Background: The Yersinia virulence regulator RovA is an intrinsic thermometer.
Results: The crystal structure of RovA is reported, and evidence is given that a thermosensing loop in the dimerization domain controls RovA activity.
Conclusion: Partial unfolding of RovA upon a temperature upshift leads to distortion of the DNA-binding domain and release from the operator sites.
Significance: Minor alterations, reflecting evolutionary changes between homologs, transform the thermotolerant regulator into a thermosensor.
Keywords: Bacterial Pathogenesis, Crystal Structure, Gene Regulation, Protein-Nucleic Acid interaction, Virulence Factors
Abstract
Pathogens often rely on thermosensing to adjust virulence gene expression. In yersiniae, important virulence-associated traits are under the control of the master regulator RovA, which uses a built-in thermosensor to control its activity. Thermal upshifts encountered upon host entry induce conformational changes in the RovA dimer that attenuate DNA binding and render the protein more susceptible to proteolysis. Here, we report the crystal structure of RovA in the free and DNA-bound forms and provide evidence that thermo-induced loss of RovA activity is promoted mainly by a thermosensing loop in the dimerization domain and residues in the adjacent C-terminal helix. These determinants allow partial unfolding of the regulator upon an upshift to 37 °C. This structural distortion is transmitted to the flexible DNA-binding domain of RovA. RovA contacts mainly the DNA backbone in a low-affinity binding mode, which allows the immediate release of RovA from its operator sites. We also show that SlyA, a close homolog of RovA from Salmonella with a very similar structure, is not a thermosensor and remains active and stable at 37 °C. Strikingly, changes in only three amino acids, reflecting evolutionary replacements in SlyA, result in a complete loss of the thermosensing properties of RovA and prevent degradation. In conclusion, only minor alterations can transform a thermotolerant regulator into a thermosensor that allows adjustment of virulence and fitness determinants to their thermal environment.
Introduction
Microbial pathogens need to adapt rapidly to milieus in warm-blooded hosts after entry from external habitats. This requires a coordinated control of large sets of genes covering a wide range of cellular processes. The thermal upshift accompanying host entry is often used by bacterial pathogens to tailor the complex regulatory network controlling virulence-associated functions (1–3).
The MarR family constitutes an important class of transcriptional regulators by which prokaryotes sense their surrounding biosphere. They control numerous biological functions important for survival, stress resistance, metabolic adaptation, and virulence (4). The SlyA/RovA family represents a subgroup of MarR-type regulators that participate in the control of virulence factors and are as such crucial for the successful establishment of an infection (5). The Salmonella SlyA protein was shown to control hemolysins and antimicrobial peptides and induces expression of SPI-2 (Salmonella pathogenicity island 2), necessary for survival in macrophages and virulence in mice (6–9). RovA, the SlyA homolog of pathogenic yersiniae, coordinates the expression of multiple genes contributing to host colonization and persistence. For instance, in enteropathogenic Yersinia species, RovA activates expression of the internalization factor invasin, allowing a more efficient colonization of gut-associated lymphatic follicles, whereas RovA of Yersinia pestis affects production of the type III secretion system and antiphagocytic Yop effector proteins, which are essential for the development of bubonic plague (10–14). In contrast to all other previously identified MarR homologs, RovA of Yersinia pseudotuberculosis, which is 100% identical to RovA of Y. pestis, represents an intrinsic proteinaceous thermometer that is able to sense temperature shifts directly through reversible alterations in its conformation. Thermo-induced conformational changes within the RovA structure modulate its DNA-binding capacity and render the regulator more susceptible to proteolytic degradation by the ATP-dependent protease Lon (11).
Use of a built-in thermosensor in the MarR-type regulator RovA to control DNA-binding activity and protein stability represents a unique regulatory strategy to adjust virulence-associated processes. To gain insight into the molecular mechanism through which RovA confers its specific biological activities, we solved the crystal structure of free and DNA-bound RovA and identified amino acids implicated in thermosensing and regulated proteolysis.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
A detailed description of the growth conditions, construction of bacterial strains and plasmids, and molecular biology procedures is provided under supplemental “Experimental Procedures.” Strains and plasmids are listed in supplemental Table S2.
