Abstract
The Atlantic bottlenose dolphin has attracted attention due to the evident impact that environmental stressors have taken on its health. In order to better understand the mechanisms linking environmental health with dolphin health, we have established cell cultures from dolphin skin as in vitro tools for molecular evaluations. The vitamin D3 pathway is one mechanism of interest because of its well established chemopreventative and immunomodulatory properties in terrestrial mammals. On the other hand, little is known of the physiological role of this molecule in aquatic animals. 1,25-dihydroxyvitamin D3 (1,25D3), the bioactive and hormonal form of vitamin D3, exerts its biological function by binding to the vitamin D receptor (VDR), a ligand-activated regulator of gene transcription. Therefore, we investigated the transcriptomic changes induced by 1,25D3 administration in dolphin skin cells. Identification of specific genes activated by 1,25D3 has provided clues to the physiological function of the vitamin D3 pathway in the dolphin. We found that exposure of the cells to 1,25D3 upregulated transactivation of a vitamin D-sensitive promoter. cDNA microarray analysis, using a novel dolphin array, identified specific gene targets within this pathway, and real-time PCR (qPCR) confirmed the enhanced expression of select genes of interest. These transcriptional changes correlated with an increase in VDR levels. This is the first report of the presence and activation of the vitamin D3 pathway in a marine mammal, and our experimental results demonstrate a number of similarities to terrestrial animals. Conservation of this pathway in the Atlantic bottlenose dolphin is consistent with the importance of nonclassic functions of vitamin D3, such as its role in innate immunity, similar to what has been demonstrated in other mammals.
Keywords: Vitamin D, Immunity, Keratinocyte, Gene expression, Transcription factors, Dolphin
1. Introduction
The health of the Atlantic bottlenose dolphin has come under scrutiny due to an increase in mortality and stranding events in correlation with observations of unusual pathologies [1–3]. Dolphin health assessments have become an indicator of detrimental changes occurring within the animal’s coastal habitat [4,5]. As a protected species, there is a need for more in vitro and molecular approaches to study the dolphin; consequently, we established cell strains and SV40-immortalized cell lines from the skin of the Atlantic bottlenose dolphin [6]. We are using these cellular models to investigate the vitamin D3 pathway and associated molecular mechanisms as biochemical platforms to correlate environmental stress with dolphin health.
Vitamin D was first identified for its classical role in calcium homeostasis, a primary function in terrestrial animals possessing calcified skeletons [7]. However, it is now well-acknowledged that the active form of vitamin D3, the secosteroid hormone 1,25D3, has additional functions of chemoprevention and immunomodulation. Chemoprevention refers to the hormone’s ability to inhibit proliferation, promote differentiation, repair DNA, and induce apoptosis in several normal and cancer cell types. 1,25D3’s title as an immunomodulator is merited by evidence of antimicrobial, antioxidant, anti-inflammatory, and wound healing capabilities [8–11]. The notion that the vitamin D pathway is functionally important beyond calcium metabolism and transport is supported by expression of VDR within several different organs and tissues [12–14] and within aquatic organisms lacking calcified skeletons, such as the sea lamprey [15] and sea squirts [16].
Vitamin D3 enters the systemic circulation either via diet or from the skin after UV-induced conversion from the sterol precursor, 7-dehydrocholesterol, which resides in the plasma membrane of skin cells [17–19]. Vitamin D3 travels to the liver where it is hydroxylated by the cytochrome p450 isoform A1 enzyme to generate 25-hydroxyvitamin D3 [20]. From here the molecule travels to the kidney where it serves as a substrate for a second cytochrome p450 enzyme, isoform B1 (CYP27B1), which hydroxylates it at position 1 to produce the biologically active and hormonal form: 1,25-dihydroxyvitamin D3 (1,25D3) [20]. Alternatively, this entire synthetic pathway for 1,25D3 can occur within the skin, which expresses all the necessary enzymes [21–24]. The skin is unique in that it may be the only organ in which the entire anabolic reaction from 7-DHC to 1,25D3 takes place [25].
1,25D3 can induce a biological response via interaction with VDR, an endocrine member of the steroid/thyroid hormone nuclear receptor superfamily that functions as a transcription factor for a large suite of genes. Upon ligand binding, VDR dimerizes with the retinoid X receptor (RXR), and this heterodimer then recognizes and binds to vitamin D response elements (VDREs) within the promoters of target genes to either activate or repress transcription. Genes identified as targets of VDR within the skin are typically associated with functions including anti-proliferation [26–28], pro-differentiation [26–28], immunomodulation [29,30], and antimicrobicity [29,31,32].
Our research effort has focused on the 1,25D3-induced transcriptional changes that occur in dolphin skin cells, using cDNA microarray analysis. The first dolphin microarray has been generated from peripheral blood leukocytes of wild bottlenose dolphins [33], providing us with the best tool available to date for transcriptomic analyses in dolphin skin. This is the first investigation on the effects of 1,25D3 in a marine mammal, and little is known about the physiological function of vitamin D in aquatic animals, in general. Transcriptomic patterns mediated by this hormone may aid in the understanding of such a function. The skin is an ideal model organ for study because it is both a source and target of endogenous 1,25D3. Within the skin, vitamin D plays a part in keratinocyte differentiation, antimicrobial peptide production, wound healing, inflammation, and hair cycling. Its link to a number of pathologies in the skin has made it a desirable therapeutic target [34]. Furthermore, the skin is directly impacted by environmental stressors, thus relying for protection on innate immune responses [35], possibly those mediated by the vitamin D3 pathway [11,36–40].
We report here the establishment of a dolphin in vitro system that has successfully allowed for the molecular evaluation of the vitamin D3 pathway in a novel species model. We found that dolphin skin cells express VDR, display sensitivity to exogenous administration of 1,25D3, and react by upregulating genes involved in proliferation, differentiation, apoptosis, wound healing, and stress responses, similarly to what has been reported in humans and other terrestrial mammals. These studies provide evidence for a nonclassical role of vitamin D3 in innate immunity, which may represent an essential mechanism in the skin for protecting the health of dolphins from environmental stressors.
2. Materials and methods
2.1. Dolphin skin cell culture
Cell strains and continuous cell lines from dolphin skin tissue have been established as previously described [6]. Skin biopsies were acquired from five wild Atlantic bottlenose dolphin individuals sampled during capture-release health assessment studies conducted in the Indian River Lagoon, FL and in Charleston Harbor, SC. Cells were cultured in Keratinocyte Serum-Free Medium (K-SFM; Gibco/Invitrogen), supplemented with 5% fetal bovine serum (FBS), 1X antibiotic-antimycotic solution (Gibco/Invitrogen), and 20 μg/ml ciprofloxacin (Bedford Laboratories). Immortalized cell lines were obtained via SV40 transformation [6]. Most experiments have been conducted in both the “cell strains” (non-transformed) and the “SV40 cell lines” (transformed). The 1,25-dihydroxyvitamin D3 (1,25D3) compound (Sigma–Aldrich, Inc.) was solubilized in ethanol and administered to the cells for the indicated dosages and times. Control cells were grown in the presence of the ethanol solvent using the same volume as 1,25D3 (0.1%, v/v). For treatment periods exceeding 48 h, culture media and 1,25D3/ethanol were replenished every 48 h from the time of initial treatment.
2.2. Western blot analysis
Cell lysates were obtained using RIPA buffer (Pierce/Thermo Fisher Scientific). 10 μg of protein was prepared with SDS loading buffer + 10% β-mercaptoethanol, boiled for 10 min, and resolved by SDS-PAGE (4–20% Tris–HCl, Pierce Ready Gels, Pierce/Thermo Fisher Scientific) at 100 V. Proteins were transferred to PVDF membranes (Millipore), and filters were blocked at room temperature for 1 h with 5% non-fat milk/0.1% Tween 20, immediately followed by overnight incubation at 4 °C with the primary antibody. Primary antibodies included anti-chicken VDR rat monoclonal, clone 9A7, IgG (Affinity BioReagents/Thermo Fisher Scientific), anti-human RXRα rabbit polyclonal IgG (Santa Cruz Biotechnology, Inc.), and anti-human β-actin mouse mono-clonal IgG (BioVision, Inc.). The next day blots were incubated with horse radish peroxidase (HRP)-conjugated IgG for 1 h at room temperature. Both primary and secondary antibodies were diluted in PBS, 1% milk, 0.2% Tween 20. After each antibody incubation, blots were washed three times in PBS, 0.2% Tween 20. Signal was detected by enhanced chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate, Pierce/Thermo Fisher Scientific) and exposure of the filters to X-ray film. Band intensities from Western blots were measured using Image J software (NIH).
2.3. MTT assay
Cells were plated in 96-well plates at 3 × 104 cells per well. Cells were treated with solvent (ethanol) or 1,25D3 (10−8 M). 5 h prior to the end of treatment, cells were incubated with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) at a 0.5 mg/ml final concentration for 5 h at 37 °C. Cells were solubilized in 200 μl DMSO, and viable cells were detected by measuring the absorbance at 570 nm with the FLUOStar Optima absorbance reader (BMG LABTECH). Absorbance was expressed as a fold change relative to the controls. A one-way ANOVA test was used to determine significant differences in absorbance between treatment groups of eight replicates each (p < 0.05).
2.4. Luciferase assay
Cells were transiently transfected with a promoter-driven reporter construct (a kind gift from John S. Adams, Cedars-Sinai Medical Center, Los Angeles, CA) containing 6 kb (from nucleotides −5500 to +455) of the promoter region of the human 25-hydroxyvitamin D3-24-hydroxylase (CYP24) gene cloned into a promoterless luciferase expression plasmid, pLUCpl, upstream of the luciferase translation start site [35,41,42]. The luciferase gene is under inducible control of the CYP24 promoter. Cells were plated in 24-well plates and transfected with 0.2 μg of the plasmid using Effectene Reagent (Qiagen) in medium (K-SFM) lacking FBS. 16 h post transfection, cells were replenished with medium containing FBS and treated with 1,25D3 or ethanol. After treatment, cells were lysed using Passive Lysis Buffer (Promega Co.) and cell lysates incubated with luciferin (Luciferase Assay System, Promega Co.) prior to measuring luciferase activity (FLUOStar Optima, BMG LABTECH). Bioluminescence was expressed as a fold change relative to control samples. Significant differences between treatment groups were determined with one-way ANOVA and student’s t-tests (p < 0.05)
2.5. RNA isolation
Total RNA was extracted from cell cultures with TRIzol reagent (Invitrogen). Chloroform was added (one-fifth volume) to isolate the RNA, which was collected in the upper, aqueous layer after centrifugation. RNA was precipitated using isopropanol followed by incubation at −20 °C, centrifugation, and washing of RNA pellets in 75% ethanol. RNA pellets were resuspended in DEPC-treated, nuclease-free water. RNA was quantified using a DU 800 UV/visible Spectrophotometer (Beckman Coulter).
