How cell polarity is determined and perpetuated is a fundamental issue in cell biology. Yamaichi et al. explore this issue in Vibrio cholera and find that HubP, a large transmembrane protein, is the organizing factor for polar localization of ParA1, ParC, and FlhG, three ParA-related ATPases involved in the recruitment of the chromosome origin, localization of chemotaxis proteins, and flagellar synthesis. The authors identify distinct regions of HubP needed to localize these proteins.
Keywords: Vibrio cholerae, cell polarity, chemotaxis, chromosome segregation, motility
Abstract
The cell poles constitute key subcellular domains that are often critical for motility, chemotaxis, and chromosome segregation in rod-shaped bacteria. However, in nearly all rods, the processes that underlie the formation, recognition, and perpetuation of the polar domains are largely unknown. Here, in Vibrio cholerae, we identified HubP (hub of the pole), a polar transmembrane protein conserved in all vibrios, that anchors three ParA-like ATPases to the cell poles and, through them, controls polar localization of the chromosome origin, the chemotactic machinery, and the flagellum. In the absence of HubP, oriCI is not targeted to the cell poles, chemotaxis is impaired, and a small but increased fraction of cells produces multiple, rather than single, flagella. Distinct cytoplasmic domains within HubP are required for polar targeting of the three ATPases, while a periplasmic portion of HubP is required for its localization. HubP partially relocalizes from the poles to the mid-cell prior to cell division, thereby enabling perpetuation of the polar domain in future daughter cells. Thus, a single polar hub is instrumental for establishing polar identity and organization.
The formation and recognition of subcellular domains is critical for numerous cellular processes, even in relatively simple unicellular organisms such as bacteria (Shapiro et al. 2009; Rudner and Losick 2010; Thanbichler 2011). Without an ability to establish intracellular landmarks, key events in the bacterial cell cycle, such as chromosome segregation and formation of the cell division plane, are instead subject to stochastic variation that can lead to detrimental results. Bacterial morphology and motility also often depend on positional recognition; e.g., for correct localization of pili and flagella. A few determinants of such subcellular patterns have been well studied, such as MinCDE, which enable a number of bacterial species to form a division plane at the mid-cell (Raskin and de Boer 1997; Lutkenhaus 2007). In Gram-positive bacteria and actinomycetes, DivIVA recruits other proteins to the cell poles and septum and thereby regulates several cell processes (Marston et al. 1998; Ben-Yehuda et al. 2003; Flärdh 2010). Furthermore, in the differentiating α-proteobacterium Caulobacter crescentus, TipN and PopZ are markers and determinants of pole formation (Huitema et al. 2006; Lam et al. 2006; Bowman et al. 2008; Ebersbach et al. 2008). However, in most bacteria, the means by which subcellular domains are generated or recognized remain unknown.
In the Gram-negative rod Vibrio cholerae, as in other rod-shaped bacteria, the cell poles are key subcellular domains, and the bacterium has the ability to distinguish poles from sides and the old pole from the new (most recently generated) pole. For example, following cell division, the origin region of the larger of V. cholerae's two chromosomes (oriC of chrI, oriCI) is always found at the old pole, as is an array of chemoreceptors with associated chemosignaling proteins, and the bacterial flagellum also develops at this site. We found that targeting of oriCI to the pole is dependent on ParA1 (Fogel and Waldor 2006), a homolog of bacterial plasmid partitioning proteins, and that the localization of the chemotactic apparati is also regulated by a ParA homolog, ParC (Ringgaard et al. 2011). Flagellar assembly is regulated by a third ParA-like protein, FlhG (Correa et al. 2005; Kusumoto et al. 2008), raising the possibility that these proteins rely on related processes to govern spatial patterning. Notably, each protein forms a focus at the old pole in newborn cells, although their distribution is not always limited to this site (Fogel and Waldor 2006; Kusumoto et al. 2008; Ringgaard et al. 2011). As the cell cycle progresses and the cell prepares for division, which will yield a “new” old pole in one of the daughter cells, ParA1, ParC, and FlhG all adopt a bipolar pattern. However, the means by which their distribution is initially determined and subsequently shifted has not been established. V. cholerae (a γ-proteobacterium) lacks apparent homologs of TipN and PopZ; in fact, these polar determinants are restricted to a subset of α-proteobacteria (Lam et al. 2006; Bowman et al. 2008; Ebersbach et al. 2008).
ParA family proteins are typically relatively small, cytoplasmic proteins that contain a deviant Walker AAA ATPase motif. They often have the ability to self-associate; e.g., into polymers that may push or pull DNA and thereby promote its equal distribution between daughter cells. ParA family proteins have also been found to govern the localization of the division site (e.g., via MinD and MipZ) (Thanbichler and Shapiro 2006; Lutkenhaus 2007), type IV pili (e.g., via TadZ/CpaE) (Viollier et al. 2002; Perez-Cheeks et al. 2012; Xu et al. 2012), and conjugative transfer machinery (via VirC1) (Atmakuri et al. 2007). They often have a dynamic distribution within the cell, and this distribution is key for proper function. In most cases, the proteins' ability to bind and/or hydrolyze ATP is critical for their subcellular localization and function (Szardenings et al. 2011; Lutkenhaus 2012). It should be noted, however, that not all ParA-like proteins, even in V. cholerae, form foci at the cell poles. The small chromosome (chrII)-associated protein ParA2 and MinD are oscillatory and more diffusely distributed throughout much of the cell (Raskin and de Boer 1999; Fogel and Waldor 2006). ParA proteins associated with plasmid segregation have also been observed to oscillate. The distinct localization patterns of these proteins are likely determined by the specific protein partners with which each interacts.
We performed a genetic screen to identify factors that contribute to the establishment of polar identity in V. cholerae and found that a single protein, VC0998 (henceforth called HubP, for hub of the pole), is required for the proper polar localization of ParA1, ParC, and FlhG. In the absence of HubP, oriCI is not targeted to the cell poles, chemotaxis is impaired due to the absence or mislocalization of polar chemotactic receptor arrays and their associated signaling proteins, and cells have an increased propensity to produce multiple, rather than single, flagella. HubP, like the three ParA family ATPases, is localized to the cell poles; however, it routinely marks both poles—rather than the single (old) pole typically observed for the other proteins in young cells—and can migrate between the two poles. Despite the homology between ParA1, ParC, and FlhG, their polar targeting by HubP appears to rely on different mechanisms, and a distinct region of HubP is needed to localize each client protein. Thus, HubP is a multifaceted scaffold necessary for the establishment of polar identity and organization; it is a junction point around which multiple polar processes are oriented.
Results
Identification of HubP, an organizer of polar features and processes
Our screen for pole-organizing factors, which was similar to a screen in C. crescentus (Huitema et al. 2006), was premised on the idea that V. cholerae lacking a polar landmark protein is likely to have impaired motility/chemotaxis, since its flagella and/or chemotaxis proteins may be mislocalized. Therefore, we focused our analysis on genes that had previously been linked to a motility defect (Cameron et al. 2008) yet did not have an obvious connection to chemotaxis or flagellar assembly. A set of 91 mutants was transformed with an expression construct for a previously described polar marker protein, ParA1[K11E]-YFP, which, in contrast to wild-type ParA1-YFP, forms discrete bipolar foci throughout the cell cycle (Ringgaard et al. 2011). Transformants were individually screened using fluorescence microscopy to identify strains in which this protein is mislocalized.