Crystallization of RovA and RovA-DNA Complexes, Data Collection, and Structure Solution
Published procedures or methods described under supplemental “Experimental Procedures” were followed for mutagenesis, expression, and purification of the RovA proteins (15, 26). RovA in the unbound and DNA-bound forms was crystallized at a concentration of 20 mg/ml with and without DNA (8 mg/ml). The crystals of RovA without DNA belong to the space group P212121, and a data set up to 2.4 Å was collected at European Synchrotron Radiation Facility beamline ID14h2. The crystals of RovA in complex with DNA belong to the space group P3 and diffract up to 1.9 Å. For details of the determination of the structure, see supplemental “Experimental Procedures.” Supplemental Table S1 summarizes data collection and refinement statistics. The final models were validated using MolProbity (27). All residues of all models fall into the allowed region of the Ramachandran plot (28). Atomic coordinates and structure factors were deposited at the Protein Data Bank (29) under accession codes 4aih (RovA), 4aik (RovA bound to an inv fragment), and 4aij (RovA bound to a rovA fragment).
Analysis of the Biochemical Properties of RovA and SlyA
CD spectroscopy of the RovA proteins, DNA retardation assays, and protein stability assays were performed as described under supplemental “Experimental Procedures” or previously (11, 30).
RESULTS AND DISCUSSION
Structure of the Thermosensor RovA
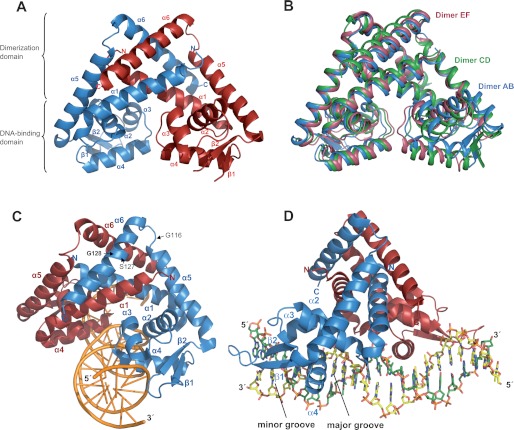
The structures of RovA of Y. pseudotuberculosis alone and in complex with DNA were solved at resolutions of 1.85 and 2.1 Å, respectively. Data collection and refinement statistics are given in supplemental Table S1. The overall structure of RovA consists of two subunits comprising six α-helices and two β-sheets and resembles other members of the MarR regulator family (Fig. 1A and supplemental Fig. S1). The two RovA monomers are tightly intertwined with the three long helices α1, α5, and α6, which form a large dimer interface of 2252.6 Å2 and a ΔG of −34.4 kcal/mol. The interface is supported mainly by hydrophobic interactions involving surface-located aliphatic and aromatic amino acids in the respective dimerization domains. The DNA-binding domain is formed by the central part of RovA and contains a winged helix-turn-helix DNA-binding motif. It comprises helices α3 (stabilizing helix) and α4 (recognition helix) and two antiparallel β-strands linked by a loop (wing) (Fig. 1A and supplemental Fig. S1).
FIGURE 1.
Structure of the thermosensing regulator RovA. A, illustration of the RovA dimer. B, superposition of three RovA dimers found in the asymmetric unit. The three different types of dimers (AB, CD, and EF) are shown in blue, green, and red. The RovA monomers could be built despite the flexible wing domain. C and D, schematic representation of a RovA dimer in complex with DNA from the front and the side, respectively. α-Helices, β-sheets, and amino acids implicated in thermosensing and RovA stability are indicated. One monomer is shown in red and the other in blue.
Three different RovA dimer variants exist in a crystallographic asymmetric unit and have the same overall structure but vary slightly in their conformation (Fig. 1B). These differences are due mainly to movements of the DNA-binding domain or the mobility of one of the long helices in the dimerization region. Similar to other structures of MarR-type regulators, the wing of the DNA-binding region is disordered in the unbound form. This indicates that the unbound form of RovA shows a considerable degree of flexibility, especially in the DNA-binding region.
