Abstract
This article serves to outline a research paradigm to investigate main effects and interactions of genes, environment, and development on behavior and psychiatric illness. We provide a historical context for candidate gene studies and genome-wide association studies, including benefits, limitations, and expected payoff. Using substance use and abuse as our driving example, we then turn to the importance of etiological psychological theory in guiding genetic, environmental, and developmental research, as well as the utility of refined phenotypic measures, such as endophenotypes, in the pursuit of etiological understanding and focused tests of genetic and environmental associations. Phenotypic measurement has received considerable attention and is informed by psychometrics, while the environment remains relatively poorly measured and is often confounded with genetic effects (i.e., gene-environment correlation). Genetically-informed designs which—thanks to ever cheaper genotyping—are no longer are limited to twin and adoption studies, are required to understand environmental influences. Finally, we outline the vast amount of individual differences in structural genomic variation, most of which remains to be leveraged in genetic association tests. While the genetic data can be burdensomely massive (tens of millions of variants per person), we argue that improved understanding of genomic structure and function will provide investigators with new tools to test specific a priori hypotheses derived from etiological psychological theory, much like current candidate gene research, but with less confusion and more payoff than candidate gene research has to date.
Over the past 50 years, twin and adoption research has revealed much about the origins of individual differences in behavior. We know that genetic factors influence individual differences in a wide range of psychological outcomes, in part because genetic factors appear to contribute substantially to both the stability of behavior as well as to behavioral change. We also know that the genetic and environmental factors that influence a specific behavioral outcome are not necessarily statistically independent nor do we expect them to be additive in their effect. The emergence of the concept of gene-environment correlation (not to be confused with gene-environment interaction) in the psychological literature arose out of a need to account for what might, at least initially, seem a paradoxical result – genetic factors appear to contribute to differences in a wide array of environmental measures including peer-group characteristics, features of the parent-child relationship, exposure to psychological stress, and becoming married as well as divorced (Kendler & Baker, 2007). The paradox is resolved by recognizing that environments are not distributed randomly; that individuals can have a major impact in shaping their experiences both directly, through the choices they make, and indirectly, through the reactions their behaviors elicit from others (Jaffee & Price, 2007; Scarr & McCartney, 1983). Alternatively, the concept that genetic and environmental factors interact in their effects has been enthusiastically embraced by psychologists; gene-environment interaction research is one of the most rapidly expanding research paradigms within psychology (Dick, 2011).
Today, the field of behavioral genetics is at a crossroads. The twin and adoption studies that helped establish that important psychological outcomes are influenced by genetic factors in the aggregate are increasingly giving way to molecular genetic investigations aimed at characterizing the nature of the contribution of specific genetic variants. This transition has not been entirely smooth. Reports of associations between specific genetic variants and behavior have been notoriously difficult to replicate, and the validity of research investigating the interaction between specific functional genetic variants and the environment has recently been called into question (Duncan & Keller, 2011; N. Risch, et al., 2009). How we think about the genetics of behavior is likely to be further challenged by recent developments in genomic science, which allow psychological researchers today to efficiently and economically interrogate vast numbers of genetic variants and, in the not-too-distant future, will allow us to sequence the entire genomes of our research participants. Genetically-minded researchers will soon be awash with genetic data and now is the time to consider the implications of this new type of genetic data and especially how it can be integrated with our knowledge of the nature of environmental influence to bring about a better understanding of individual differences in, and developmental etiology of, behavior and mental illness.
This paper is concerned with the impact of recent developments in genomics on the study of human behavior. It begins by describing those developments. Because genomics has been applied primarily with non-behavioral phenotypes, a discussion of the challenges uniquely associated with behavioral phenotypes follows. This includes a discussion of what we have learned from twin and adoption research about how best to assess the phenotype and conceptualize the nature of environmental influence and, most critically, about how genes, environment, and development be integrated to move psychology forward. Finally, we describe ongoing developments in genomics that are likely to soon have an impact on behavioral research and speculate about what the nature of what that impact is likely to be. Unfortunately, most of current genomics research on human behavior is not developmentally informed; we emphasize the importance of a developmental perspective throughout.
GWAS: Genomics in a Post-Genomic World
The heritability estimates derived from twin and adoption studies imply that aggregate differences in the DNA sequences we inherit contribute in some way to differences in our behavior. The conclusion of the Human Genome Project (see Lander [2011] for a discussion) has ushered in an era of precisely measured genomic variation. At the time of this writing it is possible to obtain your entire sequence of DNA, approximately 3.5 billion base pairs, for a couple thousand dollars. The genetic information currently used in candidate gene studies by social and behavioral scientists is a mere fraction of the total variation, and represents the tip of the genomic iceberg. This section lays the foundation for our discussion of how to incorporate a developmental perspective into modern behavioral genetic studies by briefly cataloging common forms of genetic variation, describing how behavioral scientists have typically investigated this variation, and discussing how these behavioral investigations are likely to change due to recent technological developments within genomics.
Human DNA Structure and Variability
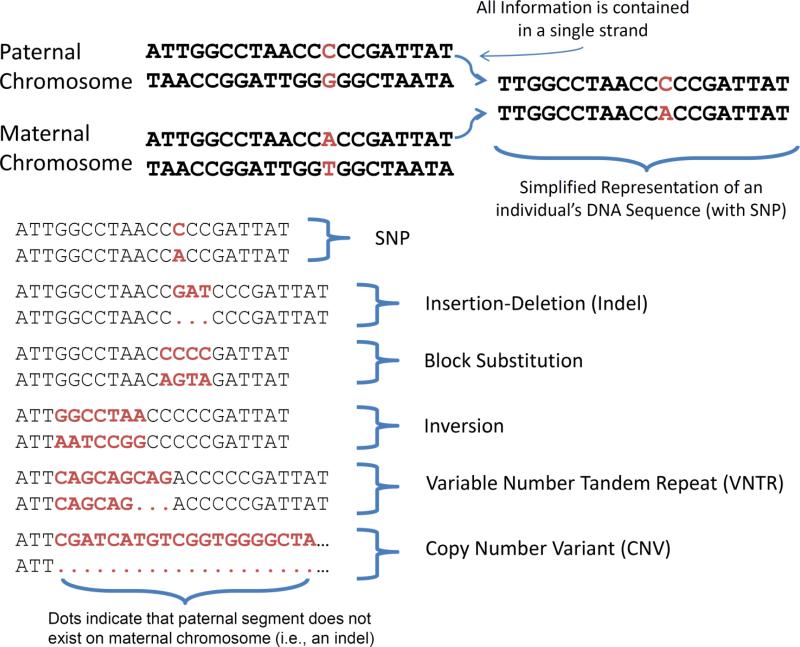
Normal human DNA is contained in 23 chromosomes—22 autosomes and a single sex chromosome. Humans also have small segments of DNA residing in cell mitochondria. We focus here on chromosomal DNA. Each autosome has two copies, one copy from the mother and one from the father. Each copy is composed of base pairs of nucleotides, the familiar A, T, C, and G (adenine, thymine, cytosine, and guanine). The nucleotides are always found in pairs, and the same nucleotides always pair with each other. An example autosomal segment, along with major types of DNA variation can be found in Figure 1.
Figure 1.
Common Forms of DNA Variation. Humans have two chromosomes, one inherited from their father (paternal) and one from their mother (maternal). Since the same bases always pair together in a single chromosomal strand (A with T; C with G), giving both pairs for each strand is redundant, and DNA sequences are therefore represented by two rows of bases (the “Simplified Representation” in the figure). The “CA” SNP represents the only difference between the maternal and paternal autosomal segments. Catalogued in this figure is the Single Nucleotide Polymorphism (SNP) as well as several common types of structural variation, including Indels, Block Substitutions, inversions, VNTRs, and CNVs. Note that these particular variants are illustrative, and that variation is not necessarily within-person. For example, this individual is heterozygous for the SNP, but other individuals may be homozygous CC or homozygous AA. The same is true for structural variants. In the Indel example, this individual has a GAT insertion on the paternal chromosome and no such insertion on the maternal chromosome. Another individual may have GAT insertions on both chromosomes; yet another individual may lack the GAT insertion altogether.
Single nucleotide polymorphisms (SNPs) represent one major source of genetic variation, with over 15 million SNPs identified in humans as of this writing (Altshuler, et al., 2010). An example SNP is given in Figure 1, where a single base pair differs between the maternal and paternal autosomal segment. Of the 15 million catalogued SNPs, only ~10 million are considered common in that they have a minor allele frequency > .05 in one or more populations (the frequency in the population of the less frequently occurring, or minor, allele). Note that the rarity, as well as the definition, of the minor allele is population-specific. A particular allele might be very common in one population (e.g., individuals of Han Chinese descent), and rare or even non-existent in another (e.g., individuals of Yoruban descent; see The International HapMap Project [2003]). Whatever racial population is under study, it is the common SNPs, and not the rare SNPs, that have been the focus of most genetics research. This is because, all else being equal, the larger the minor allele frequency the greater the power to detect an effect (Bansal, Libiger, Torkamani, & Schork, 2010). Simply put, the typical sample will not contain many individuals carrying rare SNP alleles, so a focus on common SNPs is driven in large part by pragmatics. There are many examples of studies of SNPs within psychology. Perhaps most familiar amongst these is the Val158Met (rs4680) polymorphism in catechol-O-methyltransferase (COMT) , which involves a single base pair substitution that changes the coding sequence of the gene (Meyer-Lindenberg, et al., 2006).
SNPs have been the primary focus of behavioral genetics research because they can be efficiently genotyped and have consequently been extensively mapped, but it is important to recognize that other types of genetic variants are likely also to be relevant to understanding behavior. Insertions/Deletions (“InDels”) are variants where a small number of DNA bases (usually < 100 bases) is either inserted or deleted into the DNA sequence. The most prominent In/Del in psychology involves a 44base deletion in the promoter region of the gene that codes for the serotonin transporter (5-htt; Heils, et al., 1996). Variable number of tandem repeats (VNTRs) represent another form of genetic variation. VNTRs involve a short segment of DNA that is repeated in tandem, but where the number of repeats varies from person to person. The most widely studied VNTR in psychology involve a 48-base sequence in the coding sequence of the DRD4 gene that is repeated from 2 to 10 times (Lichter, et al., 1993). Finally, copy number variants (CNVs) are relatively long sequences of DNA (100,000 bases or more) where individuals can carry other than the typical two copies. Although CNVs are generally rare, they have been implicated in multiple diseases including autism and schizophrenia (Stankiewicz & Lupski, 2010b).
While it might seem that comprehensive assessment of common SNP variation would require genotyping research participants on each of the 10 million common SNPs, in fact a small fraction of this number would suffice. This is because SNPs that are located near one another on the same chromosome tend to be correlated with one another, a phenomenon known as linkage disequilibrium (LD). Chromosomes are not transmitted across generations unaltered. Rather, during meiosis, when homologous chromosomes pair up, there is an opportunity for an exchange, or recombination, of genetic material between the maternally-inherited and the paternally-inherited chromosomes. The greater the distance between two markers, the more likely it is that the markers will recombine. But rather than occur randomly, recombination takes place most frequently at recombination hotspots along the chromosome. SNPs that are located within the same resulting chromosomal block will consequently tend to be passed along together to the next generation, and knowing one SNP in the block allows one to infer the others with a high degree of certainty. The upshot is that a minority of well-chosen SNPs (e.g., one million) contains information about nearly all the common SNPs in the genome—the well-chosen SNPs serve as proxies for neighboring SNPs in high linkage disequilibrium (Hirschhorn & Daly, 2005).
Until a few years ago, the existence of LD did little more than allow geneticists to speculate about the possible impact of comprehensively surveying common variation throughout the entire genome (Risch & Merikangas, 1996). The advent of high-throughput SNP genotyping, however, allowed speculation to become a reality. The first genome-wide association study (GWAS) was a proof-of-principle sponsored by the Wellcome Trust and involved genotyping nearly 500,000 SNPs in 14,000 cases, representing 7 different inherited disorders, and a common control sample of 3,000 (Burton, et al., 2007). This initial GWAS tentatively identified 24 SNPs as being associated with one of the seven diseases and spurred hundreds of subsequent GWAS that have resulted in the identification of several thousand SNP associations (Visscher, Brown, McCarthy, & Yang, 2012). Although GWAS posed several methodological challenges, such as how to deal with the multiple-testing burden (Hirschhorn & Daly, 2005) or how to control for subtle population differences (such as ethnicity) between cases and controls (Kang, et al., 2010; Price, et al., 2006), for the most part these issues have been addressed and a GWAS analysis today involves little more than a simple exercise of fitting an additive regression model, albeit repeated hundreds of thousands and even millions of times.