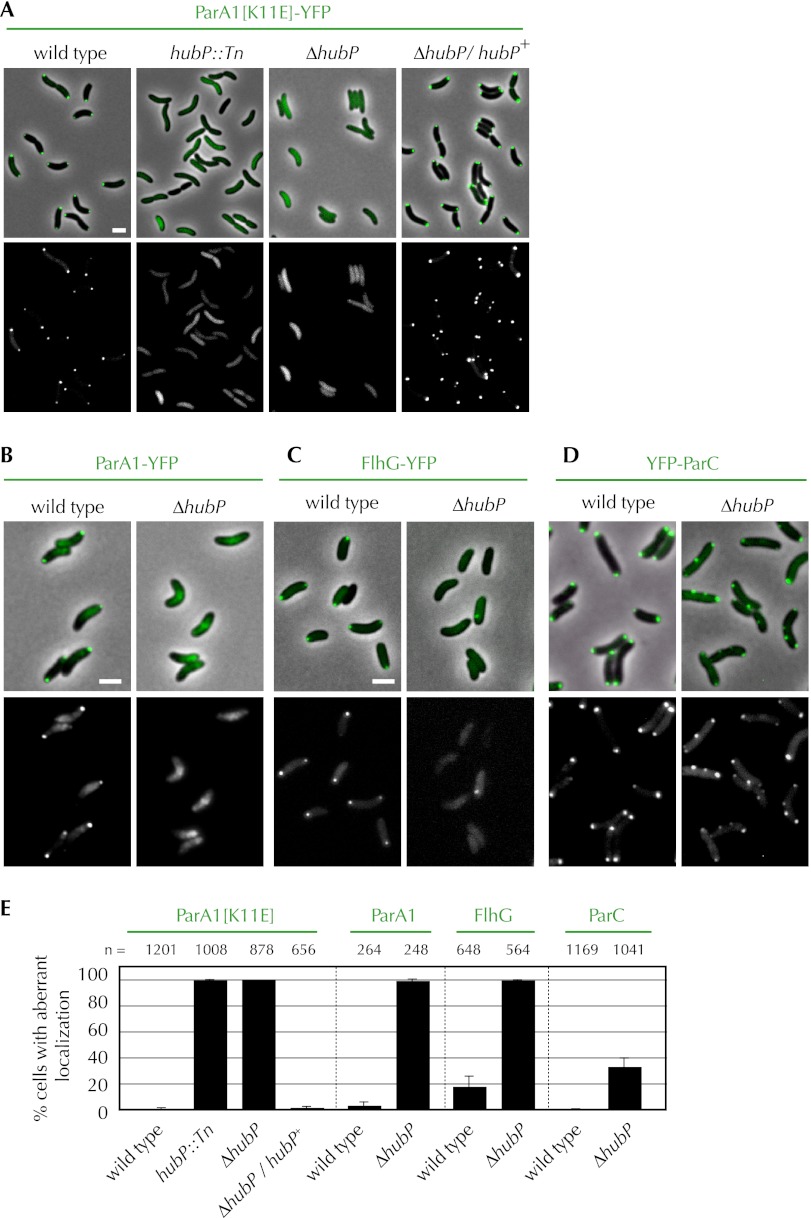
We found that disruption of a previously undescribed gene, vc0998 (hubP), caused ParA1[K11E] to be diffusely distributed throughout cells, rather than displaying its typical bipolar distribution (Fig. 1A,E). This pattern was observed in both a transposon insertion mutant (hubP∷Tn) and a strain with an in-frame deletion of hubP. Furthermore, wild-type localization of ParA1[K11E] was restored in the ΔhubP strain by expression of the protein in trans, providing strong evidence that polar targeting of ParA1[K11E] is dependent on the presence of HubP.
Figure 1.
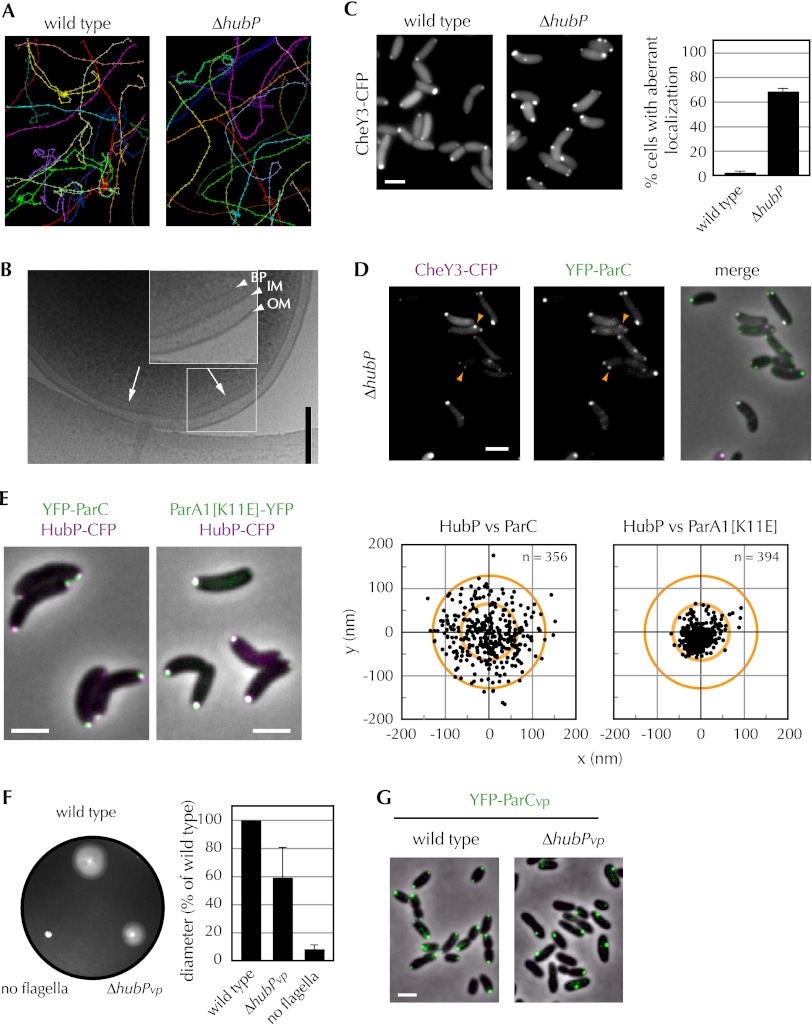
HubP is a determinant of polar protein localization. (A–D) Subcellular localization of polar proteins fused to YFP (pseudocolored in green with phase contrast in the top panels) in indicated V. cholerae strains. (A) ParA1[K11E]. (B) ParA1. (C) FlhG. (D) ParC. Pictures shown are representative fields. Bar, 2 μm. (E) Percentage of cells with mislocalized polar proteins. For ParA1[K11E], ParA1, and FlhG, mislocalization refers to cells lacking a polar focus, and for ParC, it refers to cells that contain a nonpolar focus. Means, standard deviations, and total number of cells counted (n) are shown. See also Supplemental Fig. S1.
HubP is also required for the proper cellular positioning of several additional polar factors. In wild-type cells, at least a portion of ParA1, ParC, and FlhG is found within polar foci at either one or both poles, depending on the age of the cell. However, in a strain lacking hubP, the distribution of these three proteins was markedly different than in wild-type cells (Fig. 1B–E). ParA1 was diffusely distributed throughout the cell (Fig. 1B), and FlhG was either diffusely distributed or found in a nonpolar focus (Fig. 1C); neither protein formed polar foci. ParC formed polar foci in a subset of hubP cells (Fig. 1D); however, ∼50% of cells contained nonpolar ParC foci either instead of or in addition to a polar focus. An increase in diffusely distributed ParC was also evident in the hubP cells. Thus, HubP appears to be important for the proper subcellular distribution of three ParA-type ATPases in V. cholerae. However, it is not required for the correct cellular targeting of all such paralogous proteins. The distribution of ParA2, which mediates chrII partitioning, and of MinD, which likely specifies the V. cholerae division site, was not altered in the absence of HubP (Supplemental Fig. S1). Additionally, the absence of HubP did not cause detectable changes in cell shape or cell size and did not impair cell growth.
Sequence and genomic analyses revealed several notable attributes of the HubP protein. HubP is quite large (1621 amino acids, or ∼178 kDa) and extremely acidic (pI = 3.22). Its N terminus appears to contain a signal sequence, a potential LysM peptidoglycan (PG)-binding domain, and a single transmembrane domain. Its C terminus, which is predicted to reside within the cytoplasm, contains 10 copies of an imperfect 46-amino-acid repeat, in which 19 of the consensus amino acids are acidic (Fig. 2A; Supplemental Fig. S2). The protein is fairly well conserved among vibrio and photobacteria species (Vibrionaceae/Photobacteriaceae); for example, homologs with >40% identity are present in Vibrio vulnificus and Vibrio parahaemolyticus, and Vibrio fischeri encodes a protein with 27.5% identity. Homologs appear to have similar overall structures, although the sizes and copy numbers of their acidic repeat sequences vary significantly (Supplemental Table S1; Supplemental Fig. S2). Outside of the Vibrionaceae, homologs are found principally in the γ-proteobacteria, where they are significantly less conserved, but many appear to be encoded within a similar chromosomal neighborhood. Some of these proteins (e.g., FimV of Pseudomonas aeruginosa) have been found to be associated with the assembly of type IV pili and pilus-associated twitching motility (Semmler et al. 2000; Wehbi et al. 2011).