2.6. cDNA microarray analysis
A novel dolphin microarray has been constructed comprising 3700 expressed sequence tags (ESTs) from peripheral blood lymphocytes (PBL) of wild bottlenose dolphins [33]. The ESTs were derived from libraries from LPS- and IL-2- exposed cells, and each EST was printed on the microarray in quadruplicate. In our experiments, total RNA was isolated from cells treated with ethanol or 1,25D3 (10−8 M) for 24 or 48 h. 1–2 μg of each RNA sample was used to generate aminoallyl modified (aa) RNA, using the Allyl MessageAmp II aRNA Amplification Kit (Ambion, Inc.) to allow for incorporation of a fluorescent Cy3 dye; this step was performed in triplicate for each sample treatment. Cy3-aaRNA was diluted 1:3 in hybridization buffer [50% formamide, 2.4% SDS, 4× SSPE, 2.5× Denhardt’s solution, and 1 μl of Mouse Hybloc DNA (Applied Genetics Laboratories, Inc.) blocking solution], boiled for one minute, and hybridized for 16 h at 50 °C in the dark in a hybridization oven. Prior to hybridization, the microarray slides were pre-hybridized with a pre-hybridization buffer (33.3% formamide, 1.6% SDS, 2.6× SSPE, 1.6× Denhardt’s solution and 0.1 μM salmon sperm DNA) in the dark for 1 h at 50 °C. The next day slides were washed to remove nonspecific binding. Slides were scanned with ScanArray Express and SpotArray software at 80 V PMT, and signal intensity was quantified using QuantArray Software. Data analysis was performed using a unique code developed specifically for the marinegenomics.org genomics project, as described elsewhere [33]. In brief, the data analysis entailed subtracting the background intensity from each spot for normalization; the resulting normalized intensity values were rank ordered and each value divided by the total number of valid measurements to give the within array quantile which make up a cumulative distribution plot. A value for the average differential expression (df) was derived by projecting the reference (ethanol-treated) and test (1,25D3-treated) values on the cumulative distribution plot. Values for df arbitrarily range from −1 to +1, with −1 representing those genes most downregulated and +1 those most upregulated by treatment in relation to control samples. A bottom limit was set whereby all hybridization raw intensity values less than 800 (on a scale from 1 to 65,000) were disregarded. The p-values, a measure of the consistency between triplicate arrays, were determined using the Wilcoxon signed-rank test, and only those genes with a p-value less than 0.05 were considered in further analyses.
2.7. qPCR
RNA samples (2 μg) were reverse transcribed using the Quantitect Reverse Transcription Kit (Qiagen), which incorporates a genomic DNA elimination step. Equal quantities of the resulting cDNA were subjected to real-time PCR using iQ SYBR Green master mix (Bio-Rad Laboratories). All reactions were performed in triplicate. Primers were designed with the PrimerPremier software (Bio-Rad Laboratories) against the dolphin ESTs used for the dolphin cDNA microarray that have been annotated and archived at marinegenomics.org. Primers for the vitamin D receptor (VDR) were designed against the limited cDNA sequence information we have obtained from the dolphin VDR transcript expressed in skin cells. The primers are shown in Table 1. The forward and reverse primers for each gene were pooled and diluted to a 0.3 μM final concentration. Amplification efficiency for each primer set was determined using a standard curve and use of the following equation: efficiency = [10(−1/slope)]−1, where slope was derived from the standard curve. Primer efficiencies, expressed as percentages, are displayed in Table 3. Amplification was performed with ~300 ng of cDNA template (normalized among samples) according to the following parameters: initial activation at 95 °C for 15 s, followed by 40 cycles at 95 °C for 15 s, 58 °C for 30 s, and 72 °C at 42 s, ending with a melt curve analysis at the end of the amplification. The MyiQ Single-Color Real-Time PCR Detection System, iCycler Thermal Cycler, and iQ5 Optical System Software (Bio-Rad Laboratories) were used for all PCR experiments. No template reactions and original RNA-template reactions were run as controls for primer contamination and genomic DNA contamination, respectively. Resulting threshold cycle (Ct) values were normalized to Ct values for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), the internal control. GAPDH was validated as a housekeeping gene using the BestKeeper VBA applet, version 1 [43], whereby GAPDH Ct values for control and 1,25D3-treated samples from numerous PCR reactions were compared with those of β-actin, the only other potential housekeeping gene available to us for the dolphin in this experimental system. GAPDH proved to be the more stable of the two genes for both cell strains and SV40 cell lines (cell strains: n = 57, SD = 1.37, R = 0.916, p = 0.001; SV40 cell lines: n = 56, SD = 1.30, R = 0.915, p = 0.001). Expression levels of GAPDH, using 2−ΔCt values, did not change significantly with 1,25D3 treatment (p-values > 0.05 for both cell types and both time points). Because most primer amplification efficiencies were near 100%, differential expression between treated and control samples was calculated via the 2−ΔΔCt method [44], where ΔΔCt is ΔCt1,25D3 − ΔCtEtOH, ΔCt is Cttest gene − CtGAPDH. This method does not take into account primer efficiency. Only in the case of the ARNT gene was primer efficiency significantly low (83%); thus, the Pfaffl method [45] was used to calculate differential expression for ARNT. This equation takes into account primer efficiencies of both the target and reference gene. One-way ANOVA and the Holm’s procedure for multiple testing procedures were used to determine significant differences between GAPDH-normalized Ct values of the control, 6 h, and 24 h treatment groups for each gene and each cell type (p < 0.05). Where there was significant differential expression, student’s t-tests assessed pairwise comparisons (p < 0.05).
Table 1.
Primer sets used for each gene are listed (5′–3′).
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| GAPDH | GGGAGTCCTTGCCCCAACT | GGATGGAAACCGCATGGA |
| CALM3 | TGATGGTAACGGCACAATTGA | TTCACTGTCTGTGTCTTTCATTTTTCT |
| CYC1 | AGCAGCCTCCTACCATCATCTC | CATTTCCTCTACTTTGCGTTTAGCA |
| CDC10 | GGTGTTCAGTTGCTGCTCACA | CAGGCTGCCAGCAATTACTATTATC |
| BUB3 | GGAGCTAAGATGACCGGTTCTAAC | TCCCAGGAGGAGACCAACAG |
| HINT1 | TTAATGATTGTTGGCAAGAAATGTG | CCTTCATTCACCACCATTCGA |
| MAPK6 | GGCATGTCGTTTGCACACA | CCTTGTCTTAGTGCATTTCTTTTGTC |
| AKAP10 | AGCTTGGAAGGTTGCTAAAATGA | GTGGACTTCTCTAACGGCTGATC |
| Ptpn12 | GAAAACTGGGAGTTCAACACCAA | GGAACTGTTGTGATGATCTGTGAGA |
| HSBP1 | ATGACATGAGCAGTCGCATTG | CCACCCCGGCCTGTGT |
| BRCA1 | GTCAGAGGCGATGTTGTCAATG | CTTTCTGTCCTGGGATTCTCTTG |
| MED11 | CTTCGAGGAAGGACTGTCAGATG | CAGGTTCGAGCCACCTCACT |
| APAF1 | TGGACCTGGACTCTAGCAGTCA | AAGTAGGAAGTCTGGTATGTAAAAAACCA |
| CRADD | TGACGGAAAGCCATGTTCAA | TGCTTTAGGACCCCTGGTAGGT |
| VDR | ACACTGCAGACCTACATCCG | CTTGGAGTGTTCCTCGTTCA |
Table 3.
Real-time PCR (qPCR) validation of select genes identified as upregulated by 1,25D3 from cDNA microarray analysis, presented as a table.
| Symbol | Primer efficiency (%) | qPCR result
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Cell strains
|
SV40-immortalized cells
|
||||||||
| 6h 1,25D3 Fold induction |
p-value | 24 h 1,25D3 Fold induction |
p-value | 6h 1,25P3 Fold induction |
p-value | 24h 1,25D3 Fold induction |
p-value | ||
| CALM3 | 101 | 2.49 ± 0.231 | 6.28E–04 | 2.61 ± 0.233 | 4.91E–04 | 1.95 ± 0.390 | 1.29E–02 | 1.50 ± 0.402 | p > 0.05 |
| CYC1 | 101 | 1.39 ± 0316 | NS | 1.49 ± 0.173 | NS | 4.80 ± 0.146 | 1.08E–05 | 5.97 ± 0.209 | 2.85E–05 |
| CDC10 | 101 | 2.61 ± 0.168 | 4.24E–03 | 2.53 ± 0.111 | 2.27E–03 | 1.64 ± 0.410 | NS | 1.53 ± 0.333 | NS |
| BCB3 | 101 | 2.56 ± 0.625 | 2.01E–02 | 2.04 ± 0516 | 2.60E–02 | 2.87 ± 0.200 | 1.88E–04 | 2.32 ± 0.111 | 4.84E–05 |
| HEVT1 | 97 | 2.13 ± 0.585 | 3.14E–02 | 1.35 ± 0.158 | 7.90E–03 | 1.24 ± 0.715 | NS | 1.73 ± 0.635 | NS |
| MAPK6 | 97 | 2.02 ± 0.099 | 4.97E–03 | 2.98 ± 0.031 | 9.77E–05 | 3.61 ± 0.315 | 1.80E–04 | 3.17 ± 0.175 | 5.51E–04 |
| AKAP10 | 101 | 6.38 ± 0.797 | 4.51E–04 | 1.04 ± 0.179 | p > 0.05 | 3.77 ± 0.221 | 1.13E–04 | 3.26 ± 0.355 | 1.09E–03 |
| Ptpn12 | 97 | 1.54 ± 0.121 | 8.26E–04 | 1.97 ± 0.102 | 7.85E–05 | 4.33 ± 0.280 | 1.97E–04 | 4.55 ± 0.227 | 7.43E–05 |
| HSBP1 | 104 | 1.96 ± 0.021 | 3.14E–04 | 1.15 ± 0.239 | p > 0.05 | 1.87 ± 0.120 | 1.90E–04 | 1.52 ± 0.148 | 2.11E–03 |
| ARNT | 83 | 5.09 ± 1.248 | NS | 1.70 ± 1.322 | NS | 1.94 ± 0.272 | 3.60E–03 | 2.19 ± 0.078 | 1.24E–05 |
| BRCA1 | 101 | 2.98 ± 1.065 | p>0.05 | 8.26 ± 0.688 | 1.53E–03 | 3.35 ± 0.292 | 4.94E–04 | 3.09 ± 0.099 | 9.95E–06 |
| MED11 | 97 | 1.50 ± 0.109 | 8.46E–04 | 1.20 ± 0.230 | p > 0.05 | 4.55 ± 0.227 | 1.54E–04 | 4.19 ± 0.215 | 8.17E–05 |
| APAF1 | 97 | 5.48 ± 0.705 | 3.87E–03 | 1.85 ± 0.417 | 2.18E–02 | 2.88 ± 0.122 | 2.44E–05 | 2.05 ± 0.345 | 6.11E–03 |
| CRADD | 97 | 7.33 ± 0.830 | 7.10E–03 | 2.47 ± 0.993 | 3.06E–02 | 1.53 ± 0.535 | NS | 1.10 ± 0.399 | NS |
| GAPDH | 102 | ||||||||
Listed are the amplification efficiencies for each primer set, the average fold induction × standard deviation for triplicate samples from two independent experiments, and the corresponding p-values. Significant differences among treatment groups for each gene and each cell type were determined using one-way ANOVA and the Holm’s procedure (p < 0.05). Where there were significant differences, pairwise comparisons were assessed with Student’s t-tests between control and 6 h or 24 h time points (p < 0.05). The p-values listed are those calculated by the Student’s t-tests; only significant values are shown. “NS” indicates no significance as determined by one-way ANOVA/Holm’s procedure, while “p > 0.05” indicates no significance using Student’s t-tests.
