Abstract
Antibody engineering has generally been carried out by displaying mouse or human antibodies or antibody fragments on the surface of microorganisms (phage, bacteria and yeast). We have shown that mammalian cells can be used to display single chain antibody fragments (scFvs) for affinity maturation. Using mammalian cell display one can isolate and engineer scFvs, Fabs or whole IgGs for increased affinity and other specific biological functions. Here, we describe a mammalian cell display strategy to isolate high affinity scFvs specific for CD22. Our strategy uses flow cytometry and human embryonic kidney 293T (HEK-293T) cells that are widely used for transient protein expression. Flow cytometry enhances the screen’s sensitivity thereby allowing us to isolate high affinity antibodies.
Keywords: antibody engineering, antibody affinity maturation, single chain Fv or scFv, phage display, cell display, mammalian cell display, HEK-293T, flow cytometry
1. Introduction
Creating high affinity antibodies to important biomolecules for clinical use is an important and challenging task. Twenty-one therapeutic monoclonal antibodies (mAbs) have been approved by the U.S. Food and Drug Administration, and hundreds of mAbs are in clinical trials (1). The levels of antibody affinity obtained from hybridomas are frequently not sufficient for effective clinical use, in part because of the in vivo affinity ceiling (2); therefore an improvement of antibody affinity is often required. For the past two decades phage display (3, 4) has been used for in vitro antibody affinity maturation and more recently bacterial and yeast cell surface display (5, 6) systems have been developed (7, 8). Very recently we showed that single chain antibodies can be displayed on the surface of human HEK-293T cells and used for affinity maturation. We call this method “mammalian cell display”. Our strategy is adapted from Wittrup’s yeast cell display used previously to isolate high affinity antibodies in yeast cells except we use human embryonic kidney 293T (HEK-293T) cells because these cells are already widely used for transient protein expression. Mammalian cell display is a powerful method for the isolation of scFv (9) and whole IgG (10) with high affinity and other specific biological functions. Mammalian cell display relies on the transient transfection of antibody encoded DNA to promote very high levels of antibody expression in mammalian cells. Moreover, the expressed