GWAS of Anthropometric Measures, Medical Diseases, and Behavioral Traits
While the statistical exercise is simple, reliable identification of significant GWAS associations has been challenged by two factors. First, addressing the multiple-testing burden requires use of a p-value threshold of p < 5×10-8 (Hirschhorn & Daly, 2005). Second, the effect associated with any specific SNP is very small, typically accounting for < 0.5% of the variance in the trait. Addressing these challenges requires sample sizes that are typically beyond the capabilities of any single investigator. For example, to have 80% power to detect an effect accounting for 0.1% of variance using the GWAS p-value threshold would require a sample of nearly 40,000 individuals. Detection of these small effects has motivated the establishment of large consortia, combining GWAS findings using meta-analytic methods. A consortium on height used data from over 180,000 individuals to identify 180 SNP variants that collectively accounted for approximately 10% of the variance in height (Allen, et al., 2010). Consortia for body mass index (BMI, nearly 250,000 individuals, 18 variants identified, and 1.5% of variance accounted for; Speliotes, et al., 2010), and blood lipids (more than 100,000 individuals, 95 identified variants, 10-12% of variance accounted for; Teslovich, et al., 2010) have produced similar results. Since the percent of phenotypic variance accounted for by known SNPs is far less than heritability estimates from twin and family studies, researchers have concluded that much of trait heritability remains “missing” (Manolio, et al., 2009).
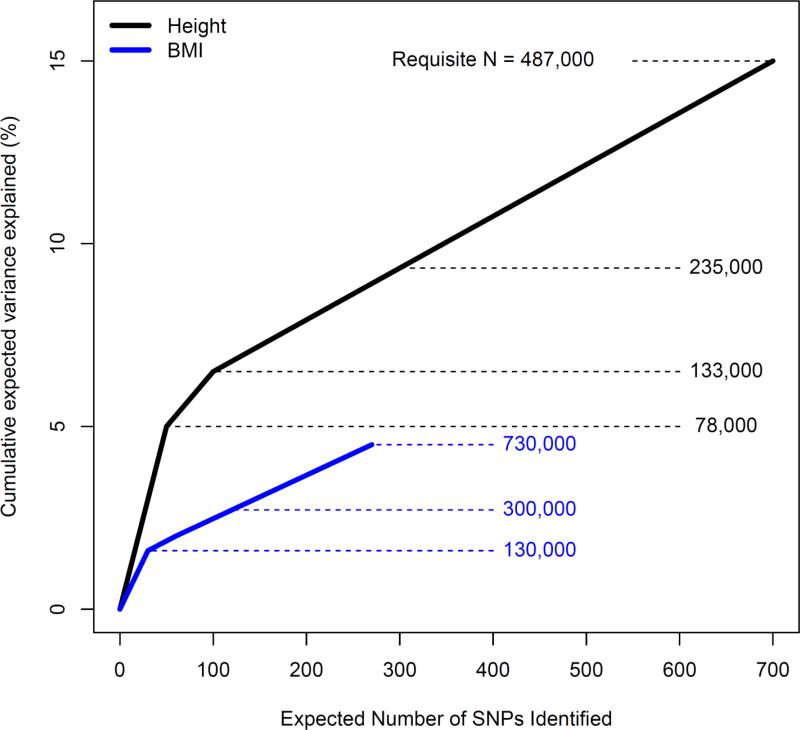
The challenge associated with finding the missing heritability is heightened in recognizing that the SNPs identified thus far are likely the low-lying fruit, those with the largest effect sizes. Accounting for greater percentages of variance will require ever larger samples to identify even smaller SNP effects. Figure 2 illustrates the projected sample sizes needed for height and BMI to increase the variance being accounted for by identified SNPs. For height, to increase from the current 10% to 15% will require a combined sample of 487,000 individuals. For BMI, going from the 1.5% that is accounted for now to a mere 5% will require a projected sample size of 730,000.
Figure 2.
Sample sizes and number of SNPs required to explain significant proportions of variance in anthropometric traits. These data are based on visual inspection of graphs provided in (Allen, et al., 2010) and (Speliotes, et al., 2010); see the original sources for full details. Note the wide difference in trend for height (estimates based on sample of ~185,000 subjects) and for BMI (estimates based on sample of ~235,000 subjects). Height is much more promising, in that it will take ~500,000 samples to obtain enough genome-wide significant SNPs to account for 15% of the variance in height. For BMI, on the other hand, it is projected that ~700,000 samples are required to account for only 5% of the variance in height. Cause of the discrepancy in genetic architecture of these traits is unknown. These values differ slightly from what is described in the text because extrapolating beyond currently available sample sizes required sample splitting and replication procedures in the original studies in order to unbiasedly estimate effect sizes.
As with anthropometric traits, large samples have been needed to reliably identify SNP effects in GWAS of behavioral and psychiatric traits. A meta-analysis (de Moor, et al., 2009) of 17,375 individuals assessed for Big 5 personality traits returned one genome-wide significant SNP for Openness, and one for Conscientiousness, neither of which subsequently replicated in a small sample (N = 3,294). A genome-wide meta-analysis of alcohol consumption (Schumann, et al., 2011) returned one SNP identified in a discovery sample of 26,316 individuals and replicated in a sample of 21,185, and a recent GWAS investigation of more than 12,000 individuals with bipolar disorder and 50,000 controls reported 18 replicated SNP effects (Sklar, et al., 2011). Not unexpectedly, most of the heritable variance for behavioral phenotypes remains missing. For example, a GWAS analysis of more than 3,000 individuals with schizophrenia and more than 3,000 controls was able to account for approximately 3% of the variance in schizophrenia liability (Purcell, et al., 2009), far less than the estimated 80% liability heritability for schizophrenia.
Accounting for missing heritability may not, however, be a simple matter of achieving ever larger pooled sample sizes, if the relevant genetic variation is not being captured by the GWAS platforms being used. Genome-wide complex trait analysis, or GCTA (Yang, Lee, Goddard, & Visscher, 2011; Yang, Manolio, et al., 2011), is a statistical method that uses the genetic relatedness among individuals, as measured by the genotyped SNPs, to estimate the variance in the phenotype accounted for by the aggregate of all of those SNPs. The method can be likened to a twin study except that instead of using twin zygosity to estimate genetic relatedness, GCTA uses the aggregate genetic similarity, as estimated by the genotyped SNPs, among each pair of research participants in a sample. Consequently, while GCTA does not tell us which of the million or so genotyped SNPs contribute to phenotypic variance, it does tell us how much variance the relevant SNPs would account for if we could sift them out. In a sample of only ~4,000 individuals, Yang, et al. (2010) found that the genotyped common SNPs accounted for 45% of the variance in height, a great deal more than the 10.5% of variance found by the Allen, et al. (2010) meta-analysis of ~185,000 individuals, but still a far cry from the 80% heritability routinely identified in twin studies. GCTA has been similarly used in large samples to estimate the variance accounted for by genotyped common SNPs to be 23% for schizophrenia (Lee, et al., 2012) and 40-50% for general cognitive ability (Davies, et al., 2011), values both well below heritability estimates for these phenotypes derived from twin and family data. Consequently, while much of the heritability of complex phenotypes could, in principle, be accounted for by the SNPs genotyped in GWAS, clearly a large portion of the heritability would remain missing even if we could achieve pooled samples in the multiple millions. In all likelihood there are sources of genetic variance in addition to common SNPs, and the additive regression model that has been the basis of GWAS analysis may need to be extended to consider non-additive effects.
While GWAS has been successful in identifying a large number of SNP variants for a large number of important traits, some have expressed disappointment that SNP effects have been uniformly small and that the vast majority of heritable variance remains missing (Gershon, Alliey-Rodriguez, & Liu, 2011). Nonetheless, GWAS has moved the field beyond a research agenda being driven by a few false positive genetic findings (Ioannidis, Castaldi, & Evangelou, 2010). We believe the question now is not so much whether GWAS has worked as it is how we can use what we have learned about the genetic architecture of complex phenotypes to better understand the origins of individual differences in behavior. We are optimistic about the future of genetically-informed behavioral research, especially if that research builds on what we have learned about the nature of heritable variation for behavioral traits; involves a consideration of environmental and developmental context; and expands to consider sources of genetic variance other than the common SNPs that have been the focus of GWAS.
The Nature of the Behavioral Phenotype
Phenotypic Definition and Measurement
Despite a long history in psychology of measurement and assessment, much work on the genetics of psychiatric disease has focused on diagnostic status or symptomatology of mental disorder categories defined by the Diagnostic and Statistical Manual of Mental Disorders (DSM; American Psychiatric Association, 2000). In part this is a practical issue, as DSM diagnoses and symptoms are often gathered as part of standard assessment protocol. This makes DSM-related measurements useful as they can easily be combined across studies in meta-analytic style, which has become standard practice in genetics.
While a categorical measurement of disease status may effectively facilitate the pooling of resources, it may not be an optimal way to measure disease and/or a trait. It is well-known that binary diagnoses throw away an immense amount of information, and have lower statistical power compared even to rough quasi-continuous measures of the same constructs, like symptom counts (Markon, Chmielewski, & Miller, 2011). It is rarely true in psychology that a binary classification is better for statistical analysis than a quasi-continuous one (Grove, 1991), but arguments favoring of the use of quantitative indicators is not based on statistical power alone. Some of the phenotypic measurements of interest to psychopathologists appear to be better represented by continuous dimensions of variation rather than as discrete entities (Krueger, Markon, Patrick, & Iacono, 2005; Vrieze, Perlman, Krueger, & Iacono, 2011; although see Vrieze, in press), a conclusion that has moreover been consistently supported in behavioral genetic research.
Used well, measurement models inform the nature and etiological structure of a phenotype for behavioral genetic investigations. For example, alcohol, nicotine, and cannabis dependence have long been considered as technically separate conditions in the DSM, but work during the last decade indicates that a significant proportion of etiology among them is shared, and is largely genetic in origin (Kendler, Jacobson, Prescott, & Neale, 2003). Factor analytic approaches give single factor solutions, suggesting similar shared causal mechanisms among nicotine, alcohol, marijuana, and illicit drugs (Hicks, Schalet, Malone, Iacono, & McGue, 2011; Krueger, et al., 2002), which begs the question of what these shared causes might be. One popular answer involves the gateway hypothesis (Kandel & Jessor, 2002), where it is proposed that the use of one drug causes use of another. Perhaps individuals use multiple drugs in their search for bigger and better highs, resulting in the strong correlations that are observed among substance dependence symptoms. Another hypothesis is that more impulsive, sensation-seeking individuals are more likely to experiment indiscriminately with drugs, and will use and become addicted to multiple drugs simultaneously, which would also result in observed correlations among different drugs. This has been termed the disinhibitory hypothesis, holding that the more disinhibited one is, the more likely one is to experiment and use indiscriminately (Iacono, Malone, & McGue, 2008).
Like many etiological theories, the gateway and disinhibitory hypotheses are developmental, in that they hypothesize the existence of etiological processes that progress, over time, to pathological behaviors such as drug abuse or addiction. Rigorous testing of these hypotheses requires developmentally informed sampling and measurement, likely not new ideas to readers of this journal. More novel, perhaps, is the utility of genetically-informative designs, even in cases where one is not explicitly interested in the role of genetic factors. For example, Irons, McGue, Iacono, & Oetting, (2007) used a genetic design called Mendelian Randomization (Smith, 2011) to test key predictions of the gateway model. Nearly 50% of individuals of East Asian ancestry inherit a variant in the ALDH2 gene that diminishes their ability to metabolize alcohol (Luczak, Glatt, & Wall, 2006). Those who inherit the gene variant are likely to experience various signs of dysphoria following ingestion of even a small amount of alcohol and consequently curb their drinking. Inheriting the ALDH2 variant is essentially random, at least among East Asians, and thus provides a natural analog to experimental randomization. The question of relevance here is whether individuals who inherit the ALDH2 variant show, in addition to reduced drinking, diminished rates of the behaviors the gateway model posits to be a consequence of use of gateway substances such as alcohol. (Irons, et al., 2007) found no evidence in support of the gateway model in that rates of non-alcohol substance use and disinhibited behavior did not vary as a function of ALDH2 status among individuals of East Asian ancestry. Nonetheless, one limitation of the (Irons, et al., 2007) study is especially relevant in the current context: participants were still in late adolescence (average age of 18.3 years) and gateway effects may not emerge until later in development. That is, there may be a gene by development interaction between ALDH2 and non-alcohol substance use. Continued follow-up of the sample is warranted.
Other research using a variety of designs, summarized by (Iacono, et al., 2008), has taken a more explicitly developmental perspective to establish that a broad risk for abusing multiple drugs is disinhibitory, at least in adolescence. Relative to adults, adolescents are more disinhibited (Steinberg, 2007), and one expects that correlations between alcohol, nicotine, and marijuana dependence to be highest at younger ages, when individual's behavior is more driven by impulsivity and sensation-seeking, and then decline over time as the youths age and become more responsible. Vrieze, Hicks, McGue, & Iacono (in press) used a large prospective longitudinal study of twins with measures of substance dependence taken at ages 11, 14, 17, 20, 24, and 29. While overall rates increased until age 20-24 and declined thereafter, the correlations among alcohol, nicotine, and marijuana dependence strictly declined from adolescence into young adulthood. In addition, by using the twins we noted a gene by environment by development interaction (GxExD). At younger ages the correlations among nicotine, alcohol, and marijuana were due largely to genetic influences. At older ages the correlations were more strongly due to non-shared environment. That is, as the individuals aged their individual life experiences increasingly influenced their propensity to use multiple drugs simultaneously.