3. Results
3.1. 1,25D3-upregulation of VDR in dolphin skin
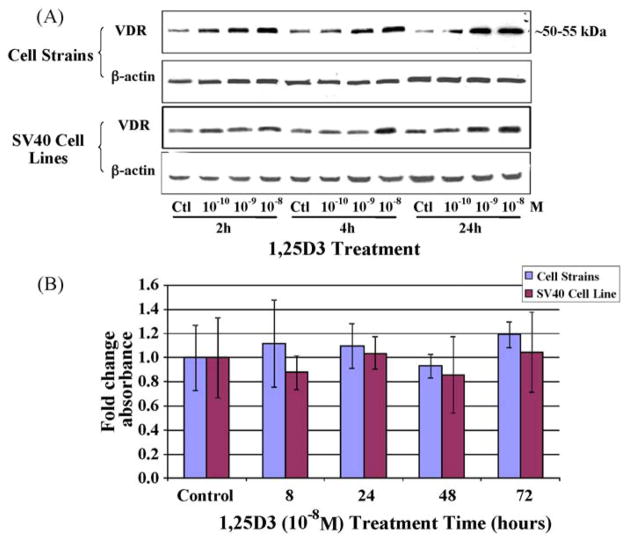
The primary route through which 1,25D3 exerts its biological function is via binding to VDR, a potent transcription factor that regulates the expression of genes involved in a number of physiological functions. It has been established that increased 1,25D3 levels upregulate VDR protein levels [46]. Therefore, our first objective was to verify that dolphin skin cells express VDR, display sensitivity to an exogenous 1,25D3 compound, and respond to 1,25D3 via upregulation of VDR. Western blot analysis was used to assess a response of the VDR protein to the compound at various dosages and time points. First, we measured the effects of three different concentrations (10−10, 10−9, and 10−8 M) of 1,25D3 on the cell strains and cell lines at multiple time points (Fig. 1A). A monoclonal antibody designed against the chicken VDR sequence was used for Western blot analysis, and it proved to cross-react with a single dolphin protein ~50–55 kDa, the same size as VDR in other species. The highest level of VDR expression was observed at a concentration of 10−8 M 1,25D3. An upregulation in VDR was apparent in both cell types after 4 h of treatment. To verify that 10−8 M levels were non-toxic, we performed an MTT assay to measure cell viability in both the cell strains and cell lines for time points up to 72 h (Fig. 1B). MTT assays showed no significant differences in absorbance between treatment groups. Consequently, we have selected a concentration of 10−8 M 1,25D3 for assessing the effects of this hormone on VDR levels and transcription in dolphin cells. This concentration of the 1,25D3 compound is comparable to levels used in other studies.
Fig. 1.
The concentration-dependent effect of 1,25D3 on VDR protein levels within dolphin skin cells. (A) VDR protein levels were assessed by Western blots after solvent (ctl), 10−10, 10−9, or 10−8 M concentrations of a 1,25D3 compound were administered to dolphin cell strains and SV40 cell lines for 2, 4, or 24 h. The same blot was stripped and reprobed with β-actin. (B) MTT assays measured cell viability as influenced by 10−8 M 1,25D3 treatment for the indicated times. Columns represent the average fold change in absorbance and bars the standard deviation among eight replicates. No significant differences in cell viability were detected within either the cell strains or the SV40 cell lines (one-way ANOVA, p < 0.05).
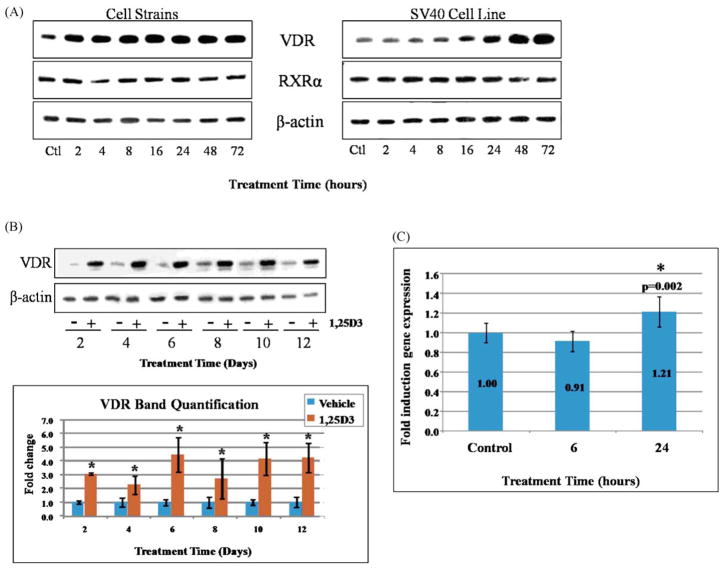
VDR protein and transcript levels were measured over time in response to 1,25D3 administration. Western blot analysis demonstrated upregulation of VDR protein in cell lysates from both dolphin cell strains and SV40-transformed cell lines treated with 1,25D3 up to 72 h (Fig. 2A). Within the cell strains, increases in VDR were observed as early as 2 h after treatment, with VDR levels appearing to plateau by 8 h. On the other hand, increased expression of VDR in the SV40 cell lines was observed later but continued to increase, even at 72 h. In addition, we measured expression of the retinoid X receptor (RXR), the heterodimeric partner for VDR necessary for transactivation. Using a polyclonal antibody designed against the human RXRα sequence, which recognized the dolphin RXRα protein, we noted very little effect on RXR levels by 1,25D3 (Fig. 2A). We also examined VDR expression over longer treatment times. SV40-transformed cell lines were treated over several days with 1,25D3 or ethanol, and protein levels were measured again by Western blot (Fig. 2B). β-actin-normalized VDR band intensities were quantified and presented graphically (Fig. 2B). At each time point measured, there was a significant upregulation in VDR relative to time-matched controls. However, the differences in VDR levels over the time course were not significant.
Fig. 2.
Time-dependent effects of 1,25D3 administration on VDR expression. (A) Western blots depict VDR, RXR, and β-actin protein levels within dolphin skin cell strains and SV40-immortalized cells before (Ctl: solvent only) and after treatment with 10−8 M 1,25D3 for the indicated times. The same blot was probed with VDR, RXR, and β-actin antibodies. (B) VDR protein levels are shown for longer treatment times (up to 12 days) by Western blot. In these experiments, SV40 cell lines were treated with solvent only (−) or 1,25D3 (+). The graph below illustrates quantified VDR band intensities after normalization to β-actin band intensities, as a fold change over time-matched controls (solvent-only). Columns represent the mean fold changes and bars the standard errors from three independent experiments. Student’s t-tests determined significant differences between controls and 1,25D3-treated samples (*, p < 0.05). No significant differences in VDR levels among the various time points were detected (one-way ANOVA, p < 0.05). (C) qPCR was used to measure VDR transcript levels in the SV40 cell lines treated with 10−8 M 1,25D3 for 6 h or 24 h. Columns represent the mean fold inductions (displayed) over the control (solvent) samples, and bars indicate the standard deviation among eleven replicate qPCR reactions. Significant changes (p < 0.05) in expression were determined by one-way ANOVA followed by pairwise comparisons using a Student’s t-test (*).
VDR contains a VDRE sequence within its own promoter [47], and 1,25D3 is known to regulate expression of its own receptor [48]; therefore, we also measured VDR transcripts. For these experiments, SV40 cell lines were treated with 1,25D3 (10−8 M) for 6 h or 24 h, and total RNA was subjected to qPCR using SYBR green. GAPDH-normalized fold changes showed a modest but significant upregulation of mRNA levels at 24 h, but not 6 h (Fig. 2C). We conclude that while 1,25D3 proves to upregulate VDR expression at the transcriptional level, post-translational effects likely play a part. This is suggested by the increase in VDR protein observed within 2 h of 1,25D3 exposure in the cell strains (Fig. 2A).
3.2. 1,25D3-transactivation of a vitamin D-sensitive promoter in dolphin skin
The observation that 1,25D3 upregulates VDR in dolphin cells prompted us to investigate its activation of a downstream transcriptional response. To assess 1,25D3-transactivation, we transfected the cells with a reporter plasmid containing a known vitamin D-sensitive promoter inserted upstream of a luciferase coding region [31,32]. The promoter within this plasmid encodes the mitochondrial cytochrome p450 24-hydroxylase (CYP24) responsible for the catabolism of 1,25D3 into its inactive byproducts. This is the most sensitive 1,25D3-inducible gene known to date [49]. Two known VDRE sequences exist within the CYP24 gene promoter, which have been shown to bind VDR-RXR heterodimers to stimulate transactivation [31,50,51]. Consequently, we selected this plasmid as the best tool for confirming 1,25D3-mediated transactivation.
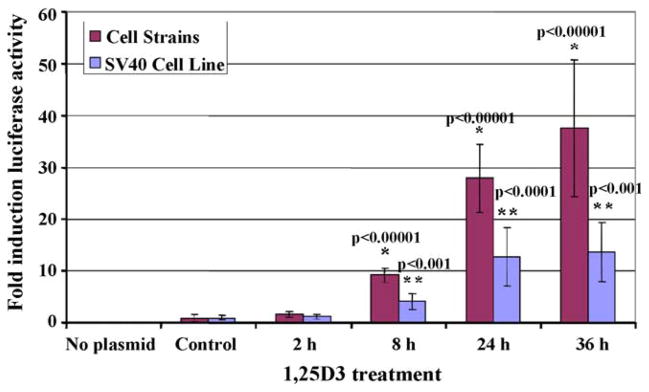
Luciferase assays were performed to measure transcriptional activity within the transfected cells after 1,25D3 (10−8 M) exposure for 2, 8, or 24 h (Fig. 3). Luciferase activity is displayed as a fold change over controls (transfected cells treated with solvent only), and it increased significantly with 8 h of 1,25D3 treatment. The dramatic increase in luciferase activity observed from these experiments demonstrates that an endogenous dolphin protein is capable of recognizing and activating the human CYP24 promoter. Interestingly, the 1,25D3-inducible expression of this promoter, as well as basal expression (not shown), were much stronger in the cell strains compared to the transformed cells. Higher activity and expression of the CYP24 enzyme have previously been reported in differentiating cells compared to proliferating cells [52]; this is consistent with the differentiation potential of our cell strains in contrast to the continuous cell line.
Fig. 3.
