Background: PRC1 silences transcription by an unknown mechanism.
Results: PRC1 can both block and dissociate PICs in general with the exception of TFIID.
Conclusion: PRC1 gene silencing may involve the ability of TFIID to remain bound to gene promoters, leaving them in a poised state.
Significance: Understanding how PRC1 regulates transcription deepens our basic understanding of key developmental processes.
Keywords: Gene Regulation, Gene Silencing, RNA Polymerase II, Transcription, Transcription Coactivators, Transcription Factors, Transcription Promoter, Transcription Regulation, Transcription Repressor
Abstract
Despite the important role of Polycomb in genome-wide silencing, little is known of the specific biochemical mechanism by which it inactivates transcription. Here we address how recombinant Polycomb repressive complex 1 (PRC1) inhibits activated RNA polymerase II preinitiation complex (PIC) assembly using immobilized H3K27-methylated chromatin templates in vitro. Recombinant PRC1 inhibited transcription, but had little effect on binding of the activator as reported previously. In contrast, Mediator and the general transcription factors were blocked during assembly or dissociated from preassembled PICs. Importantly, among the PIC components, Tata Binding Protein (TBP) was the most resistant to eviction by PRC1. Immobilized template experiments using purified PRC1, transcription factor II D (TFIID), and Mediator indicate that PRC1 blocks the recruitment of Mediator, but not TFIID. We conclude that PRC1 functions to block or dissociate PICs by interfering with Mediator, but leaves TBP and perhaps TFIID intact, highlighting a specific mechanism for PRC1 transcriptional silencing. Analysis of published genome-wide datasets from mouse embryonic stem cells revealed that the Ring1b subunit of PRC1 and TBP co-enrich at developmental genes. Further, genes enriched for Ring1b and TBP are expressed at significantly lower levels than those enriched for Mediator, TBP, and Ring1b. Collectively, the data are consistent with a model in which PRC1 and TFIID could co-occupy genes poised for activation during development.
Introduction
The Polycomb group of proteins plays a major role in transcriptional regulation during development. The first member of this group, polycomb, was identified in screens for Drosophila mutants defective in body patterning during development (1). Subsequently, other genes that fell into this group were identified that encoded proteins assembled into two distinct Polycomb Repressive Complexes (PRC)3 termed PRC1 and PRC2 (2, 3). PRC1 and PRC2 silence expression of the Hox gene network involved in development as well as the inactive X chromosome (4). The current models suggest that PRC2 methylates histone H3 at lysine 27 (H3K27) via its EZH2 subunit (5–7). This modification, in turn, provides a binding site for the chromodomain-containing Pc subunit of PRC1. Once bound, PRC1 can ubiquitinate H2AK119 via its Ring1a or Ring1b subunit (8). PRC1 binding is believed to be the major determinant in silencing, yet the precise mechanism remains unknown.
Kingston and colleagues (2) were the first to isolate and study the mechanism of PRC1 from Drosophila. They found that PRC1 comprises four subunits termed polycomb (Pc), polyhomeotic (PH), posterior sex combs (PSC), and dRING. Subsequent studies showed that mammalian counterparts of these proteins were highly conserved with chromodomain-containing Pc homologs termed CBXs, three PH homologs (PHC1–3), two dRING homologs Ring1a and Ring1b, and six human PSC homologs, the most prominent being BMI1 (9, 10).
Both native Drosophila and recombinant mouse PRC1 inhibited activated transcription on chromatin templates in mammalian in vitro systems (11). Importantly, PRC1 could only block transcription when prebound to the template. Analysis of individual subunits demonstrated that the PSC and PH subunits functioned most effectively at transcription inhibition on chromatin (11). Remarkably, PRC1 action in vitro was not dependent upon H3K27 methylation or ubiquitination. Indeed, the greatest effect was observed with PRC1 subunits lacking either the targeting or the ubiquitination functions (11). Further, a novel complex of RYBP, RING, and BMI1/MEL18 has been shown to be recruited to Polycomb-regulated genes independent of H3K27me3 (12).
The inhibition of transcription has been largely attributed to the ability of PRC1 to bind and compact chromatin. Cryo-EM analysis showed that a single molecule of PRC1 binds to three nucleosomes (13). This compaction is believed to limit access to factors necessary for transcription on chromatin. Indeed, PRC1 effectively blocked chromatin remodeling by SWI/SNF in vitro. Moreover, this inhibition occurred in the absence of ubiquitination (2).
A recent study by the Bickmore group (14) addressed the role of PRC1 and ubiquitination in gene expression and chromatin compaction in vivo. The authors found that Ring1b is necessary for silencing and chromatin compaction using fluorescence in situ hybridization (FISH) experiments on adjacent Hox genes in embryonic stem (ES) cells. Importantly, the compaction and silencing are dependent on the expression of Ring1b but independent of its ubiquitin ligase activity in agreement with the earlier in vitro work of Kingston and colleagues (13).
We were interested in understanding more specifically how PRC1 affects the assembly and function of the RNA polymerase II (Pol II) preinitiation complex. Chromatin immunoprecipitation studies performed in Drosophila showed that PRC1 co-bound to the Hox loci with the general transcription factors TBP, TFIIB, and TFIIF (15). Further, analysis of the composition of PRC1 in Drosophila identified TBP-associated factors (TAFs) as interacting proteins (16). Collectively, these studies raise the question of how PRC1 affects PIC assembly.
PICs contain Pol II, the 30-subunit Mediator co-activator complex, TFIID, the general transcription factors (GTFs) TFIIA, TFIIB, TFIIE, TFIIF, and TFIIH, and numerous chromatin-modifying and -remodeling complexes (17, 18). PIC formation in response to activator binding is a well studied and temporally regulated process. Initially, Mediator and p300 are recruited to the template directly by the activator. After p300 acetylates chromatin and itself, it dissociates from the PIC, and TFIID binds along with the GTFs (19). Mediator and TFIID binding is cooperative due to a direct interaction among the two co-activator complexes (20). The formation of the PIC also results in the recruitment of chromatin-remodeling and -modifying complexes, including CHD1, and Spt-Ada-Gcn5-acetyltransferase (SAGA) (18, 19, 21). Importantly, HeLa nuclear extracts depleted of Mediator fail to recruit most GTFs, showing that Mediator is essential for PIC formation (20).