Clearly, the same measure of a phenotype, such as symptom counts of alcohol dependence, can be the product of drastically different etiology depending on developmental stage and environmental considerations that exist when the measurement is taken. This can pose serious problems for genetic studies in samples where the genetic etiology of a phenotype is heterogeneous. An excellent example comes from recent research on obesity, where Lasky-Su, et al. (2008) found a SNP in ROBO1 was significantly associated with obesity in pediatric samples, but only marginally significant in adult samples, a GxD interaction. In work on skin cancer, Duffy, et al. (2010) identified a SNP in the IRF4 gene where the T allele was associated with high freckling in adolescents and adults, but was only associated with high nevus counts in the adolescents. High nevus counts increase risk of melanoma. The authors suggest this is likely a GxE interaction: adolescents in this cross-sectional sample sun bathe more frequently for longer periods than the adults had. The adults and the adolescents had the risk gene, but sun exposure is required to potentiate its effects.
Obvious analogues exist for substance use phenotypes. There are a host of genes that theoretically impact upon propensity for addiction, but are irrelevant unless one is exposed to the substance. This appears to be true for the nicotinic receptor gene CHRNA3, which was very strongly associated with number of cigarettes smoked per day in a sample of 74,000 current smokers (p < 2.8×10-73) but was not associated with smoking initiation in a combined sample of ~74,000 smokers and ~70,000 non-smokers (Furberg, et al., 2010). The effect of CHRNA3 is lost on those who have never smoked. Alcohol metabolism and ALDH2, the gene discussed above and responsible for flushing in many individuals of East Asian descent, also is likely not protective for alcoholism until someone starts drinking, and thereby feels the effect of that genotype.
The hypothesized existence of heterogeneity in genetic etiology, that different genes are relevant under different environmental or developmental circumstances, is in tension with the most popular current approach to evaluating common variants: combining as many samples as possible through meta-analysis. In the presence of between-sample heterogeneity of genetic associations meta-analysis can fail (DerSimonian & Kacker, 2007; Han & Eskin, 2012; Tang, 2006), at least for those SNPs that are sample-specific, e.g., due to developmental differences between some of the samples in the study. While this certainly is a problem, it is currently prohibitively difficult to amass phenotypically, developmentally, and environmentally homogenous samples powerful enough to identify common SNP interaction effects. While meta-analysis is not perfect, if used well it can identify at least those SNPs that transcend whatever heterogeneity exists, and new methods are being developed that determine which individual studies in the meta-analysis contain the SNP effect, which likely do not, and which studies are ambiguous (Han & Eskin, 2012). Such tests can provide insight into the source of genetic heterogeneity to guide future efforts. The existence of etiological heterogeneity between diverse samples is important to consider, but should not impede efforts at consortia-building and GWAS meta-analysis.
Other genotypes, such as those associated with behavioral disinhibition or impulsivity, are theoretically relevant both for initiation and maintenance of use (Zucker, Heitzeg, & Nigg, 2011). Measurements of these traits may show heterotypic continuity (Costello, Mustillo, Erkanli, Keeler, & Angold, 2003), in that the same etiological process manifests differently at different stages of development and/or in different environments. For example, problem behavior before age 15 such as tobacco use, alcohol use, trouble with police, and early sexual intercourse are predictive of age-20 psychiatric disorders like nicotine dependence, alcohol dependence, drug dependence, and antisocial personality disorder (McGue & Iacono, 2005). Studies of familial transmission also suggest a role of heterotypic continuity. Parental substance use or antisocial diagnosis is associated with increased risk for child conduct disorder, attention-deficit hyperactivity disorder, and oppositional defiant disorder (Bornovalova, Hicks, Iacono, & McGue, 2010). These studies suggest that part of the genetic predisposition to early childhood problem behavior and disorder is also relevant for adult disorders, even though the phenotypic measures can be quite different.
One can begin to see the importance of phenotypic measurement and developmental etiological theory in understanding a phenotype. Some aspects of the etiology and measurement structure of substance use phenotypes change from adolescence to young adulthood (Vrieze, et al., in press) and other aspects appear reliable over time (Bornovalova, et al., 2010; McGue & Iacono, 2005). Genes found to be associated with substance use, at whatever age, will require developmental elaboration of this kind in order to inform etiology, intervention, and measurement. The overriding goal of behavioral genetics is not to engage in statistical self-indulgence to explain variance in our pet phenotypes, but rather to test etiological developmental theories of behavior. With the ever-increasing availability of measured human genomes we expect an exciting scientific future—genetic findings will force us to discard, amend, and refine theories, leading to improved empirical measures of phenotypes and environments designed to test the refined theory, resulting in ever more genetic, biological, and environmental findings.
We expect a product of this cycle to be a pursuit of phenotypic and endophenotypic refinement. A problem facing those exploring the genetic underpinnings of DSM categories, or most other standard clinical or questionnaire measures, is that whatever utility they have in guiding research and treatment, they are not grounded in biology. This leaves open the question of how useful they can be as target phenotypes in genetic association studies. As discussed, one explanation for the problems encountered identifying replicable genetic associations with psychiatric disorders lies in the possibility that the disorders are too etiologically heterogeneous and complex to facilitate gene finding. An additional complication is that these behavioral phenotypes are far removed from the action of genes at the level of the brain. These brain processes interact with environmental context to influence various observed pathological behaviors, some constellation of which defines a DSM disorder or trait measurement thereof. An endophenotype approach that targets more directly the genetic architecture of the underlying brain processes may provide a useful complement to investigations focused on DSM categories, symptom counts, and related trait dimensions.
An endophenotype is a heritable, biologically-based, objectively quantifiable measure that is associated with a psychiatric phenotype because both share a common genetic influence. Endophenotypes hold promise for gene finding because they are believed to: a) be more proximal to the effects of genes, b) tap one of many facets of a disorder and thus be more etiologically homogeneous than the associated disorder, and c) be more highly heritable than the disorder, thus likely to produce larger effect sizes and boost GWAS power or, because the endophenotype deals with an etiologically homogenous facet of the disorder, d) be associated with fewer genes each of which may be expected to have larger individual effect sizes. To the extent that an endophenotype taps a biological mechanism associated with the development of a disorder, identifying its genetic underpinnings could provide clues regarding how the relevant gene or genes affect brain processes that underpin a disorder. It is unlikely that there are many DSM disorders with a specific neurological etiology, and likely that there are different disorders that share underlying neural processes. To the extent that a brain process (e.g., dysfunctional presynaptic prefrontal cortex) heightens risk for the development of more than one disorder (e.g., substance dependence and mood disorders; Goto, Yang, & Otani, 2010), an endophenotype has potential to provide leads to genes that affect disparate disorders as well as genes that help explain why certain disorder comorbidity combinations occur. Animal models derived from endophenotypes may be potentially useful for identifying genes, their role in neurodevelopment, and their effects on brain circuitry (Kaffman & Krystal, 2012). Many papers have been written regarding the ideal properties of an endophenotype, with a recent review paper by Iacono and Malone (2011) detailing these characteristics from a developmentally informed perspective.
Endophenotypes are by their nature developmental. Their utility for gene finding derives in part from their ability to identify genetic risk in the absence of manifest psychopathology, and a valid endophenotype should predict the subsequent development of disorder in individuals and their first-degree relatives. The ideal endophenotype would change little with development after a certain age, or change in a predictable manner. In addition, it should be relatively stable over time and affected little by changes in state, including changes associated with acute illness and remission of disorder. In their review, Iacono and Malone (2011) showed how reduced amplitude of the P300 event-related potential obtained from an oddball task (e.g., Begleiter, Porjesz, Bihari, & Kissin, 1984) shows promise as a developmental endophenotype indexing genetic risk for substance abuse and related disorders. This endophenotype is present in the pre-adolescent offspring of alcoholic fathers, is evident early in life for those who go on to develop substance use disorders, changes with development in predictable fashion (with the developmental trajectory of change itself being genetically influenced), is stable over several year intervals, and is associated with substance use disorders because of likely shared genetic influences. Molecular genetic investigations of P300 amplitude, including GWAS, are now underway.
Despite the promise of endophenotypes, skepticism has surfaced regarding the possibly idealized assumptions on which they are based and the likelihood that they will indeed facilitate gene finding (Flint & Munafo, 2007). However, it seems clear that the endophenotype strategy has yet to be fully exploited. Multivariate methods targeting the molecular genetic basis of the covariance between the endophenotype and its associated disorder have yet to be fully tested, and using endophenotypes in combination may prove fruitful. Iacono, Carlson, and Malone (2000) showed that using two endophenotypes in combination was associated with greatly increased risk for developing substance use disorders when compared to either endophenotype alone; that is, the endophenotypes each added incrementally to the prediction of who would develop a substance use disorder. Greenwood, Light, Swerdlow, Radant, and Braff (2012), obtained promising multivariate results using a novel bootstrapping approach designed to counter problems associated with multiple testing. In Greenwood, et al., (2011), they simultaneously examined the association among 12 heritable schizophrenia endophenotypes and 1536 SNPs covering 94 biologically relevant genes. They found evidence supporting the involvement of 46 genes, including those involved in neurodevelopment, and reported that eight genes affected four or more endophenotypes, suggesting pleiotropy. These studies and others (Iacono, McGue, & Krueger, 2006; Luck, et al., 2011) demonstrate that it is currently feasible to obtain endophenotype measures on samples numbering in the thousands. Although these Ns are not large by current GWAS standards, with improved statistical methods, pooling across sites, and reliance on putative endophenotypes that show strong construct validity, there is ample reason to be optimistic about the payoff from an optimally applied endophenotype study. Finally, even if endophenotypes do not help in gene identification, once disorder/trait-relevant genes are found, the association of these genes with endophenotypes may help identify relevant brain mechanisms (de Geus, 2010). This in turn would not only assist our understanding the neurobiological effects of specific genetic polymorphisms, but also enrich theoretical understanding of the etiological mechanisms contributing to the development of psychopathology.
Measuring, Selecting, and Aggregating Environments
To disentangle genetic and environmental influences on behavior and disease behavioral geneticists have traditionally used twins or adoptees and their families, which are well-suited to this task. In one test of the disinhibitory hypothesis, Keyes, Legrand, Iacono, and McGue (2008) evaluated the environmental impact of parental smoking on child tobacco, alcohol, and drug use. In biological children parental smoking was associated with increased risk for use of all drugs. In adoptive children it was only associated with a mild increase in smoking, and had no association with alcohol or other drug use. A typical family study (of biological parents and children) would have concluded that parental smoking is a risk factor for drug use, without elaboration. Having a genetically-informative design, however, elucidated the etiology. That is, parental smoking conveys risk both through an environmentally mediated pathway that appears to be specific to offspring smoking and a genetically mediated pathway that is general to offspring substance use.
Measuring and defining environments has received much less attention in clinical psychology and psychiatry than phenotypic measurement. This trend is changing, and current efforts such as the PhenX Toolkit (Hamilton, et al., 2011), devote entire measurement domains to psychosocial history and social environment, providing a total of 30 assessment protocols ranging from child maltreatment to job strain. The Toolkit is a multidisciplinary effort to standardize assessment for the genetic study of complex disease.
Studies of GxE interaction often select candidate environments on the basis of a priori hypothesis. This is more than reasonable when there exists strong a priori evidence that some environment is likely to affect the phenotype differently depending on genotype. However, measures of the environment often correlate, sometimes strongly. A standard measurement approach would model those correlations under the usual assumption that different environmental measures are imperfect measures of the same construct (Cronbach & Meehl, 1955), but this is infrequently explicitly done. For example, in the evaluation of GxE effects of externalizing behaviors, Hicks, South, DiRago, Iacono, and McGue (2009) report an average correlation of .31 among adolescent environmental adversity measures, indicating etiological overlap and the potential utility of combining environments through a factor analytic measurement model.
Evaluating defensible aggregate measures of environmental risk might be a preferable starting point for GxE studies. While such a procedure ignores the possibility of interactions at finer levels of environmental detail, the multiple testing burden of possible environmental measures times possible SNPs becomes too high too quickly for available sample sizes. Perhaps more to the point, this approach to measuring the environment directly acknowledges that environmental risk exposures seldom occur in isolation. For example, adolescents who use substances tend to have problematic relationships with their parents, to have difficulty performing at school, and to socialize with deviant peers. Rather than focusing on specific environmental risk factors, aggregating environmental measures emphasizes the accumulation of risk across domains in the etiology of problem behavior. Work from our group has demonstrated that individual risk factors can be profitably combined into broader omnibus indicators, providing valuable summaries of environmental risk (Johnson, McGue, & Iacono, 2006; Keyes, Iacono, & McGue, 2007; Legrand, McGue, & Iacono, 1999).