mouse or human antibodies can contain the posttranslational modifications that are required for antibody function. It has been suggested that mammalian cell display could be used to express the recombinant antibody fragments that cannot be expressed in E. coli (11). We have used this mammalian cell display strategy to increase the affinity of antibodies that bind CD22 (9) and to isolate human scFvs that bind mesothelin from nonimmune human libraries (Ho and Pastan, unpublished data). CD22 is an adhesion molecule expressed in B cells and overexpressed in B-cell leukemias and lymphomas. Mesothelin is highly expressed in several human solid tumors, including virtually all mesotheliomas and pancreatic adenocarcinomas, and approximately 70% of ovarian cancers and 50% of lung adenocarcinomas (reviewed in 12).
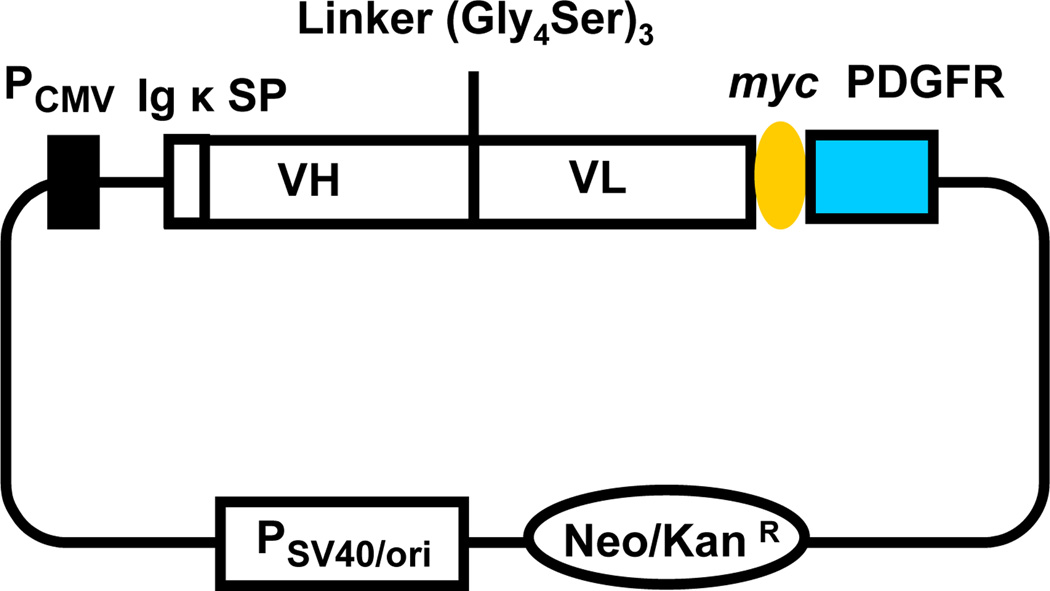
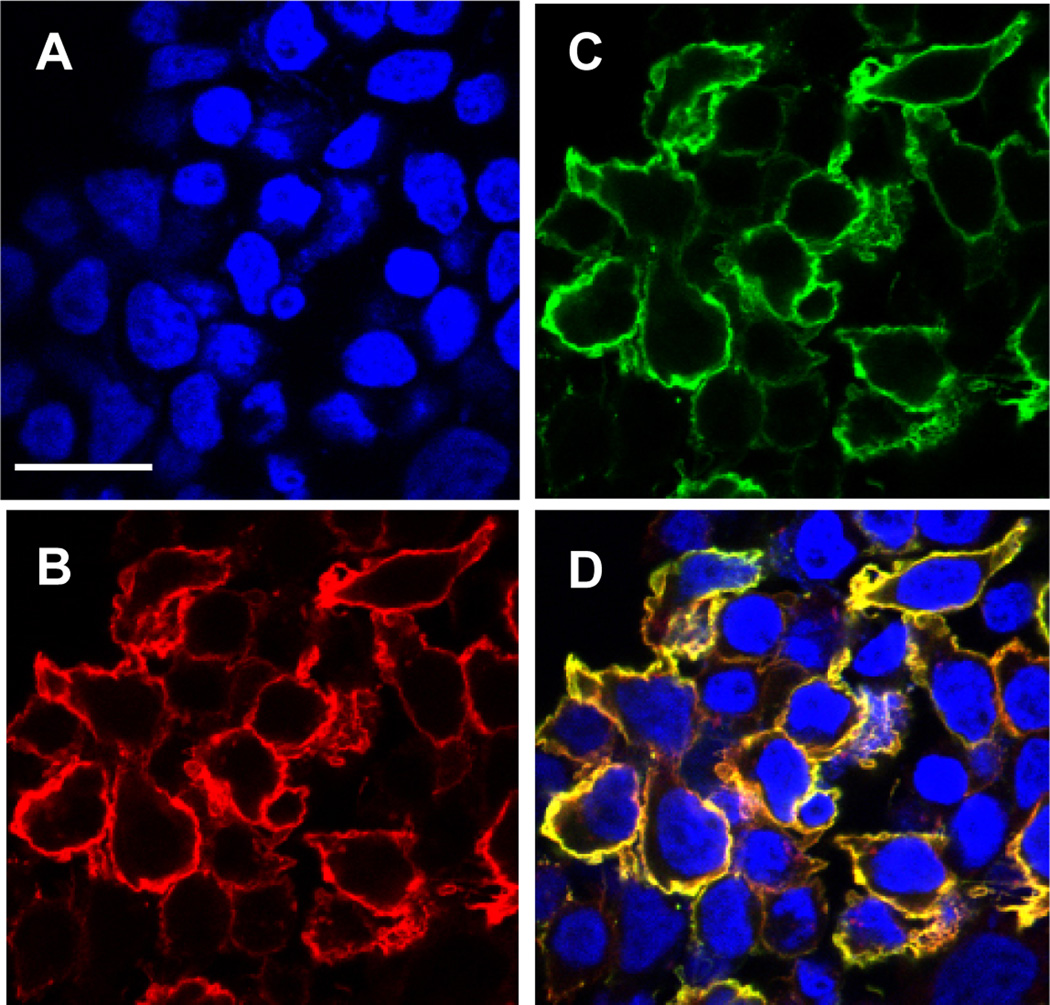
To display the Fv on the cell surface we have fused the scFv to the transmembrane domain of human platelet-derived growth factor receptor (PDGFR) (Fig. 1). The expression vector contains the cytomegalovirus promoter, the nucleotide sequence encoding the murine Igκ chain signal peptide (METDTLLLWVLLLWVPGSTGD), the scFv, a myc tag and the transmembrane domain (amino acids Ala513–Arg561) of PDGFR. We used the myc epitope tag at the carboxyl terminal of the scFv to measure the expression level. We expressed anti-CD22 (RFB4) scFv on HEK-293T cells (Fig. 2). Surface localization of the scFv-PDGFR fusion was verified by confocal fluorescence microscopy and flow cytometry. Cells labeled simultaneously with biotinylated CD22-Fc proteins and an anti-c-myc mAb were examined by laser scanning confocal microscopy (Fig. 3). Cells bearing the surface display vector expressing the scFv-PDGFR fusion protein were colabeled (Fig. 3D) by a mixture of the CD22-Fc (Fig. 3B) and the anti-c-myc antibody (Fig. 3C).
Fig. 1.
Diagram of expression plasmid for display of scFv on mammalian cells. PCMVcytomegalovirus promoter; Igκ SP, murine Igκ chain signal peptide; VH, heavy chain variable region; VL, light chain variable region; Linker, a flexible synthetic linker between VH and VL; myc, an epitope tag to measure the scFv expression level; PDGFR, the transmembrane domain of human platelet-derived growth factor receptor; PSV40/oriSV40 promoter and origin facilitating episomal replication in mammalian cells expressing SV40 large T antigen; Neo/KanRneomycin- and kanamycin-resistance gene.
