Abstract
More than 50% of myeloma cases have normal karyotypes under conventional cytogenetic analysis due to low mitotic activity and content of plasma cells in the bone marrow. We used a polymerase chain reaction (PCR)-based translocation detection assay to detect BCL1/JH t(11;14) (q13;q32) in 105 myeloma patients, and randomly selected 8 translocation positive samples for array comparative genomic hybridization (aCGH) analysis. Our findings revealed 14.3% of myeloma samples were positive for BCL1/JH t(11;14) (q13;q32) translocation (n=15 of 105). We found no significant correlation between this translocation with age (P=0.420), gender (P=0.317), ethnicity (P=0.066) or new/relapsed status of multiple myeloma (P=0.412) at 95% confidence interval level by χ2 test. In addition, aCGH results showed genomic imbalances in all samples analyzed. Frequent chromosomal gains were identified at regions 1q, 2q, 3p, 3q, 4p, 4q, 5q, 7q, 9q, 11q, 13q, 15q, 21q, 22q and Xq, while chromosomal losses were detected at 4q and 14q. Copy number variations at genetic loci that contain NAMPT, IVNS1ABP and STK17B genes are new findings that have not previously been reported in myeloma patients. Besides fluorescence in situ hybridization, PCR is another rapid, sensitive and simple technique that can be used for detecting BCL1/JH t(11;14)(q13;q32) translocation in multiple myeloma patients. Genes located in the chromosomal aberration regions in our study, such as NAMPT, IVNS1ABP, IRF2BP2, PICALM, STAT1, STK17B, FBXL5, ACSL1, LAMP2, SAMSN1 and ATP8B4 might be potential prognostic markers and therapeutic targets in the treatment and management of multiple myeloma patients positive for BCL1/JH t(11;14) (q13;q32) translocation.
Key words: immunoglobulin heavy chain, NAMPT, IVNS1ABP, STK17B, copy number variations.
Introduction
Multiple myeloma (MM) is the malignancy of differentiated B lymphocytes. It accounts for 10% of all hematologic cancers and approximately 2% of all cancer deaths.1 Translocations involving the immunoglobulin heavy chain (IGH) locus are common cytogenetic events in MM. It occurs during isotype class switching in terminally differentiated B cells.2,3 The t(11;14)(q13;q32) accounts for 15–20% of total myeloma cases and is the most common translocation found in the early stage of MM pathogenesis.4–6 In this translocation, the proto-oncogene, CCND1 at chromosome 11q13 is juxtaposed to the IGH gene at chromosome 14q32, resulting in the overexpression of the protein product, cyclin D1.7 Previous studies on t(11;14)(q13;q32) translocation in MM implied that the abnormality is associated with better prognosis when the patient is treated with traditional chemotherapy or autologous stem cell transplantation.7,8
Conventional cytogenetic analysis is routinely used for patients suspected to have chromosome imbalances. However, metaphase cytogenetic analysis of myeloma cells is difficult to perform due to low mitotic activity and content of plasma cells in the bone marrow.9 As a result, more than 50% of myeloma cases showed normal karyotypes with conventional cytogenetic analysis.7 Array comparative genomic hybridization (aCGH) is another approach currently used for genome-wide screening of chromosome imbalances.10,11 This technology is sensitive enough to detect chromosomal aberrations in resolutions of up to 200 bp simultaneously in a single experiment. Although t(11;14) (q13;q32) translocation in myeloma has been studied since 2000, most of these were carried out in Caucasian populations. Information on the frequency of t(11;14)(q13;q32) translocation in Asian myeloma patients is very limited. In this study, we aimed to study the frequency of t(11;14)(q13;q32) translocation and its correlation with the age, gender, ethnicity and new/relapsed status of MM in three major ethnic groups in Malaysia: Malays, Chinese and Indians. Chromosomal imbalances of 8 t(11;14)(q13;q32) positive cases were randomly selected for further investigation via microarray technology to identify other genomic imbalances that are closely related to t(11;14)(q13;q32) translocation. This is important to identify new prognostic markers and therapeutic targets for MM patients with t(11;14)(q13;q32) translocation.
Materials and Methods
Specimen and DNA preparation
From 2007 to 2010, DNA of 105 patients was isolated from cytogenetically fixed cell pellets using the Qiagen DNA mini kit. Ninety out of 105 patients were newly diagnosed MM whereas 15 were relapsed cases. Patient age ranged between 15 to 77 years old at the time of diagnosis (mean 57 years). Patients' characteristics are summarized in Table 1. The isolated genomic DNAs were checked on 1% denaturing agarose gel to assess the quality of the genomic DNA. The purity and concentration of the DNAs were determined by using a NanoDrop ND-1000 UV-VIS spectrophotometer. The study was approved by the Medical Research and Ethics Committee of the Ministry of Health, Malaysia, and all patients provided their informed consent.