Here we examine the biochemical effect of PRC1 on PIC assembly and function in vitro. We employed the immobilized template assay, which allows us to correlate the functional effects of PRC1 on transcription with its effects on the composition of PICs formed on H3K27me3 chromatin templates. Next, we used previously published genome-wide datasets from mouse ES cells to address the hypothesis derived from our in vitro findings.
We found that PRC1 can block assembly of the Mediator and a variety of other PIC constituents. Remarkably, the activator and TFIID were most resistant to the action of PRC1. Further, we show that purified TFIID but not Mediator is recruited to templates in the presence of PRC1. Our analysis of genome-wide binding data for TBP, Ring1b, and Mediator shows that in mouse ES cells, most Ring1b-regulated genes are also bound by TBP. Further, genes bound by Ring1b and TBP are strongly enriched for critical developmental genes. Collectively, these results suggest a more precise mechanism for PRC1-mediated transcriptional silencing and highlight the possibility that TFIID marks PRC1-regulated genes as poised for expression later in development.
EXPERIMENTAL PROCEDURES
Methyl-lysine Histone Octamer Preparation
Lysine 27 of histone H3 was mutated to a cysteine by site-directed mutagenesis of Xenopus H3.1 bearing a C110A mutation, expressed and purified from Escherichia coli inclusion bodies, and subjected to chemical alkylation by (2-bromoethyl)trimethylammonium bromide before assembly into histone octamers (22, 23). Modification of the histone tail was verified by using nano-spray mass spectrometry. Western blots were performed using standard protocols and a commercially available H3 and H3K27me3 antibody (Abcam catalog numbers ab1791 and ab6002).
Chromatin Preparation
A 602-bp biotinylated PCR fragment, which directly encompasses our GAL4-responsive promoter, was assembled into chromatin by salt dilution as described previously and validated by EMSA in native PAGE (24). Chromatin was immobilized on M280 streptavidin paramagnetic beads (Invitrogen) as described previously (25).
Immobilized Template Recruitment Assay
The 40-μl immobilized template recruitment assays contained HeLa nuclear extract and 125 fmol of a GAL4-responsive template assembled into chromatin (3 nm) as described previously (26). The template is termed G5E4T because it contains five GAL4 sites positioned upstream of the adenovirus E4 TATA box. Reactions were typically performed in the presence and absence of 250 nm PRC1. Bound protein was eluted from the immobilized templates in 10 μl of 2× Laemmli buffer, fractionated by SDS-PAGE, and immunoblotted. Antibodies used in immunoblotting included MED23 (BD Pharmingen), Pol II C-terminal domain 8WG16 (QED Bioscience), TFIIB (27), and CHD1 (Bethyl Laboratories). All other antibodies were purchased from Santa Cruz Biotechnology.
Extract and Protein Preparation
HeLa nuclear extract and GAL4-VP16 were prepared as described previously (28). Recombinant FLAG-tagged mouse BMI1 was purified from SF9 cells using a baculovirus overexpression system (Invitrogen). For the purification of the PRC1 complex, baculoviruses encoding mouse CBX2, Ring1b, and FLAG-BMI1 were co-infected into insect cells and harvested 44 h after infection. Briefly, cells were resuspended in 0.3 m Buffer F (0.3 m NaCl, 20% glycerol, 20 mm HEPES, pH 7.9, 4 mm MgCl2, 0.2% Triton X-100, and 0.1% Nonidet P-40) and sonicated. Lysates were then treated with DNase I (1 unit/ml) and heparin (12.5 μg/ml) and cleared by centrifugation at 30,000 × g. The resulting lysate was bound to M2 anti-FLAG resin (Sigma), washed extensively in 0.5 m Buffer F, and eluted using 3× FLAG peptide (0.25 mg/ml Sigma). For the analysis of proteins that interact with the recombinant PRC1 complex from HeLa nuclear extracts, MudPIT was performed as described previously (18). Mediator was purified from HeLa cells expressing FLAG-tagged human Intersex (MED29) as described previously (29). The HeLa Intersex cell line was a gift from Joan and Ron Conaway (29). TFIID was purified as described previously from a cell line expressing HA-TBP provided by Arnie Berk (30).
Genome-wide Analysis
Previous datasets for genome-wide binding of TBP, MED1, and Ring1b were obtained from the Gene Expression Omnibus (GEO) database (31, 32). The GEO accession codes for the data used are as follows: TBP, GSE22303; MED1, GSE22557; Ring1b, GSE13084; and CBX7, GSM820726. Reads were mapped to the mouse (mm9) genome using the Bowtie software (33). The mouse genome was tiled into 50-bp windows. The total counts of the input and chromatin immunoprecipitation (ChIP) samples were normalized to each other. The input sample was used to estimate the expected counts in a window. Poisson distribution analysis was used to estimate the probability of observing the ChIP counts within a window given the expected counts in the input sample window (34). In Figs. 6 and 7, all p values of enrichment are plotted to remove any bias. The expression levels for all annotated genes were determined using the GSM881355 GEO dataset.
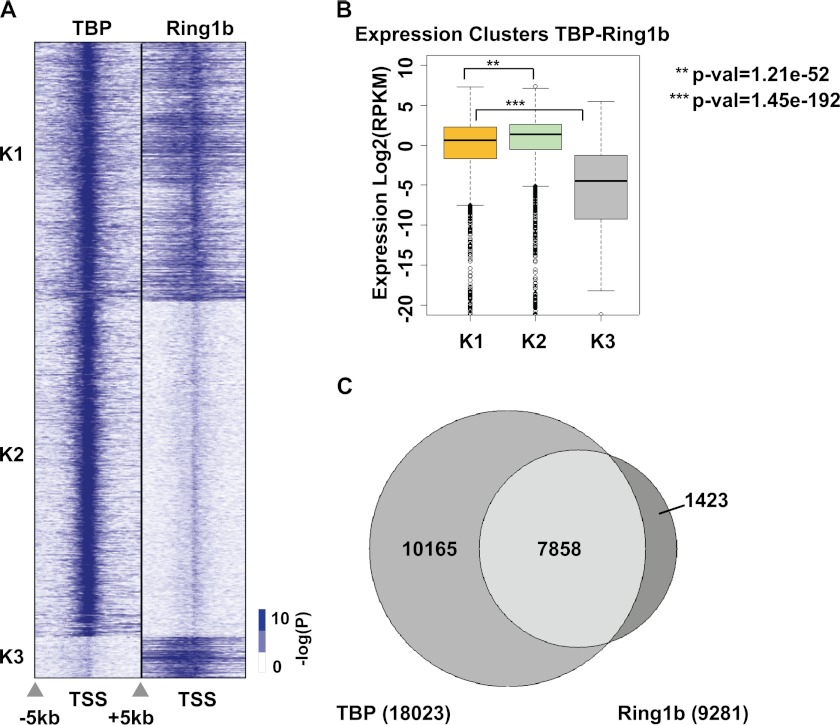
FIGURE 6.
