Abstract
In the environment, there are aquatic pollutants that disrupt androgen signaling in fish. Laboratory and field-based experiments have utilized omics technologies to characterize the molecular mechanisms underlying androgen-receptor agonism/antagonism. Transcriptomics and proteomics studies with 17β-trenbolone, a growth-promoting pharmaceutical found in water systems surrounding cattle feed lots, and androgens such as 17α-methyltestosterone and 17α-methyldihydrotestosterone, have been conducted in ovary and liver of fish that include the fathead minnow (FHM) (Pimephales promelas), common carp (Cyprinus carpio), Qurt medaka (Oryzias latipes), and zebrafish (Danio rerio). In this mini-review, we survey recent omics studies in fish and reveal that, despite the diversity of species and tissues examined, there are common cellular responses that are observed with waterborne androgenic treatments. Recurring themes in gene ontology include apoptosis, transport and oxidation of lipids, synthesis and transport of hormones, immune response, protein metabolism, and cell proliferation. However, we also discuss other mechanisms other than androgen receptor (AR) activation, such as responses to toxicant stress, estrogen receptor agonism, aromatization of androgens into estrogens, and inhibitory feedback mechanisms by high levels of androgens that may also explain molecular responses in fish. To further explore androgen-responsive protein networks, a sub-network enrichment analysis was performed on protein data collected from the livers of female FHMs exposed to 17β-trenbolone. We construct a putative AR-regulated protein/cell process network in the liver that includes B-lymphocyte differentiation, xenobiotic clearance, low-density lipoprotein oxidation, proliferation of smooth muscle cells, and permeability of blood vessels. We demonstrate that construction of protein networks can offer insight into cell processes that are potentially regulated by androgens.
Androgens and androgen mimics are a significant source of endocrine-disrupting chemicals
Historically, xeno-androgens have been given less attention in ecotoxicology compared to environmental estrogens. Recently, however, there have been a growing number of studies that have characterized the effects of exogenous androgens. The studies in teleost fishes were designed to address the toxicological and physiological impacts of androgen exposure on aquatic organisms at different levels of biological organization (i.e., gene-to-steroid physiology to secondary sex characteristics) (Jensen et al. 2006; Seki et al. 2006; Garcia-Reyero et al. 2009). As a result, adverse outcome pathways of xeno-androgenic exposures and molecular signaling pathways regulated via the androgen receptor (AR) are becoming better defined for teleost fishes.
Some of the earlier research in North America that investigated androgens in aquatic environments at a mechanistic level was spurred by studies on the masculinization of the female mosquitofish (Gambusia affinis holbrooki) in response to kraft mill effluent (KME) in FL, USA (Howell et al. 1980; Davis and Bartone 1992). Following up on these earlier studies, Cody and Bortone (1997) sampled mosquitofish in Escambia County, FL (USA) after exposure to KME. Upon visual examination of fish caught at polluted sites, there were 736 (Elevenmile Creek) and 167 (Eightmile Creek) mosquitofish that showed elongation of the anal fin, suggesting that the KME was having a masculinizing effect on female fish. The elongated fin is a gonopodium that is characteristic of maturing males and it is used for internal fertilization in this live-bearing species. Later studies confirmed using an in vitro human androgen receptor (hAR) binding assay that androgenic-like activity in the river could explain the masculinization of female fish inhabiting water downstream of plants producing KME (Parks et al. 2001). It was then concluded that the masculinization was in part related to androgenic properties of the effluent. Androstenedione was identified in the water and efforts to isolate the chemical(s) inducing elongation of the anal fin in females were employed using toxicity identification evaluations with fractionation; however, the experiment revealed that the fraction that induced the masculinization was different than the one containing cis-androstenedione (Durhan et al. 2002). Thus, the chemicals stimulating elongation of the anal fin remained elusive.
Despite the lack of a direct association between androgen and biological responses in some studies, there are known environmental pollutants that have a well documented androgenic effect by binding to the AR. Perhaps the best documented sources of environmental androgens are growth promoters used in the beef-cattle industry. Pharmaceuticals, such as 17β-trenbolone (TB), are potent growth stimulators and are used to increase tissue mass in cattle. Not all of the pharmaceutical is metabolized in the animal, and nonmetabolized steroid can be excreted from the cattle, entering the surrounding water systems via runoff from the feedlots. In a 2-year experiment conducted in feeding pens, cattle were implanted with a single hormone (i.e., estrogens, androgens, progesterone) and soil, feces, and runoff were monitored for steroids (Bartelt-Hunt et al. 2012). The concentrations of the steroids in the implants were representative of those typically used in the beef-production industry. More than 90% of runoff samples collected from the pens after rainstorm events contained androsterone and 4-androstenedione. Moreover, testosterone (T) and TB were detected in some runoff samples at concentrations as high as ∼450 and 270 ng/L, respectively. The environmental impacts of pharmaceuticals used as growth promoters in cattle have been reviewed by Kolok and Sellin (2008) and the evidence is accumulating that feedlots have the potential to be a significant source of androgens in aquatic environments.
Multiple biological endpoints are altered by exposure to androgen
Exogenous androgens have a multitude of biological effects in both male and female fish. Biological endpoints range from the morphological to the molecular level and studies have documented the presence of ovotestis (Sone et al. 2005) decreases in levels of plasma sex steroids (Ankley et al. 2003, 2004; Martinović et al. 2008), depressed vitellogenin (VTG) production (Miracle et al. 2006; Dorts et al. 2009; Hemmer et al. 2011), and increases in the expression of mRNA for AR isoforms in fin tissues (Sone et al. 2005) in female fish. In addition to the physiological disruptions reported in these studies, there can also be higher-level ecological impacts, such as skewed sex ratios biased toward males. In a study by Bogers et al. (2006), fathead minnow (Pimephales promelas) (FHM) embryos exposed to nominal concentrations of 0.10, 0.32, and 1.0 μg methyldihydrotestosterone (MDHT)/L showed increases in nuptial tubercles as well as skewed sex ratios toward males. Exposure to 0.32 μg MDHT/L resulted in 80% males and 20% mixed gonads after 114 dpf. There can be significant effects of androgens on reproductive fitness (via impaired steroidogenesis) and population viability via unbalanced sex ratios. While studies have investigated population changes in response to estrogens (Kidd et al. 2007), the long-term effects of xeno-androgens on population structure have yet to be investigated in the natural environment.