Gene-environment correlational processes, whereby measures of environmental risk come to be heritable, complicate interpretations of associations of the environment with outcome (Scarr & McCartney, 1983). For example, peer-group characteristics are substantially correlated with adolescent substance use (Hicks, et al., 2009). But rather than being simply causal, these associations may reflect that the underlying genetic liability for disinhibition can manifest in both choice of peers and decision to experiment with substances (Harden, Hill, Turkheimer, & Emery, 2008). A recent meta-analysis documented that gene-environment correlation is pervasive, with the average heritability of measures of psychosocial risk being 27% (Kendler & Baker, 2007). Nonetheless, it is important to recognize that genetically-influenced factors can still exert environmentally mediated effects on outcome (T.G. O'Connor, Deater-Deckard, Fulker, Rutter, & Plomin, 1998; Rutter, Pickles, Murray, & Eaves, 2001). That is, exposure to deviant peers may both be a manifestation of a heritable disposition and also result in increased substance misuse through the mechanisms of peer facilitation and encouragement.
A second process that is also fundamental to understanding the joint influence of genetic and environmental factors is gene-environment interaction (Thapar, Harold, Rice, Langley, & O'Donovan, 2007). Rather than being uniform across individuals, genetic influences are in many cases likely to depend on environmental context. For example, there is a growing research literature indicating that genetic influences on cognitive ability are diminished in environments that do not provide adequate opportunity for intellectual stimulation (Taylor, Roehrig, Hensler, Connor, & Schatschneider, 2010; Turkheimer, Haley, Waldron, D'Onofrio, & Gottesman, 2003). Alternatively, other research has shown that genetic influences on adolescent misbehavior are amplified in the presence of peer deviance, substance availability, and adverse family environments (Agrawal, et al., 2010; Button, Lau, Maughan, & Eley, 2008; Feinberg, Button, Neiderhiser, Reiss, & Hetherington, 2007; T. G. O'Connor, Caspi, Defries, & Plomin, 2003) and diminished in positive contexts such as being academically or prosocially engaged (Hicks, et al., 2009; Johnson, et al., 2010).
Characterizing the Nature of Environmental Influence
The goal of GxE research is, of course, to determine whether and how environmental factors modulate genetic influences. The existence of G-E correlation, however, raises the possibility that what may look like GxE may in fact be GxG (Jaffee & Price, 2007). Advancing GxE research will require not only better phenotypic or endophenotypic measurement and identification of the relevant genetic variants, it will also require proper characterization of environmental effects. Behavioral genetic methodology has a major role to play in this effort (Rutter, 2007b). We have already provided two examples where behavior genetic methodology was instrumental in resolving an ambiguous association between a putative environmental risk factor and behavioral outcome. In the first, the method of Mendelian Randomization (MR) was used to determine that adolescent alcohol use may not be a gateway to other substance abuse in adolescence (Irons, et al., 2007). But MR depends on the existence of well-characterized causal genetic variants that mimic environmental exposure (Smith & Ebrahim, 2003). Given the yield to date from large-scale GWAS, it is reasonable to expect that MR will see limited application in psychology, at least in the near-term. Nonetheless, it could be used more than it is now. For example, there are enough obesity-related genetic variants identified for MR to be used to effectively explore the psychological consequences of obesity (Timpson, et al., 2009).
Our second example involved the use of an adoptive-family design to determine that the environmental consequences of parent smoking were specific to adolescent smoking. Adoption studies are one of the most powerful methods for identifying familial environmental influences, although they have been rarely utilized within developmental psychology. This is perhaps, in part, a consequence of the logistical challenges associated with undertaking an adoption study. It is also likely a result of the belief that fundamental differences between adoptive and non-adoptive families severely limit the generalizability of adoption research (Rutter, et al., 2001). While there are certainly differences between adoptive and non-adoptive families, in general these differences are small (Rueter, Keyes, Iacono, & McGue, 2009) and when they are not can be accounted for by proper research design (McGue, et al., 2007). Adoption studies, as well as variants based on assisted reproductive technologies, are underutilized within psychology.
A cotwin control study represents a third behavioral genetic design for characterizing the nature of environmental influence. Unlike the previous two, the cotwin control design is widely used, at least by behavioral geneticists. The logic of the design derives from the counterfactual model of causality (McGue, Osler, & Christensen, 2010). Briefly, MZ twins who are discordant on exposure to some putative environmental agent represent an approximation to the ideal counterfactual design that is arguably second only to that of a randomized experiment. This is because MZ twins share a genotype and a rearing environment. So, for example, if early age of alcohol initiation is a risk factor for alcoholism in adulthood, then within MZ twin pairs discordant for early alcohol use we should observe greater alcoholism risk among the twins who are early users than those who are not (McGue, Iacono, Legrand, & Elkins, 2001; Prescott & Kendler, 1999).
An early application of the cotwin control design involved female twins discordant for childhood sexual abuse. Within discordant pairs, the abused twin was significantly more likely to develop an alcohol use disorder than the non-abused twin (Kendler, et al., 2000), indicating that the association between childhood sexual abuse and adult alcohol use disorder could not be attributed to confounding with family environment or genetic factors. Subsequently, the cotwin control design has been used to explore a broad range of putative environmental agents, including for example the impact of early cannabis abuse on drug use escalation (Grant, et al., 2010; Lynskey, et al., 2003), the impact of college attendance on drinking (Slutske, et al., 2004), and early adolescent sexual behavior on risky adult sexual behavior (Huibregtse, Bornovalova, Hicks, McGue, & Iacono, 2011).
Testing for Genetic Interactions at the Level of the Genome
GxE interactions in association studies have been even more difficult to identify than main effects, despite early promising leads (Duncan & Keller, 2011). The classic example of GxE in psychology involves the 5-HTTLPR (serotonin-transporter-linked polymorphic region) in the promoter region of SLC6A4, the serotonin transporter gene. 5-HTTLPR was found in a seminal GxE study to interact with a measure of stressful life events to predict adult depression (Caspi, et al., 2003). This result is perhaps the most well-studied and well-characterized interaction in psychiatric genetics. Yet, a decade since the 2003 study, the existence and extent of the interaction remains unclear with disagreement among experts, as well as dueling meta-analyses provided in support of very different conclusions (Caspi, Hariri, Holmes, Uher, & Moffitt, 2010; Karg, Burmeister, Shedden, & Sen, 2011; N. Risch, et al., 2009). Regardless of whether this interaction is real, the limitations of existing GxE research to clearly resolve the issue should motivate psychologists to critically evaluate existing GxE methodology.
The Caspi, et al. (2003) GxE model has provided a popular blue-print followed by investigators world-wide in testing for GxE effects. The approach is to select environments and candidate genes—usually measured by a handful of common SNPs—based on a priori hypothesis about functional importance. It has been called for this reason a candidate GxE (cGxE) approach. While theory-guided approaches are without doubt reasonable in some cases, they have yet to deliver in behavior genetics research. Indeed, much has been learned about genomic function since the Caspi study was published, including our knowledge (from GWAS) of the vanishingly small main effects for individual SNPs. It takes considerably more power to detect interaction effects compared to main effects (e.g., a common rule of thumb is four times as many subjects; Thomas, 2010a), owing in large part to the multiplicity of alternative hypotheses tested. Most candidate gene studies, including the original Caspi study, are undertaken on very small samples (the majority with N < 500), such that power to detect an effect is miniscule and Type-I errors common (Duncan & Keller, 2011). To place this in context, the average main effect for SNPs known to be associated with height account for 0.06% of the variance. To have 80% power to detect a GxE interaction of similar magnitude at alpha of .05 would require more than 10,000 unrelated participants.
From a genome-wide perspective, 80% power to detect the same effect at p < 5×10-8 would require a sample size of ~53,000 participants. Clearly, gene by environment-wide interaction studies (GEWIS) can be even more problematic than candidate GxE (Thomas, 2010a, 2010b; Thomas, Lewinger, Murcray, & Gauderman, 2011), as the many tests required in a GEWIS severely compounds the already low statistical power to detect an effect. Given the infrequency with which phenotypes and environments are consistently collected across studies, it appears infeasible to amass the sample sizes necessary to detect a GxE effect at genome-wide significant levels.
GEWIS is not feasible and, for whatever reason, the a priori selection of candidate genes based, e.g., on findings in model organisms (Caspi, et al., 2010), is not currently working. We argue that there are several ways forward.
Current atheoretical GWAS approaches can be tweaked to optimize analysis for identifying GxE interactions. These methods include A) filtering the set of common SNPs to retain only those that have promise for interaction, which we explore in the next section; and B) combining SNPs into polygenetic scores, and testing the score for interaction with the environment.
GWAS, GxE, but most importantly psychological theory, will benefit from a developmental perspective.
The standard candidate gene approach has been far from optimal, but is due to make a re-appearance new and improved. Improved phenotypic and environmental measures, along with vastly improved genomic knowledge, can revive candidate gene approaches.
Emerging Approaches to Genetic Interactions
Filtering Variants to Enrich for Interaction
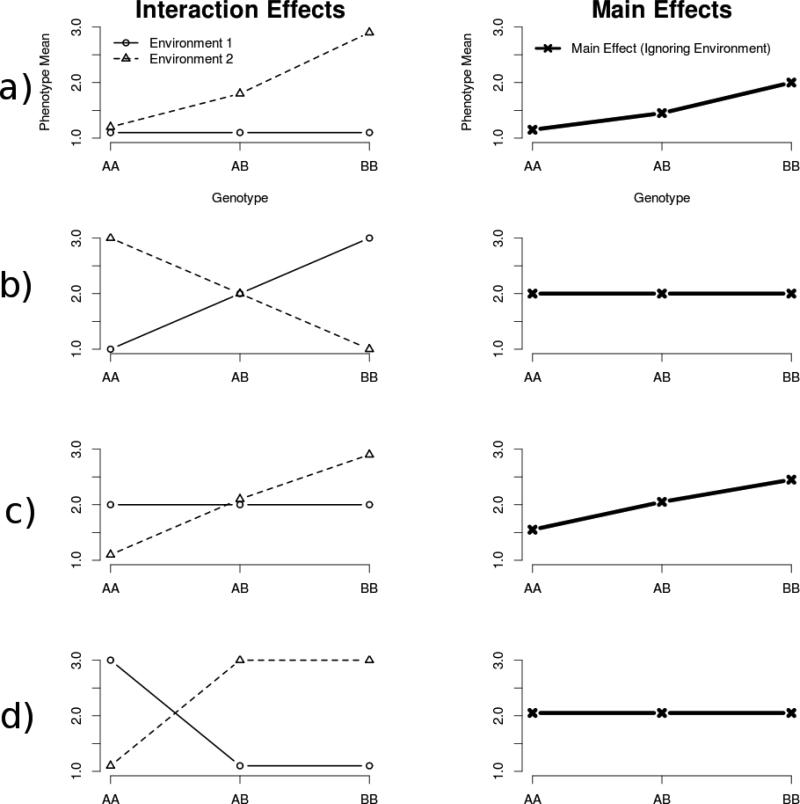
The genomically atheoretical approach of GWAS will not work for GxE because the testing burden is simply too heavy. However, we can use our general knowledge about statistical interactions to devise procedures to filter the vast number of common SNPs, resulting in a much smaller subset probabilistically enriched to show interactions (Thomas, 2010a). We briefly discuss three ways to do this. First, filter out any SNP that does not show a main effect for the phenotype. SNPs in the subset of variants with main effects are known to affect the phenotype in the first place, and are more likely to be differentially relevant depending on the environment. We can see this in Figure 3. The figure portrays a series of possible interactions between genotype, environment, and phenotype. The genotype is a single SNP with alleles A and B. On the left-hand side of the figure is a sampling of the many possible interaction effects. For each interaction we also give the corresponding main effect that would arise under that interaction. The main effects are given on the right-hand side of the figure. Notice that some interactions imply a main effect (Figure 3a & 3c), while some do not (Figure 3b and 3d). Since we generally have more power to detect main effects than interaction effects of similar size, we can use the relationship between main effects and interactions to our advantage. If we restrict tests of interaction only for those SNPs with demonstrated main effect, for example, we can greatly reduce the genome-wide testing burden by rightfully ignoring the massive number of SNPs that lack a main effect. Additionally, we expect the subset of SNPs with a main effect to be enriched for GxE interactions (Marchini, Donnelly, & Cardon, 2005; Thomas, 2010a).
Figure 3.
Association between GxE interaction effects and G main effects. Graphs are organized into rows. The left-hand graph in each row is an example GxE interaction; on the right is the corresponding main effect (assuming the environmental exposure was 50:50 in this sample). The GxE effect observed in row (a), similar to that found in (Caspi, et al., 2003), also shows a main effect when environment is ignored. The effect in row (a) would also show unequal variances across genotypes due to the increasing mean separation between environments as we move from the AA genotype to the BB genotype. The effect in row (b) shows an interaction but no main effect. Row (b) would show unequal variances across genotypes. Row (c), although similar to row (b) would show an interaction, a main effect, and unequal variances. Row (d) is unique in that the effect there would show an interaction, but would demonstrate neither a main effect nor unequal variances.