PRC1 and TBP enrichment overlaps in vivo. A, the distributions of TBP and Ring1b from −5 kb to +5 kb, centered at the transcriptional start site (TSS) for all annotated genes in mouse ES cells, are shown as heat maps of −log (p value (p-val)). Binding peaks were sorted into differential binding clusters (K1–3) comparing TBP and Ring1b across all genes in the genome. B, RNA-Seq expression analysis was used to determine the mean expression for genes in clusters K1–3. A box and whisker plot of the expression for each cluster is shown for reads per kilobase of exon per million (RPKM) fragments mapped. Asterisks indicate that the change in expression is significant between the indicated clusters as measured using a Kolmogorov-Smirnov test. Error bars indicate S.D. C, a Venn diagram comparing the enrichment of TBP alone, TBP and Ring1b, or Ring1b alone to all genes in mouse ES cells is shown.
FIGURE 7.
Genome-wide analysis of Ring1b, TBP, and Mediator in mouse ES cells. A, the distributions of TBP, Ring1b, and MED1 enrichment −5 kb to +5 kb, centered at the transcriptional start site (TSS) for all annotated genes in mouse ES cells, are shown as heat maps of −log (p value). Peaks were called and sorted into differential clusters (L1–4) based on the enrichments of TBP, Ring1b, and MED1 across all genes in the genome. Only genomic regions enriched for TBP are shown. B, mean expression analysis of genes in clusters L1–4 is shown. A box and whisker plot of the expression for each cluster indicates the mean expression for each cluster in reads per kilobase of exon per million (RPKM). The dashed line represents a 2-fold change in expression from the overall mean. The p values resultant from Kolmogorov-Smirnov test comparing clusters L1–4 are shown. Error bars indicate S.D. C–F, GO term analysis of clusters L1–4. The top six GO terms for each expression cluster are shown, and the −log of the p value of confidence for each term is graphed. G–J, the binding profile for a representative gene from each cluster is shown. Values plotted are −log (p value).
Statistical Analysis
In Figs. 2 and 3, a two-tailed Student's t test was performed comparing the quantified signals of activator-stimulated PIC formation with PIC incubated with PRC1 from Western blots using the Odyssey imaging software from LI-COR. In Figs. 6 and 7, a Kolmogorov-Smirnov test was performed to compare datasets of unequal size (35).
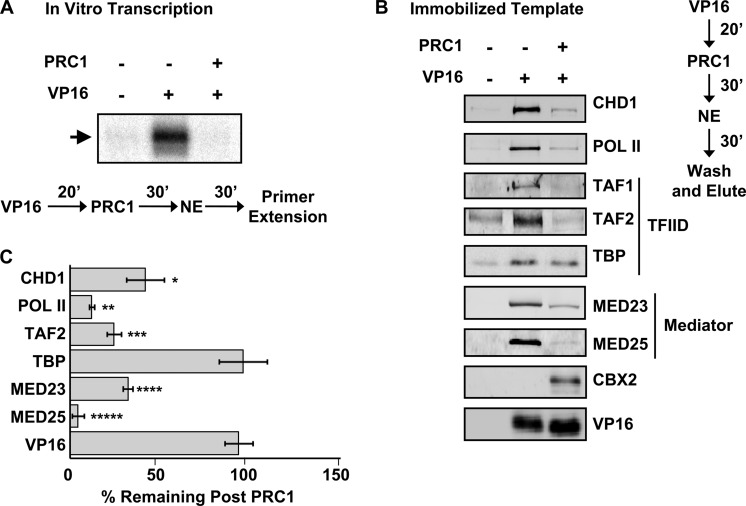
FIGURE 2.
PRC1 blocks transcription and PIC assembly. A, PRC1 blocks in vitro transcription. The flowchart indicates the order of protein addition. GAL4-VP16 was prebound to the template for 20 min, and 250 nm PRC1 was added. After 30 min, HeLa nuclear extract was added in the presence of nucleotides. After 30 min, the mRNA products were isolated and measured by primer extension. B, PRC1 blocks PIC assembly. An immobilized template assay using HeLa nuclear extract, in the presence and absence of activator and PRC1, was performed. An immunoblot of the isolated complexes comparing binding of select components of the PIC is shown. C, immunoblot signal was quantified using the LI-COR imaging software and graphed, and the statistical significance between mock- and PRC1-treated templates was calculated using a two-tailed Student's t test (n = 3). p values were determined to be *, 0.025, **, 0.03, ***, 0.015, ****, 0.022, and *****, 0.001 for the indicated signals quantified. Signals were normalized to that of activator-stimulated recruitment in lane 2 (100%). Error bars indicate S.D.
FIGURE 3.
PRC1 dissociates preassembled PICs. A, PRC1 silences in vitro transcription from preassembled PICs. The flowchart indicates the order of binding. GAL4-VP16 was prebound to the template for 20 min, after which HeLa nuclear extract was added to allow PIC assembly. After 30 min, PICs were isolated, and PRC1 was added in the presence of nucleotides. After 30 min, the mRNA products were isolated and measured by primer extension. B, PRC1 disassembles a PIC in an immobilized template assay. The flowchart indicates the order of protein addition. An immunoblot comparing binding of select components of the PIC in the presence of increasing amounts of PRC1 is shown. C, immunoblot signal was quantified using the LI-COR imaging software and graphed, and the statistical significance between recruitment of proteins to mock- and PRC1-treated templates was calculated using a two tailed Student's t test (n = 3). The p values determined were *, 0.001, **, 0.002, ***, 0.020, ****, 0.003, and *****, 0.001 for the indicated signals measured. Signals were normalized to that of activator-stimulated recruitment in lane 2 (100%). Error bars indicate S.D.
