Abstract
Sodium-dependent multivitamin transporter (SMVT) is a vital transmembrane protein responsible for translocating biotin and other essential cofactors such as pantothenate and lipoate. Unlike primary cultures of corneal and retinal pigment epithelial (RPE) cells, immortalized cells can be subcultured many times, yet maintain their physiological properties. Hence, the purpose of this study was to delineate the functional and molecular aspects of biotin uptake via SMVT on immortalized human corneal epithelial (HCEC) and RPE (D407) cells. Functional aspects of [3H] biotin uptake were studied in the presence of different concentrations of unlabeled biotin, pH, temperature, metabolic inhibitors, ions, substrates, structural analogs and biotinylated prodrug (Biotin-Acyclovir (B-ACV)). Molecular identity of SMVT was examined with reverse transcription–polymerase chain reaction. Biotin uptake was found to be saturable in HCEC and D407 cells with Km of 296.2 ± 25.9 and 863.8 ± 66.9 μM and Vmax of 77.2 ± 2.2 and 308.3 ± 10.7 pmol/mg protein/min, respectively. Uptake was found to be pH, temperature, energy, and sodium-dependent. Inhibition of biotin uptake was observed in the presence of structural analogs and specific substrates. Further, uptake was lowered in the presence of B-ACV indicating the translocation of biotinylated prodrug by SMVT. A distinct band at 774 bp confirmed the molecular existence of SMVT in both the cells. This study shows for the first time the functional and molecular presence of SMVT in HCEC and D407 cells. Therefore, these cell lines may be utilized as in vitro models to study the cellular translocation of biotin-conjugated prodrugs.
Electronic supplementary material
The online version of this article (doi:10.1208/s12248-012-9399-5) contains supplementary material, which is available to authorized users.
KEY WORDS: biotinylated prodrug., human corneal epithelial cells (HCEC), human retinal pigment epithelial cells (D407), in vitro cell culture models, SMVT
INTRODUCTION
Sodium-dependent multivitamin transporter (SMVT; product of the SLC5A6 gene) is a vital transmembrane protein responsible for translocating vitamins and other essential cofactors to the ocular tissues (1–3). This influx transporter has broad substrate specificity and can translocate biotin (a coenzyme for carboxylation reaction), pantothenic acid (a component of coenzyme A) and lipoic acid (a component in oxidative decarboxylation of pyruvate and α-ketoglutarate). Biotin (vitamin H) is an essential water-soluble vitamin required for normal cellular function, growth, and development. It cannot be synthesized by human or any other mammalian cells and generally depends on exogenous sources (4,5). It has many functions in metabolism and serves as a regulator in cell signaling pathways and gene expression (6,7). Biotin translocation is known to be either mediated by biotin transporter or SMVT. The former is a low-capacity, high-affinity transporter which regulates only the uptake of biotin (8); while the latter is a low-affinity, high-capacity transporter which can translocate biotin, pantothenic, and lipoic acid (9–13).
SMVT has been previously identified on intestine, placenta, liver, and kidney (14–17). The presence of SMVT on rabbit cornea and retina has been reported previously from our laboratory (2,3). Though excised rabbit tissues are generally utilized for various studies, the anatomical and physiological disparities between the rabbit and human eye suggest that the expression of transporters and resulting in vitro permeation data could also differ (18–21). Such differences make it very difficult to draw conclusions or predict drug absorption into human eyes (22). This generated in us the idea of studying the usefulness of human derived cells as in vitro models to predict ocular drug absorption into the human eye.
In vitro cell culture models have recently gained importance as valuable tools to predict ocular permeation of various drugs. Introduction of these cell culture models has significantly reduced the number of animal experiments, thus providing a platform for further investigations on ocular drug delivery (23–25). Primary cultures of rabbit corneal (rPCEC) and human retinal pigment epithelial (ARPE-19) cells have been used for years to evaluate drug transport into ocular tissues. Primary cultures rarely survive more than a few passages owing to their rapidly differentiating nature, especially rPCEC cells are limited to ten passages and ARPE-19 cells require 21 days of culture prior to experimentation. Moreover, these cultures frequently lose metabolic and morphological characteristics, particularly enzymatic activity, and cytoskeletal polarization (26,27). Unlike primary cultures, immortalized human corneal epithelial (HCEC) and RPE (D407) cells can be subcultured many times, yet maintain their physiological properties (26,28). Since its development, SV40-adenovirus-infected HCEC cells have become the most frequently investigated corneal cell culture model (29). These cells have been predominantly employed for studying the role of transporters in cellular translocation of the drug, bioavailability prescreening and toxicity evaluations (30–38). D407 cells, a cell line of human RPE origin are being extensively employed as an in vitro model. These cultures have been cloned from a primary culture of human RPE cells and are spontaneously transformed (26). Recent literature showcases its wide usage in identification of receptors, efflux as well as influx transporters because of its ability to preserve epithelial characteristics even after prolonged culture (39–43).
To date, no information currently exists with regard to the presence of SMVT on human corneal (HCEC) and RPE (D407) cells. Hence, the purpose of the present study is to shed light on functional and molecular aspects of biotin uptake by SMVT in HCEC and D407 cells. We also investigated the feasibility of selecting these cells as in vitro models to examine the transporter recognition and uptake of biotin-conjugated prodrugs using acyclovir (ACV) as a model drug.
MATERIALS AND METHODS
Materials
[3H] Biotin (specific activity 60 Ci/mmol) was procured from Perkin Elmer (Boston, MA, USA). Unlabeled biotin, lipoic acid, pantothenic acid, desthiobiotin, sodium azide, ouabain, 2,4-dinitrophenol, choline chloride, triton X-100, HEPES, bovine insulin, human epidermal growth factor and d-glucose were purchased from Sigma Chemical Co (St. Louis, MO, USA). NHS biotin and biocytin were acquired from ProChem, Inc (Rockford, IL, USA) and Tocris Biosciences (Ellisville, MO, USA), respectively. Dulbecco’s modified Eagle’s medium (DMEM) and Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12) were purchased from Invitrogen (Carlsbad, CA, USA). Fetal bovine serum (FBS) was obtained from Atlanta biologicals (Lawrenceville, GA, USA). Culture flasks (75 cm2 growth area) and uptake plates (3.8 cm2 growth area) were purchased from Corning Costar Corp. (Cambridge, MA, USA). The buffers for cDNA synthesis and amplification (oligodT, dNTP, MgCl2, M-MLV reverse transcriptase and Taq polymerase) were purchased from Promega Corporation (Madison, WI, USA). Light Cycler 480® SYBR I green master mix was obtained from Roche Applied Science (Indianapolis, IN, USA). Qualitative and quantitative primers used in the study were custom-designed and obtained from Invitrogen Life Technologies (Carlsbad, CA, USA). All other chemicals were obtained from Fisher Scientific Co. (Fair Lawn, NJ, USA) and utilized without further purification.
Cell Culture
HCEC and D407 cells were generous gifts from Dr. Araki-Sasaki (Kinki Central Hospital, Japan) and Dr. Richard Hunt (University or South Carolina, Columbia, SC, USA), respectively. HCEC cells were cultured following a previously published protocol (31,32,34). Cells were cultured at 37°C, humidified 5% CO2/95% air atmosphere in a culture medium containing DMEM/F-12 supplemented with 15% (v/v) FBS (heat inactivated), 15 mM HEPES, 22 mM NaHCO3, 100 mg of penicillin and streptomycin each, 5 μg/ml insulin, and 10 ng/ml of human epidermal growth factor. Cells of passage numbers between 25 and 30 were utilized for all the experiments.
D407 cells were grown at 37°C, humidified 5% CO2/95% air atmosphere in a culture medium containing DMEM supplemented with 10% (v/v) FBS (heat inactivated), 29 mM NaHCO3, 20 mM HEPES, 100 mg of penicillin and streptomycin each, and 1% nonessential amino acids at pH 7.4. Cells of passage numbers between 75 and 80 were employed for all the experiments (26,41). The growth medium was changed every other day. Both HCEC and D407 cells were cultured in flasks, harvested at 80–90% confluency with TrypLE™ Express (a superior replacement for trypsin) (Invitrogen, Carlsbad, CA, USA). Cells were then plated in 12-well uptake plates at a density of 250,000 cells/well. Cells were grown in a similar way in these plates and utilized for further studies.
