Abstract
Post-translational modification of proteins with ubiquitin regulates a variety of eukaryotic cellular processes. Ubiquitin can be conjugated to substrates either as a single moiety (monoubiquitination) or as isopeptide bond-linked chains (polyubiquitination), creating an array of ubiquitin signals. It has been established that monoubiquitination can serve important functions in many biological processes such as the regulation of gene transcription, protein trafficking, and DNA repair. Surprisingly, little is known about the mechanisms by which monoubiquitin signals are produced in the cell. Here, we discuss the potential cellular strategies for generating monoubiquitinated proteins using a few, relatively well characterized examples of monoubiquitinated proteins. These strategies include coupling ubiquitination to low affinity ubiquitin binding, using monoubiquitination-dedicated E2 conjugating enzymes, and restricting ubiquitin chain elongation. Some of these principles may be applicable to protein modifications involving ubiquitin like proteins (UBLs), which often occur in monomeric form.
Keywords: ubiquitin, monoubiquitination, ubiquitin conjugating enzymes/E2, histone, coupled monoubiquitination, ubiquitin-like proteins, ubiquitin chain, endocytosis
Ubiquitin, a 76 amino acid polypeptide, can be covalently linked to lysine (Lys), or at lesser frequencies to serine, threonine, cysteine and the N-terminal amino group for some substrates 1; 2. Ubiquitination can be classified into two major types termed monoubiquitination and polyubiquitination, respectively, depending on whether a single ubiquitin moiety or a polymerized chain is attached. Polyubiquitination may be further defined based on which Lys residue in the most proximal ubiquitin the more distal ubiquitin is attached to 3. Distinct forms of ubiquitination encode different signals, which are deciphered by a collection of ubiquitin binding domain (UBD)-containing proteins in the cell. The interplay between diverse ubiquitin signals and the corresponding receptors can change subcellular localization, stability, and protein-protein interactions for the modified polypeptides, and thus generate distinct cellular functions. For example, Lys48-linked ubiquitin chains can target proteins to the proteasome for degradation 4, whereas monoubiquitination often confers non-degradative activities when the substrate is involved in the regulation of gene transcription, the DNA damage response, and protein trafficking 5; 6.
Ubiquitination generally requires three types of enzymes: an activating enzyme (E1), a conjugating enzyme (E2), and an ubiquitin ligase (E3). The human genome encodes two E1s, approximately 40 E2s, and 600–1000 E3s 3; 7. The E1s activate ubiquitin by covalently linking it to their active site cysteine 8. Subsequently, ubiquitin is transferred to the catalytic cysteine in an E2 enzyme. Ubiquitin can be further relayed to the catalytic site of a HECT (Homologous to E6-AP Carboxyl Terminus) domain-containing E3 before being conjugated to substrate 9, or transferred directly from E2 to substrate with the assistance of a RING (Really Interesting New Gene) domain-containing E3 7. The interactions between these enzymes and the chemistry underlying ubiquitin conjugation have been subject to many reviews 1; 3; 8. However, the mechanisms that permit synthesis of different ubiquitin modifiers, particularly mono-ubiquitin signals, have not been surveyed. This is somewhat surprising considering that almost every functional aspect of monoubiquitination has been covered by one or more recent reviews 5; 6; 10–14. We therefore direct our attention specifically to the potential cellular strategies that allow mono-ubiquitinated proteins to be generated. Our goal is to summarize our current understanding of this topic, which may provide a framework for further dissecting the mechanisms of protein monoubiquitination.
Monoubiquitination of endocytic proteins: coupling ubiquitination to ubiquitin binding
A widespread function of monoubiquitination is to direct modified proteins to specific subcellular localizations, as demonstrated for many endocytosed proteins 11. Intriguingly, many endocytic regulators themselves undergo monoubiquitination 15. These endocytic regulators often use their ubiquitin binding domains to couple substrate recruitment to the ubiquitination reaction, defining a mechanism termed coupled monoubiquitination 16; 17.
E3-dependent coupled ubiquitination
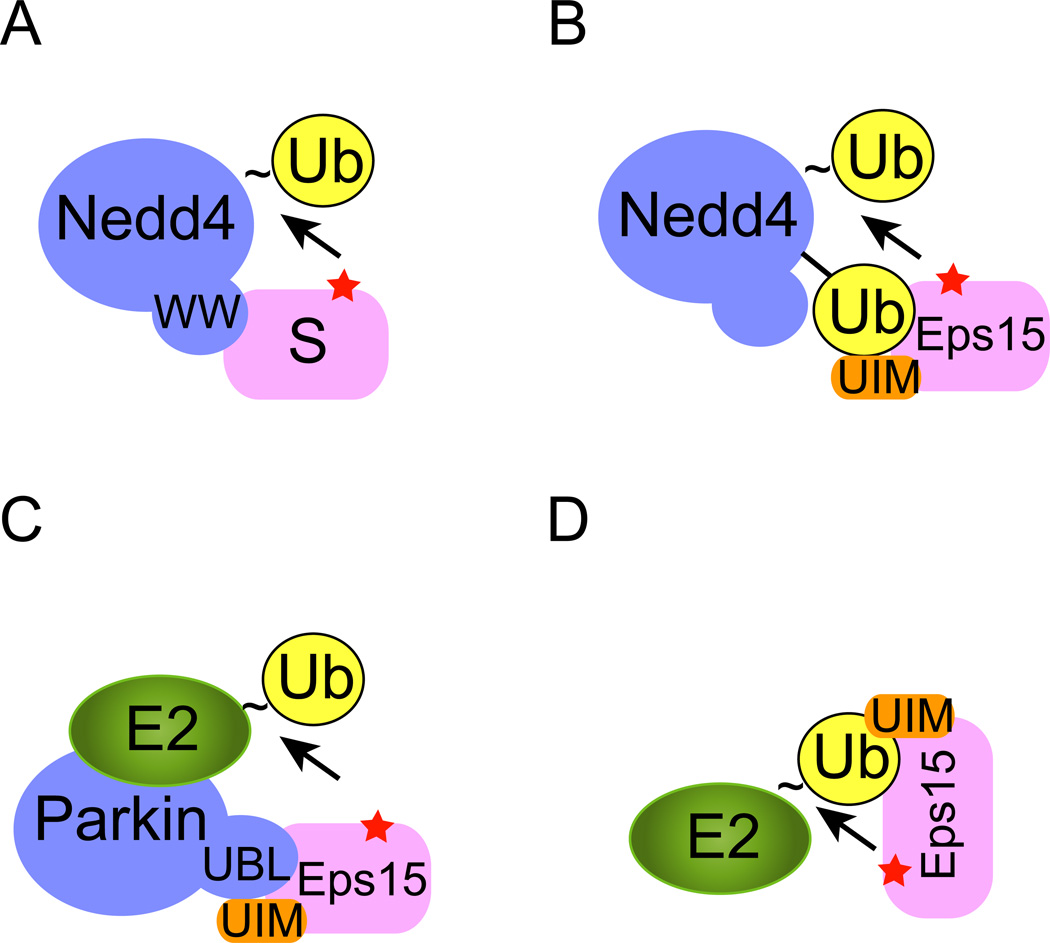
One of the best studied monoubiquitinated endocytic regulators is the epidermal growth factor (EGF) receptor pathway substrate clone 15 (Eps15). Eps15 is monoubiquitinated in cells stimulated with EGF 18. In vitro ubiquitination assays identified the carboxyl terminus of Eps15 as an essential element for ubiquitination 15; 19. This region contains two ubiquitin interacting motifs (UIM1 and UIM2). It turns out that monoubiquitin binding by UIM2 can be used to recruit the E3 ubiquitin ligase Nedd4, which leads to Eps15 monoubiquitination 17.