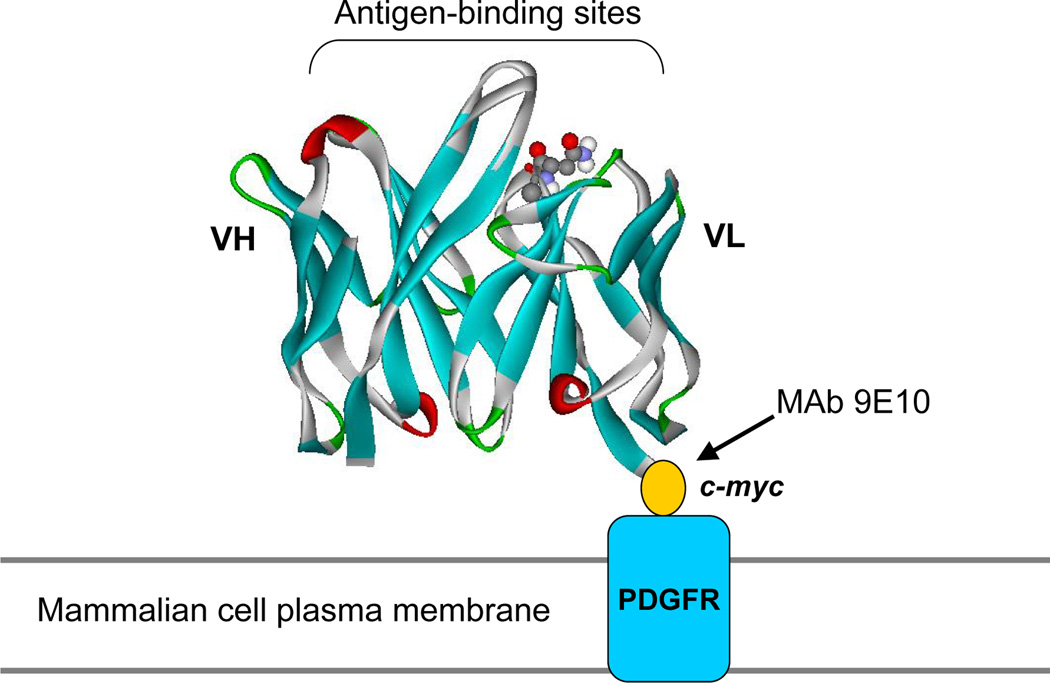
Fig. 2.
Schematic illustration of surface display on mammalian cells. An additional 10-amino acid epitope tag (c-myc) was fused to the C-terminus of the scFv (based on the anti-CD22 RFB4 Fv structural model) (13), allowing quantitation of fusion display with mAb 9E10 independent of antigen (CD22) binding. Fusion to the N-terminal portion of the PDGFR transmembrane domain was used to anchor scFv on the mammalian (HEK-293T) cell surface. (Originally published in Proceedings of the National Academy of Sciences 103(25):9637–9642, June 20, 2006); copyright 2006 National Academy of Sciences, U.S.A.
Fig. 3.
Confocal microscopic images of HEK-293T cells displaying scFv. HEK-293T cells transfected with a plasmid directing surface expression of anti-CD22 scFv were grown on cover slips. Transfected cells were fixed with 4,6-diamidino-2-phenylindole nuclear staining (A) followed by detection with biotinylated CD22-Fc (B), and anti-c-myc antibody (C) followed by Steptavidin-Alexa Fluor 594 and anti-mouse IgG-Alexa Fluor 488. (D) shows merged staining patterns. Scale bar = 25 µm. (Originally published in Proceedings of the National Academy of Sciences 103(25):9637–9642, June 20, 2006); copyright 2006 National Academy of Sciences, U.S.A.
We transfect a HEK-293T cell line with a scFv library (in the vector pDisplay). We use fluorescence-activated cell sorting (FACS) to select and collect cells which bind the targets. The library scFv DNAs are recovered and analyzed until highly enriched scFv clones with desired properties are identified.
2. Materials
2.1. Cell Culture
Growth medium for HEK-293T cells: Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen/GIBCO, Carlsbad, CA) supplemented with 10% fetal calf serum (Sigma, St. Louis, MO); 1% L-Glutamine solution (Sigma); 1% Nonessential amino acids solution (Sigma) and penicillin-streptomycin (Invitrogen, Carlsbad, CA).
Dishes for HEK-293T transfections: 100-mm tissue culture dishes (Fisher, Hampton, NH).
Cells used for mammalian cell display: HEK-293T cells. (These cells may be purchased from GenHunter, Catalog # Q401, http://genhunter.com/products/aptag-3/index.html) (see Note 1).
2.2. Transfection
An antibody library that has been directionally cloned into a mammalian expression vector pDisplay (Invitrogen) (see Note 2).
Transfection reagents: Lipofectamin reagent (Invitrogen) and PLUS reagent (Invitrogen).
Falcon 2052 tubes (Becton Dickinson, Franklin Lakes, NJ).
Solution of 0.25% trypsin and 1 mM ethylenediamine tetraacetic acid (EDTA) in PBS (Invitrogen/GIBCO).
Serum-free DMEM (DMEM plus glutamine, without antibiotics).
20% FBS in DMEM (DMEM plus 20% FBS, glutamine and antibiotics).
PBS (Invitrogen).
Ultrapure Water (Invitrogen).
2.3. Flow Cytometry
FACS Solution used as a blocking and antibody elution buffer: 5% (w/v) BSA (Bovine Serum Albumin Fraction V, heat shock, fatty acid ultra-free; Roche), 0.1% sodium azide (Sigma) in PBS.
An antibody used to bind the c-myc tag: 9E10 mAb (culture supernatant from hybridoma cell line 9E10, ATCC Catalog # CRL-1729) (see Note 3).
Fluorescein isothiocyanate (FITC) labeled goat anti-mouse (Invitrogen/Biosource) to detect the 9E10 antibody.
Streptavidin-R-phycoerythrin (Invitrogen/Molecular Probes) to visualize biotinylated-target protein (e.g., biotinylated CD22-Fc).
Target proteins (see Note 4).
Major equipment: flow cytometer (BD FACScalibur, Becton Dickinson) for analysis and BD FACSVantage SE flow cytometry (Becton Dickinson) for cell sorting (see Note 5).