Synthesis of Biotin-Acyclovir (B-ACV)
Biotin (100 mg, 0.40 mmol) was dissolved in anhydrous dimethyl formamide (DMF), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (152 mg, 0.80 mmol) was added and stirred for 1 h. In a separate reaction flask ACV (184 mg, 0.80 mmol) was dissolved in DMF, 4-dimethylaminopyridine (58 mg, 0.48 mmol) was added and stirred for 10 min at room temperature to activate the hydroxyl group of ACV. This mixture was then added dropwise into the reaction containing biotin through a syringe, and stirred continuously for 72 h under an inert atmosphere. Small portions of the reaction mixture were taken out and injected into LC/MS to ensure the complete conversion of the starting material to product. The reaction mixture is then filtered and evaporated at room temperature under reduced pressure to generate crude product. The product B-ACV was purified by silica gel column chromatography with 10% methanol/dichloromethane as eluent. The yield was approximately 78%. The synthetic scheme has been summarized in Scheme 1.
Scheme 1.
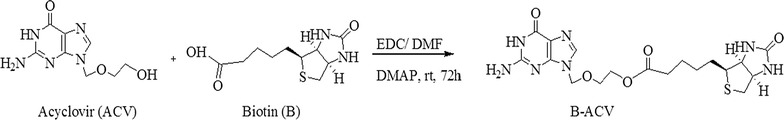
Synthesis of B-ACV. Biotin-Acyclovir (B-ACV): White solid; LC/MS (m/z): 452.1; 1HNMR(DMSO-d6): δ 1.24–1.35 (m, 2H), 1.39–1.51 (m, 3H), 1.54–1.63 (m, 1H), 2.17–2.24 (m, 2H), 2.55–2.58 (m, 1H), 2.79–2.84 (m, 1H), 3.05–3.09 (m, 1H), 3.64–3.67 (m, 2H), 4.07–4.15 (m, 3H), 4.29–4.32 (m, 1H), 5.34 (s, 2H), 6.37 (brs, 1H), 6.43 (brs, 1H), 6.55 (brs, 1H), 7.81 (s, 1H), 10.69 (brs, 1H); 13C NMR(DMSO-d6): 24.44, 27.96, 33.17, 55.38, 59.20, 61.06, 62.63, 66.59, 71.83, 116.48, 137.74, 151.45, 153.95, 156.83, 162.76, 172.79
B-ACV was characterized by 1H NMR, 13C NMR and LC/MS. 1H and 13C NMR spectra were recorded using tetra methyl silane as an internal standard on a Varian Mercury 400 Plus spectrometer. Chemical shifts (δ) were reported in parts per million relative to the NMR solvent signal (dimethyl sulfoxide (DMSO)-d6, 2.51 ppm for proton and 39.30 ppm for carbon NMR). A hybrid triple quadrupole–linear ion trap mass spectrometer (QTrap® LC/MS/MS spectrometer—Applied Biosystems) under enhanced mass mode was used for carrying out the mass analysis. Electrospray ionization was used as an ion source and operated in positive ion mode.
Uptake Studies
Uptake experiments were carried out with confluent cells 7–9 days post seeding for HCEC and 4–5 days post seeding for D407 cells. Prior to experimentation, the medium was aspirated and cells were rinsed thrice for 5 min each with 1–2 ml of Dulbecco’s phosphate-buffered saline (DPBS) containing 130 mM NaCl, 0.03 mM KCl, 7.5 mM Na2HPO4, 1.5 mM KH2PO4, 1 mM CaCl2, 0.5 mM MgSO4, 20 mM HEPES, and 5 mM glucose maintained at pH 7.4. Uptake was initiated by adding 500 μl of solution containing 0.5 μCi/ml of [3H] biotin in the presence and absence of various competing substrates. Following incubation, the solution was removed and uptake was terminated with 2 ml of ice-cold stop solution containing 200 mM KCl and 2 mM HEPES. Cells were lysed overnight at room temperature with 1 ml of lysis solution containing 0.05% (v/v) Triton X-100 in 1 N NaOH. Subsequently, the cell lysate (500 μl) from each well was transferred to scintillation vials containing 3 ml of scintillation cocktail (Fisher Scientific, Fairlawn, NJ, USA). Samples were then quantified for radioactivity using liquid scintillation spectrophotometer coulter (Beckman Instruments Inc., Fullerton, CA, USA, model LS-6500). Protein content of each sample was estimated by BioRad Protein Estimation Kit (BioRad Protein Estimation Kit, Hercules, CA, USA) using bovine serum albumin as an internal standard. The uptake rate was then normalized to its corresponding protein content.
Time and pH Dependence
Uptake of [3H] biotin on HCEC and D407 cells was assessed at different time intervals (5, 10, 15, 30, 45 and 60 min) to determine the optimal time required for uptake studies. Further, to examine the effect of extracellular pH on uptake of [3H] biotin, cells were washed and incubated with solutions of different pHs (4, 5, 6, 7.4, and 8). Also, an uptake study was performed at 4°C to differentiate the passive diffusion from the active transport in biotin uptake.
Concentration Dependence
A concentrated stock solution of biotin was prepared in DMSO. Subsequently, various concentrations (0.1–2,000 μM and 0.5–4,000 μM) of unlabeled biotin were prepared in DPBS and spiked with 0.5 μCi/ml of [3H] biotin. Then, concentration-dependent uptake of biotin was carried out in both HCEC and D407 cells. The kinetic parameters (Km and Vmax) were assessed by fitting this data into the Michaelis–Menten equation using nonlinear least squares regression analysis program (KaleidaGraph version 3.5).
Temperature and Energy Dependence
Temperature dependency of [3H] biotin uptake was determined by performing the uptake at three different temperatures (37°C, 25°C and 4°C). The uptake rate (ln(V)) was plotted against inverse temperature (1/T) to calculate activation energy (Ea). To delineate the energy requirements of the carrier system, uptake of [3H] biotin was carried out in the presence of metabolic energy inhibitors (500 μM)—ouabain, sodium azide, and 2,4-dinitrophenol (DNP).
Ion Dependence
Chloride-free buffer was prepared by replacing sodium (130 mM), potassium (0.03 mM), and calcium (1 mM) chlorides in DPBS buffer with equimolar quantities of sodium phosphate, potassium phosphate, and calcium acetate, respectively. This chloride-free buffer was then used to delineate the effect of chloride ions on cellular uptake of [3H] biotin. In a similar way, buffer solution-containing sodium chloride (130 mM) and disodium phosphate (7.5 mM) was replaced with equimolar quantities of choline chloride and dipotassium phosphate to investigate sodium involvement. This sodium-free buffer was then used to examine sodium dependency of SMVT. Uptake was also performed in the presence of amiloride, a sodium channel blocker. Further, to calculate the number of sodium ions required for translocation of one biotin molecule, uptake of [3H] biotin was carried out in the presence of different concentrations of sodium ions. The Hill coefficient was then calculated following a previous published method (44).
Substrate Specificity
Uptake of [3H] biotin was carried out in the presence of different concentrations (10, 50, and 100 μM) of competitive inhibitors for SMVT such as lipoic acid, pantothenic acid, and desthiobiotin. Structural requirements for translocation by SMVT were also delineated by performing uptake of [3H] biotin in the presence of different concentrations (10, 50, and 100 μM) of valeric acid (which possesses a free carboxylic acid group), biocytin, and NHS biotin (devoid of free carboxylic groups).
