic founder that generates mostly hypodermis and neurons; because both AB mosaics were fertile, FOG-3 is not required in AB descendants to specify the sperm fate. No mosaics were found that lost the array in D, which is a minor somatic founder that generates 20 body wall muscle cells.
Control to show that FOG-3::FLAG is expressed similarly in the mitotic zones of early L4 germlines destined for production of different gametes in adults. (a-d) Gonads dissected from early L4 hermaphrodites. Widefield images were taken at same magnification with the same settings; scale bar in (a) represents 50 μM. In each panel, gonad is outlined, empty arrowhead marks distal end and germline regions are indicated: mitotic zone (MZ), transition zone (TZ) and pachytene region (PR). (a,b) Early L4 fog-3; qIs159(FOG-3::FLAG) germlines are spermatogenic but these germlines will switch to oogenesis in adults. (c,d) Early L4 fog-3; fem-3(gf); qIs159(FOG-3::FLAG) germlines are spermatogenic and will continue spermatogenesis in adults and never switch to oogenesis. (e) Diagrams of gonads in a–b (above) or c–d (below) with squares depicting quantitated regions. (f) ImageJ quantitation of FOG-3::FLAG staining.
Brood sizes of fog-3 heterozygotes do not differ from those of wild type. p-values were generated using a two-tailed t-test.
Removal of FOG-1 and FOG-3 have opposite effects on germline proliferation in fbf-1 fbf-2 double mutants. p-values were generated using a Mann-Whitney test.
Removal of FOG-3 and FBF does not abolish germline tumors in gld-1 gld-2 double mutants. Therefore the ability of FOG-3 to promote tumor formation is context dependent.