Dissecting the transcriptional circuitry in embryonic stem cells is important to understanding self-renewal and pluripotency in these cells. Percharde et al. explore the idfentity of key coregulators that potentiate transcription in embryonic stem cells and identify Ncoa3 as a transcriptional cofactor that interacts with Esrrb and is required for the induction and maintenance of pluripotency. They also describe interactions of Ncoa3 with the Oct4–Sox2–Nanog circuitry and the role of Ncoa3 in linking Esrrb to RNA polymerase II complexes.
Keywords: Ncoa3, Esrrb, embryonic stem cells, coactivator, pluripotency, reprogramming, SRC-3
Abstract
Embryonic stem cell (ESC) pluripotency depends on a well-characterized gene regulatory network centered on Oct4, Sox2, and Nanog. In contrast, little is known about the identity of the key coregulators and the mechanisms by which they may potentiate transcription in ESCs. Alongside core transcription factors, the orphan nuclear receptor Esrrb (estrogen-related receptor β) is vital for the maintenance of ESC identity and furthermore is uniquely associated with the basal transcription machinery. Here, we show that Ncoa3, an essential coactivator, is required to mediate Esrrb function in ESCs. Ncoa3 interacts with Esrrb via its ligand-binding domain and bridges Esrrb to RNA polymerase II complexes. Functionally, Ncoa3 is critical for both the induction and maintenance of pluripotency. Through chromatin immunoprecipitation (ChIP) sequencing and microarray experiments, we further demonstrate that Ncoa3 shares overlapping gene regulatory functions with Esrrb and cooperates genome-wide with the Oct4–Sox2–Nanog circuitry at active enhancers to up-regulate genes involved in self-renewal and pluripotency. We propose an integrated model of transcriptional and coactivator control, mediated by Ncoa3, for the maintenance of ESC self-renewal and somatic cell reprogramming.
Embryonic stem cells (ESCs) are derived from the inner cell mass of early embryos and can be propagated for extended periods of time in culture while retaining their pluripotency; i.e., the capacity for multilineage specification and differentiation (Smith 2001). Coordinating with essential components of the LIF and BMP signaling pathways (Niwa et al. 1998; Matsuda et al. 1999; Ying et al. 2003), intrinsic transcription factors are crucial for specifying the undifferentiated state of mouse ESCs. Oct4 is essential for establishing and maintaining pluripotency (Nichols et al. 1998; Niwa et al. 2000; Boiani et al. 2005) and interacts with Sox2 (Avilion et al. 2003; Chew et al. 2005) in ESCs, where both factors cotarget multiple genes through cooperative binding at Oct–Sox sequence elements (Boyer et al. 2005; Loh et al. 2006). Moreover, Oct4 and Sox2, along with Klf4 and cMyc, can mediate reprogramming of mouse embryonic fibroblasts (MEFs), generating ESC-like induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka 2006). A third well-characterized core pluripotency factor is Nanog, which is notably capable of promoting ESC self-renewal even in the absence of LIF (Chambers et al. 2003; Mitsui et al. 2003).
Recent work has begun to identify new components of the ESC core transcriptional network. Along with Oct4, Sox2, and Nanog (OSN), these factors participate in auto- and cross-regulation to activate each other's expression as well as downstream self-renewal regulators while simultaneously repressing genes that promote differentiation (Chambers and Tomlinson 2009). Perturbation of these factors collapses the self-renewal circuitry and triggers specific or mixed-lineage differentiation (Ivanova et al. 2006; Loh et al. 2006). Whole-genome mapping of DNA-binding sites further revealed that the OSN triad colocalizes with a variable yet overlapping set of pluripotency-associated transcription factors at many promoters and enhancers (Chen et al. 2008). Clusters with a relatively high number of factors correlate with expression of nearby genes (Kim et al. 2008). It remains less clear, however, how this well-established network functionally interacts with basal transcription machinery complexes to activate transcription and, most importantly, which factors mediate such interactions. While the list of ESC-specific transcription factors is rapidly expanding, relatively little is known about the identity of essential coregulators (Fong et al. 2011) and the mechanisms by which they may potentiate transcription.
To gain greater insight into how transcription factors function mechanistically in ESCs, we focused here on estrogen-related receptor β (Esrrb). This orphan nuclear receptor binds many target sites that are co-occupied by OSN and Klf4 in ESCs and up-regulates the expression of these factors themselves (Chen et al. 2008). Levels of Esrrb decline rapidly upon differentiation (Xie et al. 2009), and its depletion results in a loss of ESC characteristics (Ivanova et al. 2006; Loh et al. 2006). Overexpression of Esrrb inhibits lineage commitment toward the meso- and neuro-ectodermal paths and allows robust self-renewal in the absence of LIF (Zhang et al. 2008). In addition, Esrrb can substitute for Klf4 in somatic cell reprogramming (Feng et al. 2009), further highlighting a pivotal role in both maintaining and inducing pluripotency. In somatic cells, the activity of estrogen-related receptors (ERRs) is known to be regulated through the recruitment of coactivators to the AF-2 portion of their ligand-binding domain (LBD) (Giguere 2008); for example, PGC-1 in metabolic tissues (Lin et al. 2005; Charest-Marcotte et al. 2010). These coactivators in turn act as scaffolds for other protein complexes required to bring about transcription (Rosenfeld et al. 2006). Although well studied within the context of adult somatic systems, little is known about the mechanisms by which Esrrb function is conferred in ESCs.
In this study, we show that Esrrb transcriptional and self-renewal activity is absolutely dependent on protein–protein interactions mediated via its LBD/AF-2 domain. Despite this, we found that the coactivators PGC-1α and PGC-1β commonly required for ERR activity are not expressed in ESCs and instead identify Ncoa3 (also known as SRC-3/AlB1) as an essential coactivator of Esrrb. We show that Ncoa3 expression positively correlates with the undifferentiated ESC state and itself is required for both the induction and maintenance of pluripotency. Ncoa3 binds to Esrrb specifically via its AF-2 region in ESCs and thus is corecruited to Esrrb, Klf4, Nanog, and Sox2 enhancer regions as a critical step to trigger Esrrb-dependent gene activation. Through chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) and transcriptome analysis in Ncoa3 knockdown ESCs, we further establish that Ncoa3 and Esrrb have overlapping gene regulatory functions and cooperate genome-wide with the Oct4–Sox2–Nanog circuitry at active enhancers to up-regulate genes involved in self-renewal and pluripotency. Finally, we demonstrate mechanistically that Ncoa3's presence links Esrrb to RNA polymerase II (RNApol2) complexes, identifying a crucial role for the Ncoa3–Esrrb partnership in the ESC core transcriptional network and shedding new light on how pluripotency transcription factors may be bridged to the general transcription machinery to activate transcription.