RovA Structure in the DNA-bound State
Two high-affinity RovA-binding sites (I and II) were previously detected at diverse positions in the rovA and inv promoters (supplemental Fig. S2), to which RovA bound with a similar apparent dissociation constant (Kd = 32–46 nm) (10, 11). To compare the DNA-binding mode for different target sites, RovA was crystallized in the presence of one binding site of each promoter, which contained a short consensus sequence ((A/T)ATTAT(A/T)T) shown to be important for RovA binding (10). RovA bound to both promoter fragments in a similar fashion with a root mean square deviation between common Cα atoms of the structures of 0.41 Å. The asymmetric unit contains one RovA-DNA complex. In contrast to unbound RovA, only one RovA dimer variant is present, and the wing region is well defined. This suggests that RovA binding to DNA locks the regulatory protein into one conformation (Fig. 1, C and D). The superposition of RovA in the free and DNA-bound forms shows that RovA performs a twisting motion upon DNA binding (data not shown). This conformational change increases the gap between the two DNA-binding domains in the dimer to allow the insertion of helix α4 into consecutive major grooves of the DNA (Fig. 1, C and D). Both the conformational flexibility of RovA and motions of the DNA-binding domains seem crucial to propagate thermo-induced conformational changes into an attenuation of DNA binding.
DNA-binding Mode of RovA
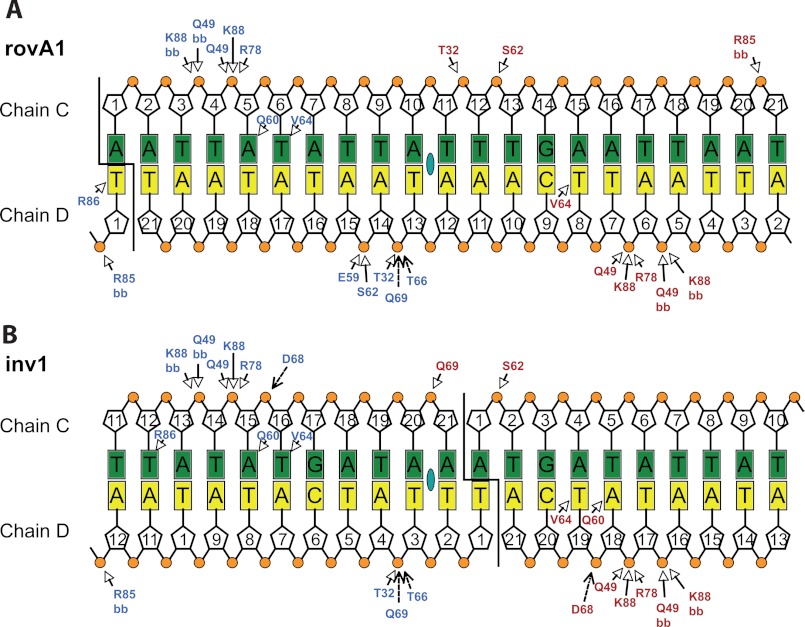
As expected, RovA bound to the previously identified DNA-binding site in the middle of the rovA promoter fragment (Fig. 2A). In the case of the inv promoter fragment, however, RovA interacted with the joint ends of the promoter fragments (Fig. 2B). The newly generated RovA-binding sequence in the pseudo-continuous DNA strand in the crystal is very similar to the originally detected binding sequence (supplemental Fig. S3), indicating that the new site represents a similar or even better target site for the RovA protein. Both RovA subunits make extensive and comparable contacts within the rovA and inv promoter regions (Fig. 2). Although helix α4 of the helix-turn-helix motif is deeply inserted into the major groove of the DNA, only very few specific interactions are visible between RovA and the DNA bases, namely Gln-60, Val-64, and Arg-86 (Fig. 2 and supplemental Fig. S4). Gln-60 of one RovA monomer is able to bind to an adenine located 6 bp from the pseudosymmetrical center in the rovA fragment (Fig. 2A). RovA binding to the original binding site in the inv fragment would have allowed a similar contact of Gln-60 by only one monomer. However, both Gln-60 residues of the RovA dimer are able to bind to adenines in the newly generated recognition site, which most probably increased the affinity for this binding sequence (supplemental Fig. S3). Notably, the importance of Gln-60 for the DNA interactions is also supported by the fact that a RovA(Q60R) variant had an impaired DNA-binding capacity and a reduced ability to stimulate inv expression (15). The other important residue, Val-64, makes hydrophobic interactions with a thymidine at position 5 from the center of pseudosymmetry in both monomers, and Arg-86 of the wing domain interacts with a thymidine residue (position 9) at the end of the pseudopalindromic sequence (Fig. 2 and supplemental Fig. S4).