In ecotoxicology, reproductive tests using small fish have been developed to detect androgenic effects of aquatic pollutants. Biological endpoints such as secondary sex characteristics and biomarkers for exposure to androgen have been adopted into the protocols of the Organization for Economic Co-operation and Development (OECD) for environmental testing for uncharacterized chemicals. Perhaps the bio-molecule best documented as an indicator of androgenic exposure is spiggin, a kidney protein that is naturally produced by male three-spined stickleback (Gasterosteus aculeatus) during nest-building during reproduction. There are now optimized androgenic/anti-androgenic bioassays for screening that are designed to assess the responses of spiggin to the test chemical (Jolly et al. 2006, 2009). These bioassays include both cell-based approaches and in vivo experiments to measure spiggin protein using polyclonal antibodies for the protein. In the FHM, the presence of nuptial tubercles (a male secondary sex character) in females following waterborne exposures to aquatic pollutants is also widely used as a bioassay for androgenic mode of action (MOA). Similarly, the gonadopodium of mosquitofish also continues to be used as a morphological bioassay for androgenic activity of polluted water, for example sewage effluent (Hou et al. 2011). Despite the continued refinement and application of bioassays for androgens in fish, molecular biomarkers at the gene and protein level (with the exception of spiggin) remain somewhat elusive in ecotoxicology. Suppression of bio-molecules typically used as indicators of exposure to xenoestrogen (VTG, zona radiata proteins) is used as a surrogate to characterize the effects of chemicals with a suspected androgenic MOA (Hemmer et al. 2008).
Omics approaches are able to address this gap in identifying potential androgen biomarkers and characterizing AR-regulated pathways in fish. Omics methods, for example transcriptomics and proteomics, can assess biological responses to exposures based upon global signature patterns of response. In this mini-review, we highlight some of the studies using omics approaches in fish to identify androgenic pathways. Anti-androgens are also important in defining androgenic pathways. There are a number of model anti-androgens that have been used to investigate AR disruption in fish at the molecular level, including flutamide and vinclozolin (Garcia-Reyero et al. 2009; Martyniuk et al. 2009; Martinović-Weigelt et al. 2011). Anti-androgenic responses of genes and proteins are compared to those evoked by androgens, and reciprocal regulation of bio-molecules is thought to be indicative of an AR-mediated effect. However, data arising from model anti-androgens are not covered here. There has been extensive transcriptomics investigation of model anti-androgens such as flutamide in the FHM (Perkins et al. 2011). Flutamide may have other modes of action in fish that may not be directly associated with AR signaling (Garcia-Reyero et al. 2009); therefore, we focus here only on those data collected with model androgens.
Transcriptomic approaches in the investigation of androgens
Microarray analyses have provided unique insight into androgen-regulated pathways in fish. The ovaries and livers of females have been the primary tissues investigated for androgenic responses at the levels of genes and proteins (Table 1). The ovary is a major producer of androgen in both female and male fish and the liver is a reproductive tissue that synthesizes VTG; thus, the importance of these tissues and how they are impacted by androgens is the focus of the majority of studies. Microarray analysis following exposures to androgens have been conducted in small-bodied and large-bodied fish species including FHM, Atlantic cod (Gadus morhua), common carp (Cyprinus carpio), Qurt medaka (Oryzias latipes), and zebrafish (Danio rerio). These studies have investigated endogenously produced androgens such as T and 11-ketotestosterone (11-KT) as well as pharmaceutical androgens (methyl-testosterone [MT] and TB). The concentrations of androgens used in the transcriptomics studies are within physiological and ecological relevance; for example T and 11-KT can fall within the range of 1–500 ng/mL blood depending upon the species and whether the fish is reproductive or not (Páll et al. 2002; Onuma et al. 2003); runoff from cattle feedlots can also be in the ng/L range for pharmaceuticals (Bartelt-Hunt et al. 2012).
Table 1.
Recent studies using transcriptomics and proteomics methods to study the action of androgen in teleost fishes
| Species | Sex and stage | Tissue | Androgenic treatment | Concentration | Duration (days) | Biological material | Gene ontology (prominent examples) | References |
|---|---|---|---|---|---|---|---|---|
| Atlantic cod (Gadus morhua) | Adult female | Oocytes (pre-vtg) | T and 11-KT | 10 and 100 μM | 5 and 10 | Transcripts | Stress (HSPs), reproduction (steroid metabolism), cell cycle (cyclin) | Kortner et al. (2008) |
| Common carp (Cyprinus carpio) | Juveniles | Liver | 17α-MT, 11-KT | 0.1, 1.0, 10 μg MT/L and 5, 50, 500 ng 11-KT/L and 5 μg MT or 11-KT/g i.p. | 1 and 4 | Transcripts | Immune response, lipid transport, oogenesis, protein metabolism, proteolysis | Moens et al. (2007) |
| Fathead minnow (Pimephales promelas) | Adult female | Liver | TB | 5.0 μg TB/L | 2 | Proteins | Mutagenesis, apoptosis, cell cycle regulation, catabolism | Martyniuk et al. (2009) |
| Fathead minnow (Pimephales promelas) | Adult female | Liver | TB | 5.0 μg TB/L | 2 | Proteins | Xenobiotic clearance, LDL oxidation, smooth muscle cell proliferation (Table 2) | Martyniuk and Denslow (this review) |
| Fathead minnow (Pimephales promelas) | Adult female | Ovary | TB | 0.5 μg TB/L | 2 | Transcripts | Secretion, embryonic development, immune response, development, regulation of apoptosis, system development, proteolysis, muscle development | Garcia-Reyero et al. (2009)a |
| Fathead minnow (Pimephales promelas) | Adult female | Ovary | TB | 1.0 μg TB/L | 4 | Transcripts | Protein ubiquitination, protein biosynthesis, lipid metabolism, transcription, DNA repair, and DNA replication | Dorts et al. (2009)b |
| Qurt medaka (Oryzias latipes) | Larvae (both) | Whole tissue | 11-KT | 100 μg 11-KT/L | 7 | Transcripts | Metabolism, stress responses, iron storage, mitotic regulation, neural networks, reproductive physiology, sex differentiation and growth | Leon et al. (2008) |
| Rainbow trout (Oncorhynchus mykiss) | Adult female | Liver | TB | 1.0 μg TB/L | 7 | Transcripts | Protein binding, transport, ATP regulation, lipid binding, copper and calcium ion binding, ribosomes, immune, cell cycle, | Hook et al. (2006)c |
| Zebrafish (Danio rerio) | Adult female | Liver | MDHT | 0.1, 0.7, 4.9 μg MDHT/L | 1 and 7 | Transcripts | Steroid metabolism, hormone transport, regulation of cell growth and proliferation polyamine biosynthesis | Hoffmann et al. (2008) |
The table includes categories of gene ontology that were prevalent in each study (i.e., major themes reported).