Nedd4 is the mammalian homologue of the yeast ubiquitin ligase Rsp5 that is responsible for ubiquitination of many endocytic proteins. Nedd4/Rsp5 belongs to the family of the HECT domain-containing E3s, most of which carry a tryptophan-tryptophan motif (WW) 20. This motif can interact with substrates bearing a PPxY (x, any amino acid) sequence, which often leads to polyubiquitination of the associated substrate 21; 22 (Fig. 1A). Eps15 does not contain a PPxY motif. Instead, it uses its UIM2 to recognize an ubiquitin moiety attached to Nedd4 as a result of Nedd4 auto-ubiquitination. This interaction must occur in a special configuration to allow a Lys residue of Eps15, positioned in proximity to the Nedd4 active site, to receive ubiquitin from Nedd4 (Fig. 1B).
Figure 1. Monoubiquitination of endocytic proteins.
To initiate ubiquitination, a ubiquitin acceptor (usually a Lys residue) in the substrate (red star) needs to be positioned in proximity to an ubiquitin carrier that contains a thioester-linked (indicated by “~”) ubiquitin at its active site. The ubiquitin carrier can be either an E2 or a HECT domain E3 such as Nedd4. A, The HECT domain E3 Nedd4 contains several WW domains that directly recognize a PPXY motif in the substrate (S), leading to the transfer of ubiquitin from Nedd4 to the substrate. B, C, D, Coupled monoubiquitination. B, Substrate recruitment is mediated by interactions of a UBD domain in the substrate (e.g. UIM in Eps15) with a ubiquitin moiety attached to Nedd4. C, The Parkin E3 ligase uses its UBL domain to recruit Eps15. D, An E3 independent mechanism of monoubiquitination. In this case, the UIM domain in Eps15 directly binds the active site-linked ubiquitin on an E2 to mediate ubiquitin transfer in the absence of an E3. Note that a color version of the figure is available online.
Why does the interaction of Eps15 with Nedd4 only produce a monoubiquitinated product, whereas Nedd4 clearly has the capacity to polyubiquitinate substrates as demonstrated for certain PPxY motif-containing proteins? One possible explanation is the self-inactivation model in which monoubiquitinated Eps15 folds back, allowing the conjugated ubiquitin moiety to bind its own UIM motif. This could prevent further rounds of association of Nedd4 with Eps15 that is required for ubiquitin chain elongation Intramolecular interactions between a UIM and ubiquitin have been demonstrated for numerous proteins, and have been proposed as a regulatory mechanism that governs the interactions of UIM domain-containing ubiquitin receptors with monoubiquitinated client proteins 23–25. However, existing experimental evidence is not fully compatible with this model. Specifically, when a mutant ubiquitin defective in UIM binding was used, Eps15 still undergoes monoubiquitination by ubiquitin-modified Nedd4 17.
In an alternative model, Woelk and colleagues proposed that the difference in substrate interaction may direct the catalytic activity of Nedd4 towards either mono- or polyubiquitination. UIMs usually bind ubiquitin with low affinities (kD in the range of 100–400 µM) and fast dissociation kinetics 16. Thus, monoubiquitinated Eps15 is expected to dissociate from Nedd4 before a second round of ubiquitin transfer occurs. This is exemplified by the observation that Nedd4 overexpression in vivo leads to increased monoubiquitination of Eps15 in the absence of any stable interactions between Eps15 and Nedd4 15. By contrast, the Nedd4/Rsp5p WW motifs can bind substrates carrying PPxY motif with higher affinities. For example, Nedd4/Rsp5p contains 4 WW motifs and it was demonstrated that the third WW motif in Nedd4 can interact with a single PPxY motif with a dissociation constant of ~4µM 26. In addition, some Nedd4 substrates contain more than one PPxY motif, which may cooperate with each other to further increase the binding affinity. High affinity interaction of E3 with substrates presumably allows the substrate to receive multiple ubiquitin moieties in a single encounter with Nedd4 and thus should increase the processivity of Nedd4-mediated ubiquitination towards certain PPxY-containing substrates. However, experimental data that correlate the Nedd4-substrate binding affinity with substrate ubiquitination status are required to prove this model.
In addition to Nedd4, the E3 ligase Parkin can also monoubiquitinate Eps15. The underlying mechanism is reminiscent of Nedd4-mediated coupled ubiquitination, but with an interesting twist. Parkin is a RING E3 ubiquitin ligase 27 and mutations in Parkin have been linked to the onset of Parkinson’s disease 28. The amino-terminus of Parkin carries an ubiquitin like domain (UBL) that can bind to some ubiquitin receptors in a similar fashion as ubiquitin. Consistent with this, Parkin can interact with Eps15 via its UBL domain and the UIMs in Eps15. Importantly, this interaction is analogous to the ubiquitin-UIM interaction, as it brings Eps15 close to a Parkin-associated E2, leading to Eps15 monoubiquitination (Fig. 1C) 25. Importantly, in the case of Parkin-mediated mono-ubiquitination, it has been demonstrated that once Eps15 is monoubiquitinated, the UIM motif can interact with the ubiquitin moiety attached to Eps15, which converts Eps15 into a closed conformation that is unable to bind Parkin (BOX1) 25. This restricts the ubiquitination reaction to only attach one ubiquitin to Eps15.
BOX 1 Potential strategies for generating monoubiquitin signals.