2.4. Construction of Mammalian Display Libraries and Rescue of scFv Plasmid DNA
pDisplay (Invitrogen) vector for library construction and serves as a negative control.
SfiI (New England Biolabs, Ipswich, MA) and SacII (New England Biolabs), 10 × digestion buffer provided with the enzymes.
Primers used to subclone an antibody library into the SfiI and SacII sites of pDisplay vector.
Glycerol (Sigma).
E. coli TOP10 (Invitrogen) at a competency of ≥109 colony forming units per µg (cfu/µg) DNA (see Note 6).
SOC medium (Invitrogen).
LB agar plates with 100 µg/mL Ampicillin (Sigma).
DpnI restriction enzyme, 10 × buffer included (New England Biolabs).
Phenol:chloroform:isoamyl alcohol (25:24:1) (Sigma).
DNA maxi- and mini-prep kits (Qiagen, Valencia, CA).
3 M sodium acetate, pH 7.0 (Sigma).
Ampicillin (Sigma).
Major equipment: 42°C water bath and tabletop centrifuge.
Hirt’s Solution (14): 0.6% SDS, 10 mM EDTA in 5 mL of ddH20 (see Note 7).
3. Methods
The following procedure describes a mammalian cell display system to identify scFv antibody proteins that bind CD22, an antigen highly expressed in B-cell leukemias and lymphomas. This method may be adapted to identify antibodies that bind other targets. We recently used this method to isolate human antibodies that bind mesothelin, a tumor antigen in solid tumors. The strategy consists of transfecting HEK-293T, a human cell line with a scFv antibody library (in this example, an anti-CD22 scFv library) and an appropriate target protein (in this example, the extracellular domain of CD22 fused to human Fc protein). The detection of cells bound to target proteins is done by FACS. The cells transfected with the scFv library are sorted by flow cytometry, and only cells in which the scFv binds to the target protein are collected. The plasmid DNAs are isolated from these cells and amplified in bacteria. These scFv DNAs are further enriched until the scFvs with increased affinity for target proteins can be isolated (see Note 8).
3.1. Construction of Mammalian Cell Display Library
An antibody scFv library should be subcloned into appropriate restriction enzyme sites of the mammalian display vector. The cloned scFvs are flanked by 5' SfiI and 3' SacII sites to facilitate in-frame cloning of scFv genes into pDisplay (Invitrogen). Oligomers with SfiI and SacII sites were used for in-frame subcloning of the scFv library into the pDisplay vector. Library scFv DNA is used as a template. The PCR product is subcloned into SfiI and SacII sites downstream of the immunoglobulinκ chain leader sequence of the pDisplay vector. The expression vector encodes a myc epitope tag and a transmembrane domain of PDGFR downstream of the scFv.
Design primers to amplify the scFv genes. The 5’ forward primer must contain a SfiI site. The 3’ reverse primer must contain a SacII site. For the anti-CD22 scFv library, we designed the following 5’ and 3' primers: Dis_SfiIF 5’-GGGGCCCAGCCGGCCATGGAAGTGCAGCTGGTG-3’, Dis_SacIIR 5’-CTGCCGCGGAGCTTTGATTTCCAG-3’, where SfiI and SacII sites are underlined.
Amplify the scFv genes. Using forward and reverse primers (final primer concentration is 10 µM) described above, use 10 ng of library DNA as the template to PCR amplify the scFv library. The template and oligonucleotides are mixed with high fidelity Tgo DNA polymerase in a 50-µL volume and then cycled using the following profile: 1 cycle at 95°C for 5 min, followed by 30 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min.
The PCR products must be digested with the SfiI and SacII, gel purified, ligated into the pDisplay (also cut with SfiI and SacII and gel purified) and transformed.
The ligation mix is used to transform E. coli TOP10 cells. Each transformation produces a cell surface display library containing ~1 × 106 independent clones. We also make a plasmid containing a wild-type scFv as a control.
3.2. Transfection
It is important to have adequate controls, as well as a sufficient number of cells for sorting and recovery of plasmid DNA. A control plate is recommended: Control Plate should be transfected with pDisplay containing a wild-type scFv antibody.
Plate five 100-mm tissue culture dishes at 2 × 106 cells per plate 24 hr before transfection (see Note 9).
Make dilution of DNA with serum-free DMEM (4 µg library DNA or control DNA in 750 µL of DMEM per dish), mix well.
Add 20 µL of PLUS reagent to the 750 µL of DNA solution, mix well, and incubate at room temperature for 15 min.
Dilute Lipofectamine reagent with serum-free DMEM in second tube (30 µL in 750 µL for 1 dish) (see Note 10).
Combine the DNA solution and the Lipofectamine diluted (1.5 mL total volume per dish). Mix well, and incubate at room temperature for more than 15 min (see Note 11).
While complexes are forming, replace the medium on cells with 5 mL of serum-free DMEM per 100 mm dish.
Gently add the DNA-lipofectamine mixture to the dishes (1.5 mL mixture per dish). Gently shake the dish several times.
Incubate the dishes in CO2-incubator for 5 hr.
Gently add 6 mL of 20% FBS DMEM to each dish and return the dish to CO2-incubator.
Add 10 mL of 10% FBS DMEM growth medium into each dish 24 hr after transfection.
After incubation for 48–72 hr, the cells are ready for sorting or analysis by flow cytometry (see Note 12).