aBiological processes shown in this table are those from Garcia-Reyero et al. (2009) that were represented by >100 genes on the FHM microarray (P < 0.03).
bBiological processes shown in this table are those from Dorts et al. (2009) that had >10 genes in the gene ontology category.
cMajor biological themes identified by Hook et al. (2006) that contained >2% genes differentially expressed compared to all regulated genes.
i.p., intraperitoneal injection.
Upon surveying these microarray and proteomics studies, common molecular themes emerge during the functional enrichment of gene ontology and during analyses of pathways (Table 1). Many of the studies reported that lipids are impacted by androgens (metabolism, transport, LDL oxidation) as well as by processes related to reproduction (steroidogenesis, hormone transport, steroid metabolism, and sex differentiation). Multiple studies also identified the immune response, protein metabolism, proteolysis, proliferation of muscle, and regulation of the cell cycle (including DNA replication) as significant biological processes affected by exposure to androgens.
There is good evidence that many of these biological processes are AR regulated in both fish and mammalian tissues, for example muscular development, lipid metabolism, and immune responses. However, there are additional considerations that are required to determine whether or not molecular responses in fish are directly related to an androgenic MOA. For example, stress-related responses are associated with different types of androgens, and molecular data suggests that there are changes in biological molecules involved in DNA repair, mutagenesis, apoptosis, xenobiotic clearance, protein ubiquitination, and heat-shock proteins (Table 1). In a recent study by Karim et al. (2011), a protein interactome for general environmental stress using data collected from fish and mammalian models revealed that energy metabolism pathways, redox reaction, mitochondrial electronic transport chain, fatty acid biosynthesis, and pathways such as interleukin signaling, delta-notch signaling, and MAPK signaling are associated with general and oxidative stress. Molecular responses that are a result of general toxicity or a result of a chemical’s MOA are difficult to tease apart, especially in the liver where one of the major functions is the detoxification of xenobiotics. Additional comparisons to other chemical stressors that do not have a defined androgenic MOA will better resolve the molecular response to xenobiotics in general compared to androgens.
A second consideration arises from the fact that androgens can be metabolized into estrogenic metabolites, including aromatization into E2. In addition, some androgens may also weakly bind to estrogen receptors, activating E2-regualted pathways (Hornung et al. 2004; Frye et al. 2008). The studies surveyed here used both nonaromatizable (TB and MDHT) and aromatizable (T) androgens and this can further complicate the interpretation of responses of genes and proteins. In male FHMs exposed to TB, there was a significant increase in plasma E2 at 50 µg TB/L and, although not significant, a variable induction of VTG (Ankley et al. 2003). An increase in VTG is an E2-mediated response and this suggests that TB may be metabolized into other estrogenic metabolites in fish. It has not been directly demonstrated in fish to the best of our knowledge, but TB may be aromatized into an estrogenic metabolite in very small quantities. This has been demonstrated in mammals, although that amount of aromatization is relatively low (Pottier et al. 1981). A second hypothesis is that the reduction of plasma E2 at lower concentrations of TB (Ankley et al. 2003) may reflect an inhibitory feedback loop involving the hypothalamic-pituitary gonadal axis, and TB is suppressing E2 levels. Perhaps in support of the hypothesis that estrogenic metabolites are affecting the transcriptome after androgen treatments, biological pathways such as immune pathways, lipid biosynthesis and metabolism pathways, DNA replication, DNA repair, and DNA metabolism (Table 1) are processes also affected by potent estrogens (e.g., 17α-ethinylestradiol) (Martyniuk et al. 2007; Garcia-Reyero et al. 2009). However, these biological processes can also be altered indirectly, due to suppression of circulating plasma E2 as a result of androgen treatments. Thus, the temporal relationship between aromatization of androgens into estrogenic metabolites and the suppression of E2 by androgens will greatly influence the estrogen/androgen ratio, resulting in transcriptional and translational responses that are not fully understood. Carefully designed experiments comparing nonaromatizable and aromatizable androgens to estrogens may help to identify transcriptomics responses that are due to direct AR binding compared to those molecular responses due to ER binding.
Proteomics and metabolomics approaches to the investigation of environmental androgens
Environmental proteomics is advancing rapidly in studies of fish ecotoxicology and there are new data being generated on chemical MOA using quantitative proteomic methods such as 2D gel electrophoresis. There are some recent reviews on the topic of proteomics in ecotoxicology (Martyniuk and Denslow 2009; Forné et al. 2010; Sanchez et al. 2011; Martyniuk et al. 2012) and this topic will not be covered here. In addition to gel-based methods, environmental proteomics has successfully implemented high throughput, non-gel-based technologies, such as isobaric tagging for relative and absolute quantitation (iTRAQ), to study the responses by fish to aquatic pollutants, including AR agonists/antagonists (Martyniuk et al. 2009). Quantitative proteomics using high-resolution mass spectrometers such as the Velos Orbitrap can generate significant amounts of information about protein changes in fish tissues. The advantages of non-gel-based over gel-based methods include increased identification and quantitation of proteins, but the cost for non-gel-based approaches with high resolution mass spectrometers can be relatively high and this approach has yet to be widely adopted in ecotoxicology.