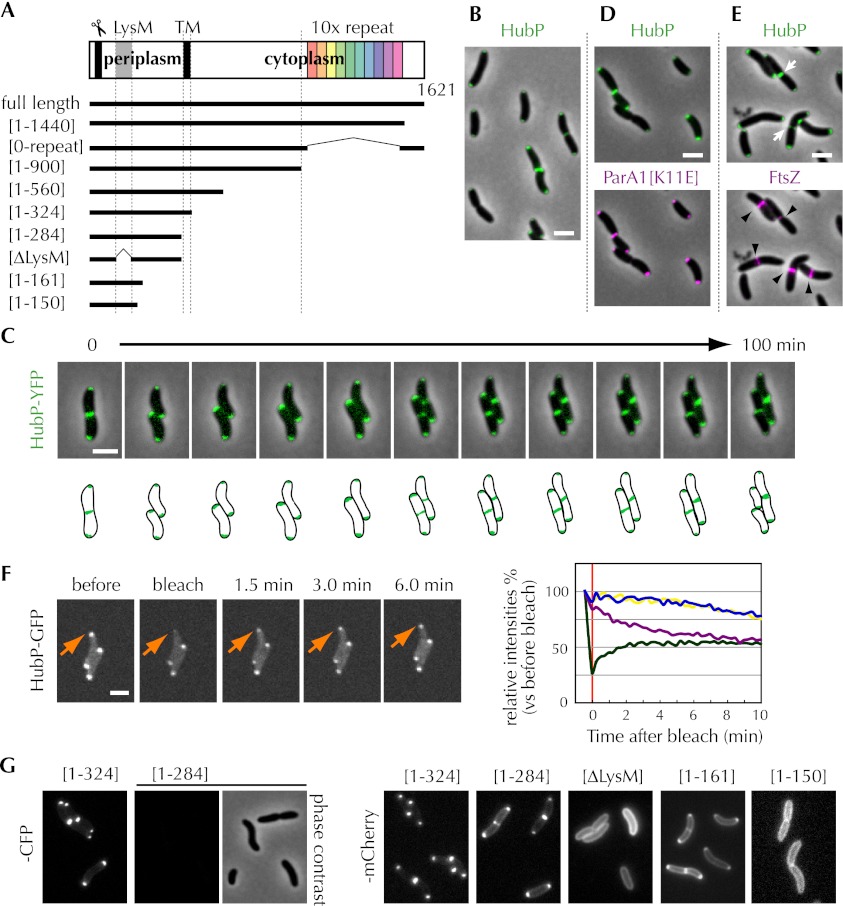
Figure 2.
HubP localization and its determinants. (A) Schematic of the HubP polypeptide (top) and truncation mutants (bottom) used in this study. (Scissors) Signal sequence; (LysM) LysM domain; (TM) transmembrane domain; (10× repeat) repeat sequence (see the text). (B) Subcellular localization of HubP-CFP in ΔhubP V. cholerae. (C, top) Time-lapse images of plasmid-borne HubP-YFP in ΔhubP V. cholerae. A schematic representation is drawn below. (D,E) Subcellular localization of HubP-CFP and ParA1[K11E]-YFP (D) and FtsZ-YFP (E) in ΔhubP V. cholerae. In E, HubP and FtsZ at mid-cell are indicated with arrowheads and arrows, respectively. (F) FRAP experiment of HubP-GFP. Pictures from a representative experiment (left) are shown, along with a graph of average signal intensities over time, based on analysis of 30 cells (right). The bleached focus is indicated by an arrow in the pictures and by the green line in the graph. The magenta line shows the average intensity of the focus at the pole opposite the bleached pole. Pale blue and yellow lines show the intensities of polar foci from unbleached (control) cells. (G) Fluorescence of -CFP and -mCherry fusions to truncated HubP in V. cholerae. Representative fields are shown. Bar, 2 μm.
HubP is directed to the V. cholerae pole and mid-cell by its periplasmic domain
Given the effect of HubP deficiency on subcellular targeting of polar proteins, we assessed whether HubP itself is present at the V. cholerae cell poles. For these experiments, we used fluorescence microscopy and visualized either ectopically or chromosomally expressed HubP-CFP (or YFP/GFP). The fusion proteins, which are functional, as they direct ParA1[K11E]-YFP to a polar position in the hubP mutant, displayed a bipolar distribution, even in small (young) cells (Fig. 2B–D). Thus, HubP appears to mark the new pole prior to the arrival of the ParA family proteins, which are typically not bipolar in young cells (Fogel and Waldor 2006; Ringgaard et al. 2011). HubP was also detectable at the mid-cell in a subset of cells, where it always colocalized with FtsZ, suggesting that it arrives at this site as cells prepare to divide (Fig. 2E). Time-lapse analyses confirmed that HubP is not present at the mid-cell in young cells, but instead arrives there as cells progress through the cell cycle, which presumably enables it to be present at newly formed poles (Fig. 2C). HubP localization was similar using plasmid- and chromosome-encoded protein, although the chromosome-encoded protein was less abundant (Supplemental Movies S1, S2).
Fluorescence microscopy was also used to explore the dynamics and determinants of HubP's subcellular localization. Notably, photobleaching of polar HubP-GFP was followed by recovery of the polar signal, revealing that HubP can be directly targeted to the pole; it does not need to arrive there via the cell division site. Recovery of fluorescence intensity at the bleached pole was accompanied by a decline of fluorescence intensity at the opposite (unbleached) pole, suggesting that there is an exchange of HubP between the two poles (Fig. 2F). Additionally, we found that polar targeting of HubP is independent of its putative cytoplasmic and membrane-spanning domains. A fusion protein consisting of the first 324 amino acids of HubP (the putative periplasmic and transmembrane sequences) fused to CFP had a cellular distribution equal to that of the full-length fusion protein (Fig. 2G). A smaller (1–284) fragment fused to CFP was not detected, consistent with the predicted localization of the HubP N-terminal domain in the periplasm (Fig. 2A), where CFP does not fluoresce. However, when smaller N-terminal fragments were fused to mCherry (which is fluorescent even in the periplasm), polar targeting was also observed (Fig. 2G, 1–284). The first 161 amino acids of HubP were sufficient to enable polar targeting of the fusion protein, although the first 150 amino acids were not (Fig. 2G, 1–161 and 1–150). We also found that polar targeting of HubP required the putative LysM domain (amino acids 90–134) (Fig. 2G, ΔLysM). In the absence of targeting sequences, HubP appeared to be diffusely distributed in the periplasm (Fig. 2G, 1–150 and ΔLysM). The requirement for the LysM domain raises the possibility that an interaction between HubP and PG contributes to its targeting and/or retention at the V. cholerae pole. However, to date, we have been unable to detect a direct interaction between HubP and PG using in vitro binding assays.