3.3. FACS Sorting
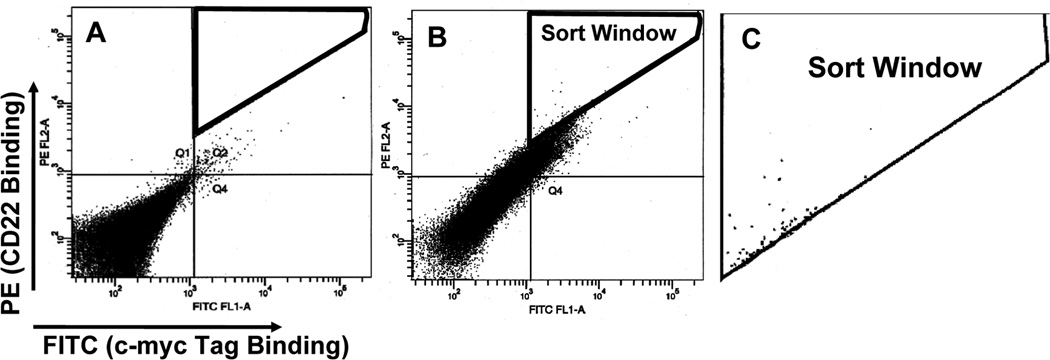
The antibodies and detection reagents described here are for the specific example of assessing binding affinity for CD22, but the method can be adapted to detect the binding affinity of any target protein. In a typical experiment, 48 hr after transfection, 1 × 107 cells (~ total HEK-293T cells collected from a 100 mm Petri dish) were used. The cells were incubated with 0.1–1 µg/mL of biotinylated CD22-Fc and 20 µg/mL of mAb 9E10 (anti-c-myc) in 500 µL of growth medium. Control cells containing wild-type anti-CD22 scFv should be separated into 4 tubes: one is stained with secondary detection conjugates only, one is stained only with 0.1–1 µg/mL of biotinylated CD22-Fc, one is stained only with 20 µg/mL of mAb 9E10 and the other one is stained with both. The single stainings are useful to determine the parameters such as compensation in FACS analysis and sorting. After incubation for 1 hr at 4°C, the cells were washed with PBS and incubated on ice with 1:200 dilution of FITC-labeled goat anti-mouse FITC and streptavidin-R-PE conjugate for 1 hr. After washing, the cells were suspended in 0.5 mL of growth medium and the cells are ready for sorting on a FACS Vantage or analysis on FACS Calibur. The top 0.1% PE-positive cells (13,300 cells) are collected (Fig. 4). Plasmids are recovered from selected HEK-293T cells and transformed into E. coli to recover plasmid scFv DNA for sequencing.
Remove the media from all plates and wash cells once with 2 mL of 1 × PBS.
Add 4 mL of trypsin EDTA buffer to the Sort Plates and the Control Plate.
Incubate for 5 min at room temperature.
Add 10 mL of complete DMEM into each dish and collect cells from Sort Plates and combine into one 50-mL tube (Sort Cells) or 15-mL tube (Control Cell).
Centrifuge the Sort Cells and the Control Cells at 300g in a tabletop centrifuge for 3 min at room temperature to pellet the cells.
Aspirate off the supernatant.
Resuspend the Sort Cells in 10 mL of complete DMEM by gentle agitation of the tubes and pipetting up and down 4 times.
Add 0.1–1 µg/mL of biotinylated antigen (for example: CD22-Fc) and 20 µg/mL of mAb 9E10 (see Note 13).
Resuspend the Control Cells (which contains wild-type scFv) from the Control Plate separately in 5 mL complete DMEM.
Aliquot 5 × 105 cells from Control Plate into 1 mL complete DMEM. Prepare 4 separate aliquots, labeling the tubes: 1A, 1B, 1C, and 1D.
Add 0.1–1 µg/mL of biotinylated antigen into 1B and 20 µg/mL of mAb 9E10 into 1C and both biotinylated antigen and 9E10 into 1D. Resuspend cells by gentle agitation of tubes.
Incubate Sort Cell samples and Control Cell samples for 60 min on ice.
Add 4 mL of complete DMEM to each control tube.
Centrifuge all samples (Control and Sort) for 5 min at 1000 rpm in a tabletop centrifuge.
Wash the cells with PBS twice.
Incubate the Sort Cells on ice with 1:200 dilution of FITC-labeled goat anti-mouse FITC and streptavidin-R-PE conjugate
Incubate the Control Cells 1A, 1B, 1C and 1D with 1:200 dilution of FITC-labeled goat anti-mouse FITC and streptavidin-R-PE conjugate.
Incubate all samples on ice for 30 min.
Wash the cells with 5 mL of complete DMEM.
Centrifuge the cells for 5 min at room temperature at 1000 rpm in a tabletop centrifuge.
Remove and discard the supernatants.
Wash the cells with 5 mL PBS twice.
Resuspend the Sort Cells in 2 mL of complete DMEM.
Resuspend Control Cell samples in 1 mL of complete DMEM.
Incubate all tubes on ice in the dark.
Set the parameters on a FACSVantage SE such that the FITC fluorescence of 2B does not overlap the PE fluorescence of 2C.
Collect the phycoerythrin- and FITC-positive Sort Cells by FACS, with the 0.1% sorting gate set as described in Fig. 4 (see Note 14).
Fig. 4.