Interaction of B-ACV with SMVT
Uptake of [3H] biotin was carried out in the presence of 50 μM of unlabeled biotin, ACV and B-ACV to delineate their interaction with SMVT in both HCEC and D407 cells. Also, cellular accumulation of B-ACV was studied on HCEC and D407 cells following a recently published procedure (45). Uptake was initiated by adding 1 ml of 50 μM concentrations of ACV and B-ACV into each well and incubated for a period of 30 min. After incubation, drug solutions were removed and uptake was terminated with ice-cold stop solution. Cells were lysed overnight at −80°C in 500 μL cremophore water (two drops of cremophore gel in 50 ml of deionized water) in each well. Samples were then analyzed with LC/MS/MS and the rate of uptake was normalized to the protein content of each well. The amount of protein in the cell lysate was estimated with BioRad protein estimation kit (BioRad, Hercules, CA) using bovine serum albumin as an internal standard.
LC/MS/MS Analysis
Stocks and stock dilutions of ACV and B-ACV were prepared similarly following a recently published procedure (45). Samples were extracted by a liquid–liquid extraction method and ganciclovir (GCV) was used as an internal standard to ensure reproducibility and reliability of the method. Prior to analysis, samples were thawed at room temperature. Two hundred microliter samples containing either ACV or B-ACV along with 20 μl of GCV (5 μg/ml) was extracted with 1 ml of organic solvent mixture containing 2:3 ratios of isopropanol and dichloromethane. Samples were vortexed vigorously for approximately 2 min and centrifuged at 12,000×g for 15 min at 4°C. The organic layer (850 μl) was transferred into Eppendorf tubes and evaporated to dryness under speed vacuum with a Speedvac (SAVANT Instruments, Inc., Holbrook, NY). The dry residue was then reconstituted in 100 μL of mobile phase, vortexed for 30 s and transferred into pre-labeled vials with silanized inserts. Subsequently, 15 μl of the resulting solution was injected onto LC/MS/MS.
A fast and sensitive LC/MS/MS method has been recently developed in multiple reaction monitoring (MRM) with electrospray positive ionization mode for the analysis of ACV and B-ACV (45). Briefly, QTrap® LC/MS/MS mass spectrometer (API 3200, Applied Biosystems/MDS Sciex, Foster City, CA, USA) was employed to analyze samples from non-radioactive cellular accumulation studies. Chromatographic separation of compounds was achieved on XTerra® RP8 Column, 5 μm, 4.6 × 50 mm (Waters Corporation, Milford, MA) with an isocratic mobile phase. The mobile phase consisted of 70% acetonitrile, 30% water, and 0.1% formic acid which were pumped at a flow rate of 0.2 mL/min. Precursor ions of the analytes as well as internal standard were determined from spectra obtained during the infusion of standard drug/prodrug solutions with an infusion pump connected directly to the electrospray ionization source. Each of these precursor ions was subjected to collision-induced dissociation to determine their respective product ions. MRM transitions at m/z [M+H]+ generated were 226.4/152.2 for ACV, 452.3/301.3 for B-ACV and 256/152 for GCV (internal standard (IS)). Appropriate calibration standards of ACV and B-ACV were prepared by spiking known analyte concentrations to blank cell homogenate (HCEC and D407) obtained from cultured cells following similar procedure. A calibration curve was generated using calibration standards. Analyst™ software was employed to integrate the peak areas for all components and the respective peak-area ratios (area of analytes to area of IS) were plotted against concentration by weighted linear regression (1/concentration).
Reverse Transcription–Polymerase Chain Reaction
Expression of SMVT on HCEC and D407 cells was determined at the molecular level with reverse transcription–polymerase chain reaction (RT-PCR) analysis. Cells were lysed with TRIzol® reagent (Invitrogen, USA) and chloroform was added to the lysate for phase separation. The aqueous phase containing RNA was separated and isopropanol was added to precipitate RNA. RNA obtained was washed twice with 75% ethanol and then resuspended in RNase-DNase free water. The concentration and purity of RNA was determined using Nanodrop (Thermo Fisher Scientific, Wilmington, DE, USA). RNA was reverse-transcribed to obtain cDNA using oligodT as a template and M-MLV reverse transcriptase. The conditions for reverse transcription were: denaturation of the template RNA for 5 min at 70°C; reverse transcription for 60 min at 42°C followed by final extension at 72°C for 5 min. cDNA obtained was then subjected to PCR for amplification of SMVT using specific primers. The primers (5′→3′) designed were forward—AGGGCTGCAGCGGTTCTATT and reverse—GCAGCTTCCAGTTTTATGGTGGAG. These primers correspond to a 774 base pair (bp) product in human SMVT cDNA (nucleotide positions 2046–2820). The conditions of PCR amplification were: denaturation for 30 s at 94°C, annealing for 1 min at 56°C, and extension for 1 min at 72°C, for 45 cycles followed by a final extension for 5 min at 72°C. PCR product obtained was analyzed by gel electrophoresis on 1.5% agarose in TAE buffer and visualized under UV.
Quantitative Real-Time PCR
Following reverse transcription, quantitative real-time PCR (qPCR) was performed using LightCycler® SYBR green technology (Roche). cDNA equivalent to 80 ng in each well was subjected to amplification using specific primers. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control to normalize the amount of cDNA in each well. The sequences of real-time primers (5′→3′) used were SMVT: forward—TACCAGTTCTGCCAGCCACAGTG and reverse—CAGGGACACCAAAACCTCCCTCT and GAPDH: forward—ATCCCTCCAAAATCAAGTGG and reverse—GTTGTCATGGATGACCTTGG. A preliminary experiment was performed to ensure that SMVT and GAPDH were amplified with equal efficiencies. The specificity of these primers was also confirmed with melting-curve analysis. The comparative threshold method was used to calculate the relative amount of SMVT in different samples (46).
Data Analysis
Radioactive Sample Analysis
Disintegrations per minute (DPM) of sample and donor solutions were used to calculate uptake of [3H] biotin as shown in Eq. 1.
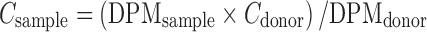 |
1 |
DPMsample and DPMdonor denote average values of DPM counts of sample and donor (n = 4) respectively; Cdonor represents the concentration of donor used and Csample represents the concentration of sample.
Calculation of Km and Vmax
Data obtained from the concentration-dependence study was fitted to the classical Michaelis–Menten equation as shown in Eq. 2:
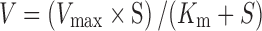 |
2 |
The uptake of [3H] biotin at a given substrate concentration [S] is represented by V. The data was fitted into a nonlinear least square regression analysis program (KaleidaGraph version 3.5, Synergy Software, Reading, PA, USA) to calculate the kinetic parameters Vmax and Km.
Calculation of Hill Ratio
The coupling ratio of [Na+] to biotin translocation was determined using the logarithmic form of the Hill equation as shown in Eq. 3:
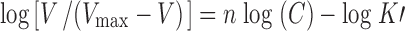 |
3 |
The uptake of [3H] biotin at a given substrate concentration [C] is represented by V and Vmax represents the maximum uptake rate. K′ is an apparent dissociation constant and n is the Hill coefficient thus calculated.
Statistical Analysis
All the experiments were conducted at least in quadruplicate (n = 4) and the results were expressed as mean ± standard deviation (SD) or mean ± standard error (SE). Student’s t test was used to calculate statistical significance and a P value of less than 0.05 was considered to be statistically significant.
RESULTS
Synthesis
The mass and NMR (both 1H NMR and 13C NMR) data for biotinylated prodrug B-ACV is summarized in Scheme 1.
Uptake Studies
Time and pH Dependence
Time-dependent [3H] biotin uptake by HCEC and D407 cells was depicted in Fig. 1. Uptake of [3H] biotin was linear until 60 min of incubation time. Hence, 30 min uptake time was selected for all subsequent uptake experiments. To examine the probable contribution of hydrogen ions on biotin uptake, incubation was carried out at pH ranging from 4.0 to 8.0. [3H] biotin uptake was highest at pH 4 and decreased with a rise in buffer pH from 4.0 to 7.4 in both HCEC and D407 cells (Fig. 2). Further, biotin uptake was significantly diminished in both HCEC and D407 cells when studies were performed at the lower temperature of 4°C instead of 37°C.