HubP modulates the localization of oriCI
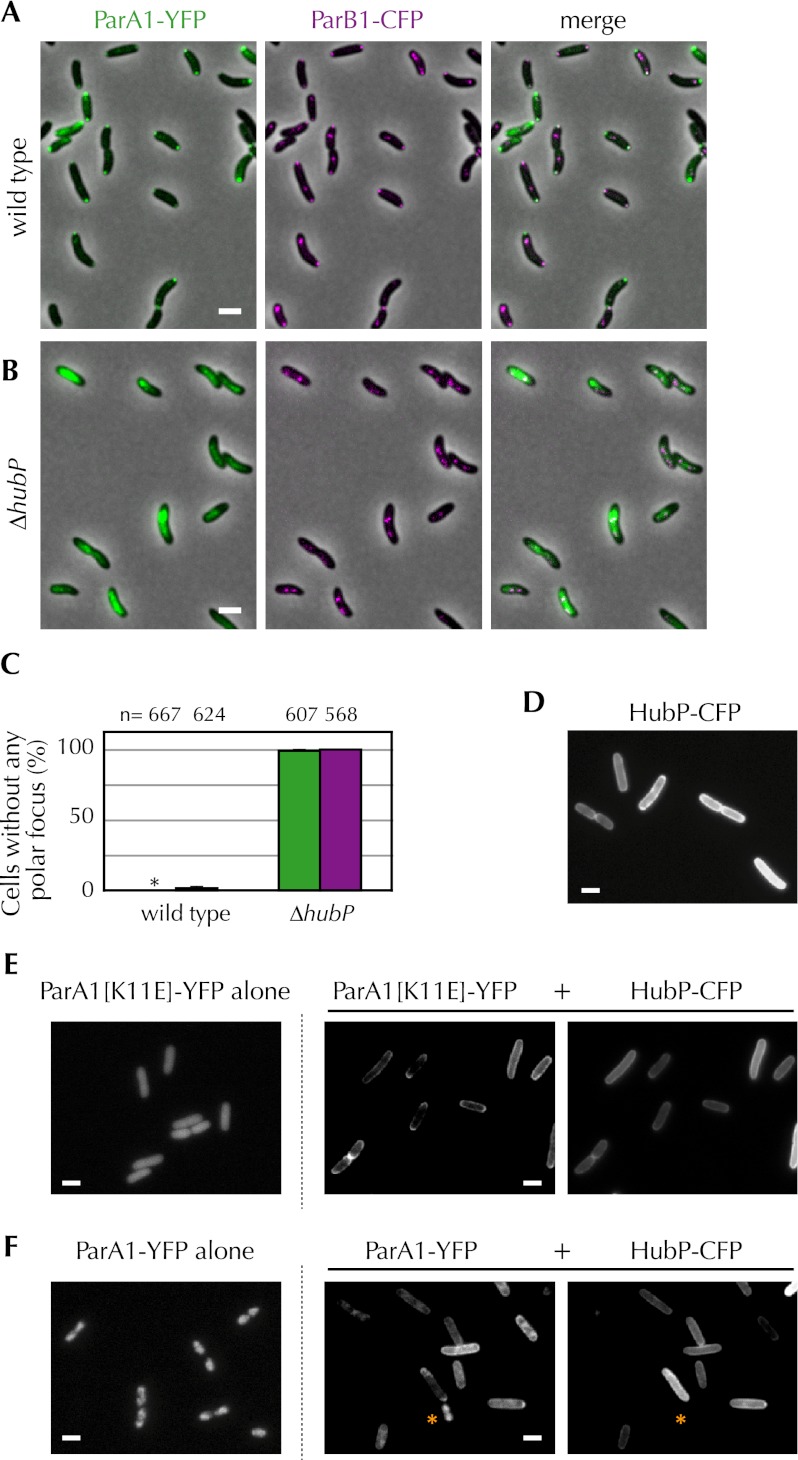
We explored whether the failure of ParA1 to localize to the cell poles in cells lacking HubP altered chrI segregation dynamics. Although ParA1 is not essential for chrI partitioning, in its absence, oriCI and the origin-associated centromere-binding protein ParB1 do not localize to the old pole in young cells, nor are they drawn to the new pole following chromosome replication (Fogel and Waldor 2006). Instead, in a parA1 deletion mutant, they are generally found at the mid-cell of young cells and near the 1/4 and 3/4 positions following replication (Fogel and Waldor 2006). We found that deletion of hubP has a similar effect on the localization of ParB1: No polar ParB1-CFP foci were detected in the hubP mutant, and cells typically contained two well-separated cytoplasmic foci (Fig. 3A–C), as observed in parA1 mutants. Thus, marking of the V. cholerae cell pole by HubP is critical for the establishment of the normal cellular distribution of the organism's major chromosome.
Figure 3.
HubP governs the subcellular distribution of ParA1. (A–C) Subcellular localization of ParA1-YFP and ParB1-CFP in wild type (A) and ΔhubP (B) V. cholerae cells. (C) The percentage of wild-type and ΔhubP cells lacking polar foci of ParA1 (green) and ParB1 (magenta) is shown, based on analysis of the number of cells (n) indicated above. Asterisk indicates 0%. (D) Fluorescence of HubP-CFP expressed in E. coli. (E,F) ParA1[K11E]-YFP (E) or ParA1-YFP (F) was expressed in E. coli in the absence (left) or presence (middle) of HubP-CFP (right). ParA1-YFP localized to the nucleoid in the cell lacking HubP-CFP expression (marked with an asterisk). Representative fields are shown. Bar, 2 μm.
HubP binds directly to ParA1 to control its subcellular distribution
Several lines of evidence suggest that HubP controls ParA1's distribution via a direct interaction between the two proteins. First, when ParA1[K11E]-YFP and HubP-CFP were coexpressed in V. cholerae, the foci they formed colocalized (Fig. 2D; see below). Second, these proteins were found to interact in a bacterial two-hybrid assay, which also showed that ParA1 can self-associate (Table 1; Supplemental Fig. S3), similar to observations for other ParA-like proteins (Leonard et al. 2005; Ebersbach et al. 2006). Finally, HubP recruited ParA1 to the cell periphery when the two proteins were coexpressed in Escherichia coli (Fig. 3D–F). When expressed alone in E. coli, ParA1[K11E]-YFP and ParA1-YFP were widely distributed within the cytoplasm or associated with the nucleoid, respectively (Fig. 3E,F). However, when expressed along with HubP-CFP, which appears to be membrane-associated but not polar in E. coli (Fig. 3D), the distribution of both wild-type ParA1 and ParA1[K11E] was shifted toward the cell membrane (Fig. 3E,F).
Table 1.
Summary of HubP's interactions
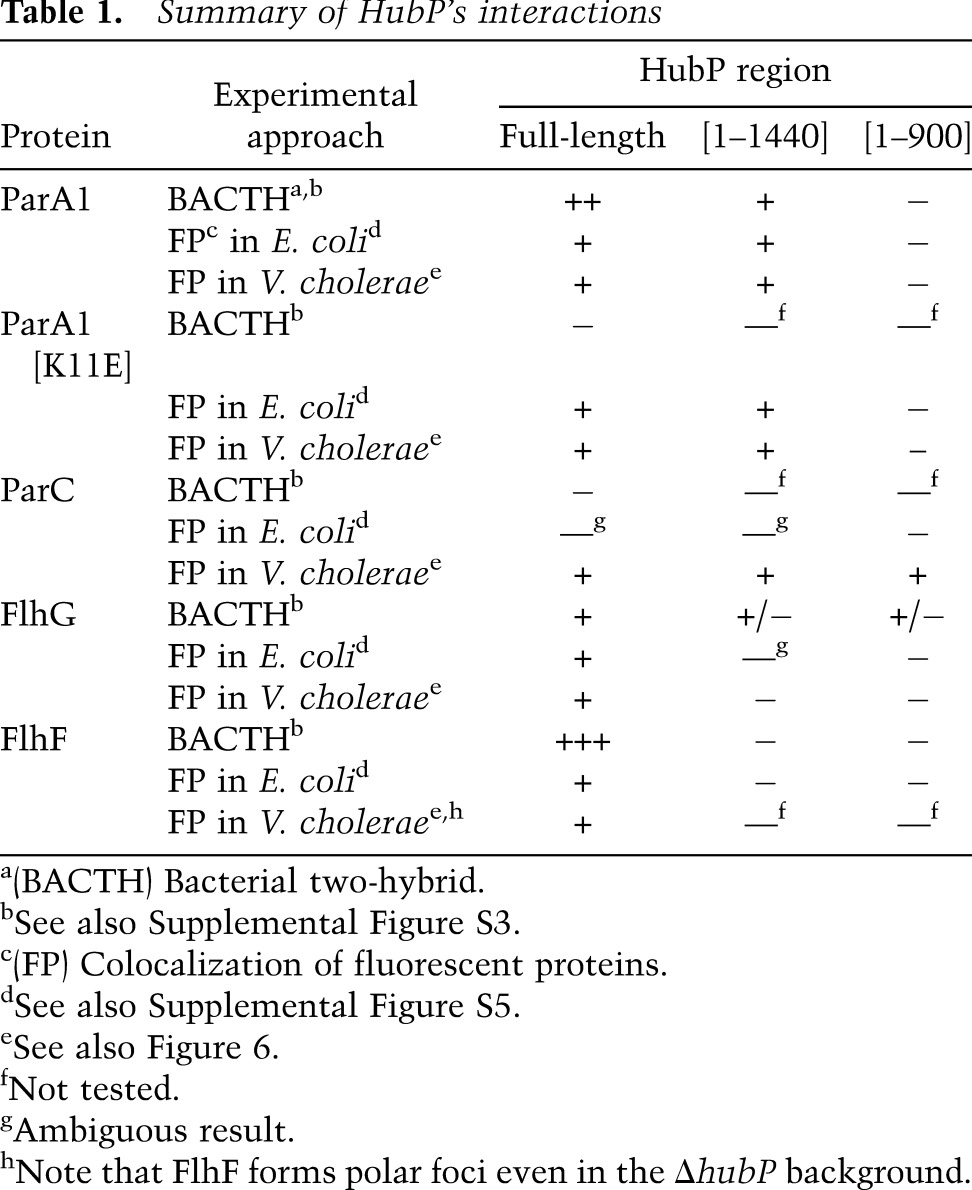
A variety of mutations within parA1 was generated in order to explore which features of the protein are needed for interaction with HubP. Based on previous analyses of ParA family members and identification of key conserved amino acids, mutations were generated to disrupt dimerization (ParA1[G12V]), DNA binding (ParA1[R189E]), ATP binding (ParA1[K16A]), and ATP hydrolysis (ParA1[D40A]), and the subcellular distribution of the mutant proteins was analyzed in E. coli and V. cholerae. Mutants predicted to be unable to dimerize or bind DNA still appeared capable of interacting with HubP in both systems; they formed HubP-dependent polar foci in V. cholerae and showed HubP-dependent membrane association in E. coli (Supplemental Fig. S4). Mutants lacking the ability to bind or hydrolyze ATP also were recruited by HubP to the cell periphery in E. coli and formed HubP-dependent polar foci in V. cholerae (Supplemental Fig. S4). However, nonpolar ParA1[K16A] and ParA1[D40A] foci were also detected in both wild-type and hubP cells regardless of the presence or absence of a chromosomal parA1+ copy, suggesting that ParA1 binding to and/or hydrolysis of ATP is not essential for, but may influence, ParA1–HubP interactions.