RESULTS
Generation of H3K27me3 Chromatin Templates
Our first goal was to recreate PRC1 silencing in vitro. To generate the chromatin docking site for PRC1, we used the methyl-lysine analog (MLA) method developed by Shokat and colleagues (22) to synthetically methylate H3K27 for biochemical analysis. The H3K27me3 MLA was validated by immunoblotting with an antibody to H3K27me3 (Fig. 1A) and quantitated by electrospray ionization mass spectrometry, which revealed that 88% of H3 was modified (data not shown). Previous studies have shown that PRC1 binding genome-wide broadly spreads across promoters; however, the high amount of modification used in our experiments may also aid in the initial recruitment of PRC1 (32). As shown in the Fig. 1B schematic, the synthetically methylated H3 was assembled into octamers and then into chromatin on biotinylated DNA templates containing a GAL4-responsive promoter for immobilized template and in vitro transcription analysis (19).
FIGURE 1.
Binding of PRC1 to H3K27me3 chromatin in vitro. A, schematic of the MLA synthesis at histone H3 lysine 27. Below the schematic is a Western blot showing the specific detection of H3K27me3 MLA by antibody against HK27me3. Unmod., unmodified. B, schematic of the immobilized chromatin template and chromatin assembly. A Coomassie Blue-stained gel of the purified recombinant Xenopus laevis histones used is shown on the left. The extent of chromatin assembly upon the addition of increasing amounts of histone octamers was monitored by EMSA; equivalent amounts of chromatin were utilized in all experiments. The positions of the free DNA and tri-nucleosome (Tri Nuc) are indicated with arrows. RT primer was used in primer extension to measure RNA during in vitro transcription. C, Coomassie Blue-stained gel of purified PRC1 showing the CBX2, BMI1, and Ring1b subunits. 2 μg of PRC1 expressed in and purified from the Bac-to-Bac baculovirus system (Invitrogen) is shown. D, immobilized template analysis of PRC1 binding to naive (unmodified) and H3K27me3 chromatin. 125 fmol of immobilized chromatin was incubated with increasing amounts of PRC1 from 62.5 to 250 nm. After washing, the bound proteins were eluted and subjected to immunoblotting. E, statistical analysis of data. Three replicates were quantitated using the LI-COR imaging package and graphed. The values were normalized to the highest point of naive chromatin used, which was set at 100%. Error bars indicate S.D.
Verification of in Vitro System to Study PRC1 Silencing
To generate a recombinant PRC1 complex, we co-expressed mouse CBX2 (Pc), Ring1b (dRING), and FLAG-BMI1 in a baculovirus expression system and isolated the complexes using FLAG antibody beads. Fig. 1C shows a Coomassie Blue-stained gel of the minimal core complex. To determine whether PRC1 displays a higher affinity for our H3K27me3 templates, we performed immobilized template analysis. Increasing concentrations of PRC1 were incubated with either naive or H3K27me3 templates immobilized on streptavidin-coated magnetic beads, and the bound proteins were captured using a magnetic particle concentrator. The immunoblot of Fig. 1D shows that recombinant PRC1 binds with higher affinity for the H3K27me3 MLA versus the naive chromatin. Approximately 5–10% of the PRC1 used in the assay is recruited to the template. Therefore, 12.5–25 nm PRC1 is binding to 3 nm H3K27me3 template in our assay. A bar graph of triplicates in Fig. 1E revealed a consistent 2-fold increase in affinity under the conditions of our binding assay, which is compatible with in vitro transcription conditions. Although previous studies with methylated histone tail peptides showed that PRC1 binds with a 4–5-fold higher affinity for H3K27me3, the use of chromatin may partially negate the effect of methylation because subunits of PRC1 other than Pc (CBX2), including BMI1, contribute strongly to its affinity (36).
As another measure of the function of the complex, we sought to confirm its ability to associate with subunits usually associated with PRC1 in vivo (9). We utilized MudPIT analysis to identify proteins from HeLa nuclear extracts able to interact with recombinant PRC1. MudPIT analysis of FLAG-PRC1 incubated with HeLa nuclear extract revealed an enrichment of numerous other PRC1 complex members as compared with FLAG beads alone (supplemental Fig. S1). Members of the PH, Pc, PSC, and dRING families associated with recombinant PRC1, showing that the complex is dynamic and can exchange subunits with those present in the nuclear extract. Using the recombinant complex in the presence of nuclear extract therefore allows for the recruitment of endogenous PRC1 members and may aid in PRC1 silencing in vitro.
Functional Consequences of PRC1 Binding
To validate the functional consequence(s) of PRC1 binding to a promoter, we carried out in vitro transcription of chromatin in HeLa nuclear extracts. We performed the assay two different ways. In one, the blocking assay, we prebound the activator GAL4-VP16 and then added PRC1 and measured transcription. In the other, the dissociation assay, we prebound GAL4-VP16, added nuclear extract, and allowed the PIC to assemble, after which we added PRC1.
PRC1 Blocks Functional PIC Formation
Our initial experiments focused on the role of PRC1 in blocking PIC formation, similar to the role that PRC1 plays in maintaining the silent state of developmental genes. The results of the in vitro transcription assay in Fig. 2A reveal stimulation by the activator GAL4-VP16 on the H3K27me3 templates. However, the addition of PRC1 completely blocked stimulation by the activator. This result demonstrated an ability to recreate, in a cell-free system, the silencing of a model reporter gene, as first reported by Kingston and colleagues (11).
We addressed the molecular basis for this inhibition by first analyzing the composition of the PICs formed in the presence of PRC1 via immobilized template assays. In this assay, the binding of our activator GAL4-VP16 to the template stimulates PIC formation. Complexes bound to the template are captured using a magnet and immunoblotted to assay the extent of PIC formation. We assayed for previously identified PIC components and chose representative proteins from each complex including Pol II, Mediator, TFIID, and the chromatin remodeler CHD1. Analyzing which PIC components are blocked will define the step in PIC formation that is blocked by PRC1. Fig. 2B is a blocking experiment showing that PRC1 does not significantly alter the amount of GAL4-VP16 bound to the template but does block the recruitment of most other PIC proteins. TBP binding was the highest for all proteins tested. Similar results were obtained using a four-member PRC1 complex that included the PH subunit (supplemental Fig. S2). Importantly, Mediator recruitment, the initial step in PIC formation, is strongly blocked. The GTFs are recruited after and dependent on Mediator during PIC formation, and we found that these were also blocked (supplemental Fig. S3). The experiment was performed in triplicate, and the percentage of protein remaining after blocking by PRC1, as measured using the LI-COR detection system, is graphed in Fig. 2C. These results reveal that PRC1 silences transcription by inhibiting the binding of the Mediator, which is integral to the formation of an activator-stimulated PIC. Further, these results suggest that TBP recruitment to a PIC is only slightly affected by PRC1 binding.