Fig. 1.
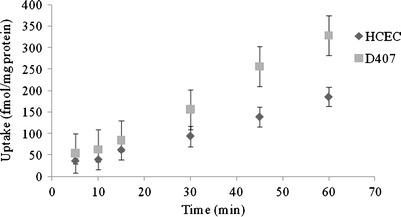
Time-dependent uptake of [3H] biotin in HCEC and D407 cells. Uptake of [3H] biotin was performed at 37°C with DPBS buffer (pH 7.4). Values represent mean ± SD (n = 4)
Fig. 2.
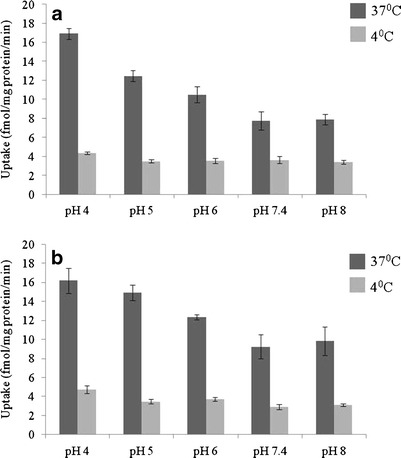
Effect of pH on [3H] biotin uptake in a HCEC and b D407 cells. Uptake of [3H] biotin was determined in the presence of different pH (4.0, 5.0, 6.0, 7.4 and 8.0) at 37°C and 4°C for 30 min. Values represent mean ± SD (n = 4)
Concentration Dependence
Saturation kinetics of biotin uptake were assessed by incubating the cells with various concentrations of unlabeled biotin for 30 min at 37°C. Biotin uptake in HCEC was found to be concentration-dependent and saturable with a Km and Vmax values of 296.2 ± 25.9 μM and 77.2 ± 2.2 pmol/mg protein/min, respectively (Fig. 3a). Also, a similar saturation kinetics plot was observed for D407 cells, but the kinetic parameters Km and Vmax values were higher than those observed with HCEC cells. The Km and Vmax values obtained from the plot were 863.8 ± 66.9 μM and 308.3 ± 10.7 pmol/mg protein/min, respectively (Fig. 3b). Lineweaver–Burk transformation of the above data obtained from both the cell lines indicate the involvement of a single carrier in the uptake process (data not shown).
Fig. 3.
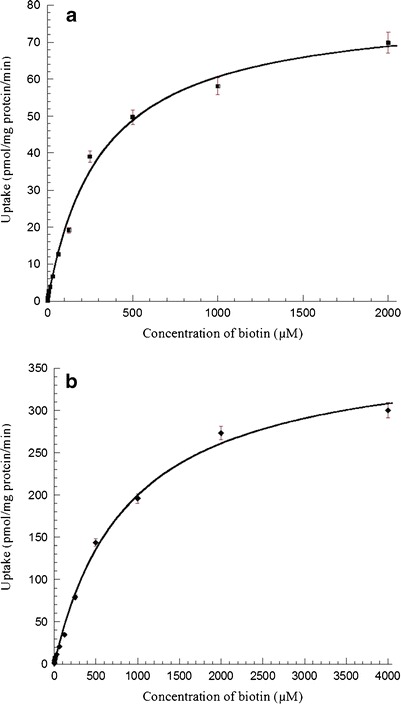
a Saturation kinetics of biotin uptake by HCEC cells at pH 7.4 in presence of different concentrations of unlabeled biotin (0.1–2,000 μM). Uptake was performed at 37°C for 30 min. Values represent mean ± SE (n = 4) of two independent experiments. b Saturation kinetics of biotin uptake by D407 cells at pH 7.4 in presence of different concentrations of unlabeled biotin (0.5–4000 μM). Uptake was performed at 37°C for 30 min. Values represent mean ± SE (n = 4) of two independent experiments
Temperature and Energy Dependence
Effect of temperature on [3H] biotin uptake by HCEC and D407 cells was studied. Cellular accumulation of biotin was inhibited to the extent of about 50% and 80% in HCEC cells at 25°C and 4°C, respectively, when compared to the uptake process at 37°C. In D407 cells, the uptake was decreased by approximately 50% and 85% when measured at 25°C and 4°C, respectively (Fig. 4a). Activation energy (Ea) was calculated to be 8.34 and 9.77 kcal/mol in HCEC and D407 cells, respectively (Fig. 4b). Effect of metabolic inhibitors on [3H] biotin uptake was studied on both HCEC and D407 cells. Biotin uptake was significantly inhibited by ouabain (Na+/K+-ATPase inhibitor), sodium azide (oxidative phosphorylation inhibitor), and 2,4-DNP (intracellular ATP reducer) (Fig. 5). This suggests that biotin uptake in both the cell lines is an active energy-dependent process and sensitive to alterations in temperature.
Fig. 4.
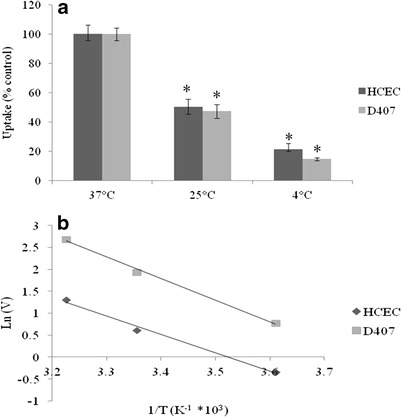
a Uptake of [3H] biotin by HCEC and D407 cells as a function of temperature (37°C, 25°C and 4°C). [3H] biotin uptake was performed at different temperatures with DPBS buffer (pH 7.4) for 30 min. Values represent mean ± SD (n = 4). A P value of < 0.05 is considered to be statistically significant and denoted by *. b Arrhenius plot of the effect of temperature on [3H] biotin uptake by HCEC and D407 cells
Fig. 5.

Uptake of [3H] biotin by HCEC and D407 cells in the presence of 500 μM concentrations of ouabain, sodium azide, and 2,4-DNP. [3H] biotin uptake was performed at 37°C with DPBS buffer (pH 7.4) for 30 min. Values represent mean ± SD (n = 4). A P value of < 0.05 is considered to be statistically significant and denoted by *
Ion Dependence
As depicted in Fig. 6, significant inhibition (>55%) in the uptake of [3H] biotin was observed in both HCEC and D407 cells in the presence of sodium-free media. Also, biotin uptake was reduced in the presence of amiloride (sodium ion transport inhibitor) indicating that this process may be sodium dependent (Fig. 6a). On the other hand, no significant difference in uptake was observed when chloride was replaced with equimolar concentrations of other monovalent cations in DPBS buffer. Since the uptake process in both the cell lines was found to be highly sodium dependent, uptake rate kinetics was investigated with various concentrations of sodium in the incubation buffer. Uptake rate increased with higher sodium concentrations and demonstrated saturation kinetics, reaching saturation at about 70 mM of sodium concentration in both HCEC and D407 cells (Fig. 6b). Hill transformation of the above data showed 1:1 molar ratio of Na+: biotin coupling in both cells (data not shown).
Fig. 6.