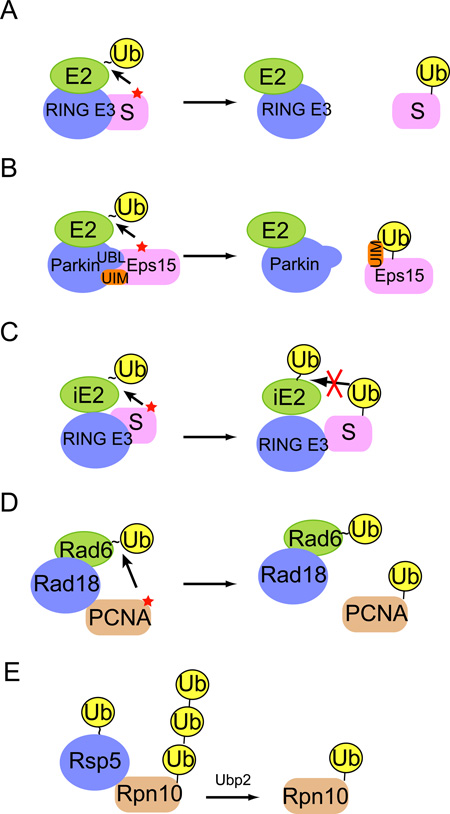
To make monoubiquitinated proteins, a ubiquitination reaction needs to be terminated after the conjugation of the first ubiquitin moiety. The schematics depict several potential mechanisms that may allow a monoubiquitinated protein to evade chain extension. A, The substrate dissociation model. If a substrate has a fast dissociation rate for E3, the substrate will be released from E3 before another E2-ubiquitin thioester joins the reaction for chain extension. The rebinding of monoubiquitinated substrate to the E3 may be unfavorable due to competitive binding of non-ubiquitinated substrates to the E3. B, The self-inactivation model. For coupled ubiquitination, rebinding of Eps15 to the E3 Parkin is inhibited by an intramolecular interaction between the Eps15 UIM and the attached monoubiquitin moiety. C, The E2 incompetent model. Ubiquitin chain formation often involves an initiation E2 (iE2) and an elongating E2 (eE2). Chain elongation usually requires interactions between the pre-attached ubiquitin molecule and an E2-ubiquitin thioester. Monoubiquitination may be achieved by using only iE2, which is intrinsically defective in E2-ubiquitin interactions and therefore lacks chain elongating activity. D, E, The chain restricting model. Certain E2s with chain-forming capability may fail to produce ubiquitin chains due to the presence of chain restricting factors. These chain-restricting regulators may inhibit chain extension by abolishing E2-ubiquitin interactions or E2-E2 cooperation. D, For PCNA, the E3 Rad18 inhibits ubiquitin binding to the backside of Rad6, which restricts chain elongation. E, Chain restriction can also be achieved by engaging a deubiquitinase that trims assembled ubiquitin chain to produce monoubiquitinated proteins. Note that a color version of the figure is available online.
E3-Independent coupled monoubiquitination
In essence, coupled ubiquitination employs weak interactions between a UBD and ubiquitin (or a UBL domain), which recruit a UBD domain-containing substrate to an ubiquitin carrier (E2 or HECT domain E3). An extension of this concept was established recently by Hoeller and colleagues who demonstrated that a UBD domain in the substrate can directly interact with ubiquitin attached to the E2 active site, and thus position the substrate in proximity to E2 for ubiquitin transfer independent of E3s 29 (Fig. 1D). In vitro ubiquitination studies using recombinant proteins carrying various UBDs demonstrated that these UBD-containing substrates can all be monoubiquitinated in a manner dependent on the UBD-ubiquitin interactions. Interestingly, only certain E2s can ubiquitinate a given substrate, indicating that specific E2 properties may be required for productive interactions between a UBD-containing substrate and an E2-ubiquitin thioester. The physiological relevance of these findings in endocytic regulation awaits further characterization, but existing evidence suggest that this mechanism may also be used to monoubiquitinate UBD-containing proteins in other cellular processes 24.
Monoubiquitination of Histone: the choice of enzyme matters
The chromatin structural protein histone H2A was the first protein identified to undergo ubiquitination. In 1977, Goldknopf et al. reported that a fraction of histone H2A exists in the cell as a fusion protein containing an isopeptide bond-linked non-histone portion 30. The identity of this non-histone adduct was later demonstrated to be ubiquitin 31, a ubiquitously expressed eukaryotic polypeptide initially discovered as a lymphocyte differentiation regulator 32. In 1980, Ciechanover and colleagues re-discovered ubiquitin as a post-translational modifier (they named it ATP-dependent proteolysis factor 1 or APF-1) that can be attached to proteins as polymerized chains to regulate protein stability 33–35. However, ubiquitination does not seem to affect the stability of histone H2A because H2A is primarily modified with a single ubiquitin moiety on Lys119. Although the function of histone ubiquitination was unclear at the time of its discovery, the observation that the level of ubiquitinated histone is modulated during cell division, DNA repair and replication had linked its putative functions to transcriptional regulation and the DNA damage response, functions that are now widely appreciated 13; 36.
In vitro ubiquitination of purified histone H2A and H2B (thereafter referred as histones) was initially established using a crude chromatographic protein preparation containing an E1 and a mixture of E2s, which showed that histone ubiquitination can occur independently of E3 37. Jentsch and colleagues later established Rad6 as the E2 that can catalyze histone monoubiquitination in vitro 38. It was subsequently found that yeast strains deficient in Rad6 have reduced levels of ubiquitinated histone H2B, establishing Rad6 as the major E2 for H2B ubiquitination in vivo 39. Although several other studies identified additional E2s that can catalyze histone ubiquitination either in the absence or presence of an E3 ligase in vitro 13; 36; 40–42, these E2s are not absolutely essential for histone ubiquitination in the cell, demonstrating some degree of E2 specificity for histone ubiquitination in the cellular context 37; 38; 43. To date, the physiologically relevant E2 for histone H2A ubiquitination is still unclear.
E3 independent histone ubiquitination does not involve any ubiquitin binding domains. Instead, the carboxyl-terminus of Rad6 and Cdc34, two E2s that ubiquitinate histones in vitro, has a stretch of acidic aspartate residues 44. This sequence is absent from E2s that fail to ubiquitinate histones such as UBC1. Because removal of this sequence from Rad6 and Cdc34 abrogates their ubiquitinating activity towards histones 41; 44; 45, and transfer of this acidic sequence to UBC1 allows the latter to gain histone monoubiquitinating activity 46, the acidic tail of these E2s may interact directly with a basic region in histones to cause their ubiquitination. This can obviate the requirement for E3 because the major function of E3s is to bring substrates into proximity of an ubiquitin-thioester (attached to the active site cysteine of an E2 or a HECT domain E3). Interestingly, although the E2 acidic sequence is critical for histone ubiquitination in vitro, it is not conserved in the higher eukaryotic Rad6 homologues. Higher eukaryotic cells may employ additional factors such as E3 ubiquitin ligases to facilitate histone recruitment to E2s 47–51.