Although not specifically focused on environmental applications, studies in fish have used gel-based proteomics methods to characterize changes in the proteome in the testes of fish, providing insight into reproductive processes that underlie maturation of sperm. This also provides a reproductive baseline for studying perturbation in the endocrine system in fish. Differential proteins, expressed in the testis of Senegalese sole (Solea senegalensis), that are correlated with poor fertilization and poor sperm production include alpha 2-macroglobulin (protease inhibitor), ferritin, cytoskeletal components such as keratin 8 (Krt8) and keratin S8 type I, glyceraldehyde 3-phosphate dehydrogenase, leukemia related protein 16 (Lrp16), chaperone gp96, and apolipoproteins A-IV 1 (Apoa4-1) (Forné et al. 2009). In general, proteins involved in cytoskeletal organization and catabolic processes, as well as in redox or antioxidant activity, were identified and it is hypothesized that these processes are important for sperm production. Physiological studies such as this one are useful for ecotoxicology and for understanding AR-mediated protein expression because plasma androgens increase during sexual development and protein changes that correspond to changes in sex steroid levels are good candidates for further exploration as bio-indicators of exposure to androgen.
Similar to proteomics, metabolomics is becoming increasingly used in ecotoxicology and has been used to study the pharmaceutical TB in FHMs. A significant advantage of using metabolomics is that it can be non-invasive, and biological material such as urine and blood can be rapidly assessed for metabolite content. Another advantage of metabolomics methods over transcriptomics and proteomics is that metabolites have known functions in the context of biochemical pathways. Using 1H NMR spectroscopy, Ekman et al. (2011) recently investigated the hepatic metabolome of FHMs after TB exposure and after 1 day, females exposed to 500 ng TB/L showed increases in the amino sulfonic acid taurine. The authors speculate this may be related to increased hepatoprotective capacity, consistent with data from genes and proteins that indicate that stress responses are activated in the livers of fish after exposure to androgens. FHM urine has also been profiled for metabolites that are associated with androgen and anti-androgens (Collette et al. 2010) and there is high potential for using fish urine as a screening tool for endocrine disruptors. However, compared to transcriptomics and metabolomics, proteomics still lacks information regarding androgen action in fish and future studies should fill in this gap if we are to utilize multiple bio-molecular endpoints to better associate early molecular changes to xeno-androgen exposure.
One challenge arising from the use of omics data is the integration of molecular data (expression and abundance of genes, proteins, metabolites) into regulatory networks that are informative and biologically meaningful. There are new algorithms to manage omics data that have greatly enabled network elucidation such as reverse engineering (Perkins et al. 2011). Protein networks can also be constructed using sub-network enrichment analysis (SNEA) that identifies key protein hubs or regulators for an enriched subset of proteins. This approach has recently been used to probe the effects of new potential hormones such as secretoneuron in the pituitary of goldfish (Trudeau et al. 2012). Therefore, we re-analyzed the protein data set from Martyniuk et al. (2009) using SNEA and constructed a putative androgen-regulated protein network in the liver. Martyniuk et al. (2009) treated female FHMs with 5 µg TB/L for 48 h and these animals showed depressed production of plasma VTG after treatment. Therefore, there was a physiological response (i.e., phenotypic anchor) that could be used as a correlate to protein responses in the liver. In order to accommodate the SNEA, three iTRAQ data sets were merged from Martyniuk et al. (2009) and TB-regulated proteins were annotated with mammalian homologs using NCBI. Proteins were imported into Pathway Studio 9.0 (Ariadne, Rockville, MD, USA). Mammalian ResNet 9.0 (∼900,000 articles) was used for extracting relationships among entities. All annotated proteins for the TB treatment are provided in Supplementary Appendix 1. Based on protein name and alias, there were 152 proteins that mapped to known homologs and 12 that did not successfully map. The objective of SNEA is to create a central seed from all functional entities in the database in order to find common protein effectors. The data from proteins were used to perform SNEA for expression targets, binding partners, and post-translation modification targets of proteins. In addition, the database was queried to determine whether or not the regulated proteins were preferentially involved in regulating a cell process or were associated with a disease.
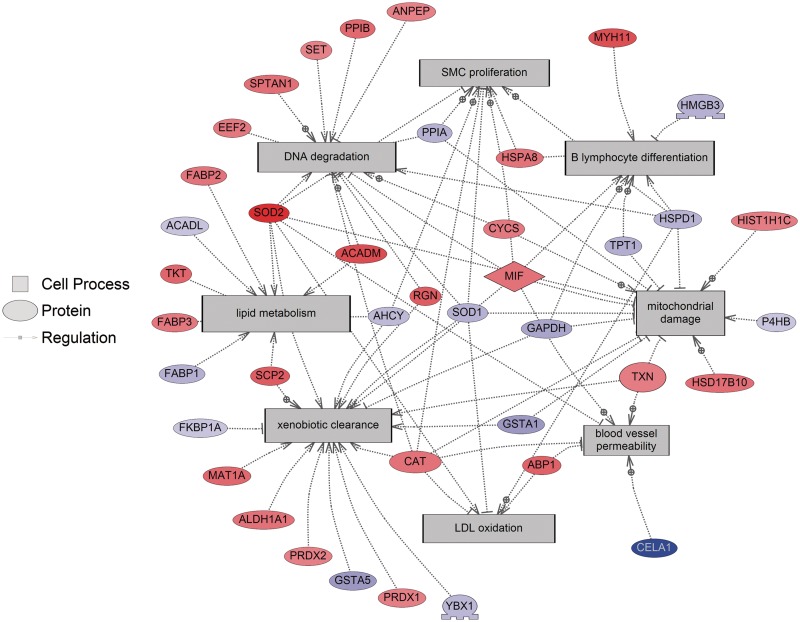
Significant cell processes that were affected by TB-regulated proteins included lymphocyte differentiation (decreasing 61%), xenobiotic clearance (increasing 12%), LDL oxidation (increasing 34%), proliferation of smooth muscle cells (increasing 33%), permeability of blood vessels (increasing 35%), and DNA degradation (increasing 34%) while disease states that involved TB-regulated proteins included hypersensitivity (increasing 34%) and genotoxicity (decreasing 67%) (Table 2). Many of the cell processes identified by SNEA have also been recently identified in a protein stress interactome for fish and mammals including xenobiotic clearance, metabolism, and effects on fatty acid biosynthesis and lipid oxidation (Karim et al. 2011). This suggests that in the FHM liver, there is a generalized proteomic response to a toxic chemical. A protein network was built using all significant sub-networks of cellular function (Figure 1). The enrichment P-value for protein seeds was set at P < 0.05 and eight protein sub-networks were included in the protein network.