Results
The LBD and AF-2 regions are essential for Esrrb transcriptional activity in ESCs
How the assembly of transcription factor clusters leads to active transcription and which essential mediating components contribute to this process are yet to be fully investigated in ESCs. Combining mutagenesis and luciferase reporter assays, we here asked whether Esrrb activity is conferred in ESCs through specific protein recruitment. Flag-tagged Esrrb mutants lacking either the entire LBD (ΔLBD) or N-terminal AF-1 (ΔAF-1) domain—regions key for mediating nuclear receptor protein–protein interactions—were generated (Supplemental Fig. S1A), and their expression was verified alongside wild-type Esrrb in ESCs (Supplemental Fig. S1B). To test these deletion mutants, we initially took advantage of previously described constructs that carry luciferase downstream from a minimal Oct4 promoter, together with ERR response element (ERRE)-containing regulatory fragments found at the Esrrb, Klf4, or Sox2 loci (Fig. 1A; Feng et al. 2009), and an additional Esrrb-dependent Nanog-Luc reporter (Rodda et al. 2005; van den Berg et al. 2008). These Enhancer-Luc vectors all show a loss of luciferase activity following knockdown of endogenous Esrrb in ESCs, an activity readily regained by coexpressing RNAi-resistant wild-type Esrrb cDNA (Supplemental Fig. S1C). As opposed to wild-type control, we found that the ΔLBD mutant failed to restore transcription from Esrrb, Klf4, Nanog, and Sox2 reporters. In contrast, deleting the AF-1 domain had no such suppressive effect on Esrrb activity (Supplemental Fig. S1C). Unlike other nuclear receptors, ERR activity is not thought to be ligand-dependent but nevertheless involves the recruitment of essential coactivators to the AF-2 region, a domain of 12 helices forming a unique protein-binding surface within the LBD (Wurtz et al. 1996). Accordingly, we repeated these experiments using Esrrb point mutants in either helix 3 (AF-2 mut1) or helix 12 (AF-2 mut2)—two analogous mutations previously found to inactivate the AF-2 region of the estrogen receptor (Danielian et al. 1992; Henttu et al. 1997)—along with a DNA-binding domain (DBD) mutant unable to bind DNA generated as a control (Fig. 1B). Despite equal expression to wild-type Esrrb in ESCs (Fig. 1C), all three point mutants were similarly unable to rescue the luciferase activity of Esrrb, Klf4, Nanog, and Sox2 reporters (Fig. 1D), unequivocally demonstrating that Esrrb in ESCs requires an active LBD/AF-2 domain to trigger transcription.
Figure 1.
The AF-2 region is essential for Esrrb activity in ESCs. (A) Schematic of Esrrb, Klf4, and Sox2 reporters used in luciferase assays. Enhancer fragments containing ERREs from the indicated genes are inserted downstream from luciferase (Feng et al. 2009). An additional Nanog-Luc reporter was also used (Rodda et al. 2005). (B) Diagram indicating Flag-tagged wild-type (Wt) and Esrrb point mutants. (DBD mut) C120G/C123G; (AF-2 mut1) K259A; (AF-2 mut2) L424A/F425A. (C) Expression of Flag-tagged wild-type Esrrb or point mutants in ESCs. (D) Luciferase assays for Esrrb-bound enhancers. ESCs were transfected with the indicated reporter plus either control (shScrambled) or shRNA to endogenous Esrrb (shEsrrb) and with or without RNAi-resistant Esrrb rescue constructs as depicted in B. Data are the mean ± SEM of three transfections. Three experiments were performed with similar results. (E) Levels of CAG-Flag-Esrrb construct expression in ESCs following stable transfection with wild-type, AF-2 mut1, or AF-2 mut2 Esrrb forms. (F,G) Ability of wild-type or Esrrb point mutants to confer LIF independence in stably transfected ESCs. Each cell line was plated at low density in normal ESC medium and, the following day, cultured in medium minus LIF for 5 d. Colonies were fixed and stained for AP and scored as undifferentiated, mixed, or differentiated. Data are the mean ± SEM of three independent experiments. Student's t-test was used to compare numbers of undifferentiated colonies in wild-type and either AF-2 mutant ESCs or empty vector control; (*) P-value < 0.05.
AF-2 region inactivation results in a loss of self-renewal in ESCs
Overexpressing Esrrb is sufficient to allow robust self-renewal in ESCs, negating the need for LIF to prevent differentiation (Zhang et al. 2008). To link the AF-2 dependence of Esrrb transcriptional activity with its self-renewal function, we next generated stable ESC lines that constitutively express Flag-tagged wild-type, AF-2 mut1, or AF-2 mut2 Esrrb (Fig. 1E). Cells were plated at low density and cultured for 5 d in the absence of LIF, then stained for alkaline phosphatase (AP) to score the number of undifferentiated, mixed, and differentiated colonies formed under these conditions (Supplemental Fig. S1D). Strikingly, and in contrast to overexpressing wild-type Esrrb cells, we observed a near-complete absence of compact, AP-positive colonies in both AF-2 mut1 and AF-2 mut2 ESC cultures (Fig. 1F,G), indicating a loss in the ability of Esrrb point mutants to confer LIF independence. To support this, we also examined the effect of treating ESCs with an ERR-specific AF-2 antagonist, diethylstilbestrol (DES). As previously reported, this compound interacts with all three ERR isoforms to block coactivator binding and thereby prevent their transcriptional activity (Supplemental Fig. S1E; Tremblay et al. 2001; Greschik et al. 2004). We demonstrated that DES treatment consistently inhibited transcription from Esrrb-dependent reporters in ESCs (Supplemental Fig. S1F) and significantly impaired the self-renewal ability of these cells in a dose-dependent fashion (Supplemental Fig. S1G,H). Taken together, these findings reveal the importance of the AF-2 region to potentiate Esrrb function and imply a crucial role for a cognate Esrrb coactivator in ESCs.
Ncoa3 is highly expressed in undifferentiated ESCs
In somatic cells, the best-characterized ERR coactivators belong to the PGC-1 family. PGC-1α and PGC-1β both potentiate ERR activity in various adult cell types to activate genes involved in processes such as cellular energy metabolism and homeostasis (Huss et al. 2002; Kamei et al. 2003; Charest-Marcotte et al. 2010). Most recently, a third member of the PGC-1 family, PRC, was identified as a regulator of NRF-1 and mitochondrial function (Vercauteren et al. 2009). Among all three PGC-1 members, only PRC was expressed in ESCs (Supplemental Fig. S2A), and thus its role as a putative Esrrb coactivator was further investigated. Knockdown of PRC in ESCs did not, however, compromise Esrrb transcriptional activity or ESC self-renewal, as seen upon depletion of Esrrb itself (Supplemental Fig. S2B–E). We next turned to the SRC/p160 family that comprises Ncoa1, Ncoa2, and Ncoa3, three alternative coactivator proteins also capable of interacting with the ERR family in mammalian cell models (Hong et al. 1999; Zhang and Teng 2000; van den Berg et al. 2010). Ncoa3 notably emerged as an interesting candidate, being highly expressed in self-renewing ESC cultures, in contrast to Ncoa1 and Ncoa2 (Fig. 2A), and promptly down-regulated alongside Esrrb and other pluripotency factors during differentiation, as assessed upon addition of retinoic acid and/or LIF withdrawal (Fig. 2B,C). A sharp decline in Ncoa3 transcript levels was also confirmed upon embryoid body (EB)-mediated differentiation, further pointing to a close association between Ncoa3 expression and the undifferentiated ESC state (Fig. 2D).