Why do histones only get monoubiquitinated in the cell? It seems that the choice of the E2-E3 partners may be a crucial factor. Although all E2s contain a core ubiquitin conjugating (UBC) domain that is highly similar in structure, the UBC cores from different E2s often contain distinct surface properties, which allow E2s to form distinct interactions with ubiquitin or cofactors (see below) 52. In particular, subtle variations in the E3 binding site allow different E2s to team up with distinct E3 ligases 3. Consequently, some E2-E3 pairs particularly excel at initiating a ubiquitination reaction (transferring ubiquitin to substrate), whereas others are specialized in elongating ubiquitin chains (catalyzing ubiquitin-ubiquitin ligation) 53. Rad6, the physiological E2 for H2B 39, seems to belong to the cohort that prefers to add only a single ubiquitin to substrates when acting with certain E3 ligases (see below).
Monoubiquitination specific conjugating systems
Recent studies have established polyubiquitination as a two-step reaction, in which a single ubiquitin moiety is first transferred from an ubiquitin carrier to the substrate. Next, in an elongation step, additional ubiquitin molecules are conjugated to the pre-existing ubiquitin molecule to form chains. Whether or not a ubiquitin molecule is conjugated to a Lys residue in the substrate or in a pre-existing ubiquitin molecule appears to be determined by the selection of E2s because E2s with distinct and dedicated roles in ubiquitination initiation and chain extension have been demonstrated 53. Hence, monoubiquitination may be achieved by only engaging an initiating E2 (Box 1). Examples of monoubiquitination carried out by initiation specific E2s include the above mentioned Rad6, and UBE2T. The former was known to monoubiquitinate histones 38; 39 and the DNA replication licensing factor PCNA 54, whereas the latter cooperates with the cognate E3 FANCL to conjugate only one ubiquitin moiety to a specific Lys residue in FANCD2 55.
Why are some E2s active in monoubiquitinating substrates, but are essentially inert in chain elongation? The answer to this question comes from comparing the chemistry of the ubiquitination initiation and elongation reactions, which appear to involve different sets of E2 interactions. The initiation reaction requires interactions between elements near a Lys acceptor in the substrate and an E2-ubiquitin thioester. Although some E2s can interact with specific sequence motifs in proximity to a Lys acceptor in the substrate 56; 57, most substrates do not seem to contain a consensus sequence for E2 recognition. Therefore, substrate-E2 interactions must be labile and relatively non-specific in nature. In principle, a stochastic collision between an E2 and a Lys-containing segment in substrate that has been positioned in proximity to an E2 active site should be sufficient to initiate ubiquitination. In contrast, chain elongation requires aligning a specific acceptor Lys in the substrate-bound ubiquitin to an E2 active site. Thus, the steric constraints for ubiquitin chain elongation likely distinguish between E2s that can add a single, versus multiple, ubiquitins to a given substrate.
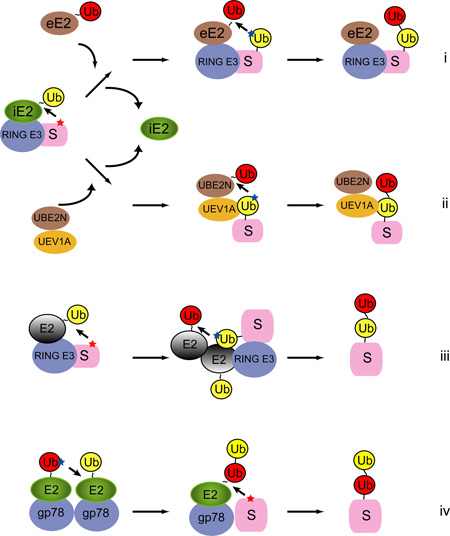
Ubiquitin chain elongation can be achieved by several mechanisms. First, direct interactions between residues surrounding an acceptor Lys in ubiquitin and an E2 active site may bring a specific Lys in ubiquitin in proximity to the E2 active site, as was initially proposed for Cdc34 58. A well illustrated example is the yeast chain forming E2 UBC1 59. Three residues (Thr84, Gln122 and Ala124) near the UBC1 active site appear to communicate with a critical Tyr residue in proximity to Lys48 in the acceptor ubiquitin, allowing UBC1 to specifically catalyze Lys48-linked ubiquitination 59. These residues are not conserved in the monoubiquitinating E2 UBC4, a member of the UBE2D family. As expected, substitution of these residues in UBC1 to the corresponding amino acids in UBC4 eliminates the chain forming capability of UBC1, converting it to a monoubiquitinating E2 59. Second, a ubiquitin binding protein capable of interacting with an E2 may serve as the matchmaker, bringing the acceptor ubiquitin close to the donor E2-ubiquitin thioester, as demonstrated by UBE2N-UEV1A-mediated assembly of Lys63-linked ubiquitin chain (BOX 2) 60; 61. In this regard, failure to evolve a strategy to effectively engage an acceptor ubiquitin might be the cause of incompetence in chain formation for some E2s.
BOX 2 E2-E2 cooperation during chain formation.
To generate polyubiquitin chains, E2s often need to cooperate with one another. The schematics depict several possible ways of E2 cooperation during ubiquitin chain assembly. It is apparent that E2-E2 cooperation needs to be avoided when monoubiquitinated proteins are the favored product. For simplicity, the schemes only show the very first step in chain extension, that is the formation of a di-ubiquitin molecule. i, After conjugation of the first ubiquitin to the substrate, an initiation E2 (iE2) dissociates from E3. An elongation E2 (eE2) carrying a ubiquitin in its active site then binds the E3-substrate complex. The elongation E2 can be a different E2 or in some cases the same as iE2. eE2 is capable of engaging a specific Lys (blue star) in the pre-attached ubiquitin (yellow) to form di-ubiquitin on the substrate. ii, The initiation step is the same as shown in (i). The elongation E2 in this case is a dimeric E2 complex. The UBE2N carries a ubiquitin in its active site, whereas UEV1A can interact with the substrate-attached ubiquitin non-covalently. This interaction positions Lys63 of ubiquitin in proximity to the active site of UBE2N for receiving the next ubiquitin. iii, Some ubiquitin E2s as well as the SUMO E2 contains a backside ubiquitin or SUMO binding site. Although un proven, these E2s may be able to form a homo-oligomer analogous to the UBE2N-UEV1A complex. The back-side ubiquitin binding by one E2 may help to orient a specific Lys in the substrate-attached ubiquitin for attacking the ubiquitin thioester on the other E2. iv, The endoplasmic reticulum-associated E3 gp78 forms homo-oligomer in cells. It contains a high affinity binding site for its cognate E2 UBE2G2, which facilitates E2-E2 self-interaction, leading to polymerization of ubiquitin chain on the active site of UBE2G2. En bloc chain transfer then allows a substrate to receive multiple ubiquitin moieties in a single encounter with the gp78-associated E2 78; 79. Note that a color version of the figure is available online.