Enrichment of mammalian cells displaying an improved scFv variant by kinetic selection and flow cytometric sorting. The dot plot shows only 50,000 cells of the total cell population (107). Each dot represents an individual observed cell. This is the 1:400 mixture of HA22:BL22-displaying cells, labeled with no antigen (A) or 1nM antigen (CD22-Fc) (B) concentration for separation. FITC fluorescence (binding to the c-myc tag) represents the number of surface expressions on an individual cell. The PE fluorescence represents binding to the antigen (CD22). A sort window (C) was drawn to include the top 0.1% of total cells in terms of ratio of PE:FITC fluorescence. Cells that fell within the window were collected. (Originally published in Proceedings of the National Academy of Sciences 103(25):9637–9642, June 20, 2006); copyright 2006 National Academy of Sciences, U.S.A.
3.4. scFv Recovery
Pipette one-half (1.5 mL) of the collected phycoerythrin- and FITC-positive Sort Cells (Collected Sort Cells) into a microcentrifuge tube.
Microcentrifuge for 5 min at 300g.
Discard the supernatant and pipette the remaining 1.5 mL of Collected Sort Cells into the same tube.
Microcentrifuge for 5 min at 1000 rpm.
Discard the supernatant (see Note 15).
Add 400 µL of Hirt’s Solution.
Incubate at room temperature for 20 min.
Add 100 µL of 5 M NaCl, mix by inversion, and leave at 4°C overnight (see Note 16).
Microcentrifuge the Collected Sort Cells tube at maximum speed for 5 min at room temperature.
Carefully transfer each supernatant into a new labeled microcentrifuge tube.
Phenol:chloroform extract each supernatant twice.
Transfer extracted supernatants to separate new microcentrifuge tubes.
Add 1 µL of glycogen to the Collected Sort Cells tube.
Fill tubes to the top with 100% ethanol to precipitate DNA.
Microcentrifuge at maximum speed for 5 min at room temperature to pellet the DNA.
Remove the supernatant and resuspend each pellet in 100 µL of ddH2O.
Add 10 µL of 3 M sodium acetate, pH 7.0 and 300 µL of 100% ethanol to each tube.
Microcentrifuge at maximum speed for 5 min at room temperature to pellet the DNA.
Remove the supernatant and resuspend each pellet in 100 µL of ddH2O.
Add 10 µL of 3 M sodium acetate, pH 7.0 and 300 µL of 100% ethanol to each tube.
Microcentrifuge the tubes for 20 min at maximum speed at 4°C.
Wash each pellet with 1 mL of 70% ethanol.
Resuspend the Collected Sort Cells pellet in 10 µL of ddH20.
(Optional) Perform a DpnI digestion using 10 µL of DNA. Incubate overnight at 37°C (see Note 17).
3.6. Transformation
Incubate each entire 10-µL isolated DNA (or after DpnI-digested) with 100 µL of highly competent TOP10 cells on ice for 20 min in Falcon 2059 tubes.
Transfer tubes to a 42°C water bath for 1 min and return to ice.
Add 1 mL of SOC Medium and incubate on the shaker at 37°C for 1 hr.
Plate entire transformation onto Amp-LB plates.
Incubate overnight at 37°C.
Count and record the number of colonies that grow (see Note 18).
Pick 10–20 individual clones after 2nd round of sorting for clone analysis and inoculate into 50 mL E. coli culture overnight.
Pick all of the rest colonies from the Collected Sort Cell plate and inoculate into 500 mL E. coli culture overnight.
Make a glycerol stock by adding 300 µL of 60% glycerol to 1 mL culture.
Store the glycerol stock at −80°C.
Prepare DNA from each round of sorting by Qiagen Maxiprep according to the manufacturer’s protocols.
Transfect the DNA isolated from previous round into HEK-293T cells and repeat the FACS sorting using the same procedure described above except that we normally reduce the antigen concentration in the following rounds to enrich high affinity binders.
3.7. Identification of Binders
At this step, the goal is to identify the scFv that bind the target protein with highest affinity in the library.
Make DNA from each individual clone (50 mL E. coli culture) by QIAgen Midi-Prep Kit.
Transfect 4 µg of each clone of DNA into the cells.
Perform analytical FACS to identify the scFv clones that produce the high affinity.
Sequence the scFv fragments with primers Dis_SfiIF and Dis_SacIIR (see Note 19).
3.8. Determination of Affinity Constants (KD) by Flow Cytometry
KD is measured by determining at what concentration of antigen is half of the scFv on the surface of the HEK-293T cell bound to antigen. Therefore measuring the MFI of the HEK-293T cells when no antigen is bound and determining the concentration of antigen that gives the maximal MFI is needed. We have shown that this is easily accomplished by setting up a series of antigen concentrations in which to label the cells with and then measuring the MFI of the antigen binding population by flow cytometry (Fig. 5).
Remove the media from all plates and wash cells once with 2 mL of 1 × PBS.
Add 4 mL of trypsin EDTA buffer to the Sort Plates and the Control Plates.
Incubate for 5 min at room temperature.
Add 10 mL of complete DMEM into each dish and collect cells into one 15-mL tube.
Centrifuge the Sort Cells and the Control Cells at 1000 rpm in a tabletop centrifuge for 3 min at room temperature to pellet the cells.
Aspirate off supernatant.
Resuspend the 1 × 105 cells in 1 mL of FACS buffer by gentle agitation of tubes and pipetting up and down for 4 times.
Add various concentrations (e.g., 0.1–100 nM) of biotinylated antigen (see Note 20).
Incubate cells for 60 min on ice.
Wash twice.
Add 1:200 streptavidin-PE and incubate on ice for 30 min.
Wash twice with PBS and analyze by flow cytometry.
Use geometric mean as MFI.