Figure 2.
Ncoa3 acts as Esrrb coactivator in ESCs. (A) Expression profiling of Ncoa1, Ncoa2, and Ncoa3 transcripts in ESCs as assessed by qRT–PCR and normalized to two housekeeping genes. Expression levels in immortalized brown adipose tissue (IMBAT) cells are shown as a control. Data are the mean ± SEM of three biological replicates. (B) Immunofluorescence staining of ESCs maintained in proliferative conditions or induced to differentiate via addition of 1 μM retinoic acid (atRA) and removal of LIF for 96 h. Bars, 50 μm. (C) Western blotting showing Ncoa3 down-regulation along with Esrrb, Nanog, and Oct4 upon LIF withdrawal. Tubulin is shown as a loading control. (D) Relative expression levels of Ncoa3 and other pluripotency factors during EB-mediated differentiation. Data are normalized to housekeeping genes and expressed relative to EB day 0 as the mean ± SEM of three independent experiments. (E) Co-IP experiments performed in COS-1 cells cotransfected with Flag-Esrrb and Ncoa3. (F) Interaction of endogenous Esrrb and Ncoa3 proteins in ESC extracts following 24 h of treatment with ethanol or 12.5 μM DES. At least two experiments were performed with similar results. (G) Loss of ESC-specific luciferase reporter activity 48 h after depletion of endogenous Esrrb or Ncoa3 in ESCs. Data are the mean ± SEM of three transfections. Three experiments were performed with similar results.
Ncoa3 acts as an Esrrb coactivator in pluripotent ESCs
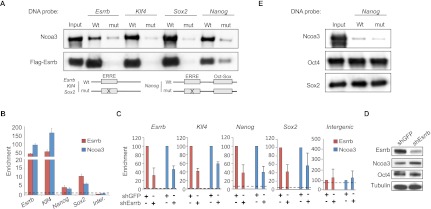
To establish a functional link between Ncoa3 and Esrrb, we first checked whether these two protein factors could indeed interact. Coimmunoprecipitation (co-IP) experiments were performed in COS-1 cells coexpressing Ncoa3 and Flag-Esrrb, readily demonstrating an Ncoa3–Esrrb interaction (Fig. 2E). As expected, this interaction was mediated via the Esrrb AF-2 region, as AF-2 mutants failed to bind to Ncoa3 (Supplemental Fig. S2F). Co-IPs were next performed using ESC extracts and, importantly, confirmed that endogenous Ncoa3 and Esrrb proteins interact in ESCs (Supplemental Fig. S2G). Moreover, Ncoa3 binding to Esrrb was abrogated in the presence of DES (Fig. 2F), a result that reflects the decreased self-renewal ability of DES-treated ESCs (Supplemental Fig. S1). To test the requirement for Ncoa3 on Esrrb-dependent enhancers, ESCs were cotransfected with a pool of siRNAs targeting Ncoa3 together with Esrrb, Klf4, Nanog, and Sox2 luciferase vectors. Ncoa3 loss closely phenocopied the suppressive effect of Esrrb depletion itself (Fig. 2G; Supplemental Fig. S2H). Furthermore, restoring Esrrb expression in depleted ESCs failed to rescue luciferase reporter activity in the absence of Ncoa3 (Supplemental Fig. S2I), indicating that Ncoa3 is indeed essential for the activation of these ESC-specific enhancers by Esrrb. We next demonstrated that Ncoa3 is recruited by Esrrb to genomic fragments containing ERREs using DNA pull-down experiments with biotinylated wild-type or ERRE mutated probes incubated together with Ncoa3 and Flag-Esrrb (Fig. 3A). These proteins both readily purified with Esrrb, Klf4, Sox2, and Nanog DNA fragments. Furthermore, point mutations in ERRE motifs that abrogated Esrrb binding also reduced Ncoa3 detection (Fig. 3A), confirming that Ncoa3 associates via Esrrb at ERREs in the vicinity of target genes. Similarly, Ncoa3 and Esrrb were corecruited to endogenous Esrrb, Klf4, Nanog, and Sox2 target genes in ESCs, as assessed by ChIP and quantitative PCR (qPCR) (Fig. 3B). Ncoa3 recruitment was also verified here to be Esrrb-dependent, as transient depletion of Esrrb protein reduced Ncoa3 enrichment at these sites (Fig. 3C). Importantly, both Ncoa3 and Oct4 protein levels remained unaltered at this time point (Fig. 3D), excluding the possibility that the loss of Ncoa3 binding is due to ESC differentiation. Given the co-occupancy of many ERRE-containing enhancers with the core pluripotency factors (Chen et al. 2008), we next examined the Oct4 and Nanog dependency of Ncoa3–Esrrb recruitment. As previously reported for the Nanog locus (van den Berg et al. 2008), removing Oct4 protein prevented the detection of Esrrb at all four ESC candidate loci, and this was strictly mirrored by a loss of Ncoa3 recruitment (Supplemental Fig. S3A,B), a trend also observed to a lesser extent upon conditional Nanog deletion (Supplemental Fig. S3C,D). However, while Ncoa3 and Esrrb readily interact (Fig. 2; Supplemental Fig. S2), we could find no evidence for an interaction between Ncoa3 and Oct4/Nanog themselves (Supplemental Fig. S3E–H), thus highlighting the specificity of the Ncoa3–Esrrb association at these sites. In line with this, DNA pull-down assays further demonstrated that although Oct4 and Sox2 bind to an Oct–Sox site also contained within the Nanog probe, they were unable alone to recruit Ncoa3 (Fig. 3E). Collectively, these results identify Ncoa3 as an important Esrrb coactivator in ESCs and point toward a critical role for the Ncoa3–Esrrb partnership in regulating pluripotency.
Figure 3.
Ncoa3 is recruited via Esrrb to ERREs at target loci. (A) DNA pull-down assays using previously described wild-type (Wt) or ERRE mutated sequences from Esrrb, Klf4, Sox2 (Feng et al. 2009), or Nanog (van den Berg et al. 2008). Biotinylated probes (40–50 base pairs [bp]) were incubated with extracts from COS-1 cells transfected with Flag-Esrrb and Ncoa3 and recovered on streptavidin beads, and DNA-associated proteins were visualized by Western blotting. (B) Esrrb and Ncoa3 enrichment levels at Esrrb, Klf4, Nanog, and Sox2 ERREs and an intergenic (Inter.) control region in ESCs as assessed by ChIP and qPCR and expressed relative to input. Data are the mean ± SEM of three biological replicates. (C) Ncoa3 and Esrrb enrichment levels at candidate loci following Esrrb depletion in ESCs, expressed relative to input. Enrichment for shGFP transfected cells is set at 100% in each individual experiment. Data are the mean ± SD of at least two independent experiments. Dotted lines in B and C indicate background enrichment by control IgGs (Santa Cruz Biotechnology). (D) Western blotting showing specific Esrrb protein depletion 48 h after transfection with shEsrrb. Note that Ncoa3 and Oct4 levels are unchanged at this time. (E) DNA pull-down assays using Nanog wild-type or ERRE mutated probe, which also contains a neighboring Oct–Sox site (van den Berg et al. 2008), and cell extracts from COS-1 transfected with Ncoa3, Oct4, and Sox2.