Table 1. Characteristics of 105 multiple myeloma patients diagnosed from 2007–2010.
| All patients n (%) | Array comparative genomic hybridization (8 cases) n (%)* | |
|---|---|---|
| Gender | ||
| Male | 58 (55.2) | 5 (62.5) |
| Female | 47 (44.8) | 3 (37.5) |
| Age at diagnosis (years) | ||
| ≤55 | 43 (40.9) | 2 (25.0) |
| >55 | 59 (56.2) | 6 (75.0) |
| Unknown | 3 (2.9) | - |
| Ethnic group | ||
| Malay | 57 (54.3) | 4 (50.0) |
| Chinese | 27 (25.7) | 1 (12.5) |
| Indian | 21 (20.0) | 3 (37.5) |
| Cytogenetic analysis | ||
| Positive° | 3 (2.9) | - |
| Negative | 56 (53.3) | 4 (50.0) |
| Unknown# | 46 (43.8) | 4 (50.0) |
| Disease type | ||
| New | 90 (85.7) | 8 (100) |
| Relapsed | 15 (14.3) | - |
Patients selected for array comparative genomic hybridization analysis;
multiple chromosomal abnormalities were seen with karyotyping;
insufficient/ short/ no chromosome spread were available for cytogenetic analysis.
Polymerase chain reaction
BCL1/JH t(11;14)(q13;q32) gene translocation was detected by IdentiClone™ assay kits. This test targets the major translocation cluster (MTC) region of the BCL1/JH translocation and amplifies genomic DNA between primers that target the BCL1 gene and the conserved joining (JH) regions of the IGH gene. PCR was carried out in 50 µL volumes containing 5 unit/µL of AmpliTaq DNA polymerase, 45 µL of PCR Master Mix and 200–400 µg of genomic DNA. Clonal control DNA and polyclonal control DNA provided in the kit were used as positive and negative control, respectively. A no-template control was included in each run to rule out cross-contamination of reagents. Amplification was performed in a thermocycler (Eppendorf) in duplicate. Cycling conditions were as follows: 95°C for 7 min followed by 35–40 cycles at 95°C for 45 s, 60°C for 45 s, 72°C for 90 s and final extension at 72°C for 10 min.
t(11;14)(q13;q32) screening
PCR products were visualized on an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) by using Agilent DNA 7500 chips according to the manufacturer's recommendations. Fragment size ranging between 150–2000 bp is positive for the detection of a BCL1/JH t(11;14) translocation. The 600 bp band is an unspecific product as it is also present in the negative control.
Cloning and sequencing
Samples which were positive for BCL1/JH t(11;14) translocation were subjected for verification via sequencing method. Polymearse chain reaction (PCR) products of interest were excised from the gel and purified according to Qiagen Gel Purification protocol (QIAquick Gel Extraction Kit). The samples were cloned into pJET1.2/blunt cloning vector (Fermentas) and transformed into Escherichia coli competent cells. Transformants were tested by PCR, and recombinant plasmids were extracted from the positive clone. The extracted plasmids were then further verified by PCR. Cycle sequencing was performed according to the procedure recommended by Bigdye Terminator v3.1 cycle sequencing kit (Applied Biosystems, USA). The insert was sequenced with vector-targeted primers on a 3730xl DNA Analyzer (Applied Biosystems, USA) in forward and reverse directions. Finally, the sequencing data were blasted with the NCBI database to confirm that they were the joining sequences of MTC of the BCL1 locus and the region of the immunoglobulin heavy chain locus.
Statistical analysis
The χ2 test was used to evaluate the correlation between BCL1/JH t(11;14)(q13;q32) translocation with the age of the patients (≤55 or >55 years), gender, ethnicity (Malay, Chinese and Indian), and status of myeloma (newly diagnosed or relapsed) at P<0.05 by IBM SPSS Statistics 19 software.
Oligonucleotide array comparative genomic hybridization and data analysis
Eight samples positive for BCL1/JH t(11;14) translocation were randomly selected for genomic imbalances analysis by using aCGH (Agilent Technologies Palo Alto, CA, USA). Reference DNAs were pooled from 10 healthy individuals from the same gender and ethnic group. All the reference and test DNAs were checked on 1% agarose gel to assess the integrity of the DNA. A total of 1.5 µg of cyanine 5-dUTP-labeled test DNA and an equal amount of cyanine 3-dUTP-labeled reference DNA were mixed and hybridized in the presence of Cot-1 DNA (Invitrogen) onto Human Genome CGH 244k array for 40 h at 65°C following Agilent's standard processing recommendations. Slides were washed (Wash Procedure B) and scanned with Agilent array scanner G2505C. Microarray images were scanned and Agilent's Feature Extraction software (version 10.7.3.1) was used to extract data from raw microarray image files in preparation for analysis. Agilent DNA Analytics software (version 4.0) was used to visualize, detect and analyze aberration patterns from CGH microarray profiles. All microarray data were analyzed according to the human reference genome hg18. Aberrations were detected with the ADM2 algorithm and the filtering options of a minimum of 3 probes and minimum average absolute log2 ratio of 0.3, with maximum aberration of 100 probes. Data were analyzed with bin size 10 and threshold 6.0. Data were filtered at a minimum of 3 probes and 37.5% or over for probe penetrance analysis. Common aberrations were performed with t-test; P threshold 0.05, overlap threshold 0.9. Aberration segments were individually reviewed with Ensembl genome browser 54.