Luciferase assay for dolphin cell cultures transfected with a 1,25D3-inducible promoter/luciferase reporter plasmid and treated with 1,25D3 (10−8 M) for 2, 8, 24, or 36 h. Luciferase activity is expressed as a fold change from the “Control” samples (solvent-treated, transfected cells). “No plasmid” samples were neither transfected nor treated. Columns represent the average fold change and bars the standard deviations for three to eight independently-transfected replicates. Significant differences (p < 0.05) were determined using one-way ANOVA followed by Student’s t-test pairwise comparisons (p-values shown) between “Control” and 1,25D3-treated cells for cell strains (*) and SV40 cell lines (**).
3.3. Specific genes targeted by 1,25D3 in dolphin skin
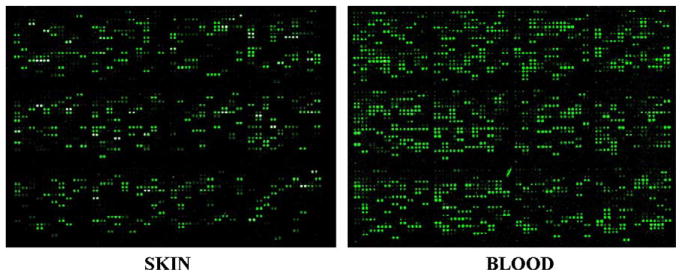
In order to identify the specific genes targeted by 1,25D3 and better understand the biological function of the vitamin D pathway in dolphin skin cells, we used the global transcriptomic approach of cDNA microarray analysis. The first dolphin microarray has recently been generated and validated [33], providing the best-available dolphin-specific transcriptomic tool to date. Of the 3700 PBL-derived ESTs on this array, 1343 have been sequenced, archived, and annotated at www.marinegenomics.org. Such sequence availability facilitated real-time PCR (qPCR) validation of the microarray results. This dolphin array has previously been validated using both dolphin PBL and our skin cells; distinct transcriptomic profiles were detected for each cell type [33]. To demonstrate hybridization efficiency in the skin cells relative to blood, we compared array hybridizations between skin RNA and blood RNA (Fig. 4). Fig. 4 depicts single analogous subarrays (out of 48 total on the microarray) from hybridizations with skin (left) and blood (right) samples. Each spot represents an individual gene (printed in duplicate per subarray and each subarray printed in duplicate per slide). The hybridization efficiency between a gene on the array to a gene expressed in the particular RNA sample is directly proportional to the observed color intensity. The degree of hybridization is noticeably less for the skin samples, as expected. However, the limitation of using a blood-derived microarray to evaluate transcriptomic profiles in skin is prevailed by the number of transcripts represented on this array that we found to also be expressed in the skin cells. The degree of hybridization appears sufficient for successful differential gene expression analysis in dolphin skin cells.
Fig. 4.
Dolphin skin cell RNA hybridization to a bottlenose dolphin blood microarray. Microarray hybridizations of RNA samples from non-treated bottlenose dolphin skin cells (left) versus bottlenose dolphin blood samples (right). One subarray (of 48 total) from the microarray chip is pictured. Fluorescent spots indicate an EST present on the array that hybridized to a gene expressed within the sample. The degree of fluorescence is directly proportional to expression of the gene.
Microarray experiments to evaluate vitamin D transcriptomic profiles in skin were conducted using RNA from cell strains treated with either ethanol or 1,25D3 (10−8 M) for 24 h or 48 h. Differential expression of genes between the control and 1,25D3-treated samples was analyzed using an original code designed specifically for the marine genomics project at marinegenomics.org [53]. This code denotes average differential expression of a gene as “df”. The value of df arbitrarily ranges from −1 to +1, with −1 being those genes most downregulated and +1 those genes most upregulated by 1,25D3 with respect to genes on the control slides. We chose to assign significant differential expression to any gene with a df value less than −0.4 or greater than +0.4 in at least one of the two time points tested (24 h and 48 h) and a p-value less than 0.05 (Wilcoxon signed-rank test). Using these parameters, 10.0% and 6.2% of the total genes represented on the array were upregulated with 1,25D3 after 24 h and 48 h, respectively. Only 1.0% and 1.9% were downregulated by 1,25D3 after 24 h and 48 h, respectively. The greatest df value observed in our datasets was 0.87 for 24 h treatment and 0.79 for 48 h treatment; the lowest df values were −0.68 and −0.79 for 24 h and 48 h treatments, respectively. Several of these significantly upregulated ESTs (among those that have been sequenced and yielded a BLAST hit against the NCBI database) were selected for further analysis. Fourteen such genes are presented in Table 2 along with the respective df and p-values for the 24 h and 48 h time points. A df value of “NA” indicates that particular EST was excluded from the final data analysis for the indicated time point. This was due to a signal intensity value less than 800; all raw intensity values less than 800, on a scale of 1 to 65,000, were automatically excluded from analysis.
Table 2.
Select genes upregulated by 1,25D3 within dolphin skin cell strains as identified by cDNA microarray analysis.
| Dolphin EST | BLAST Hit (NCBI) | Microarray results
|
|||||
|---|---|---|---|---|---|---|---|
| 24h 1,25D3 | 48h 1,25D3 | ||||||
|
|
|
||||||
| MGID# | ACC# | ACC#/description/E-value | Symbol | df | p-value | df | p-value |
| MGID140710 | DV467928 | BC120080/Bos taurus calmodulin 3 (phosphorylase kinase, delta)/0 | CALM3 | 0.80 | 1.68E–07 | 0.54 | 2.36E–07 |
| MGID140811 | DV468034 | XP001491551/PREDICTED: similar to cyclin I [Equus caballus]/5.04E-37 | CYC1 | 0.66 | 1.82E–05 | NA | NA |
| MGID140646 | DV467863 | NP001011553/cell division cycle 10 isoform 2 [Homo sapiens]/3.27E-135 | CDC10 | 0.58 | 2.17E–07 | 0.60 | 1.83E–07 |
| MGID140748 | DV467969 | NP001041371/budding uninhibited by benzimidazoles 3 homolog [Rattus norvegicus]/1.63E-19 | BUB3 | 0.58 | 1.68E–07 | 0.41 | 5.40E–07 |
| MGID140654 | DV467871 | NP071528/histidine triad nucleotide binding protein 1 [Rattus norvegicus]/1.82E-66 | HINT1 | 0.75 | 1.68E–07 | 0.62 | 1.65E–06 |
| MGID140525 | DT661425 | XM001501448/PREDICTED: Equus caballus similar to Mitogen-activated protein kinase 6 (Extracellular signal-regulated kinase 3) (ERK-3) (MAP kinase isoform p97) (p97-MAPK)/0 | MAPK6 | 0.87 | 1.68E–07 | 0.50 | 9.50E–07 |
| MGID140060 | DT660942 | ABW97719/A-kinase anchor protein 10 [Bos taurus]/0.0000006 | AKAP10 | 0.69 | 4.01E–05 | NA | NA |
| MGID141007 | DV468234 | XP610380/PREDICTED: similar to protein tyrosine phosphatase, non-receptor type 12 [Bos taurus]/4.68E-91 | Ptpn12 | 0.69 | 1.96E–04 | 0.22 | 7.40E–03 |
| MGID140751 | DV467972 | XP001112013/PREDICTED: similar to heat shock factor binding protein 1 [Macaca mulatta]/9E-34 | HSBP1 | 0.64 | 1.68E–07 | NA | NA |
| MGID139377 | DV799557 | XP540303/PREDICTED: similar to Aryl hydrocarbon receptor nuclear translocator (Dioxin receptor, nuclear translocator) (hypoxia-inducible factor 1 beta) isoform 1 [Canis familiaris]/2.87E-55 | ARNT | 0.47 | 1.01E–03 | 0.69 | 3.13E–02 |
| MGID139973 | DT660847 | AAN11299/breast and ovarian cancer susceptibility protein 1 [Canis familiaris]/3.19E-115 | BRCA1 | 0.41 | 4.63E–11 | 0.36 | 6.47E–10 |
| MGID139509 | DT660232 | XP001503009/PREDICTED: similar to mediator of RNA polymerase II transcription subunit 11 (mediator complex subunit 11) [Equus caballus]/2.08E-17 | MED11 | −0.20 | 1.40E–02 | 0.53 | 3.03E–07 |
| MGID141034 | DV468261 | NM001160/Homo sapiens apoptotic peptidase activating factor 1 (APAF1)/5E-24 | APAF1 | 0.60 | 1.22E–04 | NA | NA |
| MGID139465 | DT660182 | NP001077242/CASP2 and RIPK1 domain containing adaptor with death domain [Bos taurus]/1.14E-34 | CRADD | 0.46 | 5.91E–9 | 0.28 | 1.00E–05 |
Each dolphin EST is listed as the MGID and dolphin accession number (marinegenomics.org) along with its top BLAST hit (NCBI database). The corresponding differential expression (df) and p-values for both the 24 h and 48 h datasets are presented in the last four columns. The values for df range from −1 (most downregulated) to +1 (most upregulated). “NA” indicates exclusion of the data based on a raw intensity value less than 800.
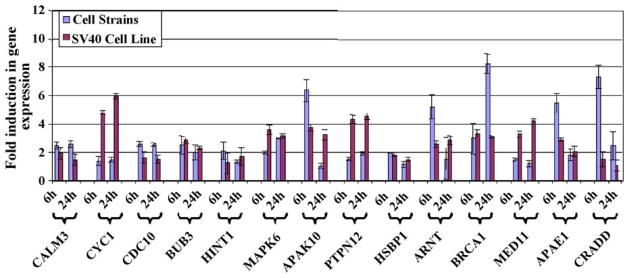
Differential expression of the genes listed in Table 2 was validated using real-time PCR (qPCR). qPCR allowed for representation of differential expression as a fold change over control samples. Validation was done in both the cell strains as well as the SV40-transformed cell lines. Total RNA from cells treated with solvent-only or 1,25D3 for 6 h or 24 h was subjected to two-step qPCR using SYBR green. Amplification efficiencies for all primer sets used are listed in Table 3. The resulting threshold cycles (Ct) were used to calculate differential expression using GAPDH as the internal, reference gene. The qPCR results are presented both as a graph (Fig. 5) and in Table 3, the latter displaying p-values. For every gene, there was significant upregulation in at least one of the cell types for at least one of the two time points. We did observe slight differences in transcriptional responses between transformed and non-transformed cells but, for the most part, 1,25D3-induced transcriptomic profiles in the SV40 cell lines were consistent with those of the cell strains.
Fig. 5.
Real-time PCR (qPCR) validation of select genes identified as upregulated by 1,25D3 from cDNA microarray analysis, presented as a chart. Differential expression between solvent- and 1,25D3-treated cells is shown as a fold induction over solvent-treated samples. Columns represent the average GAPDH-normalized fold changes, and bars represent the standard deviation for triplicate qPCR reactions from two individual experiments.
Some of the changes in expression associated with 1,25D3 were modest, yet significant, for most cases. The majority of these genes, or at least isoforms of these genes, have been implicated as 1,25D3 target genes in humans and/or mice [54], confirming the efficacy of these molecular tools for studying the bottlenose dolphin. It also supports the notion that the vitamin D pathway may be well conserved between terrestrial and marine mammals.