HubP negatively regulates polar accumulation of flagella
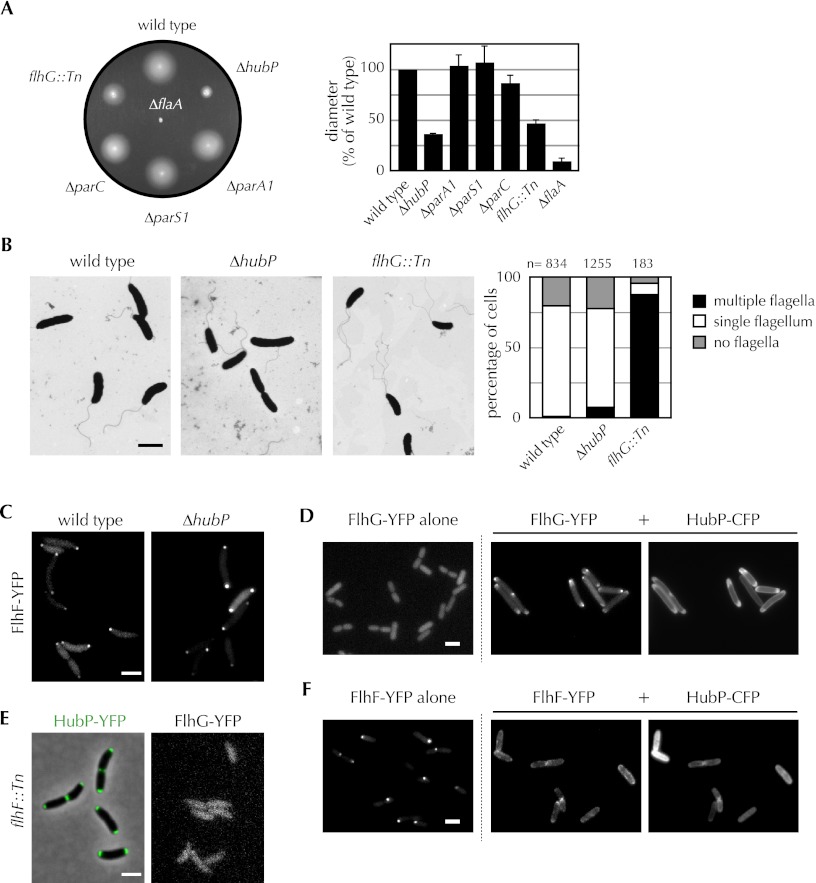
Our observation of FlhG mislocalization in the hubP mutant, coupled with the previously noted reduced motility of this mutant on soft agar plates, led us to further explore the connection between HubP and V. cholerae flagellation. The hubP mutant exhibited a >50% reduction in swarming on soft agar, comparable with the defect displayed by an flhG mutant and significantly larger than that of a parC mutant (Fig. 4A). However, deletion of hubP had a fairly modest effect on flagellation. Electron microscopy analyses revealed that a slightly increased fraction (6%) of V. cholerae lacking HubP produces more than a single polar flagellum, a feature observed in only 1% of wild-type cells (Fig. 4B). This frequency contrasts markedly with that of an flhG mutant, where ∼80% of cells had multiple flagella (Fig. 4B). In Vibrio alginolyticus, FlhG is thought to repress flagellar formation via two mechanisms: (1) by binding to FlhF (which is thought to guide polar placement of flagella) and preventing its polar recruitment, and (2) by repressing expression of flagellar genes (Kusumoto et al. 2008). In V. cholerae, deletion of hubP does not have a significant effect on the distribution of FlhF, despite its effect on FlhG localization, suggesting that polar FlhG is not a critical determinant of FlhF accumulation at this site (Fig. 4C). Furthermore, the relatively minor effect of FlhG mislocalization on flagella production in the hubP mutant suggests that mislocalization does not substantially impair its efficacy in negatively regulating flagellar production. Thus, the biological rationale for polar targeting of FlhG by HubP remains to be determined.
Figure 4.
HubP is important for V. cholerae chemotaxis and regulation of flagellar production. (A) Swarming of various V. cholerae mutants on a representative soft agar plate is pictured (left), and average swarm diameter relative to wild type, with standard deviation based on three or more experiments, is graphed (right). (B, left) Images of wild-type, hubP, and flhG V. cholerae, generated using electron microscopy, are shown. The number of flagella observed per cell for each mutant is graphed at the right. (C,E) Subcellular localization of FlhF-YFP (C), HubP-YFP (E), and FlhG-YFP (E) in the indicated V. cholerae strains. (D,F) FlhG-YFP (D) or FlhF-YFP (F) was expressed in E. coli in the absence (left) or presence (middle) of HubP-CFP (right). Representative fields are shown. Bar, 2 μm.
HubP interacts directly with FlhG and FlhF
Like ParA1, FlhG appears to be retained at the V. cholerae pole through direct interaction with HubP. Bacterial two-hybrid analyses suggest that the two proteins interact (Table 1; Supplemental Fig. S3), and coexpression of HubP along with FlhG in E. coli shifts FlhG from a diffuse cytoplasmic distribution to a pole-associated distribution that corresponds to the location of HubP (Fig. 4D). It is not clear why coexpression of these two proteins also alters the subcellular distribution of HubP (cf. Figs. 3D–F and 4D).
Unexpectedly, two-hybrid analyses suggest that FlhF also interacts with HubP (Table1), although neither protein is required for the polar targeting of the other (Fig. 4C,E). Analyses of FlhF and HubP localization in E. coli also suggest that they interact; expression of HubP shifts FlhF from a predominantly polar distribution to a pattern that matches HubP (Fig. 4F). Notably, FlhF, like HubP, is required for recruitment of FlhG to the V. cholerae pole (Fig. 4E). Thus, although the precise reason for an interaction between FlhF and HubP is unknown, it is possible that they act together to modulate the subcellular distribution of FlhG.
Deletion of hubP impairs polar localization of chemotactic signaling proteins and polar assembly of chemoreceptor arrays
In contrast to its relatively minor effect on flagellation, deletion of hubP has a far more dramatic effect on V. cholerae chemotaxis. Video tracking of swimming bacteria revealed that the hubP strain displays a significant bias toward straight swimming: Direction reversals were detected 0.16 ± 0.07 sec−1 for the mutant versus 0.57 ± 0.06 sec−1 for the wild-type strain (Fig. 5A). The effect of hubP deletion on swimming is more marked than that of a parC deletion (0.28 sec−1), consistent with its more pronounced effect on swarming on soft agar plates (Fig. 4A). Chemotaxis appears to be impaired due to mislocalization of components of chemotactic apparati in the hubP mutant. As previously shown (Briegel et al. 2009), electron cryotomography revealed that 39 of 100 wild-type cell poles examined had detectable chemoreceptor arrays (Fig. 5B). In marked contrast, only seven of 61 (11%) hubP cell poles had detectable arrays. Consistent with the reduced number of polar arrays, chemotaxis signaling protein CheY3 formed aberrant nonpolar foci in ∼60% of hubP cells (Fig. 5C). Furthermore, most of the nonpolar CheY3 foci colocalized with ParC (Fig. 5D), which we showed is also mislocalized in the hubP mutant (Fig. 1C). Together, these observations suggest that HubP (via ParC) promotes the polar localization of the entire chemoreceptor/signaling complex. Thus, at least in V. cholerae, the distribution of receptors and downstream signaling proteins is not simply the result of stochastic processes.
Figure 5.