There are many ways to filter the vast number of SNPs by main effect. Perhaps the best method is to use variants identified in the literature, preferably in large meta-samples. However, using only those variants identified as genome-wide significant (p < 5×10-8) may be too restrictive. A more exploratory GxE test might consider, perhaps, the top 100 or 1,000 SNPs with the largest estimated main effects, regardless of whether they are significant at p < 5×10-8. The multiple testing burden for 100 SNPs is p < .0005, or 5×10-4, far better than 5×10-8 incurred for testing all common SNPs. The actual number of candidate SNPs to include in such an exploratory study could easily be informed by power calculations.
Notice that filtering by main effect only works for a subset of possible interactions, as can also be seen in Figure 3. Only Figure 3a and 3c 3 show a main effect in addition to interactions. Filtering SNPs by main effect will also remove all SNPs that demonstrate interactions in the absence of main effect (Figure 3b and 3d). There are other ways to filter SNPs to enrich for interactions in the absence of main effects. For example, one might do a test of equality of variances across genotypes (Pare, Cook, Ridker, & Chasman, 2010). As can be seen in Figure 3b, for example, one would expect the variances to differ as a function of genotype. Collapsing across environment, the variance of the AA allele would be larger than the BB allele, because of the mean differences due to environment for the AA and BB genotypes that is not present for the AB genotype. In fact, every example in Figure 3 would fail a test of equality of variance (assuming sufficient statistical power) except for Figure 3d, which is a theoretically possible interaction that shows neither a main effect nor inequality of variance across alleles. Genotypes that fail a test of equality of variances would be statistically enriched for interactions and prioritized in subsequent direct tests of GxE interaction. Note however, that even Figures 3d and 3b (i.e., “cross-over” interactions) would demonstrate main effects if genotypes and environments are unequally balanced.
Combining SNPs into polygenetic SNP scores
While the individual effect of a SNP is expected to be small, rendering the expected environmental interaction effect small, the aggregate effects of SNPs can be quite large. In the meta-analytic literature on genetic main effects it has become common to sum SNPs by their univariate regression weight, resulting in a single polygenetic score for each individual (e.g., see Purcell, et al., 2009; Vrieze, McGue, et al., 2011). The resulting score can have a measureable effect on the phenotype, even in samples sizes in the thousands, allowing individual investigators to independently test interactions without need for combining studies meta-analytically.
Polygenetic scores are not as biologically informative as individual SNP (or gene) effects, but can help inform the genetic and environmental architecture of diseases and traits. For example, our group has recently conducted work on a polygenetic GxD test for cigarettes smoked per day (Vrieze, McGue, & Iacono, 2012). The score consisted of a sum of the top 91 SNPs reported in a recent GWAS meta-analysis of cigarettes smoked per day in ~75,000 smokers (Furberg, et al., 2010). In a sample of ~3,000 twins we found that the SNP score was irrelevant at ages 14 and 17, but was significantly associated with number of cigarettes smoked per day at ages 20 and 24, indicating a GxD interaction. We speculate that development here is a proxy for the environmental changes that occur between age 17 and 20, and serve to activate genetic influences on smoking. Most individuals at ages 14 and 17 have difficulty finding the opportunity to smoke more than a handful of cigarettes per day, if they smoke at all. Smoking is illegal in the U.S. at these ages, and the vast majority of youths are under teacher and/or parent supervision for most of their waking hours, limiting but by no means eliminating their opportunity to smoke. This environmental restriction on smoking is expected to temper the effect of a risky genotype. At ages 20 and 24, however, smoking is legal, and young adults often live and work independently, removing the environmental restriction on their genotypic propensity to smoke more.
Unfortunately, there does not exist for every phenotype a meta-analytic GWAS of 100,000 individuals from which to select SNPs with demonstrated main effects. Even so, several options remain. One brute force method is to obtain polygenetic scores without meta-analytic guidance, often by summing across tens of thousands of SNPs (Simonson, Wills, Keller, & McQueen, 2011). The method often involves obtaining the univariate effect of each SNP, culling SNPs in high linkage disequilibrium (LD) with more significant SNPs, then adding the remaining SNPs according to their univariate regression weight. Removing weaker SNPs in high LD with stronger SNPs removes likely LD-induced artifacts. Depending on the LD threshold, this method results in thousands to hundreds of thousands of SNPs comprising the resulting score. The method has proven useful with the usual anthropometric traits like height (Allen, et al., 2010) and BMI (Speliotes, et al., 2010), and also in schizophrenia and bipolar disorder (Purcell, et al., 2009). The score can then be incorporated in tests of environmental and developmental moderation.
Another viable method to evaluate aggregated SNP interactions is Genome-wide Complex Trait Analysis (GCTA; Yang, Lee, et al., 2011), described in the introduction. GCTA is a method by which one can estimate the variance in the phenotype accounted for by measured SNPs. The method uses the estimated genetic relatedness between individuals in the sample and models all SNPs as a random effect in a mixed effects model. Mixed effects models can include interaction terms between random and fixed effects (Pinheiro & Bates, 2000), and GCTA has a built-in facility for evaluating GxE interaction, where the G is the random effect of all measured SNPs and E is the fixed environmental measurement. At present, the software is restricted to discretely measured environments (e.g., was exposed to disease pathogen, yes/no), but quantitative measures of the environment are planned for future updates to the algorithm (Jian Yang, personal communication, February 13, 2012). It stands to reason that longitudinal repeated measures could also be implemented in a GCTA framework, allowing tests of aggregate SNP effects on developmental trajectories. There is great promise to extending GCTA to environmental and developmental interaction, as any study with genome-wide SNP data becomes immediately genetically informative, much like a twin study. Twin studies are difficult or impossible to ascertain for rare phenotype/environment combinations, and GCTA could provide a workaround in evaluating the environmental moderation of genetic effects in these situations. In the context of a rare natural event (often called a “natural experiment” (Rutter, 2007a)), such as a war or natural disaster, one could imagine genotyping survivors and comparing them to matched controls, conducting a test of GxE using GCTA to determine GxE interaction for phenotypes such as post-traumatic stress disorder. This sort of research, without using GCTA however, has been done with hurricane victims and 5-HTTLPR (Kilpatrick, et al., 2007).
Genome-wide polygenetic scores and GCTA have provided insight about the genetic architecture of disease, but they otherwise lack direct biological interpretability due to the fact that GCTA considers the genome-wide aggregate effect of hundreds of thousands of SNPs simultaneously. Such scores need not be genome-wide, however, and application of a scoring approach to a genomic region can provide improved biological relevance. Using GCTA in a sample of 12,000, Yang, Manolio, et al. (2011) found that SNPs in genes explained 38% of the variance in height, whereas SNPs in intergenic regions explained only 8%, even though the intergenic regions made up roughly half of the genome. Taking this approach one step further, Lee, et al., (2012) found that genes involved in central nervous system function accounted for ~30% of the diagnostic variance in schizophrenia, whereas they physically comprise only 20% of the genome. Other genes also accounted for ~30% of the variance, but physically comprise ~40% of the genome. As sample sizes grow, more refined subsets of genes can be evaluated. For alcoholism, for example, one might investigate the proportion of variance accounted for by SNPs in genes known to be involved in alcohol metabolism, or candidate neurotransmitter systems such as GABA, dopamine, or serotonin.
Role of GxD in evaluating GxE
GxE studies to date largely have not considered the role of development. Development can inform genetic studies in at least two ways. 1) It can serve to modify the GxE interaction, resulting in a GxExD interaction. 2) It can serve as a proxy for the environment in GxE. That is, changes in the environment can correlate strongly with changes in age or developmental stage, as illustrated in our work on cigarette smoking where the protective (or, at least, interfering) childhood environment becomes less influential as adolescents age. On a much more basic level, genomic regions have been linked to embryonic development (Woolfe, et al., 2005). These regions are often involved in regulating protein transcription from genes involved in development and cell integrity, are critical in organisms from humans to dogs to fish, and have been highly conserved across animal species (Lindblad-Toh, et al., 2005; Pennacchio, et al., 2006).
Like cGxE, candidate GxD (cGxD) studies involve the selection of a promising genomic region, such as a gene, and observation of how variants in that region are associated with a phenotype at different ages or developmental stages. An excellent example of a cGxD study involves a SNP in the gene FTO known to affect BMI in adults (Speliotes, et al., 2010) by affecting one's perception of satiation after eating. In a longitudinal meta-analysis of 8 studies Sovio, et al. (2011) demonstrated divergent developmental trajectories for different alleles of SNP rs9939609. They found that carriers of the at-risk minor allele of rs9939609 in FTO was significantly associated with a lower BMI in toddlerhood, an earlier adiposity rebound around age 5, and higher BMI in early adolescence. The study demonstrates well the potential complex interplay between genes and development that can be assessed with relatively small sample sizes – the Sovio, et al. (2011) study had, on average, ~9,500 participants at any given age of assessment. By the same token, it stresses the need for individual investigators to share existing data and collaborate on meta-analytic endeavors.
Our group at Minnesota took the cGxD paradigm in a slightly different direction (Vrieze, McGue, et al., 2011). Our goal was to understand how the Allen, et al. (2010) meta-analytic SNP findings for height play out developmentally during the pubertal growth spurt. We tested for association of 176 of those SNPs with both pre-pubertal height and the pubertal growth spurt in a sample of 3,187 twins of Caucasian ancestry from the Minnesota Center for Twin and Family Research (see Miller, et al., submitted), for a description of the study). No individual SNP was associated with pre-pubertal height, or the pubertal growth spurt, at genome-wide significance. Aggregating the SNPs into a polygenetic score—summing them together by their meta-analytic weights to form a single SNP score—resulted in a much stronger association with pre-pubertal height (p=1×10-13, r2 = 4.5%) than with the pubertal growth spurt (p=.004). The results indicated that the SNPs identified by the meta-analysis were more relevant for growth in stature between conception and 10 years of age, and largely did not explain variation in the rate at which individuals grew after the onset of puberty.
cGxD approaches based on a priori hypotheses about gene function have also proven useful in some instances. The Irons et al. (2007) study, involving the gene ALDH2 and described in detail earlier was expanded in Irons, Iacono, Oetting, and McGue (2012), where the authors tested the extent to which the protective effect of ALDH2 changed during adolescence. Recall that nearly 50% of individuals of East Asian ancestry inherit a variant in the ALDH2 gene that diminishes their ability to metabolize alcohol, and that those who inherit the deficient gene variant are likely to experience various signs of dysphoria following ingestion of even a small amount of alcohol. The deficient variant of ALDH2 was found to be protective against drinking during early adolescence (14-17) and grew increasingly protective as the individuals aged. The general results are consistent with the Vrieze, et al. (2012) findings for cigarette smoking. That is, in adolescence the genotype effect existed but was muted, likely mitigated by protective childhood environmental circumstances. As the individuals aged, their genotype increasingly exerted influence on behavior.
The Irons et al. (2012) finding is an excellent example of GxD effects for a psychiatric condition, and demonstrates how a genotype expresses differentially dependent on developmental stage or, perhaps just as likely, environmental context. As the youths develop, their environment systematically changes, often in ways that can inform knowledge of the interface between genes and environment.
Complementing GWAS: Rare and Structural Variants
Common SNPs used in GWAS studies represent a small fraction of the entirety of genetic individual differences. GWAS disregards entirely rare SNPs and structural variation. Recall the GCTA findings described in the introduction above. At best, common SNPs account for less than half of the total additive genetic variance to be explained. For the only psychiatric disease to be rigorously evaluated thusfar (schizophrenia), they account for a quarter. While nothing to sneeze at, this leaves a large proportion of the heritability of schizophrenia unexplained by common SNPs. There are many explanations for the missing heritability (Eichler, et al., 2010; Zuk, Hechter, Sunyaev, & Lander, 2012), including the existence of gene by environment interactions (Manolio, et al., 2009; Thomas, 2010a), but we focus on one: the fact that genetic variants in humans are not limited to common SNPs. Other types of variants are displayed in Figure 1, where we notice insertions, deletions, substitutions, inversions, and variable tandem number repeats, and copy number variants. Forms of genetic variation other than common SNPs are no doubt relevant for disease, and are just as likely to be environmentally and developmentally moderated as common SNPs.
Rare and structural variation is obtained by sequencing, which has not until recently become technologically or financially feasible. In current genome sequencing (called “shotgun sequencing”) the approach is to obtain several reads (or measurements) of each base pair. Because each read is relatively error-prone, it is desirable to obtain many reads (e.g., > 30×) for each base pair to correctly identify the actual nucleotide. The average number of reads for each base pair is termed the “depth” of sequencing; the deeper the coverage the higher the average number of reads and the more precise the genotype calls. Ideally, many individuals would be sequenced at high depth, which would provide accurate genotyping as well as large samples sizes (and power) for tests of phenotype-genotype associations. However, each read costs money, and there is thus a tradeoff between the number of individuals sequenced and the depth of sequencing. An optimal tradeoff can be selected, but depends on the research question (Li, Sidore, Kang, Boehnke, & Abecasis, 2011).