Results
Our results revealed 14.3% of the myeloma samples under studied were positive for BCL1/JH t(11;14) translocation (n=15 of 105). One of the samples was derived from the relapsed MM patient (RM9) while the remaining were from newly diagnosed myeloma cases (Figure 1). Sequencing results verified that all the BCL1/JH t(11;14) translocation positive samples contained the joining sequences of MTC of the BCL1 locus and the region of the immunoglobulin heavy chain locus. Secuencing result for RM9 is illustrated in Figuer 2. χ2 analysis showed that there was no significant correlation between BCL1/JH t(11;14) translocation and patient age (P=0.420), gender (P=0.317), ethnicity (P=0.066) or new/relapsed status of MM (P=0.412) at 95% confidence interval level. Interestingly, BCL1/JH t(11;14) translocation was not detected in all samples, which showed chromosomal abnormalities from conventional cytogenetic analysis.
Figure 1.
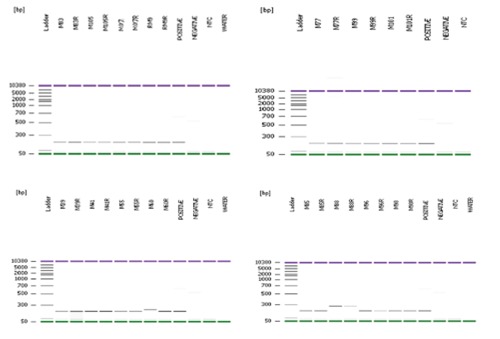
Polymerase chain reaction (PCR) products of 15 BCL1/JH t(11;14)(13q;32q) positive multiple myeloma patients on bioanalyzer. Each sample was performed in duplicate. PCR product size ranged between 200bp-300bp. Negative controls and samples showed a faint unspecific band at ∼600bp. No band was observed in the non-template controls and water controls indicated that there was no cross-contamination of reagent during the PCR preparation.
Figure 2.
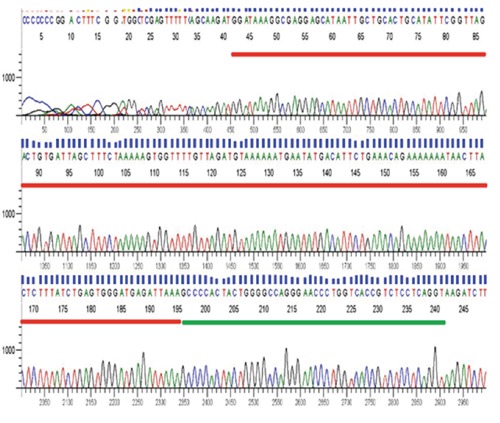
Chromatogram of RM9 in forward direction. Sequencing results showed that RM9 contained the joining sequences of G1/S-specific cyclin-D1 (red line) and IGHJ4 (green line) located at chromosome 11 and 14, respectively. The remaining sequences were the vector sequences.
Genomic imbalances were detected in 100% of BCL1/JH t(11;14) translocation positive samples selected for the aCGH analysis (M07, M39, M41, M55, M96, M98, M101 and M105). The microarray data have been submitted and approved by the Gene Expression Omnibus and assigned GEO accession numbers GSE33757.
Our findings showed that chromosomal amplification is more common than deletion and q-arm is more frequently altered than the p-arm in the MM samples under study. Chromosomal aberrations in each myeloma case are summarized in Table 2. Frequent chromosomal gains were at regions 1q, 2q, 3p, 3q, 4p, 4q, 5q, 7q, 9q, 11q, 13q, 15q, 21q, 22q and Xq, while chromosomal losses were detected at 4q and 14q. Copy number changes mentioned above were significant at P=0.05. Information on the genes included within the common aberration regions and the percentage of penetrance were identified and listed in Table 3.
Table 2. Chromosomal aberrations (gains and losses) in eight multiple myeloma patients.
| Sample ID | Gain | Loss |
|---|---|---|
| M07 | 1p, 1q, 2q, 3p, 3q, 4p, 4q, 5p, 5q, 6p, 6q, 7p, 7q, 8q, | 1q, 3q, 4q, 6q, 12p, 15q, 16p, 22q, |
| 9q, 10p, 10q, 11q, 12p, 12q, 13q, 14q, 15q, 16q, 19q, | ||
| 20p, 21q, Xq | ||
| M39 | 3q, 4q, 6p, 16q, 19q, 20p, 22q | 2p, 3q, 7q, 8p, 11p, 11q, 13q, 14q, 15q, 16p, |
| M41 | 1q, 2p, 2q, 3q, 4p, 4q, 6q, 7q, 8p, 10q, 12p, 12q, | 1q, 3q, 14q, 15q |
| 13q, 15q, 21q | ||
| M55 | 1p, 1q, 2q, 3p, 3q, 4p, 4q, 5q, 6p, 7q, 8q, 9q, 10q, | 2p, 3q, 4q, 8p, 11q, |
| 11q, 13q, 14q, 15q, 20p, 21q, 22q, Xq | ||
| M96 | 1p, 2p, 5p, 6p, 7q, 9q, 10q, 14q, 16p, 17q, 22q, Xq | 15q |
| 1p, 2p, 3q, 4q, 6q, 8p, 8q, 11q, 13q, 14q, 22q, | ||
| M98 | 2q, 4p, 7q, 11p, 11q, 13q, 21q, 22q | |
| M101 | 1q, 4q, 7q, 11q, 12p, | 1q, 2q |
| M105 | 1q, 4p, 5q, 7q, 11q, 15q, 19p, 21q, Xq | 14q |
Table 3. Gene list and the per cent penetrance of chromosomal aberration regions.