PRC1 Functionally Dissociates PICs
PRC1 regulation is dynamic during development. It silences developmental genes before differentiation and pluripotency factors such as Sox2 at the onset of differentiation. With this in mind, we asked whether PRC1 has an effect on preassembled PICs. The results of the in vitro transcription assay in Fig. 3A show that PRC1 can functionally silence a pre-existing PIC. This result was surprising because Kingston and colleagues (11) had been unable to observe this effect in previous studies. Our result may stem from the use of the immobilized template approach as it allows us to isolate PICs from crude nuclear extracts and assay the affects of PRC1 in a more purified system.
We then performed immobilized template experiments, where PICs were preformed on the template prior to PRC1 addition to assess which PIC components were dissociated by PRC1 (Fig. 3B). Preformed PICs were relatively stable up to 1 h following formation (supplemental Fig. S4), yet the addition of PRC1 to the immobilized template at 20 min following PIC formation shows that many components, including the Mediator, Pol II, and GTFs, were dissociated from the template (Fig. 3, B and C). Importantly, TBP was consistently retained on the template. In summary, these results suggest that when PRC1 silences an active gene, the mechanism is similar to PIC blocking.
PRC1 Blocks the Recruitment of Purified Mediator, but Not TFIID, in Vitro
Our results using crude nuclear extracts revealed that TBP is somewhat resistant to PRC1-mediated blocking and dissociation relative to the GTFs. Moreover, PRC1 and TFIID can interact in Drosophila, and testes-specific TAFs counteract Polycomb during development (16, 37). These findings support models where PRC1 and TFIID might either co-occupy or compete for binding to a promoter during development in higher organisms. We investigated these possibilities in vitro using purified TFIID and PRC1 in an immobilized template assay. The results in Fig. 4, A and B, show that TFIID binding is resistant to PRC1 in both blocking and dissociation assays. Further, the quantification of triplicate experiments shown in Fig. 4, C and D, highlights the lack of effect by PRC1 yet shows that CBX2 recruitment is modestly enhanced by the presence of TFIID. This result led us to question whether the recruitment of purified Mediator was affected by PRC1. We therefore performed immobilized template experiments using purified Mediator, TFIID, and PRC1. The data in Fig. 5 show that PRC1 blocks the recruitment of Mediator to the immobilized template when added either alone or with TFIID. This experiment was performed in triplicate, and amounts of binding for each protein were measured using the LI-COR detection system. The changes in Mediator binding were determined to be significant using a Student's t test comparing activator-stimulated recruitment of Mediator with PRC1-bound templates (p < 0.001). TFIID subunits were reproducibly unaffected by PRC1, similar to the results in Fig. 4. We noticed that TAFs in the extract were sensitive to PRC1, but TAFs in purified TFIID were not. To test whether an activity in extracts contributed to the effect, we also performed an immobilized template experiment where TFIID was prebound to the immobilized template followed by the addition of PRC1 and nuclear extract (supplemental Fig. S5). The results show that PRC1 has no effect on dissociating TFIID when PRC1 is recruited in the presence of nuclear extract. This result suggests that the extract does not contain an activity that dissociates the TAFs. We conclude that PRC1 blocks a critical step in PIC formation, binding of the Mediator, which is required for gene activation (38, 39).
FIGURE 4.
TFIID binds the immobilized template in the presence of PRC1. A and B, purified TFIID, PRC1, and GAL4-VP16 were incubated with 125 fmol of G5E4T template in immobilized template assays. Assays were performed to investigate both dissociation (A) and blocking (B) of TFIID binding. C and D, a quantitation of three replicate experiments for both dissociation (C) and blocking (D) assays is shown. The immunoblot signals were quantified using the LI-COR system, and values were normalized to the amount of protein bound when added alone. Error bars indicate S.D.
FIGURE 5.
TFIID but not Mediator binding is resistant to PRC1. Purified TFIID, Mediator, PRC1, and GAL4-VP16 were incubated with 125 fmol of template in an immobilized template assay. Following prebinding of the activator, PRC1 was bound to the template. TFIID or Mediator was then incubated with the template either alone or in combination. The Western blot signals for representative subunits of TFIID (TAF1, TAF2, TAF4, TBP), Mediator (MED23, MED6), and PRC1 (BMI1, CBX2) are shown.
In Vivo Association of PRC1 and TBP Correlates with Gene Repression in Mouse ES Cells
The question of whether PRC1 and TBP can co-occupy a promoter in vivo is an interesting and logical next question. We therefore sought to address this hypothesis derived from our in vitro studies in vivo. We utilized previous genome-wide data from mouse embryonic stem cells, where PRC1 is critical and required for the pluripotent state (40, 41).
ChIP-Seq datasets, deposited by investigators who previously reported the genome-wide binding of Ring1b, TBP, and MED1 in mouse embryonic stem cells, were obtained from the GEO database (31, 32). Peaks of enrichment were determined by segregating the mouse genome into 50-bp bins and comparing the ChIP and input signals in each bin. We then used Poisson distribution to calculate p values for the enrichment of ChIP signal reads in each bin. All values are plotted as −log (p value) (Figs. 6A and 7A). The mRNA expression levels for all annotated genes were assessed using a previously reported GEO dataset (GSM88135).
TBP Binding Is Retained at Ring1b-bound Promoters in Mouse ES Cells
The results of our in vitro studies implied that TFIID could bind to gene promoters regulated by PRC1. We analyzed the genome-wide binding of TBP and Ring1b in mouse ES cells from prior studies to investigate this possibility (31, 32). Fig. 6A shows a heat map of Ring1b and TBP binding distributions ±5 kb from the transcription start site for all annotated genes with each row representing one gene promoter. Clusters K1–3 were grouped based on combinatorial binding of TBP and Ring1b. For each cluster identified, the average gene expression level derived from an mRNA-Seq dataset is shown as a box plot in Fig. 6B. The Venn diagram in Fig. 6C shows the amount of overlap between TBP and Ring1b enrichments on promoters genome-wide.