4. Discussion
Dolphin molecular biology is still a nascent field, and the experimental tools that have been developed in traditional laboratory models are not yet applicable to dolphin samples. Consequently, genomic, transcriptomic, and proteomic analyses can be problematic. We are using the new tools and information we now have at our disposal to investigate the vitamin D pathway in the dolphin and its comparison to that in terrestrial mammals.
Primary cultures derived from bottlenose dolphin skin tissue have been successfully passaged to generate cell strains. We find that these cell strains tend to slow in proliferation after seven to ten passages, eventually entering senescence. To overcome the limitation of limited lifespan, one of the strains was immortalized via SV40-mediated transformation to yield continuous cell lines. This is particularly beneficial in a protected species as it eliminates the need for repeated sample acquisition. Both cellular models have provided a much-needed in vitro system for investigating dolphin health mechanisms. We are experimenting simultaneously with cell strains and SV40-immortalized cell lines to confirm results in each and to validate the cell lines as an appropriate tool in lieu of the limited cell strains. For most of the results presented here, the data from the cell lines supported what was found in the cell strains. However, the differences that were observed are noteworthy. 1,25D3, an agent affecting proliferation and differentiation, will likely have differential effects on a continuously-proliferating cell line in contrast to normal cells. For this reason, transformed cell lines should be used judiciously when compared to non-transformed cells. Nevertheless, they prove useful for confirming results found within the latter.
The luciferase assays showed that 1,25D3 transactivates a vitamin D-sensitive promoter somewhere between 2 h and 8 h. This is consistent with the qPCR results which indicated that the majority of specific genes were upregulated 6 h post-treatment. While we have not identified VDR as a direct mediator of this 1,25D3-induced transactivation within dolphin cells, we did find that VDR protein upregulation correlated with these transcriptional responses (Figs. 1A and 2A). Furthermore, it has been shown by others that ligand-bound VDR-RXR heterodimers bind and transactivate the VDRE sequences within the promoter sequence of the luciferase reporter plasmid used for our luciferase assays [31,50,51]. Yet, we do not have direct evidence that VDR is the transcriptional mediator in dolphin cells.
Microarray technology is relatively new to the marine mammal field, and the generation of the first dolphin-specific microarray is a major advancement. Still, arrays have not yet reached the complexity of those developed for organisms in which more extensive genome information is available, and this limitation should be factored into the interpretation of our results. At the time our microarray experiments were conducted, the dolphin cDNA array contained 3700 ESTs, of which only 1395 had been sequenced; the remaining 2305 are unsequenced and unidentified EST clones (i.e. clones derived from the cDNA libraries that have been printed onto the array but not yet sequenced). This pool of sequenced genes is further limited to those ESTs with significant homology to known genes/proteins (i.e. returned a BLAST hit against the NCBI database) and even further tothose also expressed in skin (see Fig. 4).While the microarray did provide an indication of general transcriptomic alterations induced by 1,25D3 in skin, we did not anticipate identification of a large number of specific genes, as with typical microarray experiments. The genes listed in Table 2 were selected from the results as key representatives of the different biological processes affected by 1,25D3 in dolphin skin cells. However, due to the skewed pool of available genes on the microarray, the genes listed are not an indication of the distribution, diversity, and totality of the biological functions affected by 1,25D3 in these cells.
Of the genes selected for further investigation (listed in Tables 1 and 2), the majority are associated in one way or another with the biological processes of proliferation, differentiation, apoptosis, or skin immunity/barrier functions. Proliferation is a key element of tumorigenesis as well as skin-associated hyperproliferative disorders and wound healing. Differentiation is a well-defined and inherent property of keratinocytes critical for skin function, wound healing, and protection of the skin against exterior pathogens. The epidermis undergoes rapid turnover to ensure proper sloughing of the exterior-most layers of the skin, and this turnover rate is particularly high in the dolphin [55]. Apoptosis is not only necessary in abnormal and/or injured cells but also for this epidermal sloughing process. Skin immunity and barrier formation are most crucial for defending the body from the external environment and for healing wounds; recent studies have begun addressing vitamin D’s position in innate immunity and wound healing within the skin [9,56].
One conspicuous indicator of the increased threat to dolphin health has been the observation of cutaneous lesions, rashes, and infections on these animals [57–62]. Such skin pathologies may be similar to those in humans, which are often associated with altered antimicrobial production, inflammation, increased proliferation with decreased differentiation, and disrupted cytokine/growth factor signaling [56,63]. All of these are well known functions of 1,25D3, the reason this pathway is implicated in several hyperproliferative and inflammatory skin conditions (e.g. atopic dermatitis, rosacea, psoriasis, and vitiligo). The genes identified in this study support such immunomodulatory properties of vitamin D in dolphin skin.
Calmodulin functions as a calcium-binding protein, and calcium’s roles in cellular differentiation, proliferation, apoptosis, and would healing within the skin have been well characterized [64]. Calmodulin itself is involved in cell cycle and wound healing and serves as a keratinocyte terminal differentiation marker [65,66]. Cyclins, cell division cycle (CDC) proteins, and the budding inhibited by benzimidazoles (BUB) proteins all are direct regulators of the cell cycle, consistent with the anti-proliferative capabilities of 1,25D3. Hyperproliferation of skin cells is not only a hallmark of skin cancers but also of autoimmune disorders such as psoriasis. Cyclin I belongs to the cyclin subgroup family that comprises cyclins G1 and G2. The biological functions of this subfamily are still obscure, but there is evidence that cyclin I is critical for cell growth arrest and is expressed in terminally differentiated tissues [67]. BUB is a mitotic checkpoint protein that regulates the alignment of chromosomes on the mitotic spindle during anaphase [68]. The CDC proteins (also known as septins) are critical for cytokinesis with mutations in the CDC10 isoform, specifically, shown to prevent cytokinesis [69]. Several genes identified from the microarray are signaling molecules associated with pathways affecting a variety of biological processes. Studies investigating the functions of HINT1 suggest it to be a modulator of apoptotic signaling, a novel tumor suppressor, and a regulator of transcription [70–72]. The MAPK family comprises phosphorylation-activated kinases responsible for a multitude of signaling pathways. MAPKs may regulate cell proliferation, differentiation, and wound healing within the epidermis [73]. A-kinase anchor proteins bind the regulatory domain of protein kinase A (PKA) to confine it within various compartments of the cell to regulate PKA signaling specificity. AKAP9 was downregulated in peripheral blood lymphocytes of patients with arsenic-induced skin lesions, suggesting a possible responsibility of AKAPs in immunity and wound healing [74]. PTPs are signaling molecules involved in differentiation, cell growth, apoptosis, mitosis and oncogenic transformation, all of which make them likely targets of the vitamin D3 pathway. Their tyrosine dephosphorylation activity serves to regulate signal transduction pathways including the MAPK cascade [75,76]. Polymorphisms of the PTPN22 isoform have been connected to several autoimmune disorders, many the same as those linked to vitamin D [77–82]. In addition, oxidative stress may inhibit PTPs [83]. More studies are addressing vitamin D’s antioxidant potential, including within the skin where it may protect keratinocytes against oxidative stress [84,85]. 1,25D3 upregulated the heat shock factor binding protein (HSBP1), which negatively regulates the heat shock response by direct inhibition of the heat shock factor [86]. We found 1,25D3 upregulation of ARNT, also known as HIF1β, particularly noteworthy due to its role in a wide array of crucial adaptive processes, as the promiscuous heterodimeric transcription factor partner for ArH, HIF1α, and MOP2 [87]. ARNT’s role in skin homeostasis and protection from environmental stress was investigated using ARNT-specific ablation in mouse skin; the authors found that ARNT was critical for epidermal sphingolipd composition, barrier function and development [88,89]. Interestingly, BRCA1 interacts with ARNT as a coactivator for the ARNT-ArH transcription complex. BRCA1 is a dominantly inherited gene that has been well-studied as a tumor suppressor for breast and ovarian cancers. Not only does BRCA negatively regulate growth of breast, ovarian, and colon cells, it may aid in DNA repair within keratinocytes [90]. Med11 is a subunit of the Mediator complex, the multi-protein coactivator required for activation of RNA polymerase II transactivation via interaction with the polymerase’s C-terminal domain [91]. Many of the mediator proteins have also been identified within the multi-subunit DRIP (VDR-interacting protein) complex, which bridges the VDR to RNA polymerase II [92,93]. It could be speculated that 1,25D3 upregulates the expression of specific subunits within this DRIP/mediator complex prior to its coactivation of VDR. Apaf1 and CRADD proteins are both directly involved in apoptotic machinery. A reduction in and/or resistance to apoptosis has been suggested as a cause of the increased cell proliferation associated with autoimmune disorders, such as psoriasis [94,95]. Apaf1 associates with the apoptosome and is involved in cytochrome c-dependent activation of caspases 3 and 9; loss of APAF1 expression has been associated with melanoma [96]. The protein encoded by CRADD (aka RAIDD) is a death domain (CARD/DD)-containing protein that serves as an adapter protein linking caspases to apoptosis signaling pathways, such as caspase 2 and the TNFR signaling complex [97,98].
Several microarray studies have been performed in various cell types treated with 1,25D3 metabolites or analogs, and a large suite of 1,25D3 target genes have been identified from them. One of the most comprehensive studies performed was done by John White’s group who compiled an extensive list of 1,25D3 target genes from humans and mice [54]. This study identified human gene targets within SCC25 cells using microarray analysis. The authors also implicated potential vitamin D targets via human and mouse genome-wide screens for VDRE consensus sequences (both the classical DR3 and ER6 elements). “Nonconsensus” sequences (single nucleotide mismatch from the classical DR3 or ER6 motifs) were included in this search. Citing these results and those from other microarray studies, we compiled a summary of homologs or homologous isoforms to each of the dolphin genes that we identified. All findings, to our knowledge, are presented in Table 4. The microarray studies conducted in the SCC25 cells identified CALML3, CCNG2, CDC2L6, MAPK13, AKAP12, PTPN1, PTPNS1, and HIF1α as upregulated by 1,25D3; CCND2 and CDC25 were downregulated. According to the human genome searches, PTPN22 contains a consensus VDRE, while, calmodulins 1, 2 and 3, CDC10, BUB1, HINT1, HSBP1, ARNT2, BRCA2, and Med8, APAF1, and several cyclin, MAPK, AKAP, and PTPN isoforms possess nonconsensus VDREs. Within the mouse genome, BUB3, HINT2, MAPK6, HIF1α and HIF3α, Med8 and several cyclin, CDC, and PTPN isoforms possess VDRE nonconsensus motifs. Beyond this study, other groups also identified 1,25D3 targets that are homologous to the dolphin genes (or their isoforms) (Table 4, far right columns). In human primary keratinocytes, 1,25D3 (12 h) upregulated cyclins G1 and G2, CDC2, and AKAP8 and downregulated BUB8 [99]. Another microarray study within breast cancer cells found that calmodulin 2 (CALM2), cyclin I (CYC1), cyclin G1 (CCNG1) and cyclin G2 (CCNG2) were upregulated by 1,25D3 after 6 h and 24 h [100]. In prostate cancer cells, cyclin K [101] and AKAP12 were upregulated [102]. 1,25D3 downregulated cyclin D1 (CCND1) and CDC6 in mouse osteoblasts (12 h) [103], BRCA2 and MAPK5 in breast cancer cells (6 h and 24 h) [100], and CDC2 in prostate cancer cells (6 h and 24 h) [102]. Others have shown via Western blot decreases in CCND1 protein levels, after treating a breast cancer cell line with a vitamin D analog for 24 h and 72 h [104].