In addition to residues close to the active site, many E2s have evolved other ways to communicate with ubiquitin, which allow them to gain chain forming capabilities. Three additional modes of E2-ubiquitin interactions have been reported. First, many chain forming-E2s such as UBE2S and UBE2G2 use a helix segment to interact with a patch of hydrophobic residues centered around Ile44 in the donor ubiquitin that has been attached to their active sites. This interaction appears to restrict the flexibility of donor ubiquitin, which may open a ‘channel’ through which an acceptor ubiquitin can attack the donor ubiquitin-E2 thioester complex 62. UBE2S also interacts with a motif termed the TEK box in the acceptor ubiquitin, which further enhances its chain forming activity 62. The E2 UbcH5c also contains a non-covalent ubiquitin binding site, but the binding site is distal from the E2 active site. This so-called backside ubiquitin binding has been shown to be required for polyubiquitination of BRCA1, a catalytic subunit of the BRCA1-BARD1 RING E3 complex 63. Hence, non-covalent E2-ubiquitin interactions can profoundly influence the chain forming capability of E2. Monoubiquitination-dedicated E2 may simply result from failure to gain ubiquitin-interacting activities or additional E2 interactions that suppress ubiquitin binding (see below).
Finally, a unique ubiquitin activating and conjugating enzyme was recently reported, which can monoubiquitinate the transcription factor Pax3 to induce its degradation by the proteasome during post-natal myogenesis 64. This ubiquitin conjugating enzyme, termed as Taf1, is a subunit/co-activator of the transcription factor TFIID complex (formerly called TafII250) 65. Taf1 is unusual in that it contains both ubiquitin-activating (E1-like) and ubiquitin-conjugating (E2-like) activities in a single polypeptide 66. It was proposed that direct interaction of substrates with Taf1 may be sufficient for monoubiquitination, but the underlying mechanism is unclear 65; 66.
Restricted ubiquitin chain formation generates monoubiquitin signals
Although E2s with dedicated roles in ubiquitin chain initiation and elongation have been reported, the boundary between the two classes is not fixed. In fact, the E2 ubiquitin conjugating activity can be modulated by various interactions, which may switch a chain-forming E2 to a monoubiquitinating E2 and vice versa. For example, the UBE2D family member UBC4 in yeast only monoubiquitinates cyclin A in conjunction with the ubiquitin ligase APC 67. Likewise, UBE2D3 monoubiquitinates IκB when acting with SCF 68. Intriguingly, UBE2D3 can form polyubiquitin chains on BRCA1 63, but the UBE2D3-BRCA1 complex predominantly catalyzes monoubiquitination of the estrogen receptor 69. Thus, conjugating partner and substrate choice can influence E2 activity and determine if a single ubiquitin is added or extended into a chain.
Another example of altered E2 activity is seen in Rad6. Rad6 has a backside ubiquitin binding site similar to the one described for UBE2D3 70. However, when Rad6 acts in conjunction with the E3 ligase Rad18, Rad18 can compete with ubiquitin for binding to Rad6. This inhibits the chain forming activity of Rad6 and thus allows only monoubiquitination to occur on PCNA 71. In light of this finding, we speculate that the interaction of certain E3s with UBE2D3 may similarly inhibit backside ubiquitin binding, and therefore restrict this E2 to only produce monoubiquitinated proteins. However, this model cannot explain why the UBE2D3-BRCA1 complex forms chains on BRCA1, but prefers to only monoubiquitinate the estrogen receptor. Further experiments are required to characterize the unknown factor(s) that influences the catalytic activity of a given E2-E3 pair towards either mono- or polyubiquitination.
E2-E3 interactions can impact the E2 chain-forming capability in several other ways. For example, RING E3-dependent polyubiquitination usually requires multiple rounds of E2-E3 interactions because an E2 generally cannot bind to E1 and E3 simultaneously 72; 73. Thus, it is conceivable that a chain-forming E2 needs to have evolved a mechanism to re-associate with E3 after each round of ubiquitin transfer, and this E3-E2 reunification should occur before the substrate leaves the complex. Indeed, the chain-forming E2 Cdc34 forms a high affinity yet dynamic interaction with its cognate E3 SCF, which is required for polyubiquitination of the substrate Sic1 58; 74. Interestingly, the chain-forming activity of Cdc34 is drastically diminished when in complex with the E3 RAG1 75. The interaction between Cdc34 and RAG1 appears to be mediated by residues not present in the conventional E2-E3 interface 75; 76. It is possible that this unusual mode of interaction does not favor E2-E3 re-association, which could explain why the ubiquitinated product of RAG1-Cdc34 is mostly a monoubiquitinated species.
The concentration of a particular E3 ligase relative to a given substrate can also dictate whether the substrate receives a single ubiquitin or a polyubiquitin chain. This was demonstrated for ubiquitination of p53 by the oncogenic E3 ligase Mdm2. Lower levels of Mdm2 activity induce p53 monoubiquitination, which does not affect the stability of p53, but results in the export of p53 from the nucleus. In contrast, high levels of Mdm2 activity, which induces polyubiquitination and degradation of p53 in the nucleus 77. What remains unclear mechanistically is how fluctuations in E3 concentrations can influence the mono- vs. polyubiquitination reaction. One possibility is that low concentrations of E3 favor substrate dissociation, which could terminate chain extension.
Another mechanism that cells use to restrict chain elongation, thus favoring monoubiquitination of substrates, is to avoid E2-E2 cooperation (BOX2). E2 cooperation can be achieved by E2 homo-oligomerization. E3-dependent E2 oligomerization has been reported for UBE2G2 and UBE2N, which is required for the polyubiquitinating activities of these E2s 78–80. In addition, UBE2D3 can be assembled into a large oligomeric complex independently of E3 by simultaneously engaging two ubiquitin molecules, one bound to the E2 active site and the other to the E2 backside 63; 81. This oligomeric assembly is required for polyubiquitination of BRCA1 63. Since many monoubiquitinating E2s do not contain this backside ubiquitin binding activity 53, lack of E2 oligomerization may contribute to the monoubiquitination preference of these E2s. Moreover, many polyubiquitination reactions require cooperation between two distinct E2s, one for chain initiation and one for elongation (BOX 2) 53; 56; 67; 68; 82; 83. Currently, there is no evidence suggesting that different E2s can cooperate in a hetero-oligomeric complex. In fact, the chain initiation and elongation reactions are independent of each other in cases examined to date. This is best illustrated by a recent study that showed that PCNA monoubiquitinated by Rad6 can serve as a substrate for the elongating E2 complex UBC13-MMS2, which assembles a Lys63-linked ubiquitin chain on PCNA 84. This finding raises an important question: How can certain monoubiquitinated substrates such as histones evade a chain elongating E2 to remain monoubiquitinated in the cell?