| Chromosome region | Molecular region | Aberration | Penetrance (%) | Gene |
|---|---|---|---|---|
| 1q25.3 | 183542641–183549823 | Gain | 75.0 | IVNS1ABP |
| 1q42.3 | 232809040–232809674 | Gain | 75.0 | IRF2BP2 |
| 2q32.2 | 191549646–191572786 | Gain | 50.0 | STAT1 |
| 2q32.3 | 196729308–196742885 | Gain | 50.0 | STK17B |
| 4p15.33 | 15222779–15249423 | Gain | 50.0 | FBXL5 |
| 4q35.1 | 185914232–185983415 | Gain | 50.0 | ACSL1 |
| 7q22.2 | 105685720–105711134 | Gain | 87.5 | NAMPT |
| 11q14.2 | 85410858–85452836 | Gain | 75.0 | PICALM |
| 15q21.2 | 47955679–48186620 | Gain | 62.5 | ATP8B4 |
| 21q11.2 | 14779624–14840578 | Gain | 62.5 | SAMSN1 |
| Xq24 | 119454654–119467091 | Gain | 50.0 | LAMP2 |
| 4q13.2 | 69069363–69166014 | Loss | 37.5 | UGT2B17 |
Discussion
Our results revealed 14.3% of myeloma patients were positive for BCL1/JH t(11;14) translocation (n=15 of 105). This agrees with the published literature which reported that 15–20% of myeloma cases carried this chromosome structural change.4 The translocation, however, is not significantly associated with patient age, gender, ethnicity or new/relapsed status of MM at 95% confidence interval level. This suggests that other causal factors might be involved in the chromosome structural abnormalities, such as disease stage and subtype, bone marrow infiltration and degree of bone lesions, which are not evaluated in this study.
Chromosomal aberrations of 8 randomly selected MM samples which harbored BCL1/JH t(11;14) translocation were further investigated with aCGH. Chromosomal alterations were detected in chromosome X and all autosomes except for chromosome 18.
The highest frequency of chromosomal gain was identified at chromosome 7q22.2 105685720–105711134 (87.5%, n=7 of 8). Nicotinamide (NAMPT) is the only gene located in this region. NAMPT, also known as pre-B colony enhancing factor (PBEF) or visfatin can function as a growth factor, cytokine and nicotinamide phosphoribosyltransferase.12–14 It functions in promoting B-cell maturation and inhibiting neutrophil apoptosis.12 No alteration in this gene has been reported in myeloma patients. However, it was described in cancers of the prostate, brain, colon and rectum.15–17 Gain of NAMPT gene could activate NAD salvage pathway. The activation raises NAD levels for cell energy production. Increased expression of NAD is needed to prolong the survival of malignant cells through DNA repair and cell survival mechanisms which are mediated by PARPs and sirtuins.18–20 It has previously been shown that inhibition of NAMPT gene expression could induce apoptosis in human liver carcinoma cells in vitro.21 Whether apoptosis of myeloma cells can be induced by down-regulating NAMPT expression is an interesting question that remains to be investigated.
P53 abnormality appears to be a crucial event in oncogenesis. P53 mediates many of its key functions by transactivation or transrepression of its target genes. Interferon regulatory factor-2-binding-protein-2 (IRF2BP2) was discovered as a new p53 target gene.22 IRF2BP2 is a transcriptional co-repressor of IRF2.23 P53 binds to the consensus binding site upstream of the promoter region of IRF2BP2 and then trans-activates its transcription. IRF2BP2 participates in the genotoxic response mediated by p53, influencing the stress response pathways of the cells.22 In vitro study of U2OS cells revealed overexpression of IRF2BP2 and delay or a reduction in apoptosis after treated with doxorubicin.22 On the other hand, overexpression of IRF2BP2 reduced S-phase population after low doses of actinomycin D treatment.22 In line with this, knockdown of IRF2BP2 leads to increasing numbers of apoptotic cells after chemotherapeutic drug treatment.22 This scenario suggested that IRF2BP2 is not only involved in apoptotic mechanism but also in maintaining a cell growth arrest state to allow the cells to repair damaged DNA. Interestingly, IRF2BP2 was later being identified as a potential immune target gene in monoclonal gammopathy of undetermined significance (MGUS) by using serological analysis of recombinant cDNA expression library.24 Gain of 1q42.3 232809040–232809674, which encodes for the IRF2BP2 gene was identified in 6 of 8 MM patients with BCL1/JH t(11;14) translocation in our study. Although we did not investigate the expression of this gene at mRNA level, consideration of the role of IRF2BP2 in the p53 pathway has highlighted its importance as a candidate gene underlying tumorigenesis of MM. The interaction between IRF2BP2 and p53 and its impact on malignant cells is a key question that remains to be answered.