Table 2.
Sub-network enrichment analysis for TB-regulated proteins in relation to cell processes, regulation of disease, expression targets, and protein-binding partners
| Relationship | Name | No. of measured neighbors | Median change | P-value |
|---|---|---|---|---|
| Proteins/chemicals-regulating cell processes | B lymphocyte differentiation | 7 | −1.61 | 0.007 |
| Xenobiotic clearance | 17 | 1.12 | 0.012 | |
| LDL oxidation | 5 | 1.34 | 0.015 | |
| Smooth muscle cell proliferation | 8 | 1.33 | 0.019 | |
| Lipid metabolism | 9 | 1.33 | 0.022 | |
| Mitochondrial damage | 14 | 1.13 | 0.029 | |
| Blood vessel permeability | 6 | 1.35 | 0.038 | |
| DNA degradation | 14 | 1.34 | 0.041 | |
| Proteins/chemicals-regulating diseases | Hypersensitivity | 5 | 1.34 | 0.018 |
| Emphysema | 5 | 1.13 | 0.022 | |
| Genotoxicity | 5 | −1.67 | 0.024 | |
| Body weight | 5 | −1.67 | 0.029 | |
| Expression targets | RAC-alpha serine/threonine–protein kinase | 6 | −1.62 | 0.024 |
| Thioredoxin | 5 | 1.35 | 0.036 | |
| Binding partner | Heat shock protein A8 | 5 | −1.39 | 0.041 |
Fig. 1.
Protein network constructed using sub-networks that are altered by waterborne exposure to 17β-trenbolone in livers of the fathead minnow liver. Abbreviations are as follows: ABP1, amiloride binding protein 1 (amine oxidase [copper-containing]); ACADL, acyl-CoA dehydrogenase, long chain; ACADM, acyl-CoA dehydrogenase, C-4 to C-12 straight chain; AHCY, adenosylhomocysteinase; ALDH1A1, aldehyde dehydrogenase 1 family, member A1; ANPEP, alanyl (membrane) aminopeptidase; CAT, catalase; CELA1, chymotrypsin-like elastase family, member 1; CYCS, cytochrome c, somatic; EEF2, eukaryotic translation elongation factor 2; FABP1, fatty acid binding protein 1, liver; FABP2, fatty acid binding protein 2, intestinal; FABP3, fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor); FKBP1A, FK506 binding protein 1A, 12kDa; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GSTA1, glutathione S-transferase alpha 1; GSTA5, glutathione S-transferase alpha 5; HIST1H1C, histone cluster 1, H1c; HMGB3, high mobility group box 3; HSD17B10, hydroxysteroid (17-beta) dehydrogenase 10; HSPA8, heat shock 70kDa protein 8; HSPD1, heat shock 60kDa protein 1 (chaperonin); MAT1A, methionine adenosyltransferase I, alpha; MIF, macrophage migration inhibitory factor (glycosylation-inhibiting factor); MYH11, myosin, heavy chain 11, smooth muscle; P4HB, prolyl 4-hydroxylase, beta polypeptide; PPIA, peptidylprolyl isomerase A (cyclophilin A); PPIB, peptidylprolyl isomerase B (cyclophilin B); PRDX1, peroxiredoxin 1; PRDX2, peroxiredoxin 2; RGN, regucalcin (senescence marker protein-30); SCP2, sterol carrier protein 2; SET, SET nuclear oncogene; SOD1, superoxide dismutase 1, soluble; SOD2, superoxide dismutase 2, mitochondrial; SPTAN1, spectrin, alpha, non-erythrocytic 1 (alpha-fodrin); TKT, transketolase; TPT1, tumor protein, translationally-controlled 1; TXN, thioredoxin; YBX1, Y box binding protein 1.
The FHM TB-regulated protein network and the cellular functions mapped onto the network are consistent with transcriptomics data. Microarray studies in fish have also found effects on the immune system (Hook et al. 2006; Moens et al. 2007), lipid metabolism (Dorts et al. 2009), and muscular development (Garcia-Reyero et al. 2009) at the transcriptomics level with androgen treatments in ovary and liver. However, as pointed out previously, the direct mechanism of TB action is not entirely understood. Other proteins that are affected by androgens include those that are related to expression targets and binding partners of thioredoxin and heat-shock protein A8, respectively, as well as those proteins involved in xenobiotic clearance. We demonstrate that construction of protein networks can offer novel insights into cell processes regulated by androgens and this approach can generate new hypotheses about potential biomarkers of androgenic activity.