Deletion of hubP indirectly alters the distribution and efficacy of vibrio chemotaxis proteins. (A) Positional tracking of swimming wild-type and ΔhubP V. cholerae. (B) Electron cryotomographs of wild-type V. cholerae. The region in the square is magnified in the inset. The arrow indicates chemoreceptor array. Arrowheads indicate outer membrane (OM), inner membrane (IM), and base plate (BP) of chemoreceptor array. (C) Foci of CheY3-CFP in wild-type and ΔhubP V. cholerae (left) and percentage of cells with mislocalized CheY3-CFP in indicated background (right). (D) CheY3-CFP and YFP-ParC foci in ΔhubP V. cholerae. Arrowheads indicate colocalization of nonpolar CheY3-CFP and YFP-ParC foci. (E) HubP-CFP (magenta) colocalizes with ParA1[K11E]-YFP (right; green) but not with YFP-ParC (left; green). Localization of YFP-ParC and ParA1[K11E]-YFP foci relative to neighboring HubP-CFP focus are plotted. Inner and outer circles correspond to diameters of 2 and 4 pixels, respectively. (F) Swarming of wild-type, ΔhubP, and flagella mutant (lacks both polar and lateral flagella) V. parahaemolyticus on a representative soft agar plate is pictured (left), and average swarm diameter relative to wild type, with standard deviation based on three or more experiments, is graphed (right). (G) Subcellular localization of YFP-ParCvp in the indicated V. parahaemolyticus strain. Representative fields are shown. Bar, 2 μm.
In contrast to ParA1 and FlhG, ParC does not appear to be anchored at the pole by directly interacting with HubP. The two proteins did not interact in a two-hybrid assay (Table 1); furthermore, there was at most a subtle change in the distribution of YFP-ParC in E. coli induced by coexpression of HubP-CFP, and the two proteins do not appear to be colocalized (Supplemental Fig. S5). Finally, although HubP-CFP and YFP-ParC are both polar proteins in V. cholerae, they do not precisely colocalize. Mapping of ParC's position relative to HubP (and ParA1[K11E]'s, for comparison) revealed that ParC was, on average, approximately two times farther from HubP than was ParA1[K11E]: 68.4 ± 40.4 nm versus 30.9 ± 16.4 nm (Student's t-test: 1.2 × 10−54) (Fig. 5E). Collectively, these observations suggest the possibility that HubP modulates the distribution of ParC indirectly, perhaps via the mediation of an as-yet-unidentified vibrio-specific factor. However, ParA and the other ParA-related ATPases do not serve this function, as previous analyses have shown that ParC localization is independent of ParA1 and FlhG as well as of FlhF (Ringgaard et al. 2011).
HubP promotes the polar localization of ParC and ParA in V. parahaemolyticus
Since the aberrant chemotaxis of the V. cholerae hubP mutant (Figs. 4A, 5A) was a particularly striking phenotype, we wondered whether hubP also modulates chemotaxis in other vibrios (e.g., V. parahaemolyticus). Like all vibrios and photobacteria, V. parahaemolyticus encodes homologs of V. cholerae HubP and ParC (69% and 86% similarity, respectively). We found that V. parahaemolyticus swarming motility, which is dependent on the organism's polar flagella, was reduced by ∼40% in a V. parahaemolyticus hubP mutant (Fig. 5F). Furthermore, as in V. cholerae, YFP-ParC routinely formed polar foci in wild-type V. parahaemolyticus. In the hubP mutant, however, YFP-ParC was often observed within nonpolar foci as well as polar foci (Fig. 5G). Thus, it seems likely that HubP's role as an organizer of chemotactic machinery is conserved, at least within the Vibrionaceae. Interestingly, there is a striking correlation in the coincidence of hubP and parC homologs among γ-proteobacteria (Supplemental Table S1), suggesting that HubP's influence over chemotaxis may extend through a broader swath of the bacteria kingdom. There is much less overlap between the distribution of HubP and ParA homologs (which are far more abundant). Nonetheless, we observed that HubP also appears to mediate the polar localization of ParA1 in V. parahaemolyticus (data not shown), indicating that HubP's role in chromosome partitioning is likewise not limited to V. cholerae.
Different HubP domains are required to mediate polar localization of ParA1, FlhG, and ParC
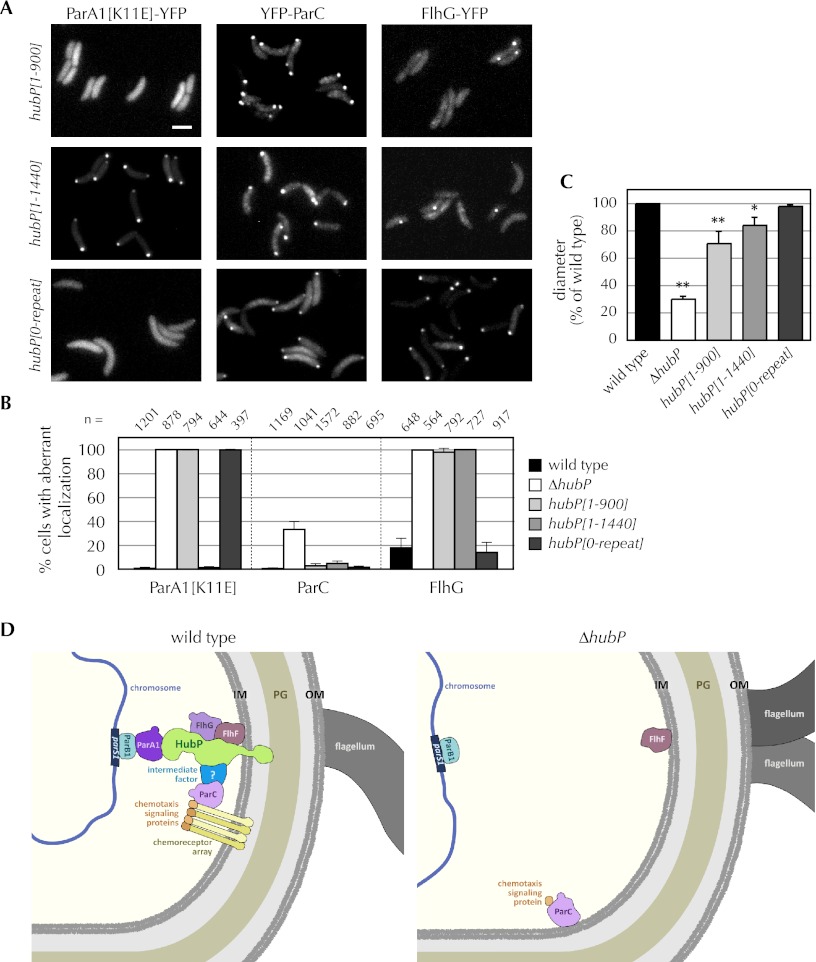
Our observation that HubP interacts directly with ParA1 and FlhG, but not with ParC, suggested that distinct regions of HubP might be important for determining the subcellular distribution of client proteins. We explored this possibility using the bacterial two-hybrid system and the fluorescence-based assays described above, except that hubP was altered by the presence of a series of deletion mutations (summarized in Table 1; data presented in Fig. 6; Supplemental Figs. S3, S5). Results from all of these systems suggest that the interaction between ParA1 and HubP requires amino acids within the repeat region of HubP. Truncation of the protein to remove the final 181 amino acids did not impair HubP's ability to interact with or guide the localization of ParA1 (hubP[1–1440] (Fig. 6A,B); however, deletion of the repeats (hubP[0-repeat]) or of the repeats along with the C terminus (hubP[1–900]) eliminated HubP's activity with respect to ParA1 (and ParA1[K11E]). In contrast, the extreme C terminus of HubP (amino acids 1441–1621) appeared to be essential for interaction between HubP and FlhG, at least based on FlhG-YFP localization in V. cholerae expressing HubP from the chromosome (Fig. 6A,B). Plasmid-based assays in E. coli suggest that additional regions of HubP may have a weak capacity to interact with FlhG (Supplemental Fig. S3); nonetheless, it is clear that amino acids 1441–1621 are important using these other assays as well. The C-terminal 181 amino acids were also critical for the interaction between HubP and FlhF (Supplemental Fig. S5F). Finally, HubP's modulation of ParC localization appears to depend on yet another domain within HubP. The first 900 amino acids of HubP were sufficient to localize ParC normally in V. cholerae (Fig. 6B), and chemotaxis (soft agar motility) by V. cholerae expressing HubP[1–900] was significantly greater than by a strain lacking the entire protein (Fig. 6C). Chemotaxis by the hubP[1–900] mutant was not equivalent to that by the wild-type strain; however, this may reflect the inadequacy of the truncated HubP for modulation of flagellar proteins. Consistent with this hypothesis, HubP[0-repeat], which interacts with both ParC and FlhG, enabled chemotaxis equivalent to that of the wild-type strain. Collectively, these results suggest that the cytoplasmic portion of HubP is likely to have multiple distinct interaction domains that mediate its spatial regulation of ParA family proteins.
Figure 6.