Apart from chromosomal gain at region 1q42.3, gain of 1q25.3 was also detected in molecular region 183542641–183549823 in 75% of patients in this study (n=6 of 8). Influenza virus NS1A binding protein (IVNS1ABP) was located in this genetic locus. Information on IVNS1ABP is very limited and its aberration has never been reported in relation with myeloma or other malignant diseases. IVNS1ABP is located within the 1q arm. Chromosome 1q gain is frequently associated with poor prognosis in myeloma.25 The amplification of partial or whole arm of chromosome 1q could induce overexpression of one or more oncogenes,25 for example, CKS1B amplification in MM patients.25,26 CKS1B influences myeloma cell growth and survival through SKP2- and p27Kip1-dependent and -independent mechanisms.27 Besides IVNS1ABP and IRF2BP2, no other gene was detected in our study located within chromosome 1q arm with high penetrance (≥37.5). Given the scarce information available about the IVNS1ABP gene, whether it is involved in myeloma pathogenesis needs further investigation.
Besides translocation involving chromosome 11q13, gain at chromosome 11q14.2 was detected in 6 of 8 t(11;14) positive patients in our study. This chromosomal gain was detected at molecular region 85410858–85452836, which contains the phosphatidylinositol-binding clathrin assembly protein (PICALM) gene. This gene was initially linked with the risk of Alzheimer's disease. The t(10;11)(p13;q14) translocation which results in the expression of the PICALM–MLLT10 fusion gene was observed in lymphoma, acute lymphoblastic leukemia and acute myeloid leukemia.28,29 Patients identified with PICALM-MLLT10 fusion gene have a poor prognosis and frequently relapse.28 The PICALM-MLLT10 fusion gene causes the elevated expression of certain HOXA cluster genes to interfere with the normal hematopoietic differentiation, resulting in a leukemogenic effect.28 The PICALM-MLLT10 fusion gene has never been reported in MM. However, aberration involving PICALM was previously reported by Largo et al. (2006) in their study of CGH and gene expression in MM cell lines.30 The role of PICALM in the pathogenesis of MM might not be associated with t(10;11)(p13;q14) rearrangement. PICALM might interact with other genes in MM predisposition.
Consistent with findings in the literature, we also found gain at chromosome region 21q11.2 14779624–14840578. This region codes for the SAM domain, SH3 domain and the nuclear localization signals 1 (SAMSN1) gene. The genomic region around the SAMSN1 locus is commonly targeted by translocation events in hematopoietic malignancies.31 SAMSN1 is up-regulated by B-cell activation signal and is involved in B-cell activation and differentiation.31 This gene is directly link to B cells and could, therefore, be altered in MM cells as well as in B cell-derived acute myeloid leukaemia.32 Apart from its involvement in hematopoietic malignancies, SAMSN1 is known to play a crucial role as a tumor suppressor gene in lung cancer.33
Two molecular regions within chromosome 2q32.2–2q32.3 were amplified in 4 of 8 cases under study. They were located in 191549646–191572786 and 196729308–196742885, which encode for signal transducers and activators of transcription 1 (STAT1) and serine/threonine kinase 17b (STK17B), respectively. Signal transducers and activators of transcription (STATs) play an important role in a wide variety of human malignancies. STATs proteins were discovered many years ago. STATs proteins transmit signals from the cell surface to the nucleus, and participate in gene regulation.34 STAT1 and STAT3 are the two most popular members of the STAT family. Deregulation of STAT1 and STAT3 were implicated in various human cancers, including myeloma, leukemia and breast cancer.35 STAT1 and STAT3 pathways are mediated by interleukin-6, a cytokine that regulates the proliferation of myeloma cells (IL-6).36,37 Overexpression of STAT1 triggers drug resistance in MM.38 A study by Fryknäs et al. on gene expression in myeloma cell line 8226/S and its doxorubicin resistant subline 8226/Dox40, revealed 17 of 50 overexpressed genes which were closely related to the STAT1 signalling pathway. Their findings indicate that increased STAT1 signalling could cause the crossresistance of myeloma cells to doxorubicin and radiation. They also found that drug resistance can be modulated by flu-arabine, a STAT1-inhibiting drug.38 Also, activation of STAT1 enhances TRAIL secretion through IFN- and triggers apoptotic cell death in the myeloma cell line, KMS-20. This further suggested that the STAT1 signalling pathway plays an important role in tumorigenesis of myeloma cells through trans-activation of its down-stream target genes.39 STAT3 is another important member of the STATs family that is responsible for the growth and survival of MM cells. Progression of MM cells occurs upon activation of the STAT3 and IL-6 pathway and up-regulated Bcl-xL gene expression, which eventually prevents apoptosis of the cancer cells.40 On the other hand, serine-threonine kinase 17B (STK17B) acts as a positive regulator for apoptosis.41 Downregulation of STK17B in non-Hodgkin's lymphoma suggested that it might play a crucial role as a tumor suppressor gene in oncogenesis.42 Instead of deletion, STK17B was amplified in the myeloma cases under study. Why a putative tumor suppressor gene could over-express in MM is unknown. A possible cause would be due to the mutational inactivation of the gene in the patients. Thus, mutational status of STK17B in myeloma is worthy of further investigation. In our study, common aberrations in chromosomes 4, 15 and X, which contain genetic codes for FBXL5 (15222779–15249423), ACSL1 (185914232–185983415), ATP8B4 (47955679–48186620) and LAMP2 (119454654–119467091), were detected by aCGH. Information on these genes is very limited and their role in hematologic malignancies is still poorly understood. Functional studies on these genes are needed to determine the clinical and genetic significance of the genes underlying the molecular pathogenesis of MM.