We found a significant number of genes with ChIP enrichment for both Ring1b and TBP (Fig. 6, A and C). Interestingly, the mean expression of genes with ChIP enrichments for PRC1 and TBP (cluster K1) was significantly lower (p = 1.21 ×10−52) than genes with enrichment for TBP alone (cluster K2) (Fig. 6B). Also, gene ontology (GO) analysis revealed that genes in clusters K1 and K3 are highly enriched for processes associated with development. This result is not surprising, but it does imply that a large portion of gene promoters bound by PRC1 in ES cells may also be bound by TBP.
To further test our hypothesis and correlate it with our biochemical analysis, we analyzed previous genome-wide binding studies for the Mediator complex and compared its binding to PRC1 and TBP (31). We generated clusters of combinatorial binding to identify TBP-bound genes based on Ring1b and MED1 binding. The results in Fig. 7A show the different gene clusters identified, L1–4, visualized as heat maps of the enrichment score. A box plot of the expression for genes in clusters L1–4 is shown in Fig. 7B. We also utilized the DAVID software to determine the top six GO terms represented for each cluster. A histogram of the −log (p value) for each GO term enriched in clusters L1–4 is shown in Fig. 7, C–F. A representative gene from each cluster is shown, respectively, in Fig. 7, G–J, with levels of binding plotted as the enrichment p value. To validate the presence of the entire PRC1 complex in cluster L1, we also analyzed the genome-wide binding of CBX7 recently reported by the Brockdorff group (12). A comparison of the normalized average enrichment for gene clusters L1–4 is shown in supplemental Fig. S6.
Our clustering reveals a number of intriguing observations. The first cluster, L1, defines a group of genes that are enriched for all proteins tested. The top six GO categories identified for this group include DNA binding and early embryonic developmental genes (Fig. 7, C and G). The L2 cluster contains genes enriched for Ring1b and TBP. Importantly, this gene cluster has a significantly lower mean expression as compared with L1, and the top GO terms include developmental proteins and homeobox genes (Fig. 7, B, D, and H). It is not a surprise to find that binding of Mediator correlates with a decreased level of Ring1b binding (Fig. 7A, cluster L1 versus L2). Cluster L3 represents highly expressed genes as there is no Ring1b bound, and this group has a significantly higher mean expression as compared with L1 and L2 (Fig. 7, A and B). Also, this cluster is enriched for constitutively active mitochondrial and RNA processing genes based on GO analysis (Fig. 7, E and I). The last cluster, L4, is enriched for TBP only and represents genes involved in protein processing and cation binding proteins by GO analysis (Fig. 7, A, F, and J).
These results collectively support the hypothesis that the Mediator is the key PIC component inhibited from recruitment to promoters by PRC1. The analysis of genome-wide binding correlated to expression levels reveals that PRC1 regulation of developmental genes in mouse ES cells may involve co-binding by TBP.
DISCUSSION
PRC1 Mechanism of Silencing
Young and colleagues (38, 39) originally postulated that the multimeric Mediator functions at the beginning of PIC formation to convey signals and efficiently recruit other machineries such as the GTFs and Pol II. The idea that Mediator coordinates these events has been verified by numerous groups including ours and is supported by the finding that Mediator recruitment is the earliest step in PIC formation in vitro using the immobilized template assay (19, 38, 39).
By utilizing a combination of the immobilized template assay and in vitro transcription, our study provides a more detailed mechanism for how PRC1 silences transcription. Our findings support a model where PRC1 blocks most PIC components with the exception of TFIID. The binding of TBP to PRC1-silenced genes in Drosophila was previously observed, and our results are consistent with that finding (42). However, our data indicate that PRC1 acts mainly by blocking Mediator, an early and integral event in activator-stimulated PIC formation (19). Curiously, we observe PRC1-mediated blocking of Pol II in vitro, whereas previous in vivo studies reported that Pol II remains on the gene (42). It is plausible that preinitiated Pol II is more sensitive to PRC1 than the paused Pol II frequently observed at promoters in vivo. We attempted but were unable to trap the paused Pol II in vitro and could not test this idea biochemically.
The finding that TFIID can dock to a promoter in the presence of PRC1 leads us to question whether genes co-bound by these two complexes may be in the poised transcriptional state. The poised state is a characteristic of bivalent promoters bearing both H3K27me3 and H3K4me3. Indeed, the TAF3 subunit of TFIID has been shown to bind to H3K4me3, and PRC1 binds to H3K27me3 (36, 43, 44). The presence of TBP, and possibly TFIID, at PRC1-regulated genes may facilitate rapid transcriptional activation when the gene is needed during development. Alternately, the binding of PRC1 to TBP may be a means of targeting PRC1 to recently inactivated genes where activators and thus Mediator have been lost. In this case, TBP or TFIID and its binding site may be acting analogous to the traditional Drosophila Polycomb repressive element. It was intriguing that in several of our experiments, PRC1 bound better when TFIID alone was present (i.e. Fig. 4) and stabilized TFIID binding even after dissociating Mediator (Fig. 5). Also, note in Fig. 6 that Ring1B binding appears centered near the transcription start site.
PIC Blocking and Dissociation by PRC1
Our in vitro transcription results verify previous studies by Kingston and colleagues (2, 11, 45) and others (42), but our immobilized template experiments provide further insight into the mechanism of PRC1 silencing, specifically, which activator-dependent PIC components are blocked from recruitment to the promoter. Specifically, we found that activator recruitment of Mediator, Pol II, and GTFs was significantly reduced, whereas TBP and GAL4-VP16 were more resistant. Using purified PRC1, TFIID, and Mediator in an immobilized template assay, we then showed that PRC1 blocks the recruitment of Mediator, whereas TFIID binding is unaffected. The ability of holo-TFIID, not just TBP, to bind in the presence of PRC1 using the purified system is likely due to a change in the binding properties of TFIID when purified as compared with nuclear extract (supplemental Fig. S5). Our results argue against an activity in the nuclear extract that affects the stability of TFIID.
Activators are known to interact directly with Mediator and TFIID. In the case of VP16, Mediator interacts via MED25 (46, 47) and TFIID via TAF9 (48). VP16 is known to recruit both complexes to DNA in vitro and in vivo (49). TFIID and Mediator also interact to form a co-activator complex, which is necessary in vitro for binding of the general transcription factors and Pol II (20). Our findings suggest a model where the Mediator interaction with VP16 is blocked, either by chromatin compaction or by inhibiting the accessibility to the template.