Let us first provide some context and motivation for whole genome sequences. The immediate goal of GWAS was to identify SNP associations with the phenotype. Because significant SNPs in GWAS are unlikely themselves to be functional (i.e., causal), but rather to be in linkage disequilibrium (LD) with functional variants, the region around a significant SNP would be subjected to further genotyping and/or sequencing to determine how that region, which the SNP has “tagged,” is causally related to the phenotype. The common SNPs thus would provide a foothold into biological investigations of etiology. There have been dramatic examples of this logical progression from GWAS hits to regional targeted sequencing and ultimate elucidation of functional variants. We discuss these to shift our attention from SNPs, which represent very minor perturbations of the genome, to genes, which are massive genomic regions integral to human biology and existence.
In a meta-analysis of GWAS on lipids, Teslovich, et al. (2010) found 95 SNPs genome-wide significant for lipid traits, such as plasma concentrations of cholesterol measures. As usual, each SNP in the GWAS had only very small effect. However, the investigators then developed mouse models to validate three genes that contained SNPs identified in the meta-analysis. In the mouse models the investigators did not simply perturb the GWAS significant SNPs; rather, they knocked out or overexpressed the entire gene, with dramatic results. The experimental mice showed marked differences in HDL cholesterol compared to controls. The results highlight the fact that SNPs identified by GWAS are “tag” SNPs, are unlikely to be functional themselves, but which can point to a gene or larger genomic region that, after further study, turns out to be phenotypically critical.
A developmentally relevant example is in human growth. Widen, et al. (2010) conducted a GWAS on growth in height during puberty. One SNP, located in LIN28B, was genome-wide significant for pubertal growth, and had previously been associated with timing of puberty (Ong, et al., 2009) and later further validated by Vrieze, McGue, et al. (2011). Similar to the work on lipids, researchers developed a mouse model where the mouse gene analog, lin28a, was overexpressed. The transgenic mice showed dramatic change in rate of growth during puberty, change in pubertal initiation, and vastly different adult height and size (Zhu, et al., 2010). Again, GWAS SNPs may only weakly point to a gene, even though the gene as a whole is crucial to phenotypic development.
Genes are the part of the genome that actually encode amino acids and proteins, and are made up of promoters, enhancers, exons, introns, and other gene units. Promoters and enhancers determine the rate of transcription of a gene – strong promoters tend to result in higher rates of transcription, and thus more of the protein(s) encoded by the gene. Weak promoters result in less gene production. The exonic regions of a gene are those regions that actually encode amino acids and proteins. Even a single mistake in the exon can completely change the protein encoded by the gene. In some cases a non-functional protein is produced, in others a chemically different but functional protein is produced. For example, a nonsense mutation can break a gene entirely, resulting in complete loss of function, similar to that discussed above for the mouse models. A good example of exomic mutations with phenotypic impact is in cystic fibrosis, where 1404 different exomic mutations have been catalogued for the CFTR gene (http://www.genet.sickkids.on.ca/cftr/Home.html). Because only a single functional copy of CFTR is required for normal function, a child inherits cystic fibrosis if both the maternal and paternal genes carries at least one non-functional mutation. In another, more sensational example, researchers used exome sequencing to make a novel diagnosis of congenital chloride diarrhea for a patient who was referred for genetic testing for diagnostic clarification (Choi, et al., 2009). The study demonstrated the potential clinical utility of genome sequences for phenotypically extreme cases that are not easily diagnostically classified.
Our knowledge of gene structure and function can be leveraged in analysis of genotype-phenotype associations. Major efforts have been conducted at characterizing variation within the exome, a term used to describe the collection of all exonic coding regions contained within the genome. The 1000 Genomes consortium sequenced 697 individuals at an average depth of 56× (very deep), identifying 12,758 SNPs with minor allele frequency > 0.1% (Altshuler, et al., 2010). Thanks to these efforts, all these exomic SNPs can now be genotyped with a relatively inexpensive exome SNP chip now commercially available.
One problem with exomic association tests is that loss-of-function mutations are likely to very rare, due to strong evolutionary selection against these variants. Since power is extremely low for rare variants (de Bakker, et al., 2005), different analytic strategies are used/devised to handle them (Bansal, et al., 2010). One attractive method is to determine the functionality of each SNP within some region, such as a gene, and then predict the extent to which that region is impaired (Cooper & Shendure, 2011; Torkamani, Scott-Van Zeeland, Topol, & Schork, 2011). For example, if an individual carries even one nonsense exonic mutation within a gene then the gene for that person would be predicted to be non-functional. This is precisely the etiology of cystic fibrosis, where there are many potential nonsense exonic mutations (among other types of mutations), all of which lead to the same phenotype. Regardless of the phenotype, one can test, for example, whether cases versus controls are more likely to have non-functional genes. Regression analysis can be used for quantitative traits, regressing the phenotype onto predicted functionality for each gene. ENCODE and other extensive databases are continuously updated with reams of genomic information. To give a few: dbSNP catalogues all identified SNPs in any organism and their population characteristics (e.g., minor allele frequency, chromosomal position); OMIM catalogues known disease-causing mutations; and the ENCODE consortium is an attempt to discover, define, and list the functional elements of the human genome (Myers, et al., 2011). Software has been developed to automatically leverage the immensity of information in these databases and apply them to individual genotypes in our studies. A list of such software is given in Cooper and Shendure (2011).
While not yet widely applied to psychiatric phenotypes, exome sequencing has shown promise in identifying genes relevant to phenotypes ranging from autism (O'Roak, et al., 2011) and schizophrenia (Xu, et al., 2011) to mental retardation (Vissers, et al., 2010). As exome sequencing becomes more affordable and the exome SNP chip more commonly used, we expect exomic studies of psychiatric disease and psychological traits to become increasingly common.
Structural variation is a substantial source of genetic differences between individuals. Structural variants include copy number variants (CNVs), insertions/deletions (indels), substitutions, and inversions, all of which are displayed in Figure 1. The extent of this variation is immense, and has only recently been well-characterized thanks to the 1000 Genomes Consortium and related projects. Altshuler, et al. (2010) identified that individuals on average have 361,669 indels, 96 of which occurred in exonic regions. Recall that one deleterious variant within an exon can disrupt a gene entirely; it is not trivial that individuals on average have 96 exonic insertions or deletions. The same study found that individuals on average have 10,000 – 11,000 non-synonymous SNPs, and around 570 in-frame structural variants (not counting CNVs) that likely have greater functional impact than single SNPs. “Non-synonomous” means the SNP disrupts the protein coding sequence, while an “in-frame” variant simply means that the variant significantly overlaps with the parts of the gene that code a protein. Of these, ~370 of the variants are likely to result in loss-of-function in the gene, rather than gain-of-function. In total these loss-of-function variants affect ~275 genes per individual in the study. In a follow-up study of copy number variants, Mills, et al. (2011) identified 28,025 CNVs, 10,995 of which overlap with genes, in many cases including the exon (1,119). Structural variants represent a significant source of genetic variation, much of which theoretically has substantial import for the genetic etiology of disease (Stankiewicz & Lupski, 2010a).
Psychiatric genetics and behavior genetics have begun to evaluate CNVs in disease, with some notable results for schizophrenia (Levinson, et al., 2011), bipolar disorder (Priebe, et al., 2011), and autism (Cook & Scherer, 2008). Promising results have also been found for neurodevelopmental conditions such as ADHD (Elia, et al., 2010; Elia, et al., 2012). Other forms of structural variation, including shorter CNVs, can now feasibly be measured with sequencing pipelines and reference genotypes provided by the 1000 Genomes consortium (Mills, et al., 2011). As more main effects of structural variants are discovered, they must be leveraged along with rare and common SNPs to assist in elucidation of genetic, environmental, and developmental etiology of disease. All of the techniques discussed above for SNPs (except perhaps GCTA) are applicable to structural variation, and we expect research in this area to begin blossoming in the near future as sequencing becomes routine.
The Return of the Candidate Gene Study
In the world after the Human Genome Project, psychiatric and behavior genetics has swung from a field dominated by the candidate gene approach, and its a priori hypotheses, to one relying on the brute force of atheoretical GWAS. With the advent of affordable whole genome sequencing and functional annotation we expect the pendulum to begin swinging back. Science has, and will, benefit from data-driven approaches like GWAS, but scientific psychology requires theory to integrate findings and focus our experimental tests (Caspi, et al., 2010). A major stumbling block of the candidate gene and cGxE approach has been the lack of theory and understanding about genomics and genetic etiology—unknown at the time, the candidates were not so good and expected effect sizes were highly optimistic. GWAS, while not curing diseases, has been an exciting learning exercise. To exaggerate for effect, we now know that testing some genetic theory of behavior using small samples, genetically un-informed phenotypes, poorly measured environments, no consideration of developmental circumstance, and a handful of SNPs in a single gene won't do the trick. GWAS undoubtedly will continue to provide leads (Visscher, et al., 2012), and point the way to relevant genes. Encouraged by past success, ever more investigators are sharing data to arrive at the required massive sample sizes. Consortia are even popping up with developmental and environmental questions—our group is contributing to a consortium evaluating adolescent cigarette smoking, for example. Identified SNPs will point to genomic regions that will be subjected to further study. Transgenic mice will be created. Identified genes will be sequenced, variants annotated and grouped, and tested in new samples.
GWAS will only get us so far. Etiological psychological theories must be proposed, tested, and amended. While biological reductionism is perhaps not always preferable or possible (Miller, 2010), theories would do well to propose, derive, and test increasingly refined (endo)phenotypes. Multidisciplinary biological, neuroscientific, and psychological research will point to chemical and structural systems relevant to psychological (endo)phenotypes, such as neurochemical systems. Gene(s) known to be involved in those systems, if not the whole genome, will be cheaply sequenced. Leveraging knowledge of gene function, the entirety of genetic variation within the gene(s) can be tested for association with the (endo)phenotype. At the same time, psychological research on environmental etiology and measurement will continue, identifying environments likely to have causal phenotypic effects. Tests of GxE and GxD can commence once plausible candidate genes have been identified.
This brief outline of candidate gene research is not prescriptive; the steps by no means must proceed in that order. Rather, it is an attempt to describe what we see as important waypoints in the path to understanding genetic and environmental etiology of behavior. In some fields many of the steps have already been taken. There is a wealth of neuroscience and biological findings for alcoholism, for example, with excellent animal models providing a wealth of biological knowledge (Spear, 2000) including metabolic and neurotransmitter systems implicated in alcohol use and abuse (Hodgkinson, et al., 2008); plausible etiological theories in psychology (Iacono, et al., 2008); well-validated endophenotypes (Iacono & Malone, 2011); environmental etiology is continuously elucidated (Irons, et al., 2007; Keyes, et al., 2008), and several studies already obtaining whole genome sequences on human samples informative for alcohol/drug use, abuse, and addiction.
Summary
Simply getting off the ground and identifying robust and replicable genetic effects has been difficult. Nonetheless, the field as a whole has learned a lot about genes, environments, and behaviors in the process. Twin, adoption, and family studies formed the foundation, and continue to be indispensible in forming and refining psychological theory. With measured genetic variation this research can be taken to a more biologically and environmentally informative level, as behaviorally relevant genetic variants are discovered and their effects characterized. While common SNPs may account for a significant proportion of heritability, there remains a very substantial source of genetic structural variation that is only beginning to be tapped. Environmental and phenotypic definitions and measurements are likely far from optimal. Better measurements, including endophenotypes, help everyone, psychologist and geneticist alike, and will only improve the science and simultaneously increase power for genetic associations. Current directions in quantitative psychiatric disease definition in DSM-5 (Krueger, et al., 2011), efforts at standardized environmental measures like the PhenX Toolkit, ever-cheaper sequencing, and improved knowledge of genomic structure and function will provide investigators with powerful tools to disentangle genomic, environmental, and developmental etiology in psychological behavior and disease. The coming decade will be an exciting one for psychologists interested in understanding the causes of human behavior.
ACKNOWLEDGEMENTS
Support was provided by grants DA05147, AA09367, AA11886, DA13240, MH66140, and DA024417. Scott Vrieze also thanks Scott Sponheim for valuable support.
References
- Agrawal A, Balasubramanian S, Smith EK, Madden PA, Bucholz KK, Heath AC, et al. Peer substance involvement modifies genetic influences on regular substance involvement in young women. Addiction. 2010;105(10):1844–1853. doi: 10.1111/j.1360-0443.2010.02993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen HL, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DL, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders. 4th ed., text rev. 2000.