PCR-based BCL1/JH t(11;14)(q13;q32) gene translocation assay is more sensitive than conventional cytogenetic analysis. Conventional cytogenetics failed to detect the chromosomal anomaly in all the 15 myeloma patients positive for BCL1/JH t(11;14)(q13;q32) translocation (8 showed normal karyotypes while 7 did not have adequate metaphases for analysis). Fluorescence in situ hybridization (FISH) is another technique used for the determination of genomic imbalances in MM.43 FISH probes for the detection of genomic alterations with prognostic importance, such as translocation t(4;14), t(11;14) and t(14;16), chromosome 13 and p53 deletions, are commercially available. However, FISH is costly and labor-intensive, requiring experienced personnel to read the slides under a fluorescent microscope. In addition, due to the low mitotic index of myeloma cells, genomic aberrations are sometimes difficult to detect via metaphase FISH. Thus, interphase FISH would be more applicable in this context. As PCR-based detection assay is limited by the availability of markers, detection for other prognostic significant markers mentioned above still need to be conducted by FISH until these markers are available.
Copy number variations were identified in all the 8 selected BCL1/JH t(11;14)(q13;q32) positive patients analyzed by aCGH. Similarly, cytogenetic analysis was unable to detect the chromosomal aberrations in these 8 patients. They either had normal karyotype (n=4) or no/insufficient chromosome spread for cytogenetic analysis (n=4). Several common chromosomal aberrations in BCL1/JH t(11;14)(q13;q32) positive patients were identified in chromosome 1q, 2q, 4p, 4q, 7q, 11q, 15, 21 and X suggest the importance of these genetic loci in the molecular pathogenesis of MM. Genes located in these genetic loci included NAMPT, IVNS1ABP, IRF2BP2, PICALM, STAT1, STK17B, FBXL5, ACSL1, LAMP2, SAMSN1 and ATP8B4. Copy number variations at genetic loci that contain NAMPT, IVNS1ABP and STK17B genes are new findings that have never been reported in MM patients.
Whether the aberration regions identified in this study represent a unique clinical and genetic subtype for BCL1/JH t(11;14)(q13;q32) translocation positive myeloma patients is an interesting question that merits further research. Although BCL1/JH t(11;14)(q13;q32) translocation alone is not clinically significant in patient prognosis, this translocation, along with other genes identified within the common chromosomal aberration regions in our present study, might be important prognostic markers that have a significant impact on MM. Theoretically, changes in copy number might change the expression levels of the genes included in the regions of variable copy number, allowing transcription levels to be higher or lower than the normal expression. However, we have not proved that these copy number changes are associated with the changes in gene expression in our present study. A larger cohort of myeloma samples is needed to assess the consistency of the copy number changes and prognostic significant of these aberrations in MM patients. Additionally, real time PCR can be used to determine whether the copy number changes have any significant impact on gene expression levels in MM patients.
Conclusions
Our study showed that PCR is another rapid, sensitive and simple technique that can be used to detect BCL1/JH t(11;14)(q13;q32) translocation in MM patients.
We suggest the importance of aCGH in the identification of copy number variations in MM patients. Genes located in the chromosomal aberration regions in our study, such as NAMPT, IVNS1ABP, IRF2BP2, PICALM, STAT1, STK17B, FBXL5, ACSL1, LAMP2, SAMSN1 and ATP8B4, might be potential prognostic markers and therapeutic targets in the treatment and management of MM patients positive for BCL1/JH t(11;14)(q13;q32) translocation.
Acknowledgments:
we would like to thank the Director of the Institute for Medical Research for support in the writing of this paper. Thanks to Ms Ten Sew Keoh for her help in editing this manuscript.