Finally, cells can make use of additional protein cofactors to restrict chain elongation, resulting in monoubiquitinated proteins. One well known example is that the presence of a ubiquitin binding domain in a polyubiquitination reaction can significantly reduce the processivity of chain formation in vitro 85; 86. The binding of a UBA domain to ubiquitinated protein might inhibit the previously mentioned ubiquitin-E2 interactions to terminate chain extension. Another class of chain restricting factors is deubiquitinating enzyme (also termed deubiquitinase), which usually acts to completely disassemble polyubiquitin chains. In this regard, it is intriguing to learn that a deubiquitinase has recently been implicated in producing monoubiquitinated protein 87. In this case, a ubiquitin chain is first assembled on the proteasome subunit Rpn10 by Rsp5, which is subsequently trimmed by the deubiquitinase (DUB) Ubp2 to yield monoubiquitinated Rpn10.
Modification with ubiquitin like proteins
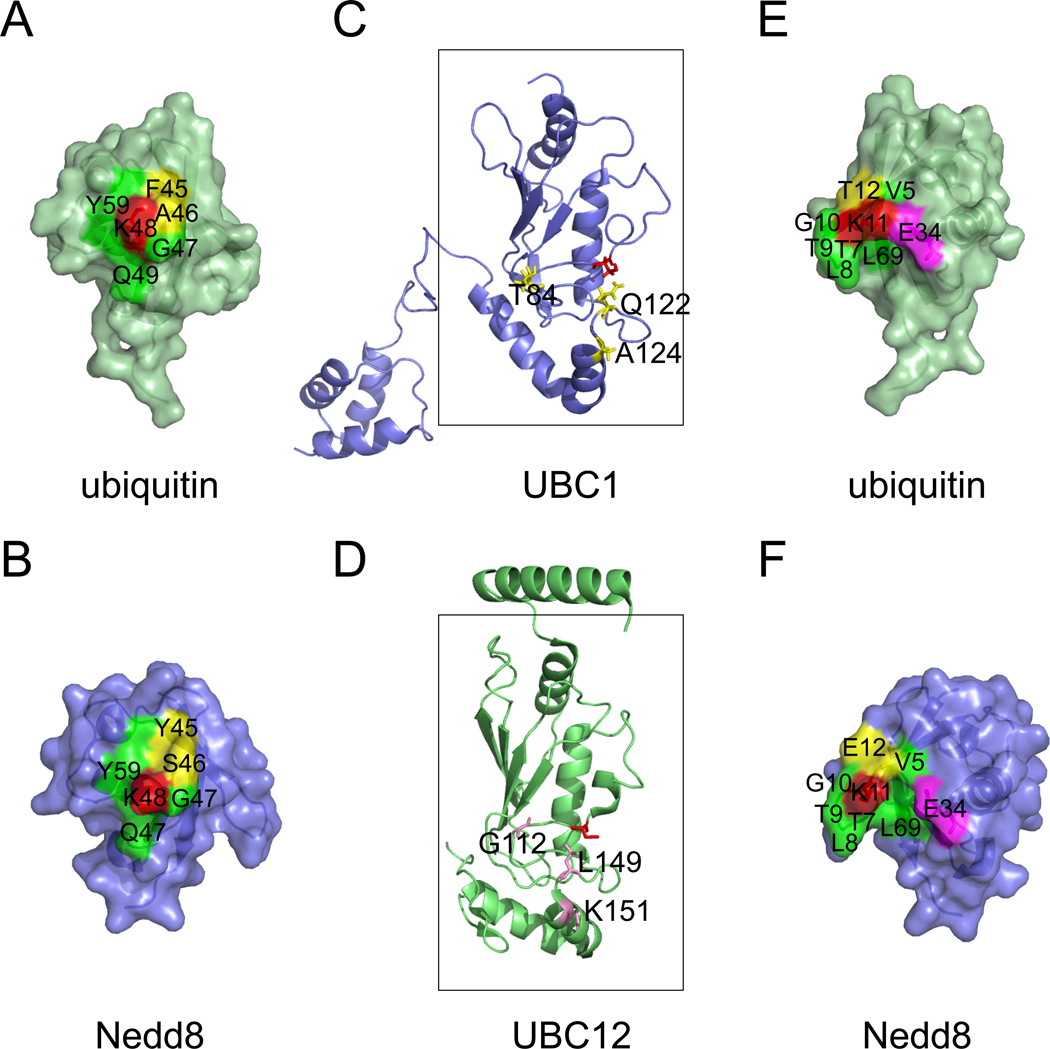
Ubiquitin like proteins (UBLs) comprise a family of small proteins that resemble ubiquitin in structure despite the lack of significant sequence similarity. Like ubiquitin, UBLs can be covalently linked to substrates by the action of an analogous E1-E2-E3 enzyme cascade. Interestingly, most UBL modifications take place in monomeric form. The lack of processivity for UBL conjugation can be attributed to several factors. First, UBLs generally do not contain all the Lys residues at positions corresponding to those in ubiquitin. Second, even when a Lys residue equivalent to that in ubiquitin is present, the environment in which the UBL Lys is located is not completely identical to that for the corresponding Lys in ubiquitin (Fig. 2 A, B, E, F). Moreover, critical residues in proximity to the E2 active sites are often variable between UBL E2s and the chain-forming ubiquitin E2s (Fig. 2C, D). These distinctions collectively may prevent the Lys residues in UBLs from attacking a corresponding E2-thioester complex, causing incompetence in chain formation.
Figure 2. A structural comparison of the ubiquitin and the Nedd8 conjugation systems.
A, B, The environment surrounding Lys48 (red) of ubiquitin and the corresponding Lys in Nedd8 is not completely identical. Note that the yellow labeled residues are not conserved between ubiquitin and Nedd8. C, D, A comparison of UBC1 and UBC12. The boxes indicate the UBC core. Active site cysteines are shown in red. Residues critical for UBC1-dependent Lys48-linked polyubiquitination are shown in yellow. Note that the corresponding residues in UBC12 are different (labeled in magenta). E, F, The environment surrounding Lys11 of ubiquitin and the corresponding residue in Nedd8. Although most residues (labeled in green) in proximity to Lys11 (red) are conserved between ubiquitin and Nedd8, the position of the Glu34 side chain is distinct. In ubiquitin, Glu34 makes close contact with Lys11 to suppress its pKa for nucleophilic attack on an E2-ubiquitin thioester 62. Note that a 7.2A gap exists between Glu34 and Lys11 in Nedd8. The following structures were used for the analyses (Protein databank code (PBD) 1UBQ, ubiquitin 92; 1NDD, Nedd8 93; 1TTE, UBC1 94; 3O2U, UBC12 95). Note that a color version of the figure is available online.