Determine equilibrium constants by using the Marquardt–Levenberg algorithm for non-linear regression with GraphPad Prism following the developer’s instruction.
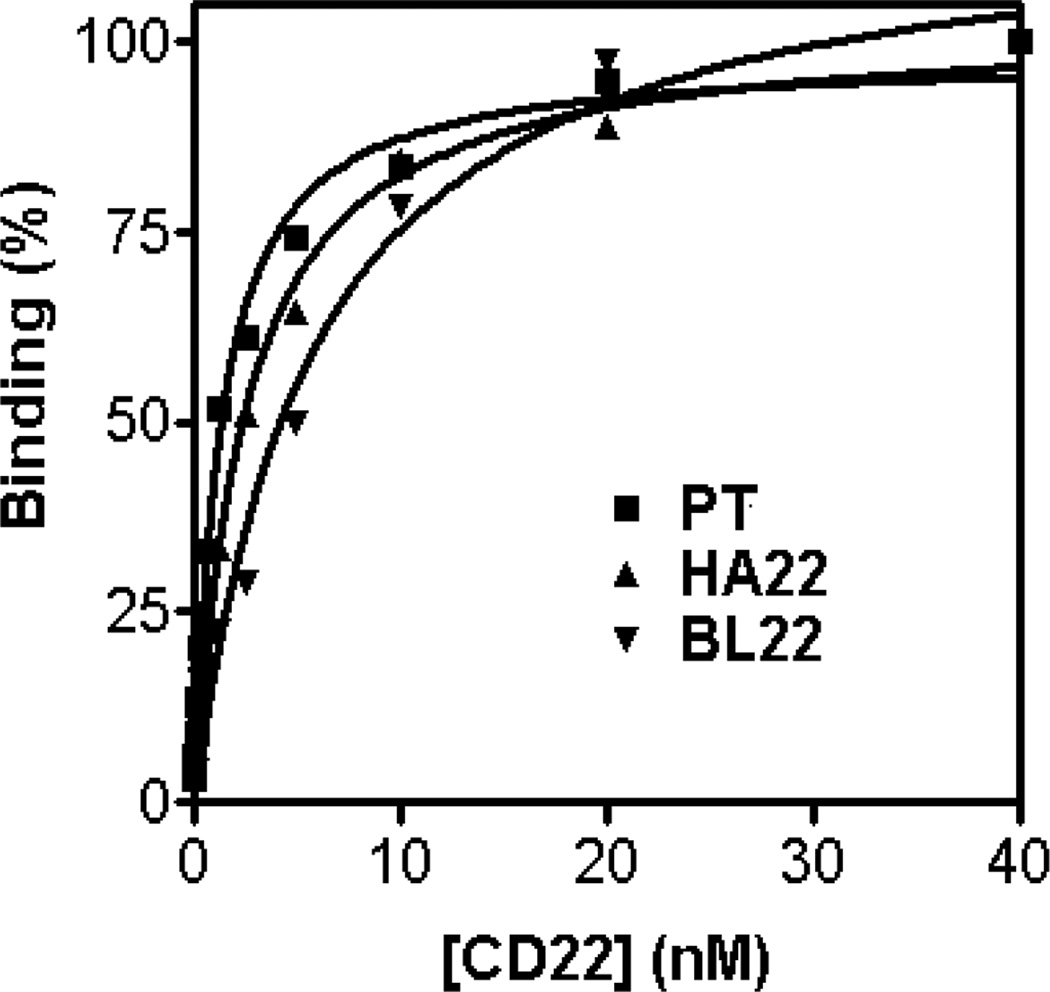
Fig. 5.
Equilibrium binding titration curves to determine dissociation constants, KD. MFI (%) of PE is plotted versus the various concentrations of biotinylated CD22-Fc used to label surface-displayed BL22 (wild-type, KD = 5.8nM, Bmax = 453), HA22 (KD = 2.5nM, Bmax = 293) or mutant PT (KD = 1.2nM, Bmax = 275) antibody. Data are fit with a nonlinear least-squares regression. (Originally published in Proceedings of the National Academy of Sciences 103(25):9637–9642, June 20, 2006); copyright 2006 National Academy of Sciences, U.S.A.
Acknowledgements
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
HEK-293T cells stably expressing the large T-antigen of SV40 (simian virus 40), allowing episomal replication of plasmids containing the SV40 origin. Any cell lines expressing SV40 large T-antigen (e.g. COS1 and COS7) may suffice. HEK-293T cells grown in 10% FBS DMEM. The cell MUST be in good condition before the transfection. HEK-293T cells should be split every 2 days to keep them growing.
The yield of pDisplay-derived plasmids from E. coli culture is low. At least 500 mL of E. coli culture is required to obtain enough DNA (200 µg of plasmid DNA) to transfect 50 dishes. Qiagen Maxi prep column-grade DNA works fine.
This antibody is used to detect expression levels of scFv.
We prefer to use a Fc fusion protein because they are often expressed in HEK-293T cells and because they can be purified in one step on a Protein A column. However, other formats of target proteins may also be suitable.
Most FACS core facilities may have these two machine; they are widely used. However, other brands of flow cytometers may suffice.
Any highly competent bacteria with ≥109 cfu/µg suitable to the pDisplay vector will suffice.
The QIAprep Spin Miniprep Kit (Qiagen) can be used as an alternative solution to recover scFv plasmid DNA from mammalian cells.