References
- 1.Bhardwaj A, Sethi G, Vadhan-Raj S, et al. Resveratrol inhibits proliferation, induces apoptosis, and overcomes chemoresistance through down-regulation of STAT3 and nuclear factor-κB–regulated antiapoptotic and cell survival gene products in human multiple myeloma cells. Blood. 2007;109:2293–302. doi: 10.1182/blood-2006-02-003988. [DOI] [PubMed] [Google Scholar]
- 2.Chesi M, Bergsagel PL, Brents LA, et al. Dysregulation of cyclin D1 by translocation into an IgH switch region in two multiple myeloma cell lines. Blood. 1996;88:674–81. [PubMed] [Google Scholar]
- 3.Bergsagel PL, Chesi M, Brents LA, et al. Translocations into IgH switch regions--the genetic hallmark of multiple myeloma. Blood. 1995;86:223–33. [Google Scholar]
- 4.Chang H, Qi XY, Stewart AK. t(11;14) does not predict long-term survival in myeloma. Leukemia. 2005;19:1078–9. doi: 10.1038/sj.leu.2403744. [DOI] [PubMed] [Google Scholar]
- 5.Fenton JAL, Pratt G, Rothwell DG, et al. Translocation t(11;14) in multiple myeloma. Analysis of translocation breakpoints on der(11) and der(14) chromosomes suggests complex molecular mechanisms of recombination. Genes, Chromosomes Cancer. 2004;39:151–5. doi: 10.1002/gcc.10304. [DOI] [PubMed] [Google Scholar]
- 6.Hoyer J, Hanson CA, Fonseca R, et al. The (11;14)(q13;q32) translocation in multiple myeloma: a morphologic and immunohistochemical study. Am J Clin Pathol. 2000;113:831–7. doi: 10.1309/4W8E-8F4K-BHUP-UBE7. [DOI] [PubMed] [Google Scholar]
- 7.Fonseca R, Blood EA, Oken MM, et al. Myeloma and t(11;14)(q13;q32): evidence for a biologically defined unique subsets of patients. Blood. 2002;99:3735–41. doi: 10.1182/blood.v99.10.3735. [DOI] [PubMed] [Google Scholar]
- 8.Moreau P, Facon T, Leleu X, et al. Recurrent 14q32 translocations determine the prognosis of multiple myeloma, especially in patients receiving intensive chemotherapy. Blood. 2002;100:1579–83. doi: 10.1182/blood-2002-03-0749. [DOI] [PubMed] [Google Scholar]
- 9.Hallek M, Bergsagel PL, Anderson KC. Multiple myeloma: increasing evidence for a multistep transformation process. Blood. 1998;91:3–21. [PMC free article] [PubMed] [Google Scholar]
- 10.Jaillard S, Drunat S, Bendavid C, et al. Identification of gene copy number variations in patients with mental retardation using array-CGH: novel syndromes in a large French series. Eur J Med Genet. 2010;53:66–75. doi: 10.1016/j.ejmg.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Shen Y, Irons M, Miller DT, et al. Development of a focused oligonucleotide-array comparative genomic hybridization chip for clinical diagnosis of genomic imbalance. Clin Chem. 2007;53:2051–9. doi: 10.1373/clinchem.2007.090290. [DOI] [PubMed] [Google Scholar]
- 12.Samal B, Sun Y, Stearns G, et al. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony enhancing factor. Mol Cell Biol. 1994;14:1431–7. doi: 10.1128/mcb.14.2.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ognjanovic S, Bao S, Yamamoto SY, et al. Genomic organization of the gene coding for human pre-B-cell colony enhancing factor and expression in human fetal membranes. J Mol Endocrinol. 2001;26:107–17. doi: 10.1677/jme.0.0260107. [DOI] [PubMed] [Google Scholar]
- 14.Martin PR, Shea RJ, Mulks M. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J Bacteriol. 2001;183:1168–74. doi: 10.1128/JB.183.4.1168-1174.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B, Hasan MK, Alvarado E, et al. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene. 2011;30:907–21. doi: 10.1038/onc.2010.468. [DOI] [PubMed] [Google Scholar]
- 16.Van Beijnum JR, Moerkerk PT, Gerbers AJ, et al. Target validation for genomics using peptide-specific phage antibodies: a study of five gene products overexpressed in colorectal cancer. Int J Cancer. 2002;101:118–27. doi: 10.1002/ijc.10584. [DOI] [PubMed] [Google Scholar]
- 17.Hufton SE, Moerkerk PT, Brandwijk R, et al. A profile of differentially expressed genes in primary colorectal cancer using suppression substractive hybridization. FEBS Lett. 1999;463:77–82. doi: 10.1016/s0014-5793(99)01578-1. [DOI] [PubMed] [Google Scholar]
- 18.Hageman GJ, Stierum RH. Niacin, poly(ADP-ribose) polymerase-1 and genomic stability. Mutat Res. 2001;475:45–56. doi: 10.1016/s0027-5107(01)00078-1. [DOI] [PubMed] [Google Scholar]
- 19.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 20.Nomura F, Yaguchi M, Togawa A, et al. Enhancement of poly-adenosine diphosphate-ribosylation in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2000;15:529–35. doi: 10.1046/j.1440-1746.2000.02193.x. [DOI] [PubMed] [Google Scholar]
- 21.Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63:7436–7436. [PubMed] [Google Scholar]