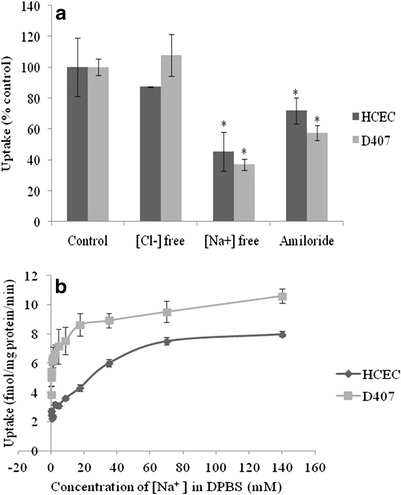
a Uptake of [3H] biotin by HCEC and D407 cells in the presence of amiloride and absence of sodium and chloride ions in DPBS buffer (pH 7.4) at 37°C. Values represent mean ± SD (n = 4). A P value of < 0.05 is considered to be statistically significant and denoted by *. b Uptake of [3H] biotin by HCEC and D407 cells as a function of sodium concentration in DPBS (pH 7.4) at 37°C. Values represent mean ± SE (n = 4) of two independent experiments
Substrate Specificity
To examine the structural requirements for interactions with the carrier system, cellular accumulation studies were carried out as described previously in the presence of various known substrates and structural analogs. Concentration-dependent inhibition of [3H] biotin uptake was observed in the presence of 50, 100, and 250 μM concentrations of lipoic acid, pantothenic acid, and desthiobiotin (Table I) in both the cell lines. A similar type of concentration-dependent inhibition of [3H] biotin uptake was found with the presence of 50, 100, and 250 μM concentrations of valeric acid, biocytin, and NHS biotin (Table II).
Table I.
Effect of Various Substrates on [3H] Biotin Uptake in HCEC and D407 Cells
| Substrates | Uptake (% of control) | |
|---|---|---|
| HCEC | D407 | |
| Control | 100 ± 23 | 100 ± 11 |
| Lipoic acid (50 μM) | 28.7 ± 0.3* | 23.8 ± 1.8* |
| Lipoic acid (100 μM) | 24.2 ± 1.8* | 21.7 ± 1.4* |
| Lipoic acid (250 μM) | 17.3 ± 0.9* | 19.9 ± 1.4* |
| Pantothenic acid (50 μM) | 44.3 ± 3.2* | 33.6 ± 4.5* |
| Pantothenic acid (100 μM) | 35.6 ± 2.6* | 31.4 ± 1.8* |
| Pantothenic acid (250 μM) | 25.2 ± 2.3* | 28.6 ± 2.6* |
| Desthiobiotin (50 μM) | 25.5 ± 1.6* | 23.7 ± 2.8* |
| Desthiobiotin (100 μM) | 15.3 ± 1.4* | 22.8 ± 1.5* |
| Desthiobiotin (250 μM) | 15.2 ± 0.7* | 18.5 ± 2.1* |
Data represents mean ± SD (n = 4)
*P value < 0.05 is considered to be statistically significant
Table II.
Effect of Various Structural Analogs on [3H] Biotin Uptake in HCEC and D407 Cells
| Structural analogs | Uptake (% of control) | |
|---|---|---|
| HCEC | D407 | |
| Control | 100 ± 5 | 100 ± 5 |
| Valeric acid (50 μM) | 20.9 ± 1.8* | 65.4 ± 9.3* |
| Valeric acid (100 μM) | 20.4 ± 1.2* | 53.8 ± 3.2* |
| Valeric acid (250 μM) | 20.3 ± 1.2* | 44.9 ± 1.3* |
| Biocytin (50 μM) | 29.9 ± 0.7* | 59.2 ± 3.1* |
| Biocytin (100 μM) | 24 ± 1.4* | 57.4 ± 3.8* |
| Biocytin (250 μM) | 23.1 ± 1.9* | 56.3 ± 6.6* |
| NHS biotin (50 μM) | 27.2 ± 1.1* | 33.9 ± 3.3* |
| NHS biotin (100 μM) | 18.6 ± 0.9* | 33.7 ± 2.5* |
| NHS biotin (250 μM) | 13.9 ± 1.3* | 20.3 ± 1.2* |
Data represents mean ± SD (n = 4)
*P value < 0.05 is considered to be statistically significant
Interaction of B-ACV with SMVT
Interaction potential of biotinylated prodrug with SMVT was tested in HCEC and D407 cells. Uptake of biotin remained unaltered in the presence of ACV. Interestingly, uptake was reduced to approximately 50% and 70% of control in the presence of 50 μM unlabeled biotin and B-ACV, respectively, in both the cells (Fig. 7a). Also, cellular accumulation of ACV and B-ACV was performed on HCEC and D407 cells. Compared to ACV, the uptake of B-ACV increased by six and five times on HCEC and D407 cells, respectively (Fig. 7b).
Fig. 7.
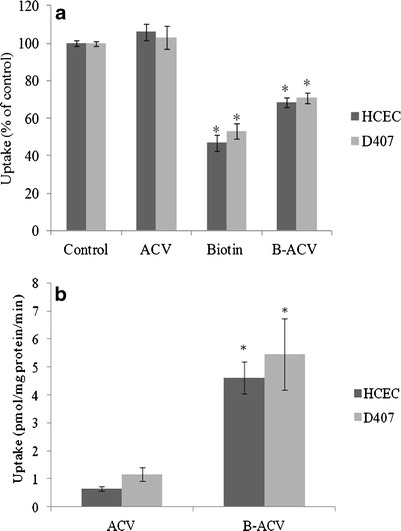
a Uptake of [3H] biotin by HCEC and D407 cells in presence of 50 μM concentration of unlabeled biotin, ACV, and B-ACV. [3H] biotin uptake was performed at 37°C with DPBS buffer (pH 7.4) for 30 min. Values represent mean ± SD (n = 4). A P value of < 0.05 is considered to be statistically significant and denoted by *. b Cellular accumulation of ACV and B-ACV (50 μM) on HCEC and D407 cells at 37°C for 30 min. Values represent mean ± SD (n = 4). A P value of <0.05 was considered to be statistically significant and denoted by *
Evaluation of the Existence of a Second Biotin-Specific High-Affinity Transporter
Uptake of [3H] biotin was studied at very low concentrations (0.3–10 nM) to evaluate the existence of a second biotin-specific high-affinity transporter. Biotin uptake was linear with increasing concentrations (see Electronic Supplementary Material (ESM), Fig. S1). Further, [3H] biotin uptake was examined in the presence of 25 nM unlabeled biotin, pantothenic acid, and lipoic acid. All the compounds at the nanomolar concentration did not show any significant inhibition (see ESM, Fig. S2).
RT-PCR Analysis
The quality of RNA extracted from HCEC, D407, and human cornea was assessed by measuring A260/A280 ratios. The ratios were found to be 1.87, 1.90, and 2.01, respectively suggesting that the RNA is pure. Molecular recognition of SMVT was confirmed with RT-PCR. The cDNA generated from total RNA isolated from either HCEC or D407 cells was PCR-amplified with the primers specific for human SMVT. Resulting PCR products were analyzed by gel electrophoresis. The band obtained at 774 bp during the gel electrophoresis confirmed the presence of SMVT. Hence, SMVT is expressed in both HCEC and D407 cells (Fig. 8).
Fig. 8.
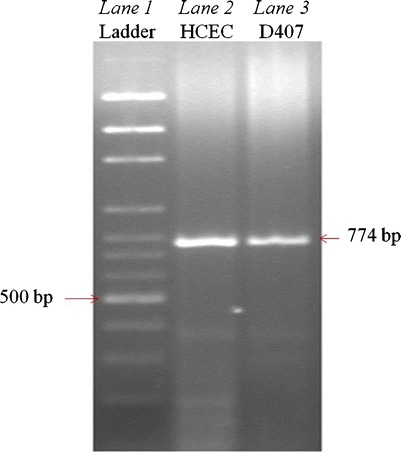
RT-PCR showing the molecular evidence of SMVT in HCEC and D407 cells. Lane 1 represents 100 bp molecular ladder and lane 2, 3 represents 774 bp PCR product obtained from HCEC and D407 cells
qPCR Analysis
Quantitative estimates of the relative abundance of SMVT mRNA were obtained with qPCR analysis. mRNA levels of SMVT were analyzed in HCEC, D407 and human cornea samples. Expression of SMVT mRNA was >5- and >3.5-fold in HCEC and D407 cells relative to the human cornea (see ESM, Fig. S3).