Ncoa3 contributes to the maintenance of pluripotency in ESCs
To explore the functional requirement for Ncoa3 in ESCs, we next knocked down Ncoa3 using two independent puromycin-selectable shRNA vectors. The extent of the knockdown was validated by qRT–PCR (Fig. 4A) and Western blot (Fig. 4B) and, strikingly, resulted in a clear and rapid loss of ESC characteristics. By day 4 post-selection, colonies displayed a flattened or spread morphology and stained negative for AP (Fig. 4C), and no stable Ncoa3 knockdown ESC clones could be established. Ncoa3 depletion was promptly accompanied by a decrease in self-renewal gene expression (Fig. 4D) and concomitant derepression of lineage-associated markers (Fig. 4E), supporting an effect of Ncoa3 on differentiation. Conversely, we asked whether, through the potentiation of Esrrb activity, elevated levels of Ncoa3 could enhance resistance to differentiation in ESCs. For this, we established a series of stable ESC lines that overexpress either Esrrb (ESCEsrrb), Ncoa3 (ESCNcoa3), or both factors together (ESCEsrrb+Ncoa3) alongside control cells (ESCcontrol) through two sequential rounds of transfection and selection, as depicted in Figure 4F. To compare the behavior of these lines, cells were plated at low density and cultured minus LIF for 5 d as previously performed (Fig. 1). The ESCEsrrb+Ncoa3 line showed a significant increase in the number of undifferentiated colonies relative to ESCEsrrb cells (Fig. 4G), demonstrating the additive effect conferred by overexpressing Esrrb and Ncoa3 together. Interestingly, a consistent increase in the number of undifferentiated and mixed colonies was also observed in ESCNcoa3 as compared with control cells (Fig. 4G,H), suggesting that overexpressing Ncoa3 alone could support the self-renewal capacity of ESCs. This was confirmed in ESCs stably expressing Ncoa3 using an alternative (pPyCAGIP) episomal vector (Supplemental Fig. S4A; Chambers et al. 2003). These cells similarly exhibited lower levels of differentiation when cultured in the absence of LIF as well as under self-renewing conditions—an effect associated with increased expression of Esrrb targets Esrrb and Klf4 (Supplemental Fig. S4B; Feng et al. 2009). Following several passages, however, Ncoa3-overexpressing ESCs cultured minus LIF eventually differentiated due to the down-regulation of endogenous Esrrb (data not shown). Furthermore, these cells were capable of EB-mediated differentiation (Supplemental Fig. S4C). Collectively, these results show that Ncoa3 plays critical roles in maintaining an intact ESC state and a self-renewal phenotype but, consistent with its role as a coactivator, is insufficient alone to maintain Esrrb levels and block differentiation.
Figure 4.
Ncoa3 plays critical roles in the maintenance of ESC self-renewal. (A,B) Specific knockdown of Ncoa3 in ESCs. ESCs were transfected with two independent puromycin-selectable shRNA vectors targeting Ncoa3, and, 48 h after transfection, Ncoa3 expression was quantified by qRT–PCR (A) and Western blotting (B). (C) AP staining following Ncoa3 knockdown. ESCs were transfected with shScrambled or shNcoa3 vectors, and, 24 h later, puromycin was added. Selection was maintained for 4 d, then cells were fixed and stained for AP. (D,E) Quantitative gene expression analysis of cells 4 d post-selection for Ncoa3 depletion. Data are the mean ± SD of at least two independent experiments. (F) Generation of stable ESC lines overexpressing combinations of Esrrb and Ncoa3. E14Tg2A (E14) ESCs were first transfected with linearized CAG-Control or CAG-Esrrb vectors and selected with puromycin. Pooled clones for each stable cell line were then transfected with linearized control vector, pCDNA4/TO-GFP, or pCDNA4/TO-Ncoa3, and overexpressing cells were selected using Zeocin. (G) ESC lines generated in F were plated at low density and cultured for 5 d without LIF. Colonies were fixed and stained for AP; shown are three independent experiments. The statistical significance of the difference in undifferentiated colonies between the indicated cell lines was calculated using Student's t-test; (*) P-value < 0.05, (**) P-value < 0.01. (H) Representative colony morphology for each cell line from G.
Ncoa3 is required for somatic cell reprogramming
Interestingly, Ncoa3 was found to be up-regulated during iPSC generation alongside Esrrb and Nanog (Supplemental Fig. S5A; Mikkelsen et al. 2008), suggesting that Ncoa3 may also be important for the induction of pluripotency by reprogramming. Detection of high levels of Ncoa3 protein was confirmed in two independently derived iPSC clones (Fig. 5A) from Oct4GFP MEFs. These MEFs carry a GFP transgene downstream from the Oct4 distal enhancer, which becomes selectively activated during successful somatic cell reprogramming (Fig. 5B; Supplemental Fig. S5B; Yeom et al. 1996). To test the functional importance of Ncoa3 in this process, we specifically depleted Ncoa3 in Oct4GFP MEFs by RNAi (Fig. 5C; Supplemental Fig. S5C) prior to transduction with Oct4, Sox2, Klf4, and cMyc (OSKM) reprogramming retroviruses (Supplemental Fig. S5D). Both scrambled and Ncoa3 knockdown MEFs showed no difference in cell proliferation (Supplemental Fig. S5E) or apoptosis (Supplemental Fig. S5F) at the time of reprogramming and infected equally highly with retroviruses (Fig. 5D). In contrast, Ncoa3 depletion induced a drastic reduction in reprogramming efficiency, as judged by the significantly lower number of GFP-positive (Fig. 5E-F) and AP-positive (Fig. 5G) colonies generated. We next asked whether Ncoa3 knockdown might inhibit reprogramming simply by inducing MEF senescence, a known impediment of reprogramming. For this, experiments were repeated in MEFs lacking p53, which allows enhanced speed and efficiency of reprogramming without cMyc (Banito et al. 2009; Kawamura et al. 2009; Marion et al. 2009). p53−/− MEFs transduced with OSK, however, showed similarly striking reductions in the number of AP-positive colonies following Ncoa3 depletion (Fig. 5H), indicating that the observed reprogramming defect was not due to p53-mediated senescence. Taken together, these findings demonstrate that as well as being critical for the maintenance of ESCs, Ncoa3 is furthermore required for the induction of pluripotency upon reprogramming.