When using mammalian cell display there are three major points that must be considered. These are (i) the size of the library to be screened; (ii) the transfection efficiency of the mammalian host cells; and (iii) episomal replication of the scFv library in these cells. An appropriate antibody library will be in a plasmid with a promoter such as CMV that drives high levels of protein expression in mammalian cells. The library sizes we have used so far range from 103 to 106although theoretically it could be as large as 109. The plasmid must also contain an origin for episomal replication, such as polyoma or SV40, which matches the elements used to drive replication (that is, polyoma large T antigen or SV40 large T antigen). We recommend using the commercially available plasmid pDisplay that contains the SV40 origin for replication suitable for simple vector rescue in cell lines expressing the large T-antigen (e.g. HEK-293T, COS1, COS7 and CHO-Tag). Episomal replication is important because it is required to amplify the transfected plasmid DNA in the mammalian host cells. This ensures that most of the plasmids can be recovered from sorted cells. Flow cytometry allows millions of cells to be screened rapidly. In a typical screen, we sort at least 1 × 107 cells and collect about 10,000 cells in the gate. Hence, mammalian cell display provides a high-throughput approach to the identification of novel high affinity antibodies for possible clinical applications.
For long-term display of scFv, we recommend that stable cell lines be cloned by transfecting with a puromycin or hygromycin-resistant plasmid, as HEK-293T is G418 resistant.
We transfect 5–10 dishes per screen and 2 control plates.
The diluted Lipofectamin reagent is not stable. Make the dilution just before the next step.
During the incubation, DNA-lipofectamine complex will be formed.
A time course experiment indicated that the expression level of the transfected scFv is at a maximum 48–72 hr after transfection.
The dilution and amount of the antibody used to measure affinity needs to be determined empirically.
We sorted more than 1 × 107 cells and collected about 10,000 cells.
All cells should be collected at the bottom of one microcentrifuge tube.
Plasmid DNA can be isolated using the QIAprep Spin Miniprep Kit following the manufacturer’s instruction (QIAGEN News, 2:11, 1995) from the sorted HEK-293T cells. We have found that as few as 100 cells are enough for successful plasmid isolation.
In theory, plasmids replicating episomally in mammalian cells due to SV40 large T are not methylated. After isolation of the plasmid DNA from the cells, the DNA is digested with DpnI, which only digests methylated DNA, thereby removing any contaminant bacterial plasmids. Thus, if the mammalian host cells contain episomal replicating library DNA (which is desired), then there will be many colonies on plates. If the host cells do not contain episomal replicating scFvs, there will be no or only a few colonies on the plates. However, we find the difference between DpnI-digested DNA and non-digested DNA from HEK-293T cells is subtle (2–fold or less). Therefore, we consider this digestion an optional step.
A typical number of colonies from a successful screen from the Collected Sort Cell plates ranged from 1000–10,000. We typically obtain about 1 clone per sorted cell.
We typically see one highly enriched clone from a small library (diversity < 2000) by only single round of FACS sorting. For a large library (diversity >106), the best enrichment is achieved at the 3rd round of FACS sorting.
The dilution and amount of the antibody used to measure affinity needs to be determine empirically. For initial measurements, we routinely use 0.1, 0.5, 1, 5, 10, 50, and 100 nM of antigen (e.g., CD22-Fc).
References
- 1.Adams GP, Weiner LM. Monoclonal antibody therapy of cancer. Nat. Biotechnol. 2005;23:1147–1157. doi: 10.1038/nbt1137. [DOI] [PubMed] [Google Scholar]
- 2.Foote J, Eisen HN. Kinetic and affinity limits on antibodies produced during immune responses. Proc. Natl. Acad. Sci. USA. 1995;92:1254–1256. doi: 10.1073/pnas.92.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 4.Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu. Rev. Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 5.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 6.Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JM, Yeung YA, Cochran JR, Heinzelman P, Colby D, Swers J, Graff C, Wiley HS, Wittrup KD. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat. Biotechnol. 2003;21:163–170. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- 7.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 8.Sergeeva A, Kolonin MG, Molldrem JJ, Pasqualini R, Arap W. Display technologies: application for the discovery of drug and gene delivery agents. Adv. Drug Deliv. Rev. 2006;58:1622–1654. doi: 10.1016/j.addr.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho M, Nagata S, Pastan I. Isolation of anti-CD22 Fv with high affinity by Fv display on human cells. Proc. Natl. Acad. Sci. USA. 2006;103:9637–9642. doi: 10.1073/pnas.0603653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akamatsu Y, Pakabunto K, Xu Z, Zhang Y, Tsurushita N. Whole IgG surface display on mammalian cells: Application to isolation of neutralizing chicken monoclonal anti-IL-12 antibodies. J. Immunol. Methods. 2007;327:40–52. doi: 10.1016/j.jim.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Schenk JA, Sellrie F, Böttger V, Menning A, Stöcklein WF, Micheel B. Generation and application of a fluorescein-specific single chain antibody. Biochimie. 2007;89:1304–1311. doi: 10.1016/j.biochi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Hassan R, Ho M. Mesothelin targeted cancer immunotherapy. Eur. J. Cancer. 2007;44:46–53. doi: 10.1016/j.ejca.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho M, Kreitman RJ, Onda M, Pastan I. In vitro antibody evolution targeting germline hot spots to increase activity of an anti-CD22 immunotoxin. J. Biol. Chem. 2005;280:607–617. doi: 10.1074/jbc.M409783200. [DOI] [PubMed] [Google Scholar]
- 14.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]