DISCUSSION
Biotin is a water-soluble vitamin essential for normal cellular function, growth, and development. Since it is not synthesized in the body, it ought to be supplemented. As a result, a specific carrier-mediated system may be involved in translocation of biotin (47). The primary intention of this study was to examine functional and molecular aspects of biotin translocation via SMVT in HCEC and D407 cells. These cell lines have been selected because of their human origin and their suitability to study active involvement of both efflux (30,32,34,40,41,43) and influx transporters (42,48–50).
Functional aspects associated with cellular translocation of biotin in HCEC and D407 cells were delineated. Time-dependency studies showed a linear increase in biotin uptake till 60 min, and therefore, a 30 min uptake time was selected for all subsequent experiments in both the cell lines utilized in this study (Fig. 1). Uptake of biotin in both HCEC and D407 cells was found to be decreasing when pH increased from acidic (pH 4.0) to physiological range (pH 7.4), but was almost similar at a pH range of 7.4–8.0 (Fig. 2). These alterations in uptake might be due to the ionic nature of biotin but may not be from the H+ gradient (51,52). The pKa of biotin is 4.65, which ensures that it exists in ionized form at physiological pH and cannot permeate across the plasma membrane by simple diffusion. At lower pH, biotin uptake may be higher because of higher availability of neutral species which usually permeates more across the lipid layer. Results from the pH-dependency study appeared to involve a partial contribution of passive diffusion in addition to active carrier-mediated process by SMVT. The uptake study performed at 4°C clearly delineated the passive diffusion component from the active biotin transport in both HCEC and D407 cells (Fig. 2). Altogether, these results suggested a pH-dependent SMVT-mediated biotin transport in the cells derived from human corneal and retinal pigment epithelia.
Concentration-dependency studies revealed saturation of carrier-mediated process at higher concentrations of unlabeled biotin. Biotin uptake was found to be saturable with a Km of 296.2 ± 25.9 μM and 863.8 ± 66.9 μM, Vmax of 77.2 ± 2.2 pmol/mg protein/min and 308.3 ± 10.7 pmol/mg protein/min in HCEC and D407 cells, respectively (Fig. 3). Km and Vmax are two important parameters that define the functional and kinetic behavior of a transporter. Km is a measure of apparent binding affinity of a substrate and Vmax is a measure of translocation capacity of the carrier-mediated system. Based on saturation kinetics, the Km value was relatively lower in corneal cells indicating a higher binding strength and affinity of biotin when compared to retinal cells. An interesting observation was that D407 cells exhibited higher transport capacity compared to HCEC cells, as evident by the higher Vmax value (308.3 ± 10.7 vs 77.2 ± 2.2 pmol/mg protein/min). These results were in accordance with previously published reports of rPCEC and ARPE-19 cells, where the corneal cells exhibited lower Km and Vmax values than retinal cells (see ESM, Table S1) (2,3). The ratio of these kinetic parameters (Vmax/Km) provides an estimate of the catalytic efficiency of a transporter. Although the Km and Vmax values were different, the transport efficiency (Vmax/Km) was observed to be almost similar in HCEC (0.26 μl/mg protein/min) and D407 (0.35 μl/mg protein/min) cells.
Significant differences in biotin uptake rates at different temperatures imply that the biotin uptake process is transporter mediated (Fig. 4). Biotin uptake in both HCEC and D407 cells diminished in the presence of ouabain, suggesting that this process is Na+/K+-ATPase dependent. In addition, sodium azide and 2,4-DNP which block oxidative phosphorylation also reduced biotin uptake. These results signify that biotin uptake in HCEC and D407 cells is an energy (ATP)-dependent active process (Fig. 5). Decreased uptake in the absence of sodium and in the presence of amiloride, a sodium ion transport inhibitor demonstrated that this transport system may be highly sodium dependent. The rate of biotin uptake remained unaffected in the absence of chloride ions in the buffer suggesting that the transport system is chloride independent (Fig. 6a).
Uphill transport of SMVT substrates was earlier reported to be energized by a transmembrane sodium gradient as well as the membrane potential (53). Therefore, we investigated whether biotin transport via SMVT in HCEC and D407 cells is found to be coupled with the electrochemical gradient of Na+ ions. Biotin uptake increased along with increasing concentrations of sodium in the uptake buffer and was saturated at higher concentrations (Fig. 6b). Our results suggested that biotin uptake is associated with the co-transport of sodium ions. The Hill ratio suggested that approximately one sodium ion (1.02 for HCEC and 0.99 for D407) is needed for translocation of each biotin molecule.
Substrate-specificity studies revealed a concentration-dependent diminution in biotin uptake in the presence of SMVT substrates (lipoic acid and pantothenic acid), and an analog of biotin (desthiobiotin) in both HCEC and D407 cells (Table I). Interestingly, significant concentration-dependent inhibition of biotin uptake was observed in the presence of structural analogs such as valeric acid, biocytin, and NHS biotin in both HCEC and D407 cells (Table II). Similar changes were not observed with rPCECs (3) and MDCK-MDR1 (51) cells as reported earlier from our laboratory. These results suggest that a free carboxylic group may or may not always be required for recognition and specific binding to SMVT.
Uptake of biotin did not alter in the presence of ACV in both HCEC and D407 cells. However, biotin-conjugated prodrug B-ACV produced significant diminution in biotin uptake indicating the recognition of prodrug by SMVT transporter (Fig. 7a). Results suggest that B-ACV competes with biotin and interacts with the SMVT carrier system. Since the prodrug may not have similar affinity pattern for SMVT relative to biotin, the inhibition of biotin uptake by B-ACV was comparatively lower. Also, cellular accumulation studies have been carried out to evaluate the rate of cellular accumulation of B-ACV for enhanced absorption of ACV. The results were consistent in both HCEC and D407 cells which demonstrated significantly higher cellular accumulation of biotinylated prodrug (B-ACV) relative to the parent drug (ACV). These observations support the hypothesis that B-ACV was transported primarily by SMVT (Fig. 7b).
The cumulative amount of [3H] biotin uptake by HCEC and D407 cells was linear as a function of biotin concentration over the nanomolar range (0.3–10 nM) (see ESM, Fig. S1). No diminution of biotin uptake was evident in the presence of 25 nM biotin, pantothenic acid, and lipoic acid suggesting that the second biotin-specific high-affinity system is not functionally active in HCEC and D407 cells (see ESM, Fig. S2).
The molecular presence of SMVT in both HCEC and D407 cells was confirmed by RT-PCR analysis (Fig. 8). We also carried out quantitative gene expression analysis on the RNA extracted from HCEC, D407 cells, and an isolated human cornea with qPCR. Expression of SMVT was relatively lower in D407 cells, although these cells exhibited a higher transport capacity than HCEC cells. Similar results were recently reported by Jwala et al. where the authors compared the expression and transporter capacity in ARPE-19 and retinoblastoma (Y-79) cells (54). Although Y-79 cells had a higher expression of SMVT, the transport capacity was relatively lower than ARPE-19 cells. Hence, the correlation between mRNA expression and transport capacity needs further investigation. Moreover, expression of SMVT in the human cornea was found to be relatively lower than that of HCEC cells. This may be due to the RNA which was extracted from a whole human cornea instead of the human corneal epithelium (see ESM, Fig. S3).
CONCLUSION
In conclusion, this study demonstrates for the first time the functional aspects and molecular expression of SMVT in HCEC, D407 cells, and an isolated human cornea. Ocular bioavailability of biotinylated conjugates (biotin-conjugated prodrugs) may be enhanced by targeting SMVT. Moreover, these cell lines may be used as in vitro cell culture models for screening the cellular accumulation of biotin-conjugated therapeutics.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
(DOCX 82 kb)
ACKNOWLEDGMENTS
This study has been supported by NIH grants R01EY09171-16 and R01EY010659-14. The authors would like to acknowledge Dr. Vadivel Ganapathy and Dr. Pamela Martin from the Department of Biochemistry and Molecular Biology at Georgia Health Sciences University for their generous help in providing human corneal RNA used in our studies. Also, the authors would like to thank Matthew Scrivner at the UMKC Writing Center for his assistance during the preparation of this manuscript.