Interestingly, among UBLs, the small ubiquitin-like modifier SUMO is unique as it can form chains on substrates. The human genome encodes three SUMO homologues that can be conjugated to substrates, but chains are preferentially formed with SUMO2 and SUMO3 because these chains are usually formed via a Lys residue that is only present at an N-terminal extension in these SUMO variants. Enzymes involved in sumoylation in human include one E1 hetero-complex comprising AOS1 and UBA2, one E2 UBE2I, and a few E3s 88. Initial in vitro sumoylation analyses revealed an E3-independent mechanism reminiscent of the Rad6-mediated histone ubiquitination. Specifically, interactions between UBE2I and a SUMO consensus motif (SCM) [I/V/L]–K–X–[D/E] (X is any amino acid) present in a SUMO substrate can dock a SUMO-charged E2 to the substrate 88; 89, positioning the Lys acceptor within the SCM for attacking the E2-SUMO thioester 90.
How can UBE2I not only initiate sumoylation but also catalyze chain elongation? Biochemical and structural studies demonstrated the existence of a SUMO binding site on the backside of UBE2I similar to the ubiquitin binding site on UBE2D3. This backside E2-SUMO interaction, when disrupted, abolishes SUMO chain formation 91. Although the precise mechanism by which this SUMO-E2 interaction governs chain formation is unclear, it has been speculated that such an interaction, if it occurs in the context of a dimeric UBE2I assembly, may bring an acceptor SUMO in proximity to a donor UBE2I-SUMO complex to facilitate SUMO-SUMO ligation analogous to UBE2N-UEV1-mediated ubiquitin chain formation (BOX 2). The E2s for other UBLs do not seem to contain this backside UBL binding activity, which would explain their preference for producing mono-UBL-modified proteins.
Perspectives
In summary, the cell has acquired a variety of strategies to produce ubiquitinated proteins. In our view, the most important factor that influences mono- versus polyubiquitination decisions is the regulation of E2 activity. Specific strategies have been evolved that allow certain E2s to form ubiquitin chains. These include E2-ubiquitin interaction, E2-E2 cooperation, and dynamic E2-E3 interactions. Meanwhile, when an E2 fails to acquire chain forming capability or their chain forming activity is negatively influenced by cofactors or deubiquitinating enzymes, it becomes a monoubiquitination-specific E2. The human genome contains ~40 E2s. It is unclear how many of these E2s are capable of producing monoubiquitinated proteins in cells. A systematic survey of E2 interactions (including E2-ubiquitin, E2-E2, and E2-E3 interactions) and the resulting ubiquitination products would provide a comprehensive view on this issue. Another major outstanding question is how a monoubiquitinated protein can evade a chain elongating E2 to remain monoubiquitinated in the cell. The key to this question may lie in how the E2 functions are regulated spatially and temporally in the cell, which may limit their access to certain monoubiquitinated proteins. The scope of deubiquitination-mediated chain restriction in monoubiquitination should also be further explored, which may reveal additional substrates that use this mechanism to achieve monoubiquitination. A key question in this model is to understand the mechanism that allows the substrate-conjugated ubiquitin to escape deubiquitinase-mediated cleavage. Investigations along these lines will no doubt give us important insights into the mechanisms underlying this crucial protein modification.
Acknowledgements
The authors thank members in the lab for stimulating discussions, and Michael Krause (NIDDK) for critical reading of the manuscript.
The authors’ research is supported by the NIH intramural AIDS Targeted Antiviral Program (IATAP) and by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
Footnotes
Declaration of Interest
The authors declare no competing financial interests.
REFERENCES
- 1.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1–3):55. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65(15):2397. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10(11):755. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243(4898):1576. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 5.Sigismund S, Polo S, Di Fiore PP. Signaling through monoubiquitination. Curr Top Microbiol Immunol. 2004;286:149. doi: 10.1007/978-3-540-69494-6_6. [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315(5809):201. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 7.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 8.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10(5):319. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10(6):398. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 10.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin binding domains-from structures to functions. Nat Rev Mol Cell Biol. 2009 doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haglund K, Di Fiore PP, Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci. 2003;28(11):598. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Hurley JH, Stenmark H. Molecular mechanisms of ubiquitin-dependent membrane traffic. Annu Rev Biophys. 2011;40:119. doi: 10.1146/annurev-biophys-042910-155404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29(6):653. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 14.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2(3):195. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 15.Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416(6879):451. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 16.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6(8):610. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 17.Woelk T, Oldrini B, Maspero E, Confalonieri S, Cavallaro E, Di Fiore PP, Polo S. Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol. 2006;8(11):1246. doi: 10.1038/ncb1484. [DOI] [PubMed] [Google Scholar]
- 18.van Delft S, Govers R, Strous GJ, Verkleij AJ, van Bergen en Henegouwen PM. Epidermal growth factor induces ubiquitination of Eps15. J Biol Chem. 1997;272(22):14013. doi: 10.1074/jbc.272.22.14013. [DOI] [PubMed] [Google Scholar]
- 19.Klapisz E, Sorokina I, Lemeer S, Pijnenburg M, Verkleij AJ, van Bergen en Henegouwen PM. A ubiquitin-interacting motif (UIM) is essential for Eps15 and Eps15R ubiquitination. J Biol Chem. 2002;277(34):30746. doi: 10.1074/jbc.M203004200. [DOI] [PubMed] [Google Scholar]
- 20.Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23(11):1972. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- 21.Staub O, Abriel H, Plant P, Ishikawa T, Kanelis V, Saleki R, Horisberger JD, Schild L, Rotin D. Regulation of the epithelial Na+ channel by Nedd4 and ubiquitination. Kidney Int. 2000;57(3):809. doi: 10.1046/j.1523-1755.2000.00919.x. [DOI] [PubMed] [Google Scholar]
- 22.Harvey KF, Shearwin-Whyatt LM, Fotia A, Parton RG, Kumar S. N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J Biol Chem. 2002;277(11):9307. doi: 10.1074/jbc.M110443200. [DOI] [PubMed] [Google Scholar]
- 23.Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, Stenmark H, Dikic I. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol. 2006;8(2):163. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- 24.Bienko M, Green CM, Sabbioneda S, Crosetto N, Matic I, Hibbert RG, Begovic T, Niimi A, Mann M, Lehmann AR, Dikic I. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol Cell. 2010;37(3):396. doi: 10.1016/j.molcel.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 25.Fallon L, Belanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, Moreau F, Voortman J, Haber M, Rouleau G, Thorarinsdottir T, Brice A, van Bergen En Henegouwen PM, Fon EA. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol. 2006;8(8):834. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- 26.Lott JS, Coddington-Lawson SJ, Teesdale-Spittle PH, McDonald FJ. A single WW domain is the predominant mediator of the interaction between the human ubiquitin-protein ligase Nedd4 and the human epithelial sodium channel. Biochem J. 2002;361(Pt 3):481. doi: 10.1042/0264-6021:3610481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25(3):302. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 28.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 29.Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26(6):891. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Goldknopf IL, Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc Natl Acad Sci U S A. 1977;74(3):864. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunt LT, Dayhoff MO. Amino-terminal sequence identity of ubiquitin and the nonhistone component of nuclear protein A24. Biochem Biophys Res Commun. 1977;74(2):650. doi: 10.1016/0006-291x(77)90352-7. [DOI] [PubMed] [Google Scholar]
- 32.Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, Boyse EA. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci U S A. 1975;72(1):11. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hershko A, Ciechanover A, Heller H, Haas AL, Rose IA. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci U S A. 1980;77(4):1783. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson KD, Urban MK, Haas AL. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J Biol Chem. 1980;255(16):7529. [PubMed] [Google Scholar]