FIGURE 2.
Interaction of RovA with the inv and rovA promoters. Shown are schematic drawings of the RovA rovA-DNA (A) and RovA inv-DNA (B) interactions. Direct interactions are indicated by arrows, and water-mediated interactions are indicated by dashed arrows. Bases of both DNA strands (chains C and D) are represented by rectangles, deoxyribose by pentagons, and phosphates by circles. The colors of the RovA residues correspond to the chains: chain A (blue) and chain B (red). The pseudosymmetrical center is indicated by a green oval in the middle of the double strand. Open arrows indicate direct interactions, and closed arrows indicate water-mediated interactions. bb, amino acid backbone.
In addition to helix α4, also helices α2 and α3 and the wing make direct contacts with DNA (Fig. 1, C and D). However, most of these interactions are nonspecific hydrogen bonds between the wing and the deoxyribose phosphate backbone of the DNA (Fig. 2). This is in line with previous results demonstrating that RovA is a global regulator that directly interacts with many promoter regions of Yersinia (10, 13, 16). The relatively small number of specific contacts between RovA and DNA allows the virulence regulator to recognize multiple non-palindromic promoter sequences in the Yersinia genome that do not share a highly conserved consensus sequence (10, 13, 16). Of the nonspecific interactions, binding of Gln-49 seems particularly important because this residue interacts with two adjacent sugar phosphate units on both sides of the bound DNA. This is supported by a previous study that showed that the Q49R mutation impairs DNA binding and renders the RovA molecule defective for transcriptional activation (15).
RovA of Yersinia and SlyA of Salmonella Display Similar DNA-binding Properties
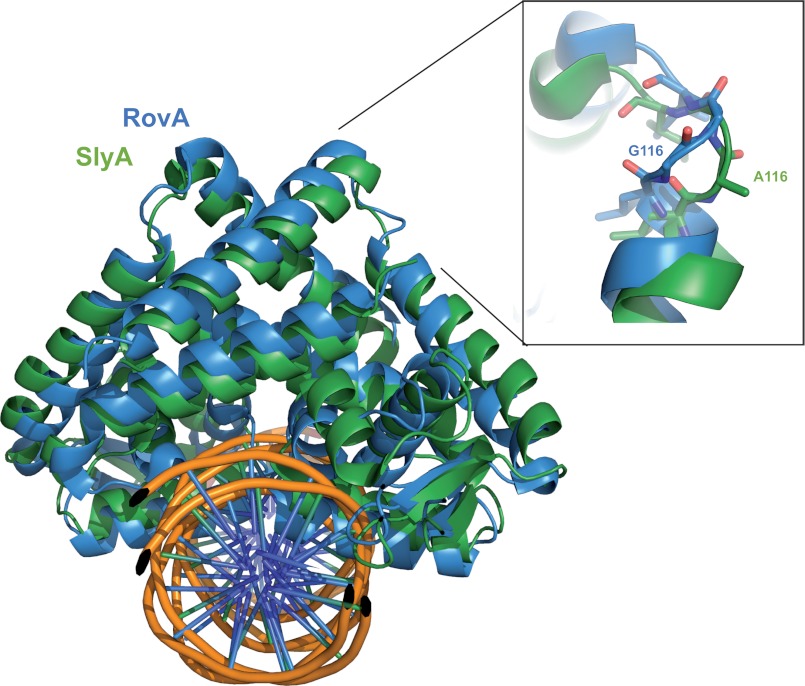
The closest homolog of RovA with a solved structure is SlyA of Salmonella enterica serovar Typhimurium, showing an amino acid sequence identity of 76% (Fig. 3 and supplemental Fig. S1) (17). In particular the DNA-binding regions are virtually identical, and both regulators show a similar promiscuity regarding their DNA-binding sites. However, in contrast to RovA, Gln-60 was not involved in DNA binding in the SlyA-DNA structure. On the other hand, Arg-65 of SlyA, which is bound to a guanine residue in the SlyA operator site (17), is not visible in the RovA structures in complex with the inv or rovA promoter fragment. This strongly suggests that RovA and SlyA recognize DNA sequences that are partially incomplete, i.e. do not contain all possible binding partners. The small number of specific contacts of RovA may be important to ensure a DNA-binding strength that allows rapid attenuation of DNA binding after a thermal upshift and counter-regulation and replacement of other competing global regulatory proteins. Previous studies showed that RovA acts mainly as an antisilencer that alleviates transcriptional repression by the small nucleoid protein H-NS, which has also little sequence specificity (10, 11, 18). This is different from other global regulators with conserved consensus sequences (e.g. the cAMP receptor protein Crp), which serve mainly to increase levels of promoter occupancy by RNA polymerase. In this case, the activator needs to be precisely positioned to ensure optimal interaction with the RNA polymerase.