HubP appears to have multiple domains for interaction with partners. (A) Fluorescent fusion proteins (as indicated) were visualized in V. cholerae in which full-length hubP was replaced by a truncated gene. Representative fields are shown. Bar, 2 μm. (B) For each protein indicated, the frequency with which the aberrantly localized fusion protein was observed is plotted as a function of hubP genotype. Protein localization was scored in the indicated number (n) of cells. For ParA1 and FlhG, aberrant localization refers to cells lacking a polar focus; for ParC, it reflects the number of cells with a nonpolar focus. Mean and standard deviations and total number of cells counted (n) are shown. Wild-type and ΔhubP data are reiterated from Figure 1E for comparison. (C) Swarming of V. cholerae hubP mutants on soft agar plates was quantitated, relative to wild type, based on swarm diameter. Truncated HubP (as indicated) was expressed from an allele inserted within the chromosome in place of the wild-type gene. Plotted values reflect the means and standard deviations from three or more experiments. (*) P < 0.05; (**) P < 0.01 by Student's t-test. (D) Schematic of HubP's role in organizing the V. cholerae old cell pole. In wild-type cells, different regions of HubP anchor three ParA-related ATPases (ParA1, ParC, and FlhG) to the old cell pole. These ATPases mediate the polar localization of the centromeric ParB1/parS1 complex and chemotaxis receptor arrays and signaling proteins and modulate flagellar production, respectively. HubP anchoring of ParC to the pole appears to be mediated through an intermediate factor. In the absence of HubP (ΔhubP), ParA1 and FlhG are diffuse, the ParB1/parS1 complex is no longer polar, ParC and chemotaxis signaling proteins form nonpolar foci (although polar foci are observed as well), and there is a small increase in cells that contain two flagella; FlhF, which also interacts with HubP, remains at the pole.
Discussion
In rod-shaped bacteria, the cell poles are critical subcellular domains, but knowledge of the mechanisms that underlie pole formation, recognition, organization, and perpetuation is restricted to very few model organisms. In V. cholerae, polarly localized ParA-related proteins are essential for mediating the polar localization of DNA/protein (ParA1 and parS1/ParB1) and protein/protein complexes (ParC and CheY3) that are required for proper segregation of oriCI and the chemotactic machinery, respectively. FlhG, a third polar ParA-like protein, modulates formation of V. cholerae's polar flagellum. Here, we found that the polar localization of ParA1, ParC, and FlhG all depend on HubP, a large transmembrane protein that is conserved in all Vibrionaceae/Photobacteriacea as well as in several other γ-proteobacteria. In the absence of this polar anchor, origin segregation and chemotaxis are impaired, and cells produce extra flagella. Polar targeting of ParA1, ParC, and FlhG requires different regions within HubP, and the ParA family proteins clearly use distinct mechanisms (including both direct and indirect interactions with HubP) for their polar tethering. Thus, the organization of the vibrio cell pole is dependent on a complex and multifaceted protein hub that anchors chromosome segregation and chemotactic machinery to this site (Fig. 6D).
Strikingly, although HubP is required for the polar placement of its ParA-related clients, there is not a perfect correspondence between their subcellular localizations. HubP is always detectable at both the old and new pole in V. cholerae and often at the future division plane at mid-cell as well. In contrast, ParC does not form detectable foci at the new pole until late in the cell cycle (Ringgaard et al. 2011), and FlhG is only bipolar in a small subset of (typically long) cells (Fig. 1). Distinct ParA1 foci are also difficult to discern at the new pole until late in the cell cycle, which corresponds to the migration of replicated oriCI to this site. However, it is presumed that some ParA1 must be anchored at the new pole at the start of chromosome segregation in order for cycles of ParA1 polymerization/depolymerization to yield a net force pulling ParB1/oriCI across the cell (Fogel and Waldor 2006). Nonetheless, there appear to be restrictions on interactions between HubP and its clients at the new pole (and at the mid-cell prior to division). What limits HubP interactions with its clients at the new pole? It is possible that additional factors must be present at this site along with HubP in order to anchor HubP's partners (as appears to be the case for ParC). Alternatively, interactions at the new pole may be limited by the presence of an additional factor that is released or inactivated later in the cell cycle. Client-specific processes, such as interactions with additional protein partners, ATP, or the nucleoid, may also regulate protein localization and may account for the apparently distinct arrival times of ParA1, FlhG, and ParC at the new pole. It remains to be seen whether there is any temporal coupling among these processes (e.g., between oriCI segregation and movement of ParC to the new pole) or whether they are fully independent.
Similarly, it is noteworthy that some HubP-dependent proteins are not exclusively targeted to either pole for at least a portion of the cell cycle. Much of ParA1 is present within the cytoplasm, although its distribution there is dynamic, and much of FlhG is diffusely localized. Polar versus cytoplasmic localization is likely modulated at least in part by ATP binding and hydrolysis. Binding of ParA-like proteins to ATP typically enables them to dimerize, and dimerization is often required for their interactions with both other proteins (e.g., ParB and homologs) and DNA (Lutkenhaus 2012). Interestingly, our analyses indicate that ParA1 dimerization is not required for its interaction with HubP. In fact, ParA1[G12V], which is thought to not dimerize, is even routinely found at new poles (Supplemental Fig. S4), suggesting that dimerization may inhibit interaction with HubP. Our analyses also suggest that ATP binding and hydrolysis are not essential for the interaction between ParA1 and HubP, since ParA1[K16A] and ParA1[D40A], which fail to bind and hydrolyze ATP, respectively, still form some polar foci, although they do have a reduced presence at the poles relative to wild-type ParA1. The ability of client proteins to bind ATP could also indirectly modulate interactions with HubP; e.g., by altering their affinity for and positioning by other interaction partners. Ultimately, the distribution of HubP's clients between the cytoplasm and the pole likely reflects the relative affinities of the client proteins for their cytoplasmic partners versus HubP.
The means by which nascent HubP is initially recruited to the mid-cell or poles warrants further investigation. Polar targeting of HubP could be due to recognition of membrane curvature, as has been proposed for DivIVA of Bacillus subtilis (Lenarcic et al. 2009; Ramamurthi and Losick 2009). However, our preliminary analyses of HubP's distribution in the spherical or oval cells formed due to mutation of rodZ (Shiomi et al. 2008; Alyahya et al. 2009; Bendezú et al. 2009) and in MreB inhibitor MP265-treated (Takacs et al. 2010) cells suggest that HubP may not simply recognize the membrane curvature associated with polar regions, as HubP retains a focal distribution even in these cases. One possibility is that HubP recognizes features of PG that are stable and specific to the poles. Polar PG is thought to be relatively inert, and pole-specific PG structures formed during cell division might serve as a “birth scar” that might be used in proper cellular targeting of HubP. There is a precedent for PG fastening other proteins to poles; e.g., C. crescentus SpmX (Radhakrishnan et al. 2008). Alternatively, HubP association with polar PG (presumably through its LysM domain) may only stabilize its association with the poles after it is recruited there by an unknown factor. Determination of the relative timing of PG synthesis at the mid-cell versus HubP recruitment there could aid in distinguishing between these possibilities.
Although HubP routinely localizes at the poles and may interact with polar PG, HubP is not stably retained at these sites; instead, it can migrate from one pole to the other, apparently bidirectionally. The significance of and mechanism underlying this movement, which has not been reported for other polar markers, remain to be determined. Additionally, it has not been investigated whether partner proteins remain associated and move with HubP or whether they are instead released and recaptured. However, the fact that HubP is detected at the mid-cell or new pole of young cells apparently without associated partners may indicate that partners are released prior to or during movement. It is also possible that HubP acquires interacting partners during its transits across the cell. If so, bipolar movement of HubP might serve as a tool for recruiting its interaction partners to polar sites.
The most profound consequences of hubP deletion that we observed relate to its regulation of chemotaxis. In the absence of HubP, polar chemotactic receptor arrays are present at a markedly reduced frequency, and chemotactic signaling complexes (marked by ParC and CheY3) are often mislocalized, although they are assumed to be structurally intact. The altered distribution of these proteins presumably is the major factor underlying the mutant's impaired chemotaxis, detectable as reduced swarming on soft agar plates and a reduced reversal frequency in liquid culture. Notably, a HubP deficiency has a more profound phenotype than does a ParC deficiency, probably both because ParC does not modulate flagellar assembly and because the frequency at which receptors and associated signaling proteins accumulate at the pole remains relatively high in the absence of ParC, so signaling is not perturbed. In contrast, in the absence of HubP, ParC is frequently mislocalized, causing mislocalization of downstream signaling proteins. However, downstream signaling proteins are also frequently mislocalized in a strain lacking hubP and parC (data not shown). Thus, although mislocalization of ParC may be more detrimental to V. cholerae than the total absence of this protein, it is possible that ParC is not the only factor through which HubP controls the localization of chemotaxis proteins. Contrasting paradigms characterize HubP's impact on FlhG and ParA1. Mislocalization of FlhG has a relatively minor effect on flagellar assembly, far less than that of FlhG's absence, perhaps because some of FlhG's regulatory roles are not dependent on its subcellular distribution. Finally, the mislocalization and absence of ParA1 have similar consequences; both result in aberrant subcellular placement of oriCI.