- 22.Koeppel M, van Heeringen SJ, Smeenk l, et al. The novel p53 target gene IRF2BP2 participates in cell survival during the p53 stress response. Nucleic Acids Res. 2009;37:322–35. doi: 10.1093/nar/gkn940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Childs KS, Goodbourn S. Identification of novel co-repressor molecules for interferon regulatory factor-2. Nucleic Acids Res. 2003;31:3016–26. doi: 10.1093/nar/gkg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blotta S, Tassone P, Prabhala RH, et al. Identification of novel antigens with induced immune response in monoclonal gammopathy of undetermined significance. Blood. 2009;114:3276–84. doi: 10.1182/blood-2009-04-219436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marzin Y, Jamet D, Douet-guilbert N, et al. Chromosome 1 abnormalities in multiple myeloma. Anticancer Res. 2006;26:953–60. [PubMed] [Google Scholar]
- 26.Sawyer JR, Tricot G, Lukacs JL, et al. Genomic instability in multiple myeloma: evidence for jumping segmental duplications of chromosome arm 1q. Genes Chromosomes Cancer. 2005;42:95–106. doi: 10.1002/gcc.20109. [DOI] [PubMed] [Google Scholar]
- 27.Zhan F, Colla S, Wu X, et al. CKS1B, overexpressed in aggressive disease, regulates multiple myeloma growth and survival through SKP2- and p27Kip1-dependent and -independent mechanisms. Blood. 2007;109:4995–5001. doi: 10.1182/blood-2006-07-038703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savage NM, Kota V, Manaloor EJ, et al. Acute leukemia with PICALM-MLLT10 fusion gene: diagnostic and treatment struggle. Cancer Genet Cytogenet. 2010;202:129–32. doi: 10.1016/j.cancergencyto.2010.07.126. [DOI] [PubMed] [Google Scholar]
- 29.Sindt A, Deau B, Brahim W, et al. Acute monocytic leukemia with coexpression of minor BCR-ABL1 and PICALM-MLLT10 fusion genes along with overexpression of HOXA9. Genes Chromosomes Cancer. 2006;45:575–82. doi: 10.1002/gcc.20320. [DOI] [PubMed] [Google Scholar]
- 30.Largo C, Alvarez S, Saez B, et al. Identification of overexpressed genes in frequently gained/amplified chromosome regions in multiple myeloma. Haematologica. 2006;91:184–91. [PubMed] [Google Scholar]
- 31.Zhu YX, Benn S, Li ZH, et al. The SHS-SAM adapter HACS1 is upregulated in B cell activation signalling cascades. J Exp Med. 2004;200:737–47. doi: 10.1084/jem.20031816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claudio JO, Zhu YX, Benn SJ, et al. HACS1 encodes a novel SH3-SAM adaptor protein differentially expressed in normal and malignant haematopoietic cells. Oncogene. 2001;20:5373–7. doi: 10.1038/sj.onc.1204698. [DOI] [PubMed] [Google Scholar]
- 33.Brandt S, Ellwanger K, Beuter-Gunia C, et al. SLy2 targets the nuclear SAP30/HDAC1 complex. Int J Biochem Cell Biol. 2010;42:1472–81. doi: 10.1016/j.biocel.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Darnell JE., Jr. STATs and gene regulation. Science. 1997;277:1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 35.Bowman T, Jove R. STAT proteins and cancer. Cancer Control. 1999;6:615–9. doi: 10.1177/107327489900600617. [DOI] [PubMed] [Google Scholar]
- 36.Ogata A, Chauhan D, Teoh G, et al. IL-6 triggers cell growth via the Ras-dependent mitogen-activated protein kinase cascade. J Immunol. 1997;159:2212–21. [PubMed] [Google Scholar]
- 37.Ishikawa H, Tsuyama N, Liu S, et al. Accelerated proliferation of myeloma cells by interleukin-6 cooperating with fibroblast growth factor receptor 3-mediated signals. Oncogene. 2005;24:6328–32. doi: 10.1038/sj.onc.1208782. [DOI] [PubMed] [Google Scholar]
- 38.Fryknäs M, Dhar S, Oberg F, et al. STAT1 signaling is associated with acquired crossresistance to doxorubicin and radiation in myeloma cell lines. Int J Cancer. 2006;120:189–95. doi: 10.1002/ijc.22291. [DOI] [PubMed] [Google Scholar]
- 39.Miura Y, Tsujioka T, Nishimura Y, et al. TRAIL expression up-regulated by interferon-gamma via phosphorylation of STAT1 induces myeloma cell death. Anticancer Res. 2006;26:4115–24. [PubMed] [Google Scholar]
- 40.Catlett-Falcone R, Landowski TH, Oshiro MM, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–15. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 41.Sanjo H, Kawai T, DRAKs AS. Novel serine/threonine kinases related to death-associated protein kinase that trigger apoptosis. J Biol Chem. 1998;273:29066–71. doi: 10.1074/jbc.273.44.29066. [DOI] [PubMed] [Google Scholar]
- 42.Capra M, Nuciforo PG, Confalonieri S, et al. Frequent alterations in the expression of serine/threonine kinases in human cancers. Cancer Res. 2006;66:8147–54. doi: 10.1158/0008-5472.CAN-05-3489. [DOI] [PubMed] [Google Scholar]
- 43.Bergsagel PL, Michael Kuehl W. Chromosome translocations in multiple myeloma. Oncogene. 2001;20:5611–22. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]