FIGURE 3.
Superposition of RovA and SlyA from S. enterica serovar Typhimurium. Shown is a schematic representation of a RovA dimer in complex with DNA. The SlyA dimer is shown in green, the RovA dimer in blue, and the DNA in orange (phosphate sugar backbone) and purple (base pairs). The flexible loop between helices α5 and α6 is enlarged in the inset. Gly-116, which is important for thermosensing, and Ala-116, the equivalent amino acid of Salmonella SlyA, are illustrated.
Flexible Loop in the Dimerization Domain Mediates Intrinsic Thermosensing of RovA
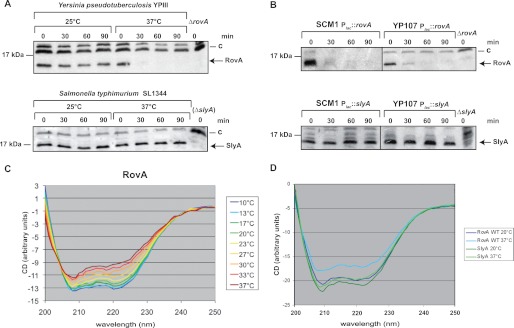
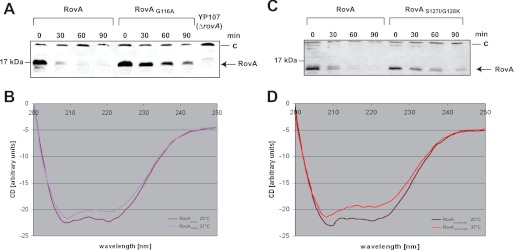
Although the structures of the Yersinia virulence factor RovA in its unbound form and in complex with DNA share many features with the Salmonella SlyA protein (Fig. 3) (17), major differences have been observed in their biochemical properties. The Yersinia RovA protein itself is a protein thermometer that uses intrinsic thermal sensing to control its DNA-binding functions and its degradation (11). In contrast, thermosensing, concomitant with a loss in stability at body temperature, has not been described for Salmonella SlyA. All previous studies on SlyA-mediated control of virulence gene expression in Salmonella were performed at and report SlyA production at 37 °C (19–22). In fact, when the stabilities of RovA and SlyA were compared in Y. pseudotuberculosis and S. enterica serovar Typhimurium, RovA was rapidly degraded, whereas the Salmonella SlyA protein remained stable at 37 °C in both pathogens (Fig. 4, A and B). However, at 25 °C, both the RovA and SlyA proteins remained stable. CD spectroscopy of the purified proteins further demonstrated that in contrast to RovA (Fig. 4C), SlyA does not undergo a temperature-dependent conformational change (Fig. 4D) and does not lose its DNA-binding capacity at body temperature (supplemental Fig. S5). This strongly indicates that SlyA is not an intrinsic thermosensor that is subjected to controlled proteolysis, although its overall structure is very similar to RovA (Fig. 3). Thermosensing has been described for only three other bacterial transcriptional regulators. However, these regulators are structurally unrelated to RovA and are most likely controlled by a different mechanism (23–25).
FIGURE 4.
Thermo-induced conformational changes and proteolysis of RovA and SlyA. The stability of RovA and SlyA was investigated at 25 and 37 °C in the original bacterial strains Y. pseudotuberculosis YPIII and S. enterica serovar Typhimurium SL1344 (A) and in the isogenic ΔslyA (SCM1) and ΔrovA (YP107) mutant strains containing either the Plac::rovA (pCM3) or Plac::slyA (pCM15) plasmid (B). A higher molecular weight protein (C) that reacted with the polyclonal antisera was used as a loading control. C and D, conformational analysis of RovA and SlyA using CD spectroscopy. CD spectra show millidegrees (m−1 cm−1) versus wavelength of RovA (0.2 mg/ml) (C) or SlyA (0.2 mg/ml) (D) as a function of temperature.