REFERENCES
- 1.Ohkura Y, Akanuma S, Tachikawa M, Hosoya K. Blood-to-retina transport of biotin via Na+-dependent multivitamin transporter (SMVT) at the inner blood-retinal barrier. Exp Eye Res. 2010;91(3):387–392. doi: 10.1016/j.exer.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Janoria KG, Boddu SH, Wang Z, Paturi DK, Samanta S, Pal D, et al. Vitreal pharmacokinetics of biotinylated ganciclovir: role of sodium-dependent multivitamin transporter expressed on retina. J Ocul Pharmacol Ther. 2009;25(1):39–49. doi: 10.1089/jop.2008.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janoria KG, Hariharan S, Paturi D, Pal D, Mitra AK. Biotin uptake by rabbit corneal epithelial cells: role of sodium-dependent multivitamin transporter (SMVT) Curr Eye Res. 2006;31(10):797–809. doi: 10.1080/02713680600900206. [DOI] [PubMed] [Google Scholar]
- 4.Bonjour JP. Biotin. In: Machlin LJ, editor. Handbook of vitamins nutritional biochemical and clinical aspects. New York: Marcel Dekker; 1984. pp. 403–435. [Google Scholar]
- 5.Sweetman L, Nyhan WL. Inheritable biotin-treatable disorders and associated phenomena. Annu Rev Nutr. 1986;6:317–343. doi: 10.1146/annurev.nu.06.070186.001533. [DOI] [PubMed] [Google Scholar]
- 6.Mall GK, Chew YC, Zempleni J. Biotin requirements are lower in human Jurkat lymphoid cells but homeostatic mechanisms are similar to those of HepG2 liver cells. J Nutr. 2010;140(6):1086–1092. doi: 10.3945/jn.110.121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dakshinamurti K, Cheah-Tan C. Liver glucokinase of the biotin deficient rat. Can J Biochem. 1968;46(1):75–80. doi: 10.1139/o68-012. [DOI] [PubMed] [Google Scholar]
- 8.Zempleni J, Mock DM. Uptake and metabolism of biotin by human peripheral blood mononuclear cells. Am J Physiol. 1998;275(2 Pt 1):C382–C388. doi: 10.1152/ajpcell.1998.275.2.C382. [DOI] [PubMed] [Google Scholar]
- 9.Said HM. Cellular uptake of biotin: mechanisms and regulation. J Nutr. 1999;129(2S Suppl):490S–493S. doi: 10.1093/jn/129.2.490S. [DOI] [PubMed] [Google Scholar]
- 10.Said HM, Ortiz A, McCloud E, Dyer D, Moyer MP, Rubin S. Biotin uptake by human colonic epithelial NCM460 cells: a carrier-mediated process shared with pantothenic acid. Am J Physiol. 1998;275(5 Pt 1):C1365–C1371. doi: 10.1152/ajpcell.1998.275.5.C1365. [DOI] [PubMed] [Google Scholar]
- 11.Said HM, Ma TY, Kamanna VS. Uptake of biotin by human hepatoma cell line, Hep G2: a carrier-mediated process similar to that of normal liver. J Cell Physiol. 1994;161(3):483–489. doi: 10.1002/jcp.1041610311. [DOI] [PubMed] [Google Scholar]
- 12.Ma TY, Dyer DL, Said HM. Human intestinal cell line Caco-2: a useful model for studying cellular and molecular regulation of biotin uptake. Biochim Biophys Acta. 1994;1189(1):81–88. doi: 10.1016/0005-2736(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 13.Said HM, Redha R, Nylander W. A carrier-mediated, Na+ gradient-dependent transport for biotin in human intestinal brush-border membrane vesicles. Am J Physiol. 1987;253(5 Pt 1):G631–G636. doi: 10.1152/ajpgi.1987.253.5.G631. [DOI] [PubMed] [Google Scholar]
- 14.Balamurugan K, Ortiz A, Said HM. Biotin uptake by human intestinal and liver epithelial cells: role of the SMVT system. Am J Physiol Gastrointest Liver Physiol. 2003;285(1):G73–G77. doi: 10.1152/ajpgi.00059.2003. [DOI] [PubMed] [Google Scholar]
- 15.Prasad PD, Wang H, Huang W, Fei YJ, Leibach FH, Devoe LD, et al. Molecular and functional characterization of the intestinal Na+-dependent multivitamin transporter. Arch Biochem Biophys. 1999;366(1):95–106. doi: 10.1006/abbi.1999.1213. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Huang W, Fei YJ, Xia H, Yang-Feng TL, Leibach FH, et al. Human placental Na+-dependent multivitamin transporter. Cloning, functional expression, gene structure, and chromosomal localization. J Biol Chem. 1999;274(21):14875–14883. doi: 10.1074/jbc.274.21.14875. [DOI] [PubMed] [Google Scholar]
- 17.Prasad PD, Wang H, Kekuda R, Fujita T, Fei YJ, Devoe LD, et al. Cloning and functional expression of a cDNA encoding a mammalian sodium-dependent vitamin transporter mediating the uptake of pantothenate, biotin, and lipoate. J Biol Chem. 1998;273(13):7501–7506. doi: 10.1074/jbc.273.13.7501. [DOI] [PubMed] [Google Scholar]
- 18.Scholz M, Schrunder S, Gartner S, Keipert S, Hartmann C, Pleyer U. Ocular drug permeation following experimental excimer laser treatment on the isolated pig eye. J Ocul Pharmacol Ther. 2002;18(2):177–183. doi: 10.1089/108076802317373923. [DOI] [PubMed] [Google Scholar]
- 19.Prausnitz MR, Noonan JS. Permeability of cornea, sclera, and conjunctiva: a literature analysis for drug delivery to the eye. J Pharm Sci. 1998;87(12):1479–1488. doi: 10.1021/js9802594. [DOI] [PubMed] [Google Scholar]
- 20.Lee VH, Robinson JR. Topical ocular drug delivery: recent developments and future challenges. J Ocul Pharmacol. 1986;2(1):67–108. doi: 10.1089/jop.1986.2.67. [DOI] [PubMed] [Google Scholar]
- 21.Maurice DM, Mishima S. Ocular Pharmakokinetics. In: Sears ML, editor. Pharmacology of the eye. Berlin: Springer; 1984. pp. 19–116. [Google Scholar]
- 22.Reichl S, Bednarz J, Muller-Goymann CC. Human corneal equivalent as cell culture model for in vitro drug permeation studies. Br J Ophthalmol. 2004;88(4):560–565. doi: 10.1136/bjo.2003.028225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reichl S. Cell culture models of the human cornea - a comparative evaluation of their usefulness to determine ocular drug absorption in-vitro. J Pharm Pharmacol. 2008;60(3):299–307. doi: 10.1211/jpp.60.3.0004. [DOI] [PubMed] [Google Scholar]
- 24.Hornof M, Toropainen E, Urtti A. Cell culture models of the ocular barriers. Eur J Pharm Biopharm. 2005;60(2):207–225. doi: 10.1016/j.ejpb.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Barar J, Asadi M, Mortazavi-Tabatabaei SA, Omidi Y. Ocular drug delivery; impact of in vitro cell culture models. J Ophthalmic Vis Res. 2009;4(4):238–252. [PMC free article] [PubMed] [Google Scholar]
- 26.Davis AA, Bernstein PS, Bok D, Turner J, Nachtigal M, Hunt RC. A human retinal pigment epithelial cell line that retains epithelial characteristics after prolonged culture. Invest Ophthalmol Vis Sci. 1995;36(5):955–964. [PubMed] [Google Scholar]
- 27.Hunt RC, Davis AA. Altered expression of keratin and vimentin in human retinal pigment epithelial cells in vivo and in vitro. J Cell Physiol. 1990;145(2):187–199. doi: 10.1002/jcp.1041450202. [DOI] [PubMed] [Google Scholar]