- 35.Ciechanover A, Elias S, Heller H, Ferber S, Hershko A. Characterization of the heat-stable polypeptide of the ATP-dependent proteolytic system from reticulocytes. J Biol Chem. 1980;255(16):7525. [PubMed] [Google Scholar]
- 36.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19(5):207. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 37.Pickart CM, Rose IA. Functional heterogeneity of ubiquitin carrier proteins. J Biol Chem. 1985;260(3):1573. [PubMed] [Google Scholar]
- 38.Jentsch S, McGrath JP, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;6135;329:131. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 39.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287(5452):501. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 40.Pickart CM, Vella AT. Ubiquitin carrier protein-catalyzed ubiquitin transfer to histones. Mechanism and specificity. J Biol Chem. 1988;263(29):15076. [PubMed] [Google Scholar]
- 41.Goebl MG, Yochem J, Jentsch S, McGrath JP, Varshavsky A, Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;241(4871):1331. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 42.Chen A, Kleiman FE, Manley JL, Ouchi T, Pan ZQ. Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase. J Biol Chem. 2002;277(24):22085. doi: 10.1074/jbc.M201252200. [DOI] [PubMed] [Google Scholar]
- 43.Haas AL, Bright PM, Jackson VE. Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. J Biol Chem. 1988;263(26):13268. [PubMed] [Google Scholar]
- 44.Morrison A, Miller EJ, Prakash L. Domain structure and functional analysis of the carboxyl-terminal polyacidic sequence of the RAD6 protein of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8(3):1179. doi: 10.1128/mcb.8.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sung P, Prakash S, Prakash L. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 1988;2(11):1476. doi: 10.1101/gad.2.11.1476. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan ML, Vierstra RD. Cloning of a 16-kDa ubiquitin carrier protein from wheat and Arabidopsis thaliana. Identification of functional domains by in vitro mutagenesis. J Biol Chem. 1991;266(35):23878. [PubMed] [Google Scholar]
- 47.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11(1):267. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 48.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell. 2005;20(4):589. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 49.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20(4):601. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 50.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11(1):261. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431(7010):873. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 52.Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem J. 2010;433(1):31. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14(10):941. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 54.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419(6903):135. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 55.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32(6):767. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 56.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133(4):653. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sadowski M, Suryadinata R, Lai X, Heierhorst J, Sarcevic B. Molecular basis for lysine specificity in the yeast ubiquitin-conjugating enzyme Cdc34. Mol Cell Biol. 2010;30(10):2316. doi: 10.1128/MCB.01094-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123(6):1107. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 59.Rodrigo-Brenni MC, Foster SA, Morgan DO. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol Cell. 2010;39(4):548. doi: 10.1016/j.molcel.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13(10):915. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 61.VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell. 2001;105(6):711. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- 62.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit e2. Cell. 2011;144(5):769. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21(6):873. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 64.Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130(2):349. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 65.Boutet SC, Biressi S, Iori K, Natu V, Rando TA. Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors. Mol Cell. 2011;40(5):749. doi: 10.1016/j.molcel.2010.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pham AD, Sauer F. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science. 2000;289(5488):2357. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- 67.Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130(1):127. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 68.Wu K, Kovacev J, Pan ZQ. Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol Cell. 2010;37(6):784. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eakin CM, Maccoss MJ, Finney GL, Klevit RE. Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase. Proc Natl Acad Sci U S A. 2007;104(14):5794. doi: 10.1073/pnas.0610887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stoll KE, Brzovic PS, Davis TN, Klevit RE. The essential Ubc4/Ubc5 function in yeast is HECT-E3 dependent and RING-E3 dependent pathways require only mono-Ub transfer by Ubc4. J Biol Chem. 2011 doi: 10.1074/jbc.M110.203968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hibbert RG, Huang A, Boelens R, Sixma TK. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1017516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1. Mol Cell. 2005;17(3):341. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 73.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12(10):933. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 74.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139(5):957. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simkus C, Bhattacharyya A, Zhou M, Veenstra TD, Jones JM. Correlation between recombinase activating gene 1 ubiquitin ligase activity and V(D)J recombination. Immunology. 2009;128(2):206. doi: 10.1111/j.1365-2567.2009.03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102(4):533. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 77.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono-versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302(5652):1972. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 78.Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446(7133):333. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- 79.Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci U S A. 2009;106(10):3722. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, Zheng L, Rich RL, Campos AD, Myszka DG, Lenardo MJ, Darnay BG, Wu H. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sakata E, Satoh T, Yamamoto S, Yamaguchi Y, Yagi-Utsumi M, Kurimoto E, Tanaka K, Wakatsuki S, Kato K. Crystal structure of UbcH5b~ubiquitin intermediate: insight into the formation of the self-assembled E2~Ub conjugates. Structure. 2010;18(1):138. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 82.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8(7):700. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 83.Summers MK, Pan B, Mukhyala K, Jackson PK. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell. 2008;31(4):544. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parker JL, Ulrich HD. Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. Embo J. 2009;28(23):3657. doi: 10.1038/emboj.2009.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ortolan TG, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K. The DNA repair protein rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol. 2000;2(9):601. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- 86.Chen L, Shinde U, Ortolan TG, Madura K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2001;2(10):933. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Isasa M, Katz EJ, Kim W, Yugo V, Gonzalez S, Kirkpatrick DS, Thomson TM, Finley D, Gygi SP, Crosas B. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol Cell. 2010;38(5):733. doi: 10.1016/j.molcel.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 89.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276(24):21664. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 90.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13(6):491. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 91.Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. Embo J. 2007;26(11):2797. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194(3):531. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 93.Whitby FG, Xia G, Pickart CM, Hill CP. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J Biol Chem. 1998;273(52):34983. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 94.Merkley N, Shaw GS. Solution structure of the flexible class II ubiquitin-conjugating enzyme Ubc1 provides insights for polyubiquitin chain assembly. J Biol Chem. 2004;279(45):47139. doi: 10.1074/jbc.M409576200. [DOI] [PubMed] [Google Scholar]
- 95.Scott DC, Monda JK, Grace CR, Duda DM, Kriwacki RW, Kurz T, Schulman BA. A dual E3 mechanism for Rub1 ligation to Cdc53. Mol Cell. 2010;39(5):784. doi: 10.1016/j.molcel.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]