Figure 5.
Essential requirement for Ncoa3 in somatic cell reprogramming. (A) Ncoa3 protein is up-regulated in fully reprogrammed iPSC lines. Western blotting depicts two independently generated iPSC clones from two different Oct4GFP genetic backgrounds (clone 1: 129/Ola; clone 2: MF1/B6) (Yeom et al. 1996) in comparison with uninfected proliferative MEFs. (B) Immunofluorescence confirming Ncoa3 and GFP expression in iPSC clone 1. Bars, 50 μm. (C) Efficient Ncoa3 knockdown in MEFs analyzed 3 d after infection with lentiviral particles expressing shScrambled or shNcoa3 shRNA. Data are the mean ± SEM of three independent experiments. (D) Equal retroviral mCherry infection of scrambled or Ncoa3 knockdown MEFs. (E) Typical GFP-positive colonies arising from transduction of shScrambled-treated Oct4GFP MEFs with OSKM reprogramming factors. Bars, 100 μm. (F) Inhibition of reprogramming following depletion of Ncoa3, analyzed by quantifying GFP-positive colonies 12–14 d after infection with OSKM vectors. The number of colonies produced from shScrambled-infected MEFs is set as 100% for each individual experiment. Three experiments were performed using different batches of MEFs and viruses. (**) P-value < 0.01, (***) P-value < 0.001, Student's t-test. (G) AP staining of iPSC colonies 14 d after infection with OSKM reprogramming factors. (H) AP staining of iPSC colonies generated in p53−/− shScrambled or shNcoa3 MEFs using OSK reprogramming factors.
Ncoa3 participates genome-wide with Esrrb and OSN at active ESC-specific enhancers
To gain greater understanding of how Ncoa3 functions in ESCs, Ncoa3 DNA-binding sites were next mapped by ChIP-seq. We first verified that Ncoa3 (this study) and Esrrb (Chen et al. 2008) data sets showed strong overlap at Esrrb, Klf4, Nanog, and Sox2 target genes and local binding profiles (Supplemental Fig. S6A). Focusing on ESC-relevant loci, we next compared all Ncoa3–Oct4–Sox2 cobound sites with Esrrb–Oct4–Sox2 and found significant overlaps between these groups (P-value = 1.11 × 10−16) (Fig. 6A). In contrast, the insulator-binding transcription factor CTCF did not show any significant overlap (P-value = 0.84). Using a de novo motif discovery algorithm (Heng et al. 2010), we identified, as expected, a canonical ERRE motif among significantly overrepresented binding sequences found at Ncoa3 peaks (Fig. 6B; Supplemental Fig. S7). Oct–Sox elements were also detected at these sites, reflecting the frequent colocalization of Esrrb with Oct4 and Sox2. GREAT gene ontology (GO) analysis at Ncoa3–Esrrb cobound sites revealed significant enrichment for developmental processes and embryonic phenotypes (Supplemental Fig. S6B). In addition, abnormal fertility and organ size were also enriched, agreeing with published mouse knockout models for Ncoa3 and/or Esrrb (Luo et al. 1997; Mitsunaga et al. 2004; Chen et al. 2010). Taken together, these data suggest a high degree of similarity between Ncoa3 and Esrrb transcriptional networks in ESCs. To look more widely at the different types of transcription factor clusters associated with Ncoa3 peaks, we next used spatial heat map analysis (Fig. 6C). This distinguished three main classes of Ncoa3 targets based on their co-occupancies by Esrrb and/or OSN as well as Nr5a2, p300, H3K27ac, and H3K4me1. Here we confirmed that ∼40% of all Ncoa3 peaks colocalized with Esrrb at active enhancers, as denoted by the presence of p300, H3K27ac, and H3K4me1 (Fig. 6C; Heintzman et al. 2007; Creyghton et al. 2010). Sites bound by both Esrrb/Ncoa3 and OSN (class Ib) represented key self-renewal regulators that are actively transcribed in ESCs, such as Esrrb, Klf2/4/5, Zfp42, Tcfcp2l1, Oct4, and Sox2, and included GO processes involving stem cells and blastocyst development (Supplemental Table S1). In contrast, sites containing Esrrb and Ncoa3 but with reduced binding for OSN (class Ia) (Fig. 6C) were uniquely associated with developmental phenotypes involving germ cell integrity and fertility (Supplemental Table S2), including Prdm14, a recently described factor critical for germ cell lineage (Yamaji et al. 2008; Gillich et al. 2012). To closely investigate transcriptional regulation by Ncoa3 and Esrrb, microarray analysis was performed on ESCs following Ncoa3 knockdown and compared with Esrrb knockdown at the same time point (Feng et al. 2009). Notably, 62.5% of the misregulated genes in Esrrb-depleted ESCs were also misregulated in Ncoa3 knockdown ESCs (Fig. 6D), most often in the same direction (Fig. 6E), showing that gene regulation by Ncoa3 and Esrrb is significantly correlated (P-value = 3.92 × 10−49). Among them, many target genes with (class Ib) or without (class Ia) high OSN enrichment were significantly down-regulated upon Ncoa3 depletion, as validated by qRT–PCR (Supplemental Fig. S8A), confirming that Ncoa3 is strictly required for Esrrb-dependent gene activation. Interestingly, at some sites, Ncoa3 also colocalized with Nr5a2—another orphan nuclear receptor previously involved in stem cells and reprogramming (Gu et al. 2005; Heng et al. 2010)—together with (class I) and without (class II) Esrrb and/or OSN (Fig. 6C). GO analysis of class II genes, however, consisted of targets associated with extraembryonic development (Supplemental Table S3), indicating that although Nr5a2 could potentially recruit Ncoa3 to a set of genes, these targets may be less relevant in ESCs. Finally, the last class identified (class III) (Supplemental Table S4) included Ncoa3-bound sites that were not co-occupied by Esrrb, Nr5a2, or OSN (Fig. 6C), suggesting that Ncoa3 may also be recruited by other factors. While marks of active enhancers—p300, H3K27ac, and H3K4me1—were absent at the majority of these sites (class IIIa), a small subset (class IIIb) colocalized with H3K4me1 alone (Fig. 6C), potentially delineating inactive/poised Ncoa3-bound enhancers in ESCs (Creyghton et al. 2010). Interestingly, microarray analysis of class IIIb revealed that out of the genes that are misregulated upon Ncoa3 knockdown, all were up-regulated (Supplemental Fig. S8B).
Figure 6.