Although the precise means by which HubP recruits and/or anchors ParC to the pole and thereby situates the remainder of the chemotactic machinery remains to be determined, bioinformatic analyses suggest that it may be a relatively conserved process. There is a strong correlation between genomes containing hubP and genomes containing parC, particularly among the γ-proteobacteria (Supplemental Table S1). Notably, since our data suggest that ParC and HubP do not interact directly, it is likely that an additional component that serves as a bridge between these two proteins will also be found to have coevolved.
HubP bears some functional similarity to TipN and PopZ, polar determinants and markers in C. crescentus, the organism in which the molecular bases for pole establishment and organization are best understood. Through their presence at the poles, all three of these proteins enable polar placement/anchoring of several additional proteins and protein complexes, such as the chromosome segregation machinery, whose proper activity depends on their polar localization. TipN, like HubP, is an early marker of new/nascent poles; both proteins are present at the mid-cell prior to cell division, thereby providing cells with cues regarding polar orientation at the start of their development. However, there are also notable differences in how these three pole-specifying proteins localize and function. In particular, HubP differs from TipN and PopZ in that it is always present at both poles, rather than marking the new pole (TipN) or primarily the old pole (PopZ). It seems possible that additional factors/processes in V. cholerae are therefore needed for the cell to distinguish between its polar regions. Additionally, there is no evidence that HubP is a primary determinant, rather than a marker, of pole formation, unlike TipN, whose overexpression can induce ectopic pole formation (Lam et al. 2006). Furthermore, HubP's clients identified to date are limited to ParA-like ATPases, whereas TipN and PopZ modulate the localization of structurally diverse proteins, although these include ParA family members (Schofield et al. 2010). The C. crescentus and V. cholerae pole-organizing proteins lack amino acid sequence similarity and clearly evolved through independent processes in α-proteobacteria and γ-proteobacteria. Despite their differences, it is striking that their core characteristics—polar proteins that anchor multiple clients to generate and perpetuate cell polarity—have so much in common. It seems likely that additional diverse hub proteins will control the activity of the growing family of ParA-related proteins that govern the polar localization of numerous subcellular structures.
Materials and methods
Plasmids and strains
Plasmids used in this study and their construction are described in the Supplemental Material (Supplemental Table S2; Supplemental Material). Oligonucleotides used for plasmid construction are listed in Supplemental Table S3. Nucleotide sequences of plasmids were confirmed whenever the construction involved PCR. Strains used in this study are listed in Supplemental Table S4. Briefly, we used El Tor O1 strains N16961 (Heidelberg et al. 2000) and C6706 (parental strain of the mapped Tn insertion library) (Cameron et al. 2008) as wild-type V. cholerae. Chromosomal deletions and insertions were introduced by allelic exchange (Donnenberg and Kaper 1991). For growth conditions and supplements including antibiotics, see the Supplemental Material.
Screening
The mapped Tn insertion library of V. cholerae contains 147 mutants showing a motility deficiency (<50% compared with wild type) (Cameron et al. 2008). Of these 147 mutants, the 56 that have Tn insertions in genes obviously linked to motility (e.g., flagellar biosynthesis genes) were discarded from the pool of candidates. A reporter plasmid encoding fluorescent fusion proteins, pBAD33 parA1[K11E]-yfp cheY3-cfp, was introduced into the remaining 91 mutants, and the subcellular localization of the fusion proteins in log-phase cells was manually examined using fluorescence microscopy.
Microscopy
For microscopy, see the Supplemental Material.
Electron microscopy
Cells were grown to O.D. ∼0.5 in LB, immobilized on grids, fixed with glutaraldehyde, and then repeatedly washed in water. Imaging was performed on a Tecnai G2 Spirit BioTWIN.
Cryomicroscopy
For cryomicroscopy, see the Supplemental Material.
Acknowledgments
We are grateful to Janet Iwasa for making Figure 6D, Paula Montero Llopis for an expert tutorial in Microbe Tracker, and John Mekalanos for providing the mapped transposon library. We also thank Richard Losick, Hubert Lam, and Waldor laboratory members for helpful discussions and comments on the manuscript. S.R. was funded with a post-doctoral fellowship from the Villum Kann Rasmussen Foundation. This work was supported by the NIAID R37 AI-042347 (to M.K.W.), NIGMS R01 GM094800B (to G.J.J), and HHMI (to M.K.W. and G.J.J.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.199869.112.
References
- Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C 2009. RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci 106: 1239–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ 2007. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J 26: 2540–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú FO, Hale CA, Bernhardt TG, de Boer PA 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532–536 [DOI] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Müller A, Iancu CV, Murphy GE, Dobro MJ, et al. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci 106: 17181–17186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DE, Urbach JM, Mekalanos JJ 2008. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci 105: 8736–8741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa NE, Peng F, Klose KE 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J Bacteriol 187: 6324–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg MS, Kaper JB 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun 59: 4310–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Ringgaard S, Møller-Jensen J, Wang Q, Sherratt DJ, Gerdes K 2006. Regular cellular distribution of plasmids by oscillating and filament-forming ParA ATPase of plasmid pB171. Mol Microbiol 61: 1428–1442 [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134: 956–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flärdh K 2010. Cell polarity and the control of apical growth in Streptomyces. Curr Opin Microbiol 13: 758–765 [DOI] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK 2006. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev 20: 3269–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, et al. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406: 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 124: 1025–1037 [DOI] [PubMed] [Google Scholar]
- Kusumoto A, Shinohara A, Terashima H, Kojima S, Yakushi T, Homma M 2008. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology 154: 1390–1399 [DOI] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C 2006. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124: 1011–1023 [DOI] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, Hamoen LW 2009. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J 28: 2272–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard TA, Butler PJ, Löwe J 2005. Bacterial chromosome segregation: Structure and DNA binding of the Soj dimer—a conserved biological switch. EMBO J 24: 270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J 2007. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem 76: 539–562 [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J 2012. The ParA/MinD family puts things in their place. Trends Microbiol 20: 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev 12: 3419–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cheeks BA, Planet PJ, Sarkar IN, Clock SA, Xu Q, Figurski DH 2012. The product of tadZ, a new member of the parA/minD superfamily, localizes to a pole in Aggregatibacter actinomycetemcomitans. Mol Microbiol 83: 694–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan SK, Thanbichler M, Viollier PH 2008. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev 22: 212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R 2009. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci 106: 13541–13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA 1997. The MinE ring: An FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91: 685–694 [DOI] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci 96: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringgaard S, Schirner K, Davis BM, Waldor MK 2011. A family of ParA-like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes Dev 25: 1544–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Losick R 2010. Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol 2: a000307 doi: 10.1101/cshperspect.a000307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield WB, Lim HC, Jacobs-Wagner C 2010. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J 29: 3068–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmler AB, Whitchurch CB, Leech AJ, Mattick JS 2000. Identification of a novel gene, fimV, involved in twitching motility in Pseudomonas aeruginosa. Microbiology 146: 1321–1332 [DOI] [PubMed] [Google Scholar]
- Shapiro L, McAdams HH, Losick R 2009. Why and how bacteria localize proteins. Science 326: 1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Sakai M, Niki H 2008. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J 27: 3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szardenings F, Guymer D, Gerdes K 2011. ParA ATPases can move and position DNA and subcellular structures. Curr Opin Microbiol 14: 712–718 [DOI] [PubMed] [Google Scholar]
- Takacs CN, Poggio S, Charbon G, Pucheault M, Vollmer W, Jacobs-Wagner C 2010. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J Bacteriol 192: 1671–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M 2011. Good things come in small packages: Subcellular organization and development in bacteria. Curr Opin Microbiol 14: 687–690 [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126: 147–162 [DOI] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L 2002. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J 21: 4420–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehbi H, Portillo E, Harvey H, Shimkoff AE, Scheurwater EM, Howell PL, Burrows LL 2011. The peptidoglycan-binding protein FimV promotes assembly of the Pseudomonas aeruginosa type IV pilus secretin. J Bacteriol 193: 540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Christen B, Chiu HJ, Jaroszewski L, Klock HE, Knuth MW, Miller MD, Elsliger MA, Deacon AM, Godzik A, et al. 2012. Structure of the pilus assembly protein TadZ from Eubacterium rectale: Implications for polar localization. Mol Microbiol 83: 712–727 [DOI] [PMC free article] [PubMed] [Google Scholar]