Table 4.
Summary of potential 1,25D3-regulated genes that are homologous to the identified dolphin genes and/or their isoforms (listed in Tables 1 and 2).
| Dolphin gene | Microarray study in human SCC25 cellsa
|
Genome-wide screens (consensus and nonconsensus VDREs)a
|
Other studies
|
|||
|---|---|---|---|---|---|---|
| Gene | Up/downregulated | Human genes | Mouse genes | Gene | Up/downregulated | |
| CALM3 | CALML3 | Up | CALM1,2,3 | CALM2 human breast cancer cellsc | Up | |
| CYC1 | CCND2 | Down | Cyclins A,B,D,E,G,H,K,M,T | Cyclins A,B,D,H | CCNK human prostate cancer cellsb | Up |
| CCNG2 | Up | CYC1, CCNG1, CCNG2 human breast cancer cellsc | Up | |||
| CCNG1, CCNG2 human keratinocytesg | Up | |||||
| CCND1 mouse osteoblastsd | Down | |||||
| CCND1 human breast cancer cellsf | Down | |||||
| CDC10 | CDC25 | Down | CDC2,6,7,10,23,25,26,27 | CDC2,7,25,26,27,45L | CDC6 mouse osteoblastsd | Down |
| CDC2L6 | Up | CDC2 human prostate cancer cellse | Down | |||
| CDC2 human keratinocytesg | Up | |||||
| BUB3 | NA | Bub1,3 | Bub3 | BUB3 human keratinocytesg | Down | |
| HINT1 | NA | HINT1,2 | HINT2 | |||
| MAPK6 | MAPK13 | Up | MAPK1,3,6,7,9,10,11,12,14 | MAPK6 | MAPK5 human breast cancer cellsc | Down |
| AKAP10 | AKAP12 | Up | AKAP1,3,7,8L,10,13 | AKAP12 human cancer and normal prostate cellse | Up | |
| AKAP8 human keratinocytesg | Up | |||||
| Ptpn12 | PTPN1 | Up | PTPN2,3,4,7,14,18,22 | PTPN2,7,14 | ||
| PTPNS1 | Up | |||||
| HSBP1 | NA | HSBP1 | ||||
| ARNT1 | HIF1α | Up | ARNT2, ARNTL | HIF1α, HIF3α | ||
| BRCA1 | NA | BRCA2 | BRCA2 human breast cancer cellsc | Down | ||
| MED11 | NA | MED8 | MED8 | |||
| APAF1 | NA | APAF1 | ||||
| CRADD | NA | CRADD | ||||
While there is ambiguity as to whether 1,25D3 upregulates or downregulates some of these genes, the discovery of their differential expression in other species supports their identities as vitamin D target genes in both terrestrial and marine mammals. Our results serve as confirmation of 1,25D3 regulation for those genes discovered to possess potential promoter VDREs in the work by Wang et al. [54]. Furthermore, similarities affirm the validity of our dolphin microarray-skin cell experimental system. Altogether, our findings presented here suggest that the vitamin D pathway may play an analogous function within the dolphin as it does in other species studied.
The study at hand contributes to the elucidation of vitamin D3, whose function depends on the cell type, organ, and organism in which it is studied. To date an extensive amount of vitamin D research has been conducted in terrestrial animal models; however, nothing is known of the pathway in marine mammals. Originally discovered for its role in calcium metabolism, today 1,25D3 is additionally acknowledged for its chemopreventative and immunomodulatory properties. An increasing body of experimental evidence corroborates the role of the vitamin D pathway in enhancing both innate and adaptive immunity in humans [36,40]. The similarities we have detected between the bottlenose dolphin and man suggest that vitamin D enhances innate immunity in this marine mammal as well. Therefore, dolphin skin cells will be used to assess 1,25D3’s effects on vitamin D-related phenotypes including antimicrobial peptide production, wound healing, cell proliferation, apoptosis, and oxidative stress. Any effects can be correlated with upregulation of the genes identified in this study, most of which associate directly or indirectly with one or more of these functions. We are currently testing the protective effects of 1,25D3 in vitro on dolphin skin cells after exposure to harmful agents. The ultimate objective is to evaluate the effects of environmental stressors on 1,25D3-induced transcriptomic profiles and phenotypes in order to understand how the vitamin D3 pathway might protect marine mammals from the negative impact of stressors.
Acknowledgments
This work was partially supported by a National Science Foundation EPSCoR grant (MSK, EPS-0132573 and EPS-0447660), Veterans Administration Merit Review (MSK) and a National Oceanic and Atmospheric Administration grant to Center for Coastal Environmental Health and Biomolecular Research (NOAA Biotech Contract, MOA-2004-168/1259), SC Sea Grant (MOA-2006-025/7182) and the Marine Biomedicine and Environmental Sciences Center, Oceans and Human Health Training Program.
Abbreviations
- 1,25D3
1,25-dihydroxyvitamin D3
- VDR
vitamin D receptor
- qPCR
real-time polymerase chain reaction
- CYP27B1
cytochrome p450 enzyme, isoform B1
- 7-DHC
7-dihydroxycholesterol
- RXR
retinoid X receptor
References
- 1.Reif JS, Mazzoil MS, McCulloch SD, Varela RA, Goldstein JD, Fair PA, et al. Lobomycosis in Atlantic bottlenose dolphins from the Indian River Lagoon, Florida. J Am Vet Med Assoc. 2006;228:104–8. doi: 10.2460/javma.228.1.104. [DOI] [PubMed] [Google Scholar]
- 2.Taubenberger JK, Tsai M, Krafft AE, Lichy JH, Reid AH, Schulman FY, et al. Two morbilliviruses implicated in bottlenose dolphin epizootics. Emerg Infect Dis. 1996;2:213–6. doi: 10.3201/eid0203.960308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipscomb TP, Schulman FY, Moffett D, Kennedy S. Morbilliviral disease in Atlantic bottlenose dolphins (Tursiops truncatus) from the 1987–1988 epizootic. J Wildl Dis. 1994;30:567–71. doi: 10.7589/0090-3558-30.4.567. [DOI] [PubMed] [Google Scholar]
- 4.Dolphins Klinowska M. IUCN. The IUCN Red Data Book. Gland; Switzerland: Cambridge; U.K: 1991. porpoises and whales of the world; p. 429. [Google Scholar]
- 5.Gubbins CM. Behavioral ecology and social structure of coastal bottlenose dolphins in South Carolina. Reno, NV: University of Nevada; 2000. p. 162. [Google Scholar]
- 6.Yu J, Kindy MS, Ellis BC, Baatz JE, Peden-Adams M, Ellingham TJ, et al. Establishment of epidermal cell lines derived from the skin of the Atlantic bottlenose dolphin (Tursiops truncatus) Anat Rec A Discov Mol Cell Evol Biol. 2005;287:1246–55. doi: 10.1002/ar.a.20266. [DOI] [PubMed] [Google Scholar]
- 7.Lin R, White JH. The pleiotropic actions of vitamin D. Bioessays. 2004;26:21–8. doi: 10.1002/bies.10368. [DOI] [PubMed] [Google Scholar]
- 8.van Etten E, Mathieu C. Immunoregulation by 1,25-dihydroxyvitamin D3: basic concepts. J Steroid Biochem Mol Biol. 2005;97:93–101. doi: 10.1016/j.jsbmb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Schauber J, Gallo RL. The vitamin D pathway: a new target for control of the skin’s immune response? Exp Dermatol. 2008;17:633–9. doi: 10.1111/j.1600-0625.2008.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuritzky LA, Finlay-Jones JJ, Hart PH. The controversial role of vitamin D in the skin: immunosuppression vs. photoprotection. Clin Exp Dermatol. 2008;33:167–70. doi: 10.1111/j.1365-2230.2007.02632.x. [DOI] [PubMed] [Google Scholar]
- 11.Bikle DD. Vitamin D receptor, UVR, and skin cancer: a potential protective mechanism. J Invest Dermatol. 2008;128:2357–61. doi: 10.1038/jid.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitfield GK, Dang HT, Schluter SF, Bernstein RM, Bunag T, Manzon LA, et al. Cloning of a functional vitamin D receptor from the lamprey (Petromyzon marinus), an ancient vertebrate lacking a calcified skeleton and teeth. Endocrinology. 2003;144:2704–16. doi: 10.1210/en.2002-221101. [DOI] [PubMed] [Google Scholar]
- 13.Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–67. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 14.Nagpal S, Na S, Rathnachalam R. Noncalcemic actions of vitamin D receptor ligands. Endocr Rev. 2005;26:662–87. doi: 10.1210/er.2004-0002. [DOI] [PubMed] [Google Scholar]
- 15.Dusso AS, Brown AJ, Slatopolsky E. Vitamin D Am J Physiol Renal Physiol. 2005;289:F8–28. doi: 10.1152/ajprenal.00336.2004. [DOI] [PubMed] [Google Scholar]
- 16.Reschly EJ, Krasowski MD. Evolution and function of the NR1I nuclear hormone receptor subfamily (VDR, PXR, and CAR) with respect to metabolism of xenobiotics and endogenous compounds. Curr Drug Metab. 2006;7:349–65. doi: 10.2174/138920006776873526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holick MF. McCollum Award Lecture 1994: vitamin D—new horizons for the 21st century. Am J Clin Nutr. 1994;60:619–30. doi: 10.1093/ajcn/60.4.619. [DOI] [PubMed] [Google Scholar]
- 18.Tian XQ, Chen TC, Matsuoka LY, Wortsman J, Holick MF. Kinetic and thermodynamic studies of the conversion of previtamin D3 to vitamin D3 in human skin. J Biol Chem. 1993;268:14888–92. [PubMed] [Google Scholar]
- 19.Holick MF, Tian XQ, Allen M. Evolutionary importance for the membrane enhancement of the production of vitamin D3 in the skin of poikilothermic animals. Proc Natl Acad Sci USA. 1995;92:3124–6. doi: 10.1073/pnas.92.8.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haussler MR. Vitamin D receptors: nature and function. Annu Rev Nutr. 1986;6:527–62. doi: 10.1146/annurev.nu.06.070186.002523. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann B, Genehr T, Knuschke P, Pietzsch J, Meurer M. UVB-induced conversion of 7-dehydrocholesterol to 1alpha,25-dihydroxyvitamin D3 in an in vitro human skin equivalent model. J Invest Dermatol. 2001;117:1179–85. doi: 10.1046/j.0022-202x.2001.01538.x. [DOI] [PubMed] [Google Scholar]