- Bansal V, Libiger O, Torkamani A, Schork NJ. Statistical analysis strategies for association studies involving rare variants. Nature Reviews Genetics. 2010;11(11):773–785. doi: 10.1038/nrg2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B, Bihari B, Kissin B. Event-related brain potentials in boys at risk for alcoholism. Science. 1984;225(4669):1493–1496. doi: 10.1126/science.6474187. [DOI] [PubMed] [Google Scholar]
- Bornovalova MA, Hicks BM, Iacono WG, McGue M. Familial transmission and heritability of childhood disruptive disorders. American Journal of Psychiatry. 2010;167(9):1066–1074. doi: 10.1176/appi.ajp.2010.09091272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. [Review]. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button TM, Lau JY, Maughan B, Eley TC. Parental punitive discipline, negative life events and gene-environment interplay in the development of externalizing behavior. Psychological Medicine. 2008;38(1):29–39. doi: 10.1017/S0033291707001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Hariri AR, Holmes A, Uher R, Moffitt TE. Genetic Sensitivity to the Environment: The Case of the Serotonin Transporter Gene and Its Implications for Studying Complex Diseases and Traits. American Journal of Psychiatry. 2010;167(5):509–527. doi: 10.1176/appi.ajp.2010.09101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science. 2003;297:851–854. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Choi M, Scholl UI, Ji WZ, Liu TW, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(45):19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455(7215):919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nature Reviews Genetics. 2011;12(9):628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- Costello EJ, Mustillo S, Erkanli A, Keeler G, Angold A. Prevalence and development of psychiatric disorders in childhood and adolescence. Archives of General Psychiatry. 2003;60(8):837–844. doi: 10.1001/archpsyc.60.8.837. [DOI] [PubMed] [Google Scholar]
- Cronbach LJ, Meehl PE. Construct validity in psychological tests. Psychological Bulletin. 1955;52(4):281–302. doi: 10.1037/h0040957. [DOI] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Molecular Psychiatry. 2011;16(10):996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PIW, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nature Genetics. 2005;37(11):1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- de Geus EJ. From genotype to EEG endophenotype: a route for post-genomic understanding of complex psychiatric disease? Genome Medicine. 2010;2(9):63. doi: 10.1186/gm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor MHM, Boomsma DI, de Geus EJC, Willemsen G, Hottenga JJ, Distel MA, et al. Meta-analysis of genome-wide association results in > 10.000 individuals for the big five personality traits. Behavior Genetics. 2009;39(6):643–643. [Google Scholar]
- DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemperary Clinical Trials. 2007;28(2):105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Dick DM. Gene-environment interaction in psychological traits and disorders. Annual Review of Clinical Psychology. 2011;7:383–409. doi: 10.1146/annurev-clinpsy-032210-104518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DL, Iles MM, Glass D, Zhu G, Barrett JH, Hoiom V, et al. IRF4 variants have age-specific effects on nevus count and predispose to melanoma. American Journal of Human Genetics. 2010;87(1):6–16. doi: 10.1016/j.ajhg.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. A Critical Review of the First 10 Years of Candidate Gene-by-Environment Interaction Research in Psychiatry. American Journal of Psychiatry. 2011;168(10):1041–1049. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nature Reviews Genetics. 2010;11(6):446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Molecular Psychiatry. 2010;15(6):637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia J, Glessner JT, Wang K, Takahashi N, Shtir CJ, Hadley D, et al. Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nature Genetics. 2012;44(1):78–U113. doi: 10.1038/ng.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg ME, Button TM, Neiderhiser JM, Reiss D, Hetherington EM. Parenting and adolescent antisocial behavior and depression: evidence of genotype × parenting environment interaction. Archives of General Psychiatry. 2007;64(4):457–465. doi: 10.1001/archpsyc.64.4.457. [DOI] [PubMed] [Google Scholar]
- Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychological Medicine. 2007;37(2):163–180. doi: 10.1017/S0033291706008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furberg H, Kim Y, Dackor J, Boerwinkle E, Franceschini N, Ardissino D, et al. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature Genetics. 2010;42(5):441–U134. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon ES, Alliey-Rodriguez N, Liu CY. After GWAS: Searching for Genetic Risk for Schizophrenia and Bipolar Disorder. American Journal of Psychiatry. 2011;168(3):253–256. doi: 10.1176/appi.ajp.2010.10091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Yang CR, Otani S. Functional and dysfunctional synaptic plasticity in prefrontal cortex: roles in psychiatric disorders. Biological Psychiatry. 2010;67(3):199–207. doi: 10.1016/j.biopsych.2009.08.026. [DOI] [PubMed] [Google Scholar]
- Grant JD, Lynskey MT, Scherrer JF, Agrawal A, Heath AC, Bucholz KK. A cotwin-control analysis of drug use and abuse/dependence risk associated with early-onset cannabis use. Addictive Behaviors. 2010;35(1):35–41. doi: 10.1016/j.addbeh.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood TA, Lazzeroni LC, Murray SS, Cadenhead KS, Calkins ME, Dobie DJ, et al. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. American Journal of Psychiatry. 2011;168(9):930–946. doi: 10.1176/appi.ajp.2011.10050723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood TA, Light GA, Swerdlow NR, Radant AD, Braff DL. Association analysis of 94 candidate genes and schizophrenia-related endophenotypes. PLoS One. 2012;7(1):e29630. doi: 10.1371/journal.pone.0029630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove WM. When Is a Diagnosis Worth Making - a Statistical Comparison of 2 Prediction Strategies. Psychological Reports. 1991;69(1):3–17. [PubMed] [Google Scholar]
- Hamilton CM, Strader LC, Pratt JG, Maiese D, Hendershot T, Kwok RK, et al. The PhenX Toolkit: Get the Most From Your Measures. American Journal of Epidemiology. 2011;174(3):253–260. doi: 10.1093/aje/kwr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Eskin E. Interpreting meta-analyses of genome-wide association studies. PLoS Genetics. 2012;8(3) doi: 10.1371/journal.pgen.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden KP, Hill JE, Turkheimer E, Emery RE. Gene-environment correlation and interaction in peer effects on adolescent alcohol and tobacco use. Behavior Genetics. 2008;38(4):339–347. doi: 10.1007/s10519-008-9202-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heils A, Teufel A, Petri S, Stober G, Riederer P, Bengel D, et al. Allelic variation of human serotonin transporter gene expression. Journal of Neurochemistry. 1996;66(6):2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- Hicks BM, Schalet BD, Malone SM, Iacono WG, McGue M. Psychometric and genetic architecture of substance use disorder and behavioral disinhibition measures for gene association studies. Behavior Genetics. 2011;41(4):459–475. doi: 10.1007/s10519-010-9417-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks BM, South SC, DiRago AC, Iacono WG, McGue M. Environmental Adversity and Increasing Genetic Risk for Externalizing Disorders. Archives of General Psychiatry. 2009;66(6):640–648. doi: 10.1001/archgenpsychiatry.2008.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nature Reviews Genetics. 2005;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Yuan QP, Xu K, Shen PH, Heinz E, Lobos EA, et al. Addictions biology: Haplotype-based analysis for 130 candidate genes on a single array. Alcohol and Alcoholism. 2008;43(5):505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse BM, Bornovalova MA, Hicks BM, McGue M, Iacono W. Testing the Role of Adolescent Sexual Initiation in Later-Life Sexual Risk Behavior: A Longitudinal Twin Design. Psychological Science. 2011;22(7):924–933. doi: 10.1177/0956797611410982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Carlson SR, Malone SM. Identifying a multivariate endophenotype for substance use disorders using psychophysiological measures. International Journal of Psychophysiology. 2000;38(1):81–96. doi: 10.1016/s0167-8760(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Malone SM. Developmental Endophenotypes: Indexing Genetic Risk for Substance Abuse With the P300 Brain Event-Related Potential. Child Development Perspectives. 2011;5(4):239–247. doi: 10.1111/j.1750-8606.2011.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, McGue M. Behavioral disinhibition and the development of early-onset addiction: Common and specific influences. Annual Review of Clinical Psychology. 2008;4:325–348. doi: 10.1146/annurev.clinpsy.4.022007.141157. [DOI] [PubMed] [Google Scholar]
- Iacono WG, McGue M, Krueger RF. Minnesota Center for Twin and Family Research. Twin Research and Human Genetics. 2006;9(6):978–984. doi: 10.1375/183242706779462642. [DOI] [PubMed] [Google Scholar]
- The International HapMap Project Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Ioannidis JPA, Castaldi P, Evangelou E. A Compendium of Genome-Wide Associations for Cancer: Critical Synopsis and Reappraisal. [Review]. Journal of the National Cancer Institute. 2010;102(12):846–858. doi: 10.1093/jnci/djq173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons DE, Iacono WG, Oetting WS, McGue M. Developmental trajectory and environmental moderation of the effect of ALDH2 polymporphism on alcohol use. Alcoholism: Clinical and Experimental Research. 2012 doi: 10.1111/j.1530-0277.2012.01809.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons DE, McGue M, Iacono WG, Oetting WS. Mendelian randomization: A novel test of the gateway hypothesis and models of gene-environment interplay. Development and Psychopathology. 2007;19(4):1181–1195. doi: 10.1017/S0954579407000612. [DOI] [PubMed] [Google Scholar]
- Jaffee SR, Price TS. Gene-environment correlations: a review of the evidence and implications for prevention of mental illness. [Review]. Molecular Psychiatry. 2007;12(5):432–442. doi: 10.1038/sj.mp.4001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W, Kyvik KO, Mortensen EL, Skytthe A, Batty GD, Deary IJ. Education reduces the effects of genetic susceptibilities to poor physical health. International Journal of Epidemiology. 2010;39(2):406–414. doi: 10.1093/ije/dyp314. [DOI] [PubMed] [Google Scholar]
- Johnson W, McGue M, Iacono WG. Genetic and environmental influences on academic achievement trajectories during adolescence. Developmental Psychologyl. 2006;42(3):514–532. doi: 10.1037/0012-1649.42.3.514. [DOI] [PubMed] [Google Scholar]
- Kaffman A, Krystal JJ. New frontiers in animal research of psychiatric illness. Methods in Molecular Biology. 2012;829:3–30. doi: 10.1007/978-1-61779-458-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel DB, Jessor R. The gateway hypothesis revisited. In: Kandel DB, editor. Stages and Pathways of Drug Involvement. Cambridge University Press; New York: 2002. [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, et al. Variance component model to account for sample structure in genome-wide association studies. Nature Genetics. 2010;42(4):348–110. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Archives of General Psychiatry. 2011;68(5):444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Baker JH. Genetic influences on measures of the environment: a systematic review. [Review]. Psychological Medicine. 2007;37(5):615–626. doi: 10.1017/S0033291706009524. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Bulik CM, Silberg J, Hettema JM, Myers J, Prescott CA. Childhood sexual abuse and adult psychiatric and substance use disorders in women - An epidemiological and Cotwin control analysis. Archives of General Psychiatry. 2000;57(10):953–959. doi: 10.1001/archpsyc.57.10.953. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. American Journal of Psychiatry. 2003;160(4):687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- Keyes MA, Iacono WG, McGue M. Early onset problem behavior, young adult psychopathology, and contextual risk. Twin Research and Human Genetics. 2007 doi: 10.1375/twin.10.1.45. [DOI] [PubMed] [Google Scholar]
- Keyes MA, Legrand LN, Iacono WG, McGue M. Parental smoking and adolescent problem behavior: An adoption study of general and specific effects. American Journal of Psychiatry. 2008;165(10):1338–1344. doi: 10.1176/appi.ajp.2008.08010125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick DG, Koenen KC, Ruggiero KJ, Acierno R, Galea S, Resnick HS, et al. The serotonin transporter genotype and social support and moderation of posttraumatic stress disorder and depression in hurricane-exposed adults. [Article; Proceedings Paper]. American Journal of Psychiatry. 2007;164(11):1693–1699. doi: 10.1176/appi.ajp.2007.06122007. [DOI] [PubMed] [Google Scholar]
- Krueger RF, Eaton NR, Clark LA, Watson D, Markon KE, Derringer J, et al. Deriving an empirical structure of personality pathology for DSM-5. Journal of Personality Disorders. 2011;25(2):170–191. doi: 10.1521/pedi.2011.25.2.170. [DOI] [PubMed] [Google Scholar]
- Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WG, McGue M. Etiologic connections among substance dependence, antisocial behavior, and personality: Modeling the externalizing spectrum. Journal of Abnormal Psychology. 2002;111(3):411–424. [PubMed] [Google Scholar]
- Krueger RF, Markon KE, Patrick CJ, Iacono WG. Externalizing psychopathology in adulthood: A dimensional-spectrum conceptualization and its implications for DSM-V. Journal of Abnormal Psychology. 2005;114(4):537–550. doi: 10.1037/0021-843X.114.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470(7333):187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- Lasky-Su J, Lyon HN, Emilsson V, Heid IM, Molony C, Raby BA, et al. On the replication of genetic associations: Timing can be everything! American Journal of Human Genetics. 2008;82(4):849–858. doi: 10.1016/j.ajhg.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Decandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nature Genetics. 2012 doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand LN, McGue M, Iacono WG. Searching for interactive effects in the etiology of early-onset substance use. Behavior Genetics. 1999;29(6):433–444. doi: 10.1023/a:1021627021553. [DOI] [PubMed] [Google Scholar]