Ncoa3 is an integrated component of the ESC transcriptional network. (A) Significant overlap between Esrrb and Ncoa3 peaks within 200 bp at Oct4–Sox2-bound sites. The intersection with CTCF is shown for comparison. (B) MEME de novo motif discovery analysis of top Ncoa3-bound sequences. (C) Spatial heat map depicting binding peak intensity of indicated factors, centered on the peak of maximum Ncoa3 binding for each site. The two-dimensional matrix was provided as input for the K-mean clustering program in Matlab to determine the classes of each position. (D) Venn diagram showing the overlap of genes that are significantly regulated by Ncoa3 and Esrrb, as determined by microarray following Ncoa3 (this study) and Esrrb (Feng et al. 2009) knockdown in ESCs (fold change >1.5). The Fisher's exact test P-value for the correlation of fold change is indicated. See also Supplemental Table 5. (E) Heat map of class I genes bound by Ncoa3 and Esrrb. First, all of the genes regulated by Esrrb within 100 kb of the class I region were taken, then their fold change upon Esrrb (left) or Ncoa3 (right) knockdown was plotted according to Esrrb knockdown. Shades of green and red indicate down-regulation and up-regulation, respectively. Note that the majority of genes is down-regulated. (F) Co-IP of Esrrb in ESCs treated for 24 h with ethanol or 12.5 μM DES, immunoblotted for the indicated proteins. (G) Co-IP in ESCs showing the interaction of endogenous Ncoa3 and RNApol2. Two representative blots are shown. (H) Co-IP of Esrrb immunoblotted for RNApol2 in ESCs following 48 h of transfection with siControl or siNcoa3 oligos. The square brackets in F–H indicate bands corresponding to RNApol2. All co-IPs were performed at least twice with similar results.
Ncoa3 bridges Esrrb to the general transcription machinery
Recently, Esrrb, among other pluripotency factors, was identified as being uniquely associated with RNApol2 complexes (van den Berg et al. 2010), suggesting a critical function for Esrrb, distinct from OSN, in activating transcription. Thus, we investigated the importance of the Esrrb AF-2 domain and Ncoa3 for this function. We first demonstrated by co-IP that RNApol2 does indeed interact with endogenous Esrrb in ESCs (Fig. 6F, top left). Strikingly, however, upon short treatment with DES, we observed a reduction of the Esrrb–RNApol2 interaction (Fig. 6F, top left). This appeared to be specifically associated with loss of Ncoa3 binding (Fig. 6F, top right), as neither the interaction between Esrrb and Oct4 (van den Berg et al. 2008) nor the efficiency of anti-Esrrb immunoprecipitation itself was affected (Fig. 6F, bottom). These results suggested that Ncoa3 could mediate the interaction between Esrrb and RNApol2. To test this further, we performed co-IPs between RNApol2 and Ncoa3 itself and demonstrated an interaction between these proteins in ESCs (Fig. 6G). Finally, to confirm that Ncoa3 is required for the association between Esrrb and RNApol2, co-IPs were repeated in cells treated with siControl or siNcoa3 oligos to deplete endogenous Ncoa3 protein. Transfection with siNcoa3 specifically reduced the interaction between endogenous Esrrb and RNApol2 (Fig. 6H; Supplemental Fig. S8C–E). Taken together, these findings reveal Ncoa3 as a key limiting factor that bridges Esrrb to the general transcription machinery in ESCs.
Discussion
Here, we present the first detailed characterization of a nuclear receptor coactivator in the maintenance of ESC pluripotency. Although it is not a tissue-specific factor, we observed that elevated Ncoa3 expression is a defining property and essential requirement of ESCs and iPSCs. Numerous transcription factors have been well characterized in this context. In contrast, there exist few matching studies for critical coactivators. Our work notably complements recent findings by Fong et al. (2011) identifying a novel Oct4/Sox2 coactivator, the XPC nucleotide excision repair complex, which is essential to activate Nanog and support the ESC state while also safeguarding genome integrity.
In this study, we provide significant data to suggest that Ncoa3 action in ESCs is primarily mediated through Esrrb function. We show that Ncoa3 binds to Esrrb specifically via its AF-2 domain and that Ncoa3 recruitment is strictly required to potentiate Esrrb activity in ESCs. AF-2 domain inactivation in ESCs or depletion of Ncoa3 itself both suppress transcription from Esrrb-dependent gene reporters and result in a loss of ESC self-renewal. Conversely, overexpressing Ncoa3 in ESCs enhances Esrrb-mediated resistance to differentiation but is not sufficient alone to block differentiation, in agreement with Ncoa3 acting as an Esrrb coactivator and not a transcription factor. Through genome-wide ChIP-seq analysis, we demonstrate the presence of Ncoa3 at a large number of active Esrrb-bound enhancers that are shared with the OSN triad and enriched for p300, H3K27ac, and H3K4me1 (Heintzman et al. 2007; Creyghton et al. 2010). GO analysis at this set of targets shows enrichment for key self-renewal regulators whose expression is significantly reduced in Ncoa3 knockdown ESCs, as determined by microarray and qRT–PCR, identifying Ncoa3 as an integrated component of the ESC transcriptional network.
A distinct group of Ncoa3- and Esrrb-bound loci showing reduced OSN occupancies is exclusively associated with germ cell and fertility-related phenotypes. This corroborates with a primordial germ cell (PGC)-specific defect observed in vivo in the absence of Esrrb (Mitsunaga et al. 2004) and an infertility phenotype in Ncoa3 knockout mice (Xu et al. 2000). It furthermore suggests that Esrrb and Ncoa3 might also act in synergy during PGC reprogramming. Consistently, we found that germ cell-relevant targets such as Prdm14 are significantly down-regulated upon Ncoa3 knockdown (Supplemental Fig. S8A), supporting a putative role for the Ncoa3–Esrrb partnership in this process. Aside from Esrrb, it is still possible that other nuclear receptors or transcription factors may also recruit Ncoa3 to some genes. Nr5a2, for example, maps in ESCs to many Esrrb-, Ncoa3-, and OSN-bound targets (Heng et al. 2010; this study) and could potentially interact with Ncoa3 via its LBD, although this remains to be formally demonstrated. In contrast, we could find no evidence to support a direct role for OSN in recruiting Ncoa3 at these sites, although these core factors most likely stabilize the Esrrb–Ncoa3 complex through protein–protein interactions with Esrrb at multitranscription factor clusters (Supplemental Fig. S3; van den Berg et al. 2008).
Critically, we shed new light on how the Ncoa3–Esrrb self-renewal function is conferred in ESCs. A recent study reported the Oct4 interactome of 166 proteins, including many transcription factors and chromatin-modifying complexes not previously known to associate with the ESC network (van den Berg et al. 2010). Among these Oct4 partners, Esrrb was uniquely found to be associated with the basal transcription machinery. An association between Ncoa3 and Esrrb was also observed there. Our mechanistic work on Ncoa3 thus elucidates the functional importance of this interaction and further highlights the relevance of Oct4's broad range of interaction partners. Here, we confirm that endogenous Esrrb and RNApol2 are physically associated in ESCs and demonstrate that Ncoa3 mediates this association. We propose that Ncoa3 functionally bridges Esrrb and RNApol2 at loci co-occupied by Oct4 as well as Sox2, Nanog, and additional pluripotency factors (see Fig. 7), highlighting a distinct role for the Ncoa3–Esrrb partnership to the core ESC transcription factors. Ncoa3 is also established in this study as being required for efficient somatic cell reprogramming—a property shared with other Oct4/Esrrb partners, including core members of Trithorax/MLL complexes (Ang et al. 2011). These chromatin-modifying complexes are essential for open chromatin and pluripotency and may therefore open the path to the reprogramming process by facilitating Oct4 occupancy at genomic loci encoding self-renewal genes. Esrrb–Ncoa3–RNApol2 bridging would in turn perpetuate strong transcriptional activation at Oct4/Esrrb-bound sites, allowing for the full establishment of iPSCs or the maintenance of ESC self-renewal.