It was rather interesting to find that PRC1 can dissociate functional PICs. Previous studies implicate a role for PRC1 in silencing genes active in stem cells, such as Sox2, following differentiation (50). In our immobilized template assays, PRC1 binding to a template post-PIC formation resulted in the demolition of PICs. The majority of proteins assayed for were significantly dissociated from the PICs, except for TBP and activator, similar to results obtained for PIC blocking. Our results reveal the ability of PRC1 to silence a gene by disrupting pre-existing PIC components in vitro.
Genome-wide Analysis of PRC1 Regulated Genes
We tested our hypothesis that TFIID is the most resistant to PIC inhibition by PRC1 by analyzing the genome-wide binding of TBP and Ring1b in mouse ES cells. Our analysis indicated that the majority of Ring1b-bound genes are also bound by TBP (Fig. 6, A and C). Further, the genes bound by both Ring1b and TBP have a significantly lower expression level as compared with those bound by TBP alone, suggesting that they are indeed targets of PRC1 silencing (Fig. 6B, clusters K1–3).
Our further analysis of genes bound by MED1, TBP, and Ring1b revealed a number of interesting binding clusters (Fig. 7A). Clusters L1 and L2 are bound by both PRC1 and TBP, whereas Mediator binds in cluster L1 only. It was interesting that MED1 is bound to genes that are also bound by Ring1b. Based on GO analysis, the genes in L1 are early differentiation genes that may be lowly expressed due to heterogeneity in the ES cultures (Fig. 7C). It is also possible that the Ring1b present at genes in the L1 cluster may not be sufficient for silencing or may be acting in a function not related to PRC1. The genes located in cluster L2 highlight an area where TBP and PRC1 cross-talk may occur. Importantly, the enrichment of Ring1b is increased in this cluster. Additionally, cluster L2 has the highest average enrichment for CBX7 (supplemental Fig. S6). Note that Ring1b enrichment appears to spread more broadly across the genes in this category and that MED1 binding is greatly reduced at these genes, possibly due to the higher levels of PRC1 present (Fig. 7A, supplemental Fig. S6). The GO terms associated with this category are homeobox genes and developmental proteins, potentially implicating TBP in helping to poise critical developmental genes for expression upon differentiation.
The ability of PRC1 to block Mediator in vitro is reminiscent of our previous study with a different silencing protein HP1. In that study, HP1 also blocked most of Mediator and the TAF subunits of TBP, but unlike PRC1, some components of Mediator, namely MED6, MED23, and MED25, were unaffected (51). This suggests that the two complexes have similar yet distinct mechanisms of silencing. This study, along with previous studies from our laboratory, supports the idea that transcriptional silencers work by targeting critical steps in PIC assembly (51).
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM074701 (to M. C.).
This article was selected as a Paper of the Week.

This article contains supplemental Figs. S1–S6.
- PRC
- Polycomb repressive complex
- PSC
- posterior sex combs
- PIC
- preinitiation complex
- TBP
- Tata Binding Protein
- TFII
- transcription factor II
- GTF
- general transcription factor
- TAF
- TBP-associated factor
- Pol II
- RNA polymerase II
- MLA
- methyl-lysine analog
- CBX
- Chromobox protein homolog
- GO
- gene ontology.
REFERENCES
- 1. Lewis E. B. (1978) A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 [DOI] [PubMed] [Google Scholar]
- 2. Shao Z., Raible F., Mollaaghababa R., Guyon J. R., Wu C. T., Bender W., Kingston R. E. (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98, 37–46 [DOI] [PubMed] [Google Scholar]
- 3. Ng J., Hart C. M., Morgan K., Simon J. A. (2000) A Drosophila ESC-E(Z) protein complex is distinct from other Polycomb group complexes and contains covalently modified ESC. Mol. Cell Biol. 20, 3069–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Plath K., Fang J., Mlynarczyk-Evans S. K., Cao R., Worringer K. A., Wang H., de la Cruz C. C., Otte A. P., Panning B., Zhang Y. (2003) Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135 [DOI] [PubMed] [Google Scholar]
- 5. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 6. Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V. (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196 [DOI] [PubMed] [Google Scholar]
- 7. Müller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A., Wild B., Miller E. L., O'Connor M. B., Kingston R. E., Simon J. A. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208 [DOI] [PubMed] [Google Scholar]
- 8. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 9. Levine S. S., Weiss A., Erdjument-Bromage H., Shao Z., Tempst P., Kingston R. E. (2002) The core of the Polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell Biol. 22, 6070–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simon J. A., Kingston R. E. (2009) Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697–708 [DOI] [PubMed] [Google Scholar]
- 11. King I. F., Francis N. J., Kingston R. E. (2002) Native and recombinant Polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol. Cell Biol. 22, 7919–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tavares L., Dimitrova E., Oxley D., Webster J., Poot R., Demmers J., Bezstarosti K., Taylor S., Ura H., Koide H., Wutz A., Vidal M., Elderkin S., Brockdorff N. (2012) RYBP-PRC1 complexes mediate H2A ubiquitylation at Polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Francis N. J., Kingston R. E., Woodcock C. L. (2004) Chromatin compaction by a Polycomb group protein complex. Science 306, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 14. Eskeland R., Leeb M., Grimes G. R., Kress C., Boyle S., Sproul D., Gilbert N., Fan Y., Skoultchi A. I., Wutz A., Bickmore W. A. (2010) Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 38, 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Breiling A., Turner B. M., Bianchi M. E., Orlando V. (2001) General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412, 651–655 [DOI] [PubMed] [Google Scholar]
- 16. Saurin A. J., Shao Z., Erdjument-Bromage H., Tempst P., Kingston R. E. (2001) A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412, 655–660 [DOI] [PubMed] [Google Scholar]
- 17. Thomas M. C., Chiang C. M. (2006) The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 [DOI] [PubMed] [Google Scholar]