- 22.Lehmann B, Knuschke P, Meurer M. UVB-induced conversion of 7-dehydrocholesterol to 1 alpha,25-dihydroxyvitamin D3 (calcitriol) in the human keratinocyte line HaCaT. Photochem Photobiol. 2000;72:803–9. doi: 10.1562/0031-8655(2000)072<0803:uicodt>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 23.Schuessler M, Astecker N, Herzig G, Vorisek G, Schuster I. Skin is an autonomous organ in synthesis, two-step activation and degradation of vitamin D(3): CYP27 in epidermis completes the set of essential vitamin D(3)-hydroxylases. Steroids. 2001;66:399–408. doi: 10.1016/s0039-128x(00)00229-4. [DOI] [PubMed] [Google Scholar]
- 24.Vantieghem K, Overbergh L, Carmeliet G, De Haes P, Bouillon R, Segaert S. UVB-induced 1,25(OH)2D3 production and vitamin D activity in intestinal CaCo-2 cells and in THP-1 macrophages pretreated with a sterol Delta7-reductase inhibitor. J Cell Biochem. 2006;99:229–40. doi: 10.1002/jcb.20910. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann B. The vitamin D3 pathway in human skin and its role for regulation of biological processes. Photochem Photobiol. 2005;81:1246–51. doi: 10.1562/2005-02-02-IR-430. [DOI] [PubMed] [Google Scholar]
- 26.Banerjee P, Chatterjee M. Antiproliferative role of vitamin D and its analogs –a brief overview. Mol Cell Biochem. 2003;253:247–54. doi: 10.1023/a:1026072118217. [DOI] [PubMed] [Google Scholar]
- 27.Bikle DD. Vitamin D and skin cancer. J Nutr. 2004;134:3472S–8. doi: 10.1093/jn/134.12.3472S. [DOI] [PubMed] [Google Scholar]
- 28.Bikle DD, Oda Y, Xie Z. Calcium and 1,25(OH)2D: interacting drivers of epidermal differentiation. J Steroid Biochem Mol Biol. 2004;89–90:355–60. doi: 10.1016/j.jsbmb.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Zasloff M. Sunlight, vitamin D, and the innate immune defenses of the human skin. J Invest Dermatol. 2005;125:xvi–i. doi: 10.1111/j.0022-202X.2005.23924.x. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann B, Querings K, Reichrath J. Vitamin D and skin: new aspects for dermatology. Exp Dermatol. 2004;13:11–5. doi: 10.1111/j.1600-0625.2004.00257.x. [DOI] [PubMed] [Google Scholar]
- 31.Mallbris L, Edstrom DW, Sundblad L, Granath F, Stahle M. UVB upregulates the antimicrobial protein hCAP18 mRNA in human skin. J Invest Dermatol. 2005;125:1072–4. doi: 10.1111/j.0022-202X.2005.23872.x. [DOI] [PubMed] [Google Scholar]
- 32.Weber G, Heilborn JD, Chamorro Jimenez CI, Hammarsjo A, Torma H, Stahle M. Vitamin D induces the antimicrobial protein hCAP18 in human skin. J Invest Dermatol. 2005;124:1080–2. doi: 10.1111/j.0022-202X.2005.23687.x. [DOI] [PubMed] [Google Scholar]
- 33.Mancia A, Lundqvist ML, Romano TA, Peden-Adams MM, Fair PA, Kindy MS, et al. A dolphin peripheral blood leukocyte cDNA microarray for studies of immune function and stress reactions. Dev Comp Immunol. 2007;31:520–9. doi: 10.1016/j.dci.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Kragballe K. Treatment of psoriasis with calcipotriol and other vitamin D analogues. J Am Acad Dermatol. 1992;27:1001–8. doi: 10.1016/0190-9622(92)70302-v. [DOI] [PubMed] [Google Scholar]
- 35.Katiyar S, Mukhtar H. Immunotoxicity of environmental agents in the skin. In: Fuchs PLJ, editor. Environmental stressors in health and disease. New York: Marcel Dekker, Inc; 2001. pp. 345–364. [Google Scholar]
- 36.Adorini L, Penna G. Control of autoimmune diseases by the vitamin D endocrine system. Nat Clin Pract Rheumatol. 2008;4:404–12. doi: 10.1038/ncprheum0855. [DOI] [PubMed] [Google Scholar]
- 37.Adams JS, Liu PT, Chun R, Modlin RL, Hewison M. Vitamin D in defense of the human immune response. Ann N Y Acad Sci. 2007;1117:94–105. doi: 10.1196/annals.1402.036. [DOI] [PubMed] [Google Scholar]
- 38.Birlea SA, Costin GE, Norris DA. Cellular and molecular mechanisms involved in the action of vitamin D analogs targeting vitiligo depigmentation. Curr Drug Targets. 2008;9:345–59. doi: 10.2174/138945008783954970. [DOI] [PubMed] [Google Scholar]
- 39.Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–3. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 40.Bikle D. Nonclassic actions of vitamin D. J Clin Endocrinol Metab. 2009;94:26–34. doi: 10.1210/jc.2008-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pike JW, Kerner SA, Jin CH, Allegretto EA, Elgort MG. Direct activation of the human 25-(OH)2D3 and PMA: Identification of cis elements and transactivators. J Bone Miner Res. 1994;9:S144. [Google Scholar]
- 42.Allegretto EA, Shevde N, Zou A, Howell SR, Boehm MF, Hollis BW, et al. Retinoid X receptor acts as a hormone receptor in vivo to induce a key metabolic enzyme for 1,25-dihydroxyvitamin D3. J Biol Chem. 1995;270:23906–9. doi: 10.1074/jbc.270.41.23906. [DOI] [PubMed] [Google Scholar]
- 43.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–15. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krishnan AV, Feldman D. Regulation of vitamin D receptor abundance. In: Feldman D, Glorieux FH, Pike JW, editors. Vitamin D. San Diego: Academic Press; 1997. pp. 179–200. [Google Scholar]
- 47.Zella LA, Kim S, Shevde NK, Pike JW. Enhancers located in the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol. 2007;103:435–9. doi: 10.1016/j.jsbmb.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 48.Estebana LM, Eismana JA, Gardiner EM. Vitamin D receptor promoter and regulation of receptor expression. In: Feldman D, Pike JW, Glorieux FH, editors. Vitamin D. Vol. 2. New York: Elsevier/Academic Press; 2005. pp. 193–217. [Google Scholar]
- 49.Dwivedi PP, Omdahl JL, Kola I, Hume DA, May BK. Regulation of rat cytochrome P450C24 (CYP24) gene expression. Evidence for functional cooperation of Ras-activated Ets transcription factors with the vitamin D receptor in 1,25-dihydroxyvitamin D(3)-mediated induction. J Biol Chem. 2000;275:47–55. doi: 10.1074/jbc.275.1.47. [DOI] [PubMed] [Google Scholar]
- 50.Zou A, Elgort MG, Allegretto EA. Retinoid X receptor (RXR) ligands activate the human 25-hydroxyvitamin D3-24-hydroxylase promoter via RXR heterodimer binding to two vitamin D-responsive elements and elicit additive effects with 1,25-dihydroxyvitamin D3. J Biol Chem. 1997;272:19027–34. doi: 10.1074/jbc.272.30.19027. [DOI] [PubMed] [Google Scholar]
- 51.Chen KS, DeLuca HF. Cloning of the human 1 alpha,25-dihydroxyvitamin D-3 24-hydroxylase gene promoter and identification of two vitamin D-responsive elements. Biochim Biophys Acta. 1995;1263:1–9. doi: 10.1016/0167-4781(95)00060-t. [DOI] [PubMed] [Google Scholar]
- 52.Bikle DD. In: Vitamin D: Role in Skin and Hair Vitamin D. David Feldman FG, Wesley Pike J, editors. San Diego: Elsevier; 2005. pp. 609–20. [Google Scholar]
- 53.McKillen DJ, Chen YA, Chen C, Jenny MJ, Trent HF, 3rd, et al. Marine genomics: a clearing-house for genomic and transcriptomic data of marine organisms. BMC Genomics. 2005;6:34. doi: 10.1186/1471-2164-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang TT, Tavera-Mendoza LE, Laperriere D, Libby E, MacLeod NB, Nagai Y, et al. Large-scale in silico and microarray-based identification of direct 1,25-dihydroxyvitamin D3 target genes. Mol Endocrinol. 2005;19:2685–95. doi: 10.1210/me.2005-0106. [DOI] [PubMed] [Google Scholar]
- 55.Hicks BD, St Aubin DJ, Geraci JR, Brown WR. Epidermal growth in the bottlenose dolphin, Tursiops truncates. J Invest Dermatol. 1985;85:60–3. doi: 10.1111/1523-1747.ep12275348. [DOI] [PubMed] [Google Scholar]
- 56.Segaert S, Simonart T. The epidermal vitamin D system and innate immunity: some more light shed on this unique photoendocrine system? Dermatology. 2008;217:7–11. doi: 10.1159/000118506. [DOI] [PubMed] [Google Scholar]
- 57.Van Bressem MF, Van Waerebeek K, Montes D, Kennedy S, Reyes JC, Garcia-Godos IA, et al. Diseases, lesions and malformations in the long-beaked common dolphin Delphinus capensis from the Southeast Pacific. Dis Aquat Organ. 2006;68:149–65. doi: 10.3354/dao068149. [DOI] [PubMed] [Google Scholar]
- 58.Manire CA, Smolarek KA, Romero CH, Kinsel MJ, Clauss TM, Byrd L. Proliferative dermatitis associated with a novel alphaherpesvirus in an Atlantic bottlenose dolphin (Tursiops truncatus) J Zoo Wildl Med. 2006;37:174–81. doi: 10.1638/05-006.1. [DOI] [PubMed] [Google Scholar]
- 59.Bonar CJ, Boede EO, Hartmann MG, Lowenstein-Whaley J, Mujica-Jorquera E, Parish SV, et al. A retrospective study of pathologic findings in the Amazon and Orinoco river dolphin (Inia geoffrensis) in captivity. J Zoo Wildl Med. 2007;38:177–91. doi: 10.1638/1042-7260(2007)038[0177:ARSOPF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 60.Cowan DF. Lobo’s disease in a bottlenose dolphin (Tursiops truncatus) from Matagorda Bay, Texas. J Wildl Dis. 1993;29:488–9. doi: 10.7589/0090-3558-29.3.488. [DOI] [PubMed] [Google Scholar]
- 61.Schulman FY, Lipscomb TP. Dermatitis with invasive ciliated protozoa in dolphins that died during the 1987–1988 Atlantic Bottlenose Dolphin morbilliviral epizootic. Vet Pathol. 1999;36:171–4. doi: 10.1354/vp.36-2-171. [DOI] [PubMed] [Google Scholar]
- 62.Van Bressem MF, Gaspar R, Aznar FJ. Epidemiology of tattoo skin disease in bottlenose dolphins Tursiops truncatus from the Sado estuary. Portugal Dis Aquat Organ. 2003;56:171–9. doi: 10.3354/dao056171. [DOI] [PubMed] [Google Scholar]
- 63.Gurlek A, Pittelkow MR, Kumar R. Modulation of growth factor/cytokine synthesis and signaling by 1alpha, 25-dihydroxyvitamin D(3): implications in cell growth and differentiation. Endocr Rev. 2002;23:763–86. doi: 10.1210/er.2001-0044. [DOI] [PubMed] [Google Scholar]