Figure 7.
Proposed model. Esrrb interacts with Nanog and Oct4/Sox2 at ESC enhancers and recruits Ncoa3 via its AF-2 region. Ncoa3 interacts with RNApol2 and bridges Esrrb at enhancers to the general transcriptional machinery. These factors would thus act, together with other factors such as chromatin proteins and Mediator (Kagey et al. 2010), to bring about transcription of ESC-associated genes.
In summary, our work elucidates a previously unrecognized functional link between the core ESC transcriptional network and a nuclear receptor coactivator and, importantly, provides novel mechanistic insights into the role of Ncoa3–Esrrb in sustaining both ESC self-renewal and reprogramming. It would be of interest in future studies to elucidate whether coactivator recruitment is indeed a general requirement for many ESC pluripotency factors. Moreover, acting as “sensors,” these cofactors could also be capable of integrating stimuli from multiple signaling pathways, thus adding another layer of control to the complex network governing pluripotency and differentiation.
Materials and methods
Cell culture and RNAi experiments
Mouse E14Tg2A (E14) ESCs were cultured on 0.1% gelatin as previously described (Azuara et al. 2006). COS-1 and 293T were grown in DMEM supplemented with 10% FCS, penicillin/streptomycin, and L-glutamine. MEFs and iPSCs were cultured as previously described (Feng et al. 2009). RNAi experiments followed by puromycin selection were performed using pLKO.1 or pSuper vectors. Transfection of ESCs was performed with Lipofectamine 2000 according to the manufacturer's protocols. See Supplemental Table S6 for shRNA sequences.
Antibodies
The primary antibodies used in this study for immunofluorescence, Western blotting, and ChIP were as follows: anti-Esrrb (R&D Systems, H6705 and H6707), anti-Nanog (Cosmo, RCAB0002P), anti-Ncoa3 (Santa Cruz Biotechnology, M-397; BD Biosciences, #611105), anti-Oct4 (Santa Cruz Biotechnology, C-10), anti-Sox2 (Abcam, ab97959), anti-Flag (Sigma, M2), anti-RNApol2 (Santa Cruz Biotechnology, N-20), anti-Gapdh (Millipore), anti-Tubulin (Sigma), anti-cleaved Caspase-3 (Cell Signaling, #9664), and anti-BrdU (Abcam, ab6326).
Co-IP and ChIP
For co-IP experiments performed with ectopically expressed proteins, COS-1 cells in 10-cm dishes were transfected with 3–5 μg of each expression vector using Fugene HD (Roche), according to the manufacturer's guidelines. Cells were collected 48 h later and lysed in RIPA buffer. Eight hundred micrograms of protein extract was precleared with protein A Sepharose beads (Sigma, P3391) for 2 h at 4°C, then incubated overnight at 4°C with the indicated antibodies. Protein A beads were added for 2 h, then washed with RIPA and TSE buffer (2mM EDTA, 20 mM Tris at pH 8, 150 mM NaCl) three times and loaded on 7% acrylamide gels. Detection of endogenous interactions was performed using 1-μg extracts similarly prepared from untransfected E14 ESCs. Anti-Ncoa3 (#611105; mouse monoclonal) was used to coimmunoprecipitate Ncoa3 expressed in COS-1 cells when examining interactions with Nanog (rabbit polyclonal); all other co-IP, immunofluorescence, and ChIP experiments were performed with M-397. ChIP experiments were carried out with chromatin prepared from E14 cells as previously described (Stock et al. 2007) using 8–10 μg of primary antibody and 600 μg of precleared chromatin per immunoprecipitation. Immunoprecipitated DNA fragments were purified and analyzed via SYBR Green qPCR.
DNA pull-down assays
COS-1 cells were transfected with Ncoa3 together with Flag-Esrrb or Oct4 and Sox2 expression vectors. Forty-eight hours after transfection, cells were lysed in HKMG buffer (10 mM HEPES at pH 7.9, 100 mM KCl, 5 mM MgCl2, 10% glycerol, 0.5% NP-40, 1 mM DTT, protease inhibitors [Complete EDTA-free; Roche]) and lysates precleared for 1 h with pre-equilibrated streptavidin-coupled Dynabeads (Invitrogen, M-280). Pairs of complementary DNA oligonucleotides with the sense oligonucleotide biotinylated at the 5′ end (Sigma) were annealed and incubated for 2 h at 4°C with precleared cell lysates. DNA-bound proteins were collected by incubation for 1 h at 4°C with streptavidin-coupled Dynabeads (Invitrogen), washed four times in HKMG buffer, and separated by SDS-PAGE. Ncoa3, Flag-Esrrb, Oct4, and Sox2 were detected by Western blot. See Supplemental Table S6 for probe sequences.
Luciferase assays
ESCs were plated on gelatin-coated 96-well plates at a density of 2 × 104 cells per well and, 5 h later, transfected with the indicated luciferase reporters and expression constructs using Lipofectamine 2000. For knockdown experiments, either 30 ng of shRNA vector or 2–3 pmol of siRNA was cotransfected per well. pCmv.Renilla or pCDNA4/TO.eGFP was used to normalize luciferase values in each well. Firefly and Renilla luciferase activities were measured 48 h post-transfection using the SteadyLite Plus kit (PerkinElmer).
Retroviral production
For reprogramming experiments, PlatE cells were used to produce pMXs-based retroviruses containing the cDNA for Pou5f1/Oct4, Sox2, Klf4, or cMyc, as previously described (Feng et al. 2009). PlatE-generated mCherry retrovirus was used to monitor MEF infection efficiency in each experiment.
iPSC generation
Embryonic day 13.5 (E13.5) Oct4-GFP MEFs were derived from embryos produced via the intercross between wild-type MF1 females and homozygous Oct4-GFP B6 males (Yeom et al. 1996). For iPSC induction, 2 × 104 Oct4-GFP MEFs were seeded per well of a 24-well plate and, 5 h later, infections were performed using equal amounts of each indicated retrovirus in the presence of 8 μg/mL polybrene. Medium was exchanged for MEF medium 24 h post-infection, and, 48 h after infection, MEFs were collected and reseeded onto inactivated feeders in iPSC medium. Emerging iPSC colonies were monitored until day 12–14 when cells were harvested or individual colonies were picked for further analysis.
ChIP-seq and microarray analysis
ChIP-seq and microarray analysis were performed on mouse ESCs as previously described (Chen et al. 2008; Feng et al. 2009). Detection of Ncoa3 sequenced peaks was carried out using MACS with a P-value cut-off of 1.0 × 10−5.