- 18. Lin J. J., Lehmann L. W., Bonora G., Sridharan R., Vashisht A. A., Tran N. (2011) Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev. 25, 2198–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Black J. C., Choi J. E., Lombardo S. R., Carey M. (2006) A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell 23, 809–818 [DOI] [PubMed] [Google Scholar]
- 20. Johnson K. M., Wang J., Smallwood A., Arayata C., Carey M. (2002) TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 16, 1852–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krebs A. R., Karmodiya K., Lindahl-Allen M., Struhl K., Tora L. (2011) SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes, and ATAC defines a class of p300-independent enhancers. Mol. Cell 44, 410–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simon M. D., Chu F., Racki L. R., de la Cruz C. C., Burlingame A. L., Panning B., Narlikar G. J., Shokat K. M. (2007) The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luger K., Rechsteiner T. J., Flaus A. J., Waye M. M., Richmond T. J. (1997) Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272, 301–311 [DOI] [PubMed] [Google Scholar]
- 24. Steger D. J., Owen-Hughes T., John S., Workman J. L. (1997) Analysis of transcription factor-mediated remodeling of nucleosomal arrays in a purified system. Methods 12, 276–285 [DOI] [PubMed] [Google Scholar]
- 25. Lin J. J., Carey M. (2012) In vitro transcription and immobilized template analysis of preinitiation complexes. Curr. Protoc. Mol. Biol. Chapter 12, Unit 12.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ikeda K., Steger D. J., Eberharter A., Workman J. L. (1999) Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell Biol. 19, 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tantin D., Chi T., Hori R., Pyo S., Carey M. (1996) Biochemical mechanism of transcriptional activation by GAL4-VP16. Methods Enzymol. 274, 133–149 [DOI] [PubMed] [Google Scholar]
- 28. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sato S., Tomomori-Sato C., Banks C. A., Parmely T. J., Sorokina I., Brower C. S., Conaway R. C., Conaway J. W. (2003) A mammalian homolog of Drosophila melanogaster transcriptional coactivator intersex is a subunit of the mammalian Mediator complex. J. Biol. Chem. 278, 49671–49674 [DOI] [PubMed] [Google Scholar]
- 30. Zhou Q., Lieberman P. M., Boyer T. G., Berk A. J. (1992) Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 6, 1964–1974 [DOI] [PubMed] [Google Scholar]
- 31. Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., van Berkum N. L., Ebmeier C. C., Goossens J., Rahl P. B., Levine S. S., Taatjes D. J., Dekker J., Young R. A. (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ku M., Koche R. P., Rheinbay E., Mendenhall E. M., Endoh M., Mikkelsen T. S., Presser A., Nusbaum C., Xie X., Chi A. S., Adli M., Kasif S., Ptaszek L. M., Cowan C. A., Lander E. S., Koseki H., Bernstein B. E. (2008) Genome-wide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4, e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ferrari R., Su T., Li B., Bonora G., Oberai A., Chan Y., Sasidharan R., Berk A. J., Pellegrini M., Kurdistani S. K. (2012) Reorganization of the host epigenome by a viral oncogene. Genome Res. 22, 1212–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eadie W. T. (1971) Statistical Methods in Experimental Physics, pp. 313–318, American Elsevier Publishing Co., New York [Google Scholar]
- 36. Fischle W., Wang Y., Jacobs S. A., Kim Y., Allis C. D., Khorasanizadeh S. (2003) Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17, 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen X., Hiller M., Sancak Y., Fuller M. T. (2005) Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science 310, 869–872 [DOI] [PubMed] [Google Scholar]
- 38. Koleske A. J., Young R. A. (1994) An RNA polymerase II holoenzyme responsive to activators. Nature 368, 466–469 [DOI] [PubMed] [Google Scholar]
- 39. Chao D. M., Gadbois E. L., Murray P. J., Anderson S. F., Sonu M. S., Parvin J. D., Young R. A. (1996) A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature 380, 82–85 [DOI] [PubMed] [Google Scholar]
- 40. Endoh M., Endo T. A., Endoh T., Fujimura Y., Ohara O., Toyoda T., Otte A. P., Okano M., Brockdorff N., Vidal M., Koseki H. (2008) Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135, 1513–1524 [DOI] [PubMed] [Google Scholar]
- 41. Stock J. K., Giadrossi S., Casanova M., Brookes E., Vidal M., Koseki H., Brockdorff N., Fisher A. G., Pombo A. (2007) Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 9, 1428–1435 [DOI] [PubMed] [Google Scholar]
- 42. Dellino G. I., Schwartz Y. B., Farkas G., McCabe D., Elgin S. C., Pirrotta V. (2004) Polycomb silencing blocks transcription initiation. Mol. Cell 13, 887–893 [DOI] [PubMed] [Google Scholar]
- 43. Vermeulen M., Mulder K. W., Denissov S., Pijnappel W. W., van Schaik F. M., Varier R. A., Baltissen M. P., Stunnenberg H. G., Mann M., Timmers H. T. (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 [DOI] [PubMed] [Google Scholar]
- 44. Min J., Zhang Y., Xu R. M. (2003) Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys-27. Genes Dev. 17, 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Francis N. J., Saurin A. J., Shao Z., Kingston R. E. (2001) Reconstitution of a functional core Polycomb repressive complex. Mol. Cell 8, 545–556 [DOI] [PubMed] [Google Scholar]
- 46. Uhlmann T., Boeing S., Lehmbacher M., Meisterernst M. (2007) The VP16 activation domain establishes an active mediator lacking CDK8 in vivo. J. Biol. Chem. 282, 2163–2173 [DOI] [PubMed] [Google Scholar]
- 47. Yang F., DeBeaumont R., Zhou S., Näär A. M. (2004) The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. U.S.A. 101, 2339–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goodrich J. A., Hoey T., Thut C. J., Admon A., Tjian R. (1993) Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75, 519–530 [DOI] [PubMed] [Google Scholar]
- 49. Berk A. J., Boyer T. G., Kapanidis A. N., Ebright R. H., Kobayashi N. N., Horn P. J., Sullivan S. M., Koop R., Surby M. A., Triezenberg S. J. (1998) Mechanisms of viral activators. Cold Spring Harb. Symp. Quant. Biol. 63, 243–252 [DOI] [PubMed] [Google Scholar]
- 50. Hawkins R. D., Hon G. C., Lee L. K., Ngo Q., Lister R., Pelizzola M., Edsall L. E., Kuan S., Luu Y., Klugman S., Antosiewicz-Bourget J., Ye Z., Espinoza C., Agarwahl S., Shen L., Ruotti V., Wang W., Stewart R., Thomson J. A., Ecker J. R., Ren B. (2010) Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6, 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smallwood A., Black J. C., Tanese N., Pradhan S., Carey M. (2008) HP1-mediated silencing targets Pol II coactivator complexes. Nat. Struct. Mol. Biol. 15, 318–320 [DOI] [PubMed] [Google Scholar]