- 64.Lansdown AB. Calcium: a potential central regulator in wound healing in the skin. Wound Repair Regen. 2002;10:271–85. doi: 10.1046/j.1524-475x.2002.10502.x. [DOI] [PubMed] [Google Scholar]
- 65.Dlugosz AA, Yuspa SH. Protein kinase C regulates keratinocyte transglutaminase (TGK) gene expression in cultured primary mouse epidermal kera-tinocytes induced to terminally differentiate by calcium. J Invest Dermatol. 1994;102:409–14. doi: 10.1111/1523-1747.ep12372171. [DOI] [PubMed] [Google Scholar]
- 66.Vicanova J, Boelsma E, Mommaas AM, Kempenaar JA, Forslind B, Pallon J, et al. Normalization of epidermal calcium distribution profile in reconstructed human epidermis is related to improvement of terminal differentiation and stratum corneum barrier formation. J Invest Dermatol. 1998;111:97–106. doi: 10.1046/j.1523-1747.1998.00251.x. [DOI] [PubMed] [Google Scholar]
- 67.Liu Y, Tang MK, Cai DQ, Li M, Wong WM, Chow PH, et al. Cyclin I and p53 are differentially expressed during the terminal differentiation of the postnatal mouse heart. Proteomics. 2007;7:23–32. doi: 10.1002/pmic.200600456. [DOI] [PubMed] [Google Scholar]
- 68.Chan GK, Yen TJ. The mitotic checkpoint: a signaling pathway that allows a single unattached kinetochore to inhibit mitotic exit. Prog Cell Cycle Res. 2003;5:431–9. [PubMed] [Google Scholar]
- 69.Cooper JA, Kiehart DP. Septins may form a ubiquitous family of cytoskeletal filaments. J Cell Biol. 1996;134:1345–8. doi: 10.1083/jcb.134.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weiske J, Huber O. The histidine triad protein Hint1 triggers apoptosis independent of its enzymatic activity. J Biol Chem. 2006;281:27356–6. doi: 10.1074/jbc.M513452200. [DOI] [PubMed] [Google Scholar]
- 71.Wang L, Zhang Y, Li H, Xu Z, Santella RM, Weinstein IB. Hint1 inhibits growth and activator protein-1 activity in human colon cancer cells. Cancer Res. 2007;67:4700–8. doi: 10.1158/0008-5472.CAN-06-4645. [DOI] [PubMed] [Google Scholar]
- 72.Weinstein IB, Li H. Mast cells provide a “HINT” to the function of an exotic nucleotide. Immunity. 2004;20:119–20. doi: 10.1016/s1074-7613(04)00029-9. [DOI] [PubMed] [Google Scholar]
- 73.Geilen CC, Wieprecht M, Orfanos CE. The mitogen-activated protein kinases system (MAP kinase cascade): its role in skin signal transduction. A review J Dermatol Sci. 1996;12:255–62. doi: 10.1016/0923-1811(95)00481-5. [DOI] [PubMed] [Google Scholar]
- 74.Argos M, Kibriya MG, Parvez F, Jasmine F, Rakibuz-Zaman M, Ahsan H. Gene expression profiles in peripheral lymphocytes by arsenic exposure and skin lesion status in a Bangladeshi population. Cancer Epidemiol Biomarkers Prev. 2006;15:1367–75. doi: 10.1158/1055-9965.EPI-06-0106. [DOI] [PubMed] [Google Scholar]
- 75.Paul S, Lombroso PJ. Receptor and nonreceptor protein tyrosine phosphatases in the nervous system. Cell Mol Life Sci. 2003;60:2465–82. doi: 10.1007/s00018-003-3123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Denu JM, Dixon JE. Protein tyrosine phosphatases: mechanisms of catalysis and regulation. Curr Opin Chem Biol. 1998;2:633–41. doi: 10.1016/s1367-5931(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 77.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–8. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 78.Kyogoku C, Langefeld CD, Ortmann WA, et al. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet. 2004;75:504–7. doi: 10.1086/423790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Begovich AB, Carlton VE, Honigberg LA, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–7. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Orozco G, Sanchez E, Gonzalez-Gay MA, Lopez-Nevot MA, Torres B, Caliz R, et al. Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum. 2005;52:219–24. doi: 10.1002/art.20771. [DOI] [PubMed] [Google Scholar]
- 81.Velaga MR, Wilson V, Jennings CE, Owen CJ, Herington S, Donaldson PT, et al. The codon 620 tryptophan allele of the lymphoid tyrosine phosphatase (LYP) gene is a major determinant of Graves’ disease. J Clin Endocrinol Metab. 2004;89:5862–5. doi: 10.1210/jc.2004-1108. [DOI] [PubMed] [Google Scholar]
- 82.Szodoray P, Nakken B, Gaal J, Jonsson R, Szegedi A, Zold E, et al. The complex role of vitamin D in autoimmune diseases. Scand J Immunol. 2008;68:261–9. doi: 10.1111/j.1365-3083.2008.02127.x. [DOI] [PubMed] [Google Scholar]
- 83.Krejsa CM, Schieven GL. Impact of oxidative stress on signal transduction control by phosphotyrosine phosphatases. Environ Health Perspect. 1998;106 (Suppl 5):1179–84. doi: 10.1289/ehp.98106s51179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ravid A, Rubinstein E, Gamady A, Rotem C, Liberman UA, Koren R. Vitamin D inhibits the activation of stress-activated protein kinases by physiological and environmental stresses in keratinocytes. J Endocrinol. 2002;173:525–32. doi: 10.1677/joe.0.1730525. [DOI] [PubMed] [Google Scholar]
- 85.Diker-Cohen T, Koren R, Liberman UA, Ravid A. Vitamin D protects keratinocytes from apoptosis induced by osmotic shock, oxidative stress, and tumor necrosis factor. Ann N Y Acad Sci. 2003;1010:350–3. doi: 10.1196/annals.1299.064. [DOI] [PubMed] [Google Scholar]
- 86.Satyal SH, Chen D, Fox SG, Kramer JM, Morimoto RI. Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev. 1998;12:1962–74. doi: 10.1101/gad.12.13.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hogenesch JB, Chan WK, Jackiw VH, Brown RC, Gu YZ, Pray-Grant M, et al. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J Biol Chem. 1997;272:8581–93. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 88.Geng S, Mezentsev A, Kalachikov S, Raith K, Roop DR, Panteleyev AA. Targeted ablation of Arnt in mouse epidermis results in profound defects in desquamation and epidermal barrier function. J Cell Sci. 2006;119:4901–12. doi: 10.1242/jcs.03282. [DOI] [PubMed] [Google Scholar]
- 89.Takagi S, Tojo H, Tomita S, Sano S, Itami S, Hara M, et al. Alteration of the 4-sphingenine scaffolds of ceramides in keratinocyte-specific Arnt-deficient mice affects skin barrier function. J Clin Invest. 2003;112:1372–82. doi: 10.1172/JCI18513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yarosh DB, Boumakis S, Brown AB, Canning MT, Galvin JW, Both DM, et al. Measurement of UVB-Induced DNA damage and its consequences in models of immunosuppression. Methods. 2002;28:55–62. doi: 10.1016/s1046-2023(02)00209-8. [DOI] [PubMed] [Google Scholar]
- 91.Myers LC, Gustafsson CM, Bushnell DA, Lui M, Erdjument-Bromage H, Tempst P, et al. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–9. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 93.Rachez C, Suldan Z, Ward J, Chang CP, Burakov D, Erdjument-Bromage H, et al. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang J, Li Y, Liu YQ, Long JW, Tian F, Dong J, Shen GX, Tu YT, Tao J. Expression of antiapoptotic protein c-FLIP is upregulated in psoriasis epidermis. Eur J Dermatol. 2009 Jan-Feb;19(1):29–33. doi: 10.1684/ejd.2008.0571. [DOI] [PubMed] [Google Scholar]
- 95.Ozawa M, Aiba S. Immunopathogenesis of psoriasis. Curr Drug Targets Inflamm Allergy. 2004;3:137–44. doi: 10.2174/1568010043343868. [DOI] [PubMed] [Google Scholar]
- 96.Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–11. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 97.Duan H, Dixit VM. RAIDD is a new ‘death’ adaptor molecule. Nature. 1997;385:86–9. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- 98.Ahmad M, Srinivasula SM, Wang L, Talanian RV, Litwack G, Fernandes-Alnemri T, et al. CRADD, a novel human apoptotic adaptor molecule for caspase-2, and FasL/tumor necrosis factor receptor-interacting protein RIP. Cancer Res. 1997;57:615–9. [PubMed] [Google Scholar]
- 99.Moll PR, Sander V, Frischauf AM, Richter K. Expression profiling of vitamin D treated primary human keratinocytes. J Cell Biochem. 2007;100:574–92. doi: 10.1002/jcb.21061. [DOI] [PubMed] [Google Scholar]
- 100.Swami S, Raghavachari N, Muller UR, Bao YP, Feldman D. Vitamin D growth inhibition of breast cancer cells: gene expression patterns assessed by cDNA microarray. Breast Cancer Res Treat. 2003;80:49–62. doi: 10.1023/A:1024487118457. [DOI] [PubMed] [Google Scholar]
- 101.Khanim FL, Gommersall LM, Wood VH, Smith KL, Montalvo L, O’Neill LP, et al. Altered SMRT levels disrupt vitamin D3 receptor signalling in prostate cancer cells. Oncogene. 2004;23:6712–25. doi: 10.1038/sj.onc.1207772. [DOI] [PubMed] [Google Scholar]
- 102.Peehl DM, Shinghal R, Nonn L, Seto E, Krishnan AV, Brooks JD, et al. Molecular activity of 1,25-dihydroxyvitamin D3 in primary cultures of human prostatic epithelial cells revealed by cDNA microarray analysis. J Steroid Biochem Mol Biol. 2004;92:131–41. doi: 10.1016/j.jsbmb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 103.Eelen G, Verlinden L, Van Camp M, Mathieu C, Carmeliet G, Bouillon R, et al. Microarray analysis of 1alpha,25-dihydroxyvitamin D3-treated MC3T3-E1 cells. J Steroid Biochem Mol Biol. 2004;89–90:405–7. doi: 10.1016/j.jsbmb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 104.Hussain-Hakimjee EA, Peng X, Mehta RR, Mehta RG. Growth inhibition of carcinogen-transformed MCF-12F breast epithelial cells and hormone-sensitive BT-474 breast cancer cells by 1alpha-hydroxyvitamin D5. Carcinogenesis. 2006;27:551–9. doi: 10.1093/carcin/bgi231. [DOI] [PubMed] [Google Scholar]