Accession numbers
ChIP-seq and microarray data are accessible at the Gene Expression Omnibus database under accession number GSE40193.
Detailed Materials and Methods, antibodies, and sequences are available in the Supplemental Material.
Acknowledgments
We are grateful to Paul Robson, Vincent Cavaillès, and Mark Christian for providing plasmids, and to Eleni Daniel and Anthony Okolo for IMBAT cDNA. Thanks to Rachel Amoroux for E13.5 Oct4GFP embryos; Sadaf Khan for p53−/− MEFs; Matias Autio for iMEFs; Hitoshi Niwa and Ian Chambers for the ZHBTc4 and RCNβHB ESC lines, respectively; and Ana Banito and Lai-Ping Yaw for their technical assistance. Thanks also to Mark Christian, Helle Jorgensen, and Tristan Rodriguez, as well as members of the Epigenetics and Development Group and Molecular Endocrinology Group for discussions and/or critical reading of the manuscript. This work was supported by the BBSRC, Genesis Research Trust, FCT, MRC, Genome Institute of Singapore, and Imperial College London.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.195545.112.
References
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. 2011. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17: 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. 2006. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. 2009. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 23: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiani M, Gentile L, Gambles VV, Cavaleri F, Redi CA, Scholer HR 2005. Variable reprogramming of the pluripotent stem cell marker Oct4 in mouse clones: Distinct developmental potentials in different culture environments. Stem Cells 23: 1089–1104 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Tomlinson SR 2009. The transcriptional foundation of pluripotency. Development 136: 2311–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113: 643–655 [DOI] [PubMed] [Google Scholar]
- Charest-Marcotte A, Dufour CR, Wilson BJ, Tremblay AM, Eichner LJ, Arlow DH, Mootha VK, Giguere V 2010. The homeobox protein Prox1 is a negative modulator of ERRα/PGC-1α bioenergetic functions. Genes Dev 24: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Chen X, Liu Z, Xu J 2010. The cooperative function of nuclear receptor coactivator 1 (NCOA1) and NCOA3 in placental development and embryo survival. Mol Endocrinol 24: 1917–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, et al. 2005. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol 25: 6031–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, White R, Lees JA, Parker MG 1992. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J 11: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, et al. 2009. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol 11: 197–203 [DOI] [PubMed] [Google Scholar]
- Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R 2011. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell 147: 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere V 2008. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev 29: 677–696 [DOI] [PubMed] [Google Scholar]
- Gillich A, Bao S, Grabole N, Hayashi K, Trotter MW, Pasque V, Magnusdottir E, Surani MA 2012. Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell 10: 425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greschik H, Flaig R, Renaud JP, Moras D 2004. Structural basis for the deactivation of the estrogen-related receptor γ by diethylstilbestrol or 4-hydroxytamoxifen and determinants of selectivity. J Biol Chem 279: 33639–33646 [DOI] [PubMed] [Google Scholar]
- Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, et al. 2005. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol 25: 3492–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, et al. 2010. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell 6: 167–174 [DOI] [PubMed] [Google Scholar]
- Henttu PM, Kalkhoven E, Parker MG 1997. AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol Cell Biol 17: 1832–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Yang L, Stallcup MR 1999. Hormone-independent transcriptional activation and coactivator binding by novel orphan nuclear receptor ERR3. J Biol Chem 274: 22618–22626 [DOI] [PubMed] [Google Scholar]
- Huss JM, Kopp RP, Kelly DP 2002. Peroxisome proliferator-activated receptor coactivator-1α (PGC-1α) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. Identification of novel leucine-rich interaction motif within PGC-1α. J Biol Chem 277: 40265–40274 [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR 2006. Dissecting self-renewal in stem cells with RNA interference. Nature 442: 533–538 [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Ohizumi H, Fujitani Y, Nemoto T, Tanaka T, Takahashi N, Kawada T, Miyoshi M, Ezaki O, Kakizuka A 2003. PPARγ coactivator 1β/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci 100: 12378–12383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1: 361–370 [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Luo J, Sladek R, Bader JA, Matthyssen A, Rossant J, Giguere V 1997. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-β. Nature 388: 778–782 [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA 2009. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T 1999. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J 18: 4261–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature 454: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642 [DOI] [PubMed] [Google Scholar]
- Mitsunaga K, Araki K, Mizusaki H, Morohashi K, Haruna K, Nakagata N, Giguere V, Yamamura K, Abe K 2004. Loss of PGC-specific expression of the orphan nuclear receptor ERR-β results in reduction of germ cell number in mouse embryos. Mech Dev 121: 237–246 [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391 [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev 12: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24: 372–376 [DOI] [PubMed] [Google Scholar]
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P 2005. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem 280: 24731–24737 [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK 2006. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20: 1405–1428 [DOI] [PubMed] [Google Scholar]
- Smith AG 2001. Embryo-derived stem cells: Of mice and men. Annu Rev Cell Dev Biol 17: 435–462 [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Tremblay GB, Kunath T, Bergeron D, Lapointe L, Champigny C, Bader JA, Rossant J, Giguere V 2001. Diethylstilbestrol regulates trophoblast stem cell differentiation as a ligand of orphan nuclear receptor ERRβ. Genes Dev 15: 833–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg DL, Zhang W, Yates A, Engelen E, Takacs K, Bezstarosti K, Demmers J, Chambers I, Poot RA 2008. Estrogen-related receptor β interacts with Oct4 to positively regulate Nanog gene expression. Mol Cell Biol 28: 5986–5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA 2010. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercauteren K, Gleyzer N, Scarpulla RC 2009. Short hairpin RNA-mediated silencing of PRC (PGC-1-related coactivator) results in a severe respiratory chain deficiency associated with the proliferation of aberrant mitochondria. J Biol Chem 284: 2307–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, Moras D, Gronemeyer H 1996. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol 3: 206. [DOI] [PubMed] [Google Scholar]
- Xie CQ, Jeong Y, Fu M, Bookout AL, Garcia-Barrio MT, Sun T, Kim BH, Xie Y, Root S, Zhang J, et al. 2009. Expression profiling of nuclear receptors in human and mouse embryonic stem cells. Mol Endocrinol 23: 724–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O'Malley BW 2000. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci 97: 6379–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M 2008. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet 40: 1016–1022 [DOI] [PubMed] [Google Scholar]
- Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR 1996. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122: 881–894 [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115: 281–292 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Teng CT 2000. Estrogen receptor-related receptor α1 interacts with coactivator and constitutively activates the estrogen response elements of the human lactoferrin gene. J Biol Chem 275: 20837–20846 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang J, Wang T, Esteban MA, Pei D 2008. Esrrb activates Oct4 transcription and sustains self-renewal and pluripotency in embryonic stem cells. J Biol Chem 283: 35825–35833 [DOI] [PubMed] [Google Scholar]