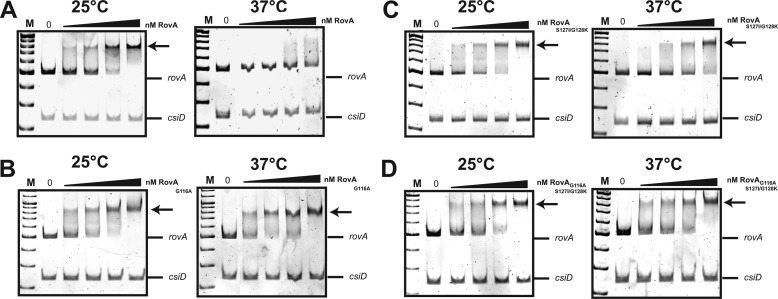
Superposition of the SlyA and RovA structures to identify variations between the two regulatory proteins revealed differences in a small loop between helices α5 and α6 (Fig. 3). This loop, including Gly-116 of RovA, has a different conformation in SlyA, in which position 116 is occupied by an alanine (Fig. 3 and supplemental Fig. S1). We introduced a corresponding substitution into RovA and found that the mutant protein (RovA(G116A)) showed no major thermally induced loss of structural integrity (Fig. 5B) and did not display a thermo-induced reduction in DNA binding (Fig. 6B). DNA binding of the RovA(G116A) variant was comparable at 25 and 37 °C. This demonstrated that RovA(G116A) lost most of its intrinsic thermosensitivity observed for wild-type RovA. Consequently, RovA(G116A) did not reduce its DNA-binding property at 37 °C, which was concomitant with an enhanced stability of RovA (Fig. 5A). It is likely that the recognition sites for the Lon protease are occluded in the active DNA-bound RovA(G116A) variant even at 37 °C. In fact, previous studies with chimeric RovA proteins indicated that amino acids in the vicinity of the DNA-binding region are important for proteolytic susceptibility (11). We conclude that the specific intrinsic thermosensing ability of RovA involves Gly-116 located in a flexible loop between helices α5 and α6 implicated in RovA dimerization. This represents a novel thermosensing mechanism. Of the three known thermosensing regulators, the heat-sensing domain is known only for CtsR, a master regulator of protein quality control of low-GC, Gram-positive bacteria (25). Strikingly, in CtsR, intrinsic heat sensing occurs through a tetraglycine loop connecting the two β-sheets of its winged helix-turn-helix domain. Although both regions are structurally and functionally unrelated, they possess a low thermal stability due to a flexible loop structure.
FIGURE 5.
Thermo-induced conformational changes and proteolysis of RovA mutant proteins. The stability and thermo-induced conformational changes of the RovA mutants RovA(G116A) (A) and RovA(S127I/G128K) (C) were investigated at 37 °C. A higher molecular weight protein (C) that reacted with the polyclonal antisera was used as a loading control. Thermo-induced conformational changes in RovA(G116A) (B) and RovA(S127I/G128K) (D) were detected by CD spectroscopy. CS spectra, degrees (m−1 cm−1) versus wavelength of RovA(G116A) (0.2 mg/ml) or RovA(S127I/G128K) (0.2 mg/ml) at 20 and 37 °C, are presented.
FIGURE 6.
Interaction of RovA, RovA(G116A), RovA(S127I/G128K), or RovA(G116A/S127I/G128K) with the rovA regulatory region at 25 or 37 °C. Double-stranded promoter fragments of the rovA regulatory region harboring RovA-binding site I (10) were incubated without or with increasing amounts (25, 36, 48, and 65 nm) of purified RovA (A) and its variants (RovA(G116A) (B), RovA(S127I/G128K) (C), or RovA(G116A/S127I/G128K) (D)) at 25 or 37 °C. Lanes in which lower concentrations of RovA (<25 nm) were added to the DNA fragments are not shown. A nonspecific probe containing an unrelated sequence (csiD promoter of Escherichia coli) was included as a negative control. The RovA-DNA complexes are indicated by arrows. M, DNA size marker–100 bp DNA ladder.