Conclusions
There are now experimental data collected from all three of the major omics methods used in ecotoxicology; transcriptomics (León et al. 2008; Dorts et al. 2009; Garcia-Reyero 2009), proteomics (Martyniuk et al. 2009), and metabolomics (Collette et al. 2010; Ekman et al. 2011) after androgenic exposures in fish. These studies include both endogenous hormones (11-KT) and pharmaceuticals that are androgen-receptor agonists (e.g., 17β-trenbolone). These studies have contributed significantly to our understanding of AR-mediated pathways in teleost tissues and have identified some of the most commonly observed responses to xeno-androgens, although in many cases the direct mechanism of action for androgens remains elusive.
With increased characterization of pathways using transcriptomics, proteomics, and metabolomics, novel bioassays will continue to be developed for the chemical screening of androgens. There are already well-utilized bioassays in fish that include the androgen-sensitive MDA-kb2 cell line (Blake et al. 2010) and androgen-sensitive bioassays based on cultures of primary cells and tissue slices from the three-spined stickleback (G. aculeatus) (Björkblom et al. 2007). Programs such as the European Union Registration, Evaluation and Authorisation of Chemicals (REACH) have focused on the development of rapid, toxicological screening assays and omics technologies, while currently cost-prohibitive for high throughput screening, can provide a significant amount of information on chemical MOA.
Funding
This work was supported by a Canada Research Chair and Natural Sciences and Engineering Research Council Discovery Grant (C.J.M.) and a grant from the Superfund Basic Research Program from the National Institute of Environmental Health Sciences (R01 ES015449) (N.D.D.).
Supplementary Data
Supplementary Data are available at ICB online.
Acknowledgments
The authors would like to thank Dr L. Tomanek for organizing the Environmental Proteomics Symposium at the 2011 Society for Integrative and Comparative Biology conference.
References
- Ankley GT, Defoe DL, Kahl MD, Jensen KM, Makynen EA, Miracle A, Hartig P, Gray LE, Cardon M, Wilson V. Evaluation of the model anti-androgen flutamide for assessing the mechanistic basis of responses to an androgen in the fathead minnow (Pimephales promelas) Environ Sci Technol. 2004;38:6322–7. doi: 10.1021/es040022b. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Jensen KM, Makynen EA, Kahl MD, Korte JJ, Hornung MW, Henry TR, Denny JS, Leino RL, Wilson VS, et al. Effects of the androgenic growth promoter 17β-trenbolone on fecundity and reproductive endocrinology of the fathead minnow. Environ Toxicol Chem. 2003;22:1350–60. [PubMed] [Google Scholar]
- Bartelt-Hunt SL, Snow DD, Kranz WL, Mader TL, Shapiro CA, Donk SJ, Shelton DP, Tarkalson DD, Zhang TC. Effect of growth promotants on the occurrence of endogenous and synthetic steroid hormones on feedlot soils and in runoff from beef cattle feeding operations. Environ Sci Technol. 2012;46:1352–60. doi: 10.1021/es202680q. [DOI] [PubMed] [Google Scholar]
- Björkblom C, Olsson PE, Katsiadaki I, Wiklund T. Estrogen- and androgen-sensitive bioassays based on primary cell and tissue slice cultures from three-spined stickleback (Gasterosteus aculeatus) Comp Biochem Physiol C Toxicol Pharmacol. 2007;146:431–42. doi: 10.1016/j.cbpc.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Blake LS, Martinović D, Gray LE, Jr, Wilson VS, Regal RR, Villeneuve DL, Ankley GT. Characterization of the androgen-sensitive MDA-kb2 cell line for assessing complex environmental mixtures. Environ Toxicol Chem. 2010;29:1367–76. doi: 10.1002/etc.166. [DOI] [PubMed] [Google Scholar]
- Bogers R, De Vries-Buitenweg S, Van Gils M, Baltussen E, Hargreaves A, van de Waart B, De Roode D, Legler J, Murk A. Development of chronic tests for endocrine active chemicals. Part 2: an extended fish early-life stage test with an androgenic chemical in the fathead minnow (Pimephales promelas) Aquat Toxicol. 2006;80:119–30. doi: 10.1016/j.aquatox.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Cody RP, Bortone SA. Masculinization of mosquitofish as an indicator of exposure to kraft mill effluent. Bull Environ Contam Toxicol. 1997;58:429–36. doi: 10.1007/s001289900352. [DOI] [PubMed] [Google Scholar]
- Collette TW, Teng Q, Jensen KM, Kahl MD, Makynen EA, Durhan EJ, Villeneuve DL, Martinović-Weigelt D, Ankley GT, Ekman DR. Impacts of an anti-androgen and an androgen/anti-androgen mixture on the metabolite profile of male fathead minnow urine. Environ Sci Technol. 2010;44:6881–6. doi: 10.1021/es1011884. [DOI] [PubMed] [Google Scholar]
- Davis WP, Bortone SA. Effects of kraft mill effluent on the sexuality of fishes: an environmental early warning? In: Colborn T, Clement C, editors. Chemically-induced alterations in sexual and functional development: the wildlife/human connection. Vol. 435. Princeton, NJ: Princeton Scientific Publishing Company; 1992. pp. 113–27. [Google Scholar]
- Dorts J, Richter CA, Wright-Osment MK, Ellersieck MR, Carter BJ, Tillitt DE. The genomic transcriptional response of female fathead minnows (Pimephales promelas) to an acute exposure to the androgen, 17beta-trenbolone. Aquat Toxicol. 2009;91:44–53. doi: 10.1016/j.aquatox.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durhan EJ, Lambright C, Wilson V, Butterworth BC, Kuehl DW, Orlando EF, Guillette LJ, Jr, Gray LE, Ankley GT. Evaluation of androstenedione as an androgenic component of river water downstream of a pulp and paper mill effluent. Environ Toxicol Chem. 2002;21:1973–6. [PubMed] [Google Scholar]