- 28.Notara M, Daniels JT. Characterisation and functional features of a spontaneously immortalised human corneal epithelial cell line with progenitor-like characteristics. Brain Res Bull. 2010;81(2–3):279–286. doi: 10.1016/j.brainresbull.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Araki-Sasaki K, Ohashi Y, Sasabe T, Hayashi K, Watanabe H, Tano Y, et al. An SV40-immortalized human corneal epithelial cell line and its characterization. Invest Ophthalmol Vis Sci. 1995;36(3):614–621. [PubMed] [Google Scholar]
- 30.Vellonen KS, Mannermaa E, Turner H, Hakli M, Wolosin JM, Tervo T, et al. Effluxing ABC transporters in human corneal epithelium. J Pharm Sci. 2010;99(2):1087–1098. doi: 10.1002/jps.21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karla PK, Quinn TL, Herndon BL, Thomas P, Pal D, Mitra A. Expression of multidrug resistance associated protein 5 (MRP5) on cornea and its role in drug efflux. J Ocul Pharmacol Ther. 2009;25(2):121–132. doi: 10.1089/jop.2008.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karla PK, Earla R, Boddu SH, Johnston TP, Pal D, Mitra A. Molecular expression and functional evidence of a drug efflux pump (BCRP) in human corneal epithelial cells. Curr Eye Res. 2009;34(1):1–9. doi: 10.1080/02713680802518251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becker U, Ehrhardt C, Daum N, Baldes C, Schaefer UF, Ruprecht KW, et al. Expression of ABC-transporters in human corneal tissue and the transformed cell line, HCE-T. J Ocul Pharmacol Ther. 2007;23(2):172–181. doi: 10.1089/jop.2006.0095. [DOI] [PubMed] [Google Scholar]
- 34.Karla PK, Pal D, Quinn T, Mitra AK. Molecular evidence and functional expression of a novel drug efflux pump (ABCC2) in human corneal epithelium and rabbit cornea and its role in ocular drug efflux. Int J Pharm. 2007;336(1):12–21. doi: 10.1016/j.ijpharm.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranta VP, Laavola M, Toropainen E, Vellonen KS, Talvitie A, Urtti A. Ocular pharmacokinetic modeling using corneal absorption and desorption rates from in vitro permeation experiments with cultured corneal epithelial cells. Pharm Res. 2003;20(9):1409–1416. doi: 10.1023/A:1025754026449. [DOI] [PubMed] [Google Scholar]
- 36.Toropainen E, Ranta VP, Vellonen KS, Palmgren J, Talvitie A, Laavola M, et al. Paracellular and passive transcellular permeability in immortalized human corneal epithelial cell culture model. Eur J Pharm Sci. 2003;20(1):99–106. doi: 10.1016/S0928-0987(03)00173-8. [DOI] [PubMed] [Google Scholar]
- 37.Toropainen E, Ranta VP, Talvitie A, Suhonen P, Urtti A. Culture model of human corneal epithelium for prediction of ocular drug absorption. Invest Ophthalmol Vis Sci. 2001;42(12):2942–2948. [PubMed] [Google Scholar]
- 38.Saarinen-Savolainen P, Jarvinen T, Araki-Sasaki K, Watanabe H, Urtti A. Evaluation of cytotoxicity of various ophthalmic drugs, eye drop excipients and cyclodextrins in an immortalized human corneal epithelial cell line. Pharm Res. 1998;15(8):1275–1280. doi: 10.1023/A:1011956327987. [DOI] [PubMed] [Google Scholar]
- 39.Wan WJ, Cui DM, Yang X, Hu JM, Li CX, Hu SL, et al. Expression of adenosine receptors in human retinal pigment epithelium cells in vitro. Chin Med J (Engl) 2011;124(8):1139–1144. [PubMed] [Google Scholar]
- 40.Mannermaa E, Vellonen KS, Ryhanen T, Kokkonen K, Ranta VP, Kaarniranta K, et al. Efflux protein expression in human retinal pigment epithelium cell lines. Pharm Res. 2009;26(7):1785–1791. doi: 10.1007/s11095-009-9890-6. [DOI] [PubMed] [Google Scholar]
- 41.Constable PA, Lawrenson JG, Dolman DE, Arden GB, Abbott NJ. P-Glycoprotein expression in human retinal pigment epithelium cell lines. Exp Eye Res. 2006;83(1):24–30. doi: 10.1016/j.exer.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 42.Maenpaa H, Gegelashvili G, Tahti H. Expression of glutamate transporter subtypes in cultured retinal pigment epithelial and retinoblastoma cells. Curr Eye Res. 2004;28(3):159–165. doi: 10.1076/ceyr.28.3.159.26244. [DOI] [PubMed] [Google Scholar]
- 43.Kennedy BG, Mangini NJ. P-glycoprotein expression in human retinal pigment epithelium. Mol Vis. 2002;8:422–430. [PubMed] [Google Scholar]
- 44.Talluri RS, Katragadda S, Pal D, Mitra AK. Mechanism of l-ascorbic acid uptake by rabbit corneal epithelial cells: evidence for the involvement of sodium-dependent vitamin C transporter 2. Curr Eye Res. 2006;31(6):481–489. doi: 10.1080/02713680600693629. [DOI] [PubMed] [Google Scholar]
- 45.Vadlapudi AD, Vadlapatla RK, Kwatra D, Earla R, Samanta SK, Pal D, et al. Targeted lipid based drug conjugates: a novel strategy for drug delivery. Int J Pharm. 2012;434(1–2):315–324. doi: 10.1016/j.ijpharm.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vadlapudi AD, Vadlapatla RK, Mitra AK. Sodium dependent multivitamin transporter (SMVT): a potential target for drug delivery. Curr Drug Targets. 2012;13(7):994–1003. doi: 10.2174/138945012800675650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vellonen KS, Hakli M, Merezhinskaya N, Tervo T, Honkakoski P, Urtti A. Monocarboxylate transport in human corneal epithelium and cell lines. Eur J Pharm Sci. 2010;39(4):241–247. doi: 10.1016/j.ejps.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 49.Maenpaa H, Mannerstrom M, Toimela T, Salminen L, Saransaari P, Tahti H. Glutamate uptake is inhibited by tamoxifen and toremifene in cultured retinal pigment epithelial cells. Pharmacol Toxicol. 2002;91(3):116–122. doi: 10.1034/j.1600-0773.2002.910305.x. [DOI] [PubMed] [Google Scholar]
- 50.Toimela TA, Tahti H. Effects of mercuric chloride exposure on the glutamate uptake by cultured retinal pigment epithelial cells. Toxicol In Vitro. 2001;15(1):7–12. doi: 10.1016/S0887-2333(00)00057-6. [DOI] [PubMed] [Google Scholar]
- 51.Luo S, Kansara VS, Zhu X, Mandava NK, Pal D, Mitra AK. Functional characterization of sodium-dependent multivitamin transporter in MDCK-MDR1 cells and its utilization as a target for drug delivery. Mol Pharm. 2006;3(3):329–339. doi: 10.1021/mp0500768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Said HM, Redha R, Nylander W. Biotin transport in the human intestine: site of maximum transport and effect of pH. Gastroenterology. 1988;95(5):1312–1317. doi: 10.1016/0016-5085(88)90366-6. [DOI] [PubMed] [Google Scholar]
- 53.Prasad PD, Ganapathy V. Structure and function of mammalian sodium-dependent multivitamin transporter. Curr Opin Clin Nutr Metab Care. 2000;3(4):263–266. doi: 10.1097/00075197-200007000-00004. [DOI] [PubMed] [Google Scholar]
- 54.Jwala J, Vadlapatla RK, Vadlapudi AD, Boddu SH, Pal D, Mitra AK. Differential expression of folate receptor-alpha, sodium-dependent multivitamin transporter, and amino acid transporter (B ((0, +))) in human retinoblastoma (Y-79) and retinal pigment epithelial (ARPE-19) cell lines. J Ocul Pharmacol Ther Off J Assoc Ocul Pharmacol Ther. 2012;28(3):237–244. doi: 10.1089/jop.2011.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 82 kb)