- Levinson DF, Duan JB, Oh S, Wang K, Sanders AR, Shi JX, et al. Copy Number Variants in Schizophrenia: Confirmation of Five Previous Findings and New Evidence for 3q29 Microdeletions and VIPR2 Duplications. American Journal of Psychiatry. 2011;168(3):302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: Implications for design of complex trait association studies. Genome Research. 2011;21(6):940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter JB, Barr CL, Kennedy JL, Vantol HHM, Kidd KK, Livak KJ. A hypervariable segment in the human dopamine receptor D4 (DRD4) gene. Human Molecular Genetics. 1993;2(6):767–773. doi: 10.1093/hmg/2.6.767. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438(7069):803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Luck SJ, Mathalon DH, O'Donnell BF, Hamalainen MS, Spencer KM, Javitt DC, et al. A roadmap for the development and validation of event-related potential biomarkers in schizophrenia research. Biological Psychiatry. 2011;70(1):28–34. doi: 10.1016/j.biopsych.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczak SE, Glatt SJ, Wall TL. Meta-analyses of ALDH2 and ADH1B with alcohol dependence in Asians. Psychological Bulletin. 2006;132(4):607–621. doi: 10.1037/0033-2909.132.4.607. [DOI] [PubMed] [Google Scholar]
- Lynskey MT, Heath AC, Bucholz KK, Slutske WS, Madden PAF, Nelson EC, et al. Escalation of drug use in early-onset cannabis users vs co-twin controls. Journal of the American Medical Association. 2003;289(4):427–433. doi: 10.1001/jama.289.4.427. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nature Genetics. 2005;37(4):413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- Markon KE, Chmielewski M, Miller CJ. The Reliability and Validity of Discrete and Continuous Measures of Psychopathology: A Quantitative Review (vol 137, pg 856, 2011). Psychological Bulletin. 2011;137(6):1093–1093. doi: 10.1037/a0023678. [DOI] [PubMed] [Google Scholar]
- McGue M, Iacono WG. The association of early adolescent problem behavior with adult psychopathology. American Journal of Psychiatry. 2005;162(6):1118–1124. doi: 10.1176/appi.ajp.162.6.1118. [DOI] [PubMed] [Google Scholar]
- McGue M, Iacono WG, Legrand LN, Elkins I. The origins and consequences of age at first drink. I. Associations with substance-use disorders, disinhibitory behavior and psychopathology, and P3 amplitude. Alcoholism: Clinical and Experimental Research. 2001;25:1156–1165. [PubMed] [Google Scholar]
- McGue M, Keyes M, Sharma A, Elkins I, Legrand L, Johnson W, et al. The environments of adopted and non-adopted youth: Evidence on range restriction from the Sibling Interaction and Behavior Study (SIBS). Behavior Genetics. 2007;37(3):449–462. doi: 10.1007/s10519-007-9142-7. [DOI] [PubMed] [Google Scholar]
- McGue M, Osler M, Christensen K. Causal inference and observational research: The utility of twins. Perspectives on Psychological Science. 2010;5(5):546–556. doi: 10.1177/1745691610383511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Nichols T, Callicott JH, Ding J, Kolachana B, Buckholtz J, et al. Impact of complex genetic variation in COMT on human brain function. Molecular Psychiatry. 2006;11(9):867–877. doi: 10.1038/sj.mp.4001860. [DOI] [PubMed] [Google Scholar]
- Miller GA. Mistreating Psychology in the Decades of the Brain. Perspectives on Psychological Science. 2010;5(6):716–743. doi: 10.1177/1745691610388774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Basu S, Cunningham J, Oetting W, Schork NJ, Iacono WG, et al. The Minnesota Center for Twin and Family Research Genome-Wide Association Study. submitted. [DOI] [PMC free article] [PubMed]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470(7332):59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, et al. A User's Guide to the Encyclopedia of DNA Elements (ENCODE). Plos Biology. 2011;9(4) doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor TG, Caspi A, Defries JC, Plomin R. Genotype-environment interaction in children's adjustment to parental separation. Journal of Child Psychology and Psychiatry. 2003;44(6):849–856. doi: 10.1111/1469-7610.00169. [DOI] [PubMed] [Google Scholar]
- O'Connor TG, Deater-Deckard K, Fulker DW, Rutter M, Plomin R. Genotype-environment correlation in late childhood and early adolscence: Antisocial behavior problems and coercive parenting. Developmental Psychology. 1998;34:970–981. doi: 10.1037//0012-1649.34.5.970. [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature Genetics. 2011;43(6):585–125. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KK, Elks CE, Li SX, Zhao JH, Luan J, Andersen LB, et al. Genetic variation in LIN28B is associated with the timing of puberty. Nature Genetics. 2009;41(6):729–733. doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare G, Cook NR, Ridker PM, Chasman DI. On the Use of Variance per Genotype as a Tool to Identify Quantitative Trait Interaction Effects: A Report from the Women's Genome Health Study. Plos Genetics. 2010;6(6) doi: 10.1371/journal.pgen.1000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444(7118):499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pinheiro JC, Bates DM. Mixed-effects models in S and S-PLUS. Springer; New York: 2000. [Google Scholar]
- Prescott CA, Kendler KS. Age at first drink and risk for alcoholism: A noncausal association. Alcoholism: Clinical and Experimental Research. 1999;23:101–107. [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Priebe L, Degenhardt FA, Herms S, Haenisch B, Mattheisen M, Nieratschker V, et al. Genome-wide survey implicates the influence of copy number variants (CNVs) in the development of early-onset bipolar disorder. Molecular Psychiatry. 2011 doi: 10.1038/mp.2011.8. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: A meta-analysis. Journal of the American Medical Association. 2009;301(23):2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch NJ, Merikangas KR. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Rueter MA, Keyes MA, Iacono WG, McGue M. Family interactions in adoptive compared to nonadoptive families. Journal of Family Psychology. 2009;23(1):58–66. doi: 10.1037/a0014091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M. Proceeding From Observed Correlation to Causal Inference The Use of Natural Experiments. Perspectives on Psychological Science. 2007a;2(4):377–395. doi: 10.1111/j.1745-6916.2007.00050.x. [DOI] [PubMed] [Google Scholar]
- Rutter M. Proceeding from observed correlation to causal inference: The use of natural experiments. Perspectives on Psychological Science. 2007b;2(4):377–395. doi: 10.1111/j.1745-6916.2007.00050.x. [DOI] [PubMed] [Google Scholar]
- Rutter M, Pickles A, Murray R, Eaves LJ. Testing hypotheses on specific causal effects on behavior. Psychological Bulletin. 2001;127:291–324. doi: 10.1037/0033-2909.127.3.291. [DOI] [PubMed] [Google Scholar]
- Scarr S, McCartney K. How people make their own environments: A theory of genotype => environment effects. Child Development. 1983;54:424–435. doi: 10.1111/j.1467-8624.1983.tb03884.x. [DOI] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption (vol 108, pg 7119, 2011). Proceedings of the National Academy of Sciences of the United States of America. 2011;108(22):9316–9316. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson MA, Wills AG, Keller MC, McQueen MB. Recent methods for polygenic analysis of genome-wide data implicate an important effect of common variants on cardiovascular disease risk. BMC Medical Genetics. 2011;12:146. doi: 10.1186/1471-2350-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature Genetics. 2011;43(10):977–U162. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutske WS, Hunt-Carter EE, Nabors-Oberg R,E, Sher KJ, Bucholz KK, Madden PAF, et al. Do college students drink more than their non-college-attending peers? Evidence from a population-based longitudinal female twin study. Journal of Abnormal Psychology. 2004;113(4):530–540. doi: 10.1037/0021-843X.113.4.530. [DOI] [PubMed] [Google Scholar]
- Smith GD. Mendelian randomization for strengthening causal inference in observational studies: Application to gene X environment interactions. Perspectives on Psychological Science. 2011;6(3):314–314. doi: 10.1177/1745691610383505. [DOI] [PubMed] [Google Scholar]
- Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? International Journal of Epidemiology. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- Sovio U, Mook-Kanamori DO, Warrington NM, Lawrence R, Briollais L, Palmer CNA, et al. Association between Common Variation at the FTO Locus and Changes in Body Mass Index from Infancy to Late Childhood: The Complex Nature of Genetic Association through Growth and Development. Plos Genetics. 2011;7(2) doi: 10.1371/journal.pgen.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear LP. The adolescent brain and age-related behavioral manifestations. Neuroscience and Biobehavioral Reviews. 2000;24(4):417–463. doi: 10.1016/s0149-7634(00)00014-2. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42(11):937–953. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Structural Variation in the Human Genome and its Role in Disease. Annual Review of Medicine. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- Steinberg L. Risk taking in adolescence - New perspectives from brain and behavioral science. Current Directions in Psychological Science. 2007;16(2):55–59. [Google Scholar]
- Tang H. Confronting ethnicity-specific disease risk. Nature Genetics. 2006;38(1):13–15. doi: 10.1038/ng0106-13. [DOI] [PubMed] [Google Scholar]
- Taylor J, Roehrig AD, Hensler BS, Connor CM, Schatschneider C. Teacher Quality Moderates the Genetic Effects on Early Reading. Science. 2010;328(5977):512–514. doi: 10.1126/science.1186149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapar A, Harold G, Rice F, Langley K, O'Donovan M. The contribution of gene-environment interaction to psychopathology. Development and Psychopathology. 2007;19(4):989–1004. doi: 10.1017/S0954579407000491. [DOI] [PubMed] [Google Scholar]
- Thomas DC. Gene-environment-wide association studies: emerging approaches. Nature Reviews Genetics. 2010a;11(4):259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DC. Methods for Investigating Gene-Environment Interactions in Candidate Pathway and Genome-Wide Association Studies. Annual Review of Public Health. 2010b;31:21–36. doi: 10.1146/annurev.publhealth.012809.103619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DC, Lewinger JP, Murcray CE, Gauderman WJ. Invited Commentary: GE-Whiz! Ratcheting Gene-Environment Studies up to the Whole Genome and the Whole Exposome. American Journal of Epidemiology. 2011 doi: 10.1093/aje/kwr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson NJ, Harbord R, Smith GD, Zacho J, Tybjaerg-Hansen A, Nordestgaard BG. Does Greater Adiposity Increase Blood Pressure and Hypertension Risk? Mendelian Randomization Using the FTO/MC4R Genotype. Hypertension. 2009;54(1):84–U131. doi: 10.1161/HYPERTENSIONAHA.109.130005. [DOI] [PubMed] [Google Scholar]
- Torkamani A, Scott-Van Zeeland AA, Topol EJ, Schork NJ. Annotating individual human genomes. Genomics. 2011;98(4):233–241. doi: 10.1016/j.ygeno.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkheimer E, Haley A, Waldron M, D'Onofrio B, Gottesman II. Socioeconomic status modifies heritability of IQ in young children. Psychological Science. 2003;14(6):623–628. doi: 10.1046/j.0956-7976.2003.psci_1475.x. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. American Journal of Human Genetics. 2012;90(1):7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LELM, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, et al. A de novo paradigm for mental retardation. Nature Genetics. 2010;42(12):1109–+. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- Vrieze SI. Model selection and psychological theory: A discussion of the differences between the Akaike Information Criterion (AIC) and the Bayesian Information Criterion (BIC). Psychological Methods. doi: 10.1037/a0027127. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Hicks BM, McGue M, Iacono WG. Genetic Influence on the Co-Occurrence of Alcohol, Marijuana, and Nicotine Dependence Symptoms Declines from Age 14 to 29. American Journal of Psychiatry. doi: 10.1176/appi.ajp.2012.11081268. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, McGue M, Iacono WG. The interplay of genes and adolescent development in substance use disorders: Leveraging findings from GWAS meta-analyses to test developmental hypotheses about nicotine consumption. Human Genetics. 2012;131:791–801. doi: 10.1007/s00439-012-1167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, McGue M, Miller MB, Legrand LN, Schork NJ, Iacono WG. An assessment of the individual and collective effects of variants on height using twins and a developmentally informative study design. PLoS Genetics. 2011;7(12):e1002413. doi: 10.1371/journal.pgen.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Perlman G, Krueger RF, Iacono WG. Is the Continuity of Externalizing Psychopathology the Same in Adolescents and Middle-Aged Adults? A Test of the Externalizing Spectrum's Developmental Coherence. Journal of Abnormal Child Psychology. 2011 doi: 10.1007/s10802-011-9571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widen E, Ripatti S, Cousminer DL, Surakka I, Lappalainen T, Jarvelin MR, et al. Distinct Variants at LIN28B Influence Growth in Height from Birth to Adulthood. American Journal of Human Genetics. 2010;86(5):773–782. doi: 10.1016/j.ajhg.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, et al. Highly conserved non-coding sequences are associated with vertebrate development. Plos Biology. 2005;3(1):116–130. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nature Genetics. 2011;43(9):864–U872. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JA, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nature Genetics. 2010;42(7):565–U131. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JA, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JA, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nature Genetics. 2011;43(6):519–U544. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nature Genetics. 2010;42(7):626–U106. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker RA, Heitzeg MM, Nigg JT. Parsing the Undercontrol/Disinhibition Pathway to Substance Use Disorders: A Multilevel Developmental Problem. Child Development Perspectives. 2011;5(4):248–255. doi: 10.1111/j.1750-8606.2011.00172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proceedings of the National Academy of Sciences U S A. 2012 doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]