- Ekman DR, Villeneuve DL, Teng Q, Ralston-Hooper KJ, Martinović-Weigelt D, Kahl MD, Jensen KM, Durhan EJ, Makynen EA, Ankley GT, et al. Use of gene expression, biochemical and metabolite profiles to enhance exposure and effects assessment of the model androgen 17β-trenbolone in fish. Environ Toxicol Chem. 2011;30:319–29. doi: 10.1002/etc.406. [DOI] [PubMed] [Google Scholar]
- Forné I, Abián J, Cerdà J. Fish proteome analysis: model organisms and non-sequenced species. Proteomics. 2010;10:858–72. doi: 10.1002/pmic.200900609. [DOI] [PubMed] [Google Scholar]
- Forné I, Agulleiro MJ, Asensio E, Abián J, Cerdà J. 2-D DIGE analysis of Senegalese sole (Solea senegalensis) testis proteome in wild-caught and hormone-treated F1 fish. Proteomics. 2009;9:2171–81. doi: 10.1002/pmic.200800696. [DOI] [PubMed] [Google Scholar]
- Frye CA, Koonce CJ, Edinger KL, Osborne DM, Walf AA. Androgens with activity at estrogen receptor beta have anxiolytic and cognitive-enhancing effects in male rats and mice. Horm Behav. 2008;54:726–34. doi: 10.1016/j.yhbeh.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyero N, Kroll KJ, Liu L, Orlando EF, Watanabe KH, Sepúlveda MS, Villeneuve DL, Perkins EJ, Ankley GT, Denslow ND. Gene expression responses in male fathead minnows exposed to binary mixtures of an estrogen and antiestrogen. BMC Genomics. 2009;10:308. doi: 10.1186/1471-2164-10-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyero N, Villeneuve DL, Kroll KJ, Liu L, Orlando EF, Watanabe KH, Sepúlveda MS, Ankley GT, Denslow ND. Expression signatures for a model androgen and antiandrogen in the fathead minnow (Pimephales promelas) ovary. Environ Sci Technol. 2009;43:2614–9. doi: 10.1021/es8024484. [DOI] [PubMed] [Google Scholar]
- Hemmer MJ, Cripe GM, Hemmer BL, Goodman LR, Salinas KA, Fournie JW, Walker CC. Comparison of estrogen-responsive plasma protein biomarkers and reproductive endpoints in sheepshead minnows exposed to 17beta-trenbolone. Aquat Toxicol. 2008;88:128–36. doi: 10.1016/j.aquatox.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Hemmer MJ, Salinas KA, Harris PS. Application of protein expression profiling to screen chemicals for androgenic activity. Aquat Toxicol. 2011;103:71–8. doi: 10.1016/j.aquatox.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Hoffmann JL, Thomason RG, Lee DM, Brill JL, Price BB, Carr GJ, Versteeg DJ. Hepatic gene expression profiling using GeneChips in zebrafish exposed to 17alpha-methyldihydrotestosterone. Aquat Toxicol. 2008;87:69–80. doi: 10.1016/j.aquatox.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Hook SE, Skillman AD, Small JA, Schultz IR. Gene expression patterns in rainbow trout, Oncorhynchus mykiss, exposed to a suite of model toxicants. Aquat Toxicol. 2006;77:372–85. doi: 10.1016/j.aquatox.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung MW, Jensen KM, Korte JJ, Kahl MD, Durhan EJ, Denny JS, Henry TR, Ankley GT. Mechanistic basis for estrogeniceffects in fathead minnow (Pimephales promelas) following exposureto the androgen 17a-methyltestosterone: conversion of 17a-methyltestosterone to 17a-methylestradiol. Aquat Toxicol. 2004;66:15–23. doi: 10.1016/j.aquatox.2003.06.004. [DOI] [PubMed] [Google Scholar]
- Hou L, Xie Y, Ying G, Fang Z. Developmental and reproductive characteristics of western mosquitofish (Gambusia affinis) exposed to paper mill effluent in the Dengcun River, Sihui, South China. Aquat Toxicol. 2011;103:140–9. doi: 10.1016/j.aquatox.2011.02.018. [DOI] [PubMed] [Google Scholar]
- Howell WM, Black DA, Bortone SA. Abnormal expression of secondary sex characters in a population of mosquitofish, Gambusia affinis holbrooki: evidence for environmentally induced masculinization. Copeia. 1980;1980:676–81. [Google Scholar]
- Jensen KM, Makynen EA, Kahl MD, Ankley GT. Effects of the feedlot contaminant 17alpha-trenbolone on reproductive endocrinology of the fathead minnow. Environ Sci Technol. 2006;40:3112–7. doi: 10.1021/es052174s. [DOI] [PubMed] [Google Scholar]
- Jolly C, Katsiadaki I, Le Belle N, Mayer I, Dufour S. Development of a stickleback kidney cell culture assay for the screening of androgenic and anti-androgenic endocrine disrupters. Aquat Toxicol. 2006;79:158–66. doi: 10.1016/j.aquatox.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Jolly C, Katsiadaki I, Morris S, Le Belle N, Dufour S, Mayer I, Pottinger TG, Scott AP. Detection of the anti-androgenic effect of endocrine disrupting environmental contaminants using in vivo and in vitro assays in the three-spined stickleback. Aquat Toxicol. 2009;92:228–39. doi: 10.1016/j.aquatox.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Karim M, Puiseux-Dao S, Edery M. Toxins and stress in fish: proteomic analyses and response network. Toxicon. 2011;57:959–69. doi: 10.1016/j.toxicon.2011.03.018. [DOI] [PubMed] [Google Scholar]
- Kidd KA, Blanchfield PJ, Mills KH, Palace VP, Evans RE, Lazorchak JM, Flick RW. Collapse of a fish population after exposure to a synthetic estrogen. Proc Natl Acad Sci USA. 2007;104:8897–901. doi: 10.1073/pnas.0609568104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolok AS, Sellin MK. The environmental impact of growth-promoting compounds employed by the United States beef cattle industry: history, current knowledge, and future directions. Rev Environ Contam Toxicol. 2008;195:1–30. doi: 10.1007/978-0-387-77030-7_1. [DOI] [PubMed] [Google Scholar]
- Kortner TM, Rocha E, Silva P, Castro LF, Arukwe A. Genomic approach in evaluating the role of androgens on the growth of Atlantic cod (Gadus morhua) previtellogenic oocytes. Comp Biochem Physiol Part D Genomics Proteomics. 2008;3:205–18. doi: 10.1016/j.cbd.2008.04.001. [DOI] [PubMed] [Google Scholar]
- León A, Wu PS, Hall LC, Johnson ML, Teh SJ. Global gene expression profiling of androgen disruption in Qurt strain medaka. Environ Sci Technol. 2008;42:962–9. doi: 10.1021/es071785c. [DOI] [PubMed] [Google Scholar]
- Martinović D, Blake LS, Durhan EJ, Greene KJ, Kahl MD, Jensen KM, Makynen EA, Villeneuve DL, Ankley GT. Reproductive toxicity of vinclozolin in the fathead minnow: confirming an anti-androgenic mode of action. Environ Toxicol Chem. 2008;27:478–88. doi: 10.1897/07-206R.1. [DOI] [PubMed] [Google Scholar]
- Martinović-Weigelt D, Wang RL, Villeneuve DL, Bencic DC, Lazorchak J, Ankley GT. Gene expression profiling of the androgen receptor antagonists flutamide and vinclozolin in zebrafish (Danio rerio) gonads. Aquat Toxicol. 2011;101:447–58. doi: 10.1016/j.aquatox.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Alvarez S, Denslow ND. DIGE and iTRAQ as biomarker discovery tools in aquatic toxicology. Ecotoxicol Environ Saf. 2012;76:3–10. doi: 10.1016/j.ecoenv.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyniuk CJ, Alvarez S, McClung S, Villeneuve DL, Ankley GT, Denslow ND. Quantitative proteomic profiles of androgen receptor signaling in the liver of fathead minnows (Pimephales promelas) J Proteome Res. 2009;8:2186–200. doi: 10.1021/pr800627n. [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Denslow ND. Towards functional genomics in fish using quantitative proteomics. Gen Comp Endocrinol. 2009;164:135–41. doi: 10.1016/j.ygcen.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Gerrie ER, Popesku JT, Ekker M, Trudeau VL. Microarray analysis in the zebrafish (Danio rerio) liver and telencephalon after exposure to low concentration of 17alpha-ethinylestradiol. Aquat Toxicol. 2007;84:38–49. doi: 10.1016/j.aquatox.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Miracle A, Ankley G, Lattier D. Expression of two vitellogenin genes (vg1 and vg3) in fathead minnow (Pimephales promelas) liver in response to exposure to steroidal estrogens and androgens. Ecotoxicol Environ Saf. 2006;63:337–42. doi: 10.1016/j.ecoenv.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Moens LN, van der Ven K, Van Remortel P, Del-Favero J, De Coen WM. Gene expression analysis of estrogenic compounds in the liver of common carp (Cyprinus carpio) using a custom cDNA microarray. J Biochem Mol Toxicol. 2007;21:299–311. doi: 10.1002/jbt.20190. [DOI] [PubMed] [Google Scholar]
- Onuma T, Higashi Y, Ando H, Ban M, Ueda H, Urano A. Year-to-year differences in plasma levels of steroid hormones in pre-spawning chum salmon. Gen Comp Endocrinol. 2003;133:199–215. doi: 10.1016/s0016-6480(03)00171-0. [DOI] [PubMed] [Google Scholar]
- Páll MK, Mayer I, Borg B. Androgen and behavior in the male three-spined stickleback, Gasterosteus aculeatus I.–changes in 11-ketotestosterone levels during the nesting cycle. Horm Behav. 2002;41:377–83. doi: 10.1006/hbeh.2002.1777. [DOI] [PubMed] [Google Scholar]
- Parks LG, Lambright CS, Orlando EF, Guillette LJ, Jr, Ankley GT, Gray LE., Jr Masculinization of female mosquitofish in Kraft mill effluent-contaminated Fenholloway River water is associated with androgen receptor agonist activity. Toxicol Sci. 2001;62:257–67. doi: 10.1093/toxsci/62.2.257. [DOI] [PubMed] [Google Scholar]
- Perkins EJ, Chipman JK, Edwards S, Habib T, Falciani F, Taylor R, Van Aggelen G, Vulpe C, Antczak P, Loguinov A. Reverse engineering adverse outcome pathways. Environ Toxicol Chem. 2011;30:22–38. doi: 10.1002/etc.374. [DOI] [PubMed] [Google Scholar]
- Pottier J, Cousty C, Heitzman RJ, Reynolds IP. Differences in the biotransformation of a 17 beta-hydroxylated steroid trenbolone acetate, in rat and cow. Xenobiotica. 1981;11:489–500. doi: 10.3109/00498258109045859. [DOI] [PubMed] [Google Scholar]
- Sanchez BC, Ralston-Hooper K, Sepúlveda MS. Review of recent proteomic applications in aquatic toxicology. Environ Toxicol Chem. 2011;30:274–82. doi: 10.1002/etc.402. [DOI] [PubMed] [Google Scholar]
- Seki M, Fujishima S, Nozaka T, Maeda M, Kobayashi K. Comparison of response to 17beta-estradiol and 17 beta-trenbolone among three small fish species. Environ Toxicol Chem. 2006;25:2742–52. doi: 10.1897/05-647r.1. [DOI] [PubMed] [Google Scholar]
- Sone K, Hinago M, Itamoto M, Katsu Y, Watanabe H, Urushitani H, Tooi O, Guillette LJ, Jr, Iguchi T. Effects of an androgenic growth promoter 17beta-trenbolone on masculinization of Mosquitofish (Gambusia affinis affinis) Gen Comp Endocrinol. 2005;143:151–60. doi: 10.1016/j.ygcen.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Trudeau VL, Martyniuk CJ, Zhao E, Hu H, Volkoff H, Decatur WA, Basak A. Is secretoneurin a new hormone? Gen Comp Endocrinol. 2012;175:10–8. doi: 10.1016/j.ygcen.2011.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.