Additional Residues Important for Thermo-induced Degradation of RovA
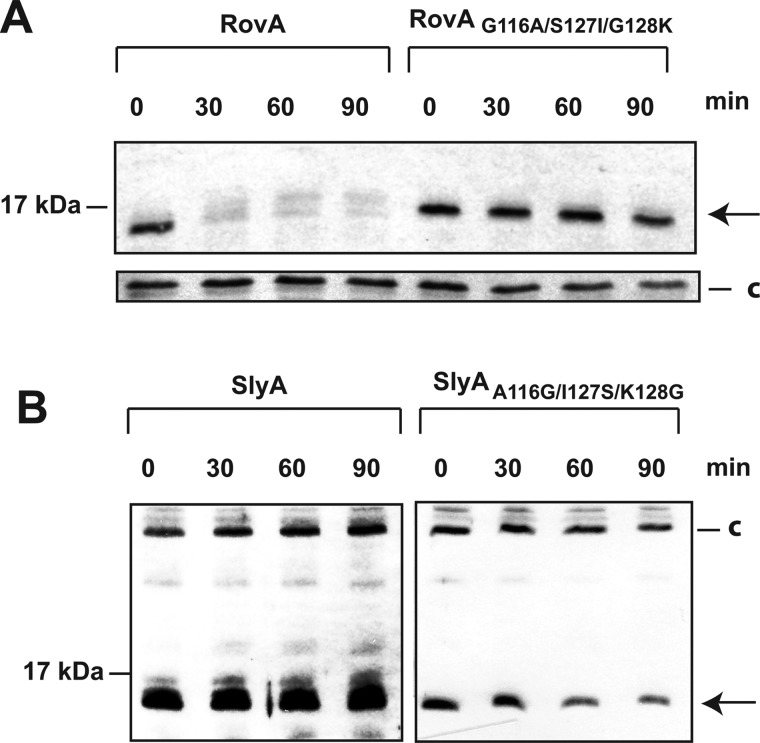
Although thermosensing and degradation of RovA(G116A) were strongly reduced, degradation was not entirely abolished at 37 °C (Fig. 5A). Compared with SlyA, slightly less of the RovA protein was detectable 90 min after blockage of protein synthesis. This indicates that additional amino acids contribute to the thermostabilization of SlyA. We replaced other amino acids of RovA with the equivalent amino acids of SlyA and found that a substitution of S127I/G128K in addition to G116A (RovA(G116A/S127I/G128K)) further increased the stability of RovA to the level of SlyA (Fig. 7A). DNA binding studies further showed that RovA(G116A/S127I/G128K) did not bind DNA in a temperature-dependent manner (Fig. 6D), similar to the SlyA protein (supplemental Fig. S5). We also included both mutations in RovA without the G116A substitution and found that the RovA(S127I/G128K) protein was more stable than the wild-type RovA protein, although it was still degraded at 37 °C (Fig. 5C). In contrast to RovA(G116A), only a slight reduction in the thermosensing capacity was apparent in the RovA(S127I/G128K) protein (Fig. 5D). In agreement with this observation, DNA binding of this variant was increased at 37 °C but was still lower relative to RovA(G116A) (Fig. 6, B and C). Interestingly, thermal denaturation measured by far-UV CD spectroscopy revealed two unfolding transitions for RovA and its variants, whereas SlyA exhibited only a single transition at a higher temperature (supplemental Fig. S6). The midpoints of both unfolding transitions were lowest for RovA, considerably higher for RovA(G116A), and highest for RovA(G116A/S127I/G128K) (supplemental Fig. S6). The in vitro thermal denaturation behavior is therefore in full agreement with the thermosensing capacities of the variants observed in vivo. To confirm these observations, we also included the reverse mutations in SlyA to determine whether introduction of the reciprocal substitutions leads to its destabilization at body temperature. In fact, we observed a considerable degradation of SlyA(A116G/I127S/K128G) in contrast to wild-type SlyA, which remained completely stable during the test period (Fig. 7B). All other changes designed to convert RovA into SlyA resulted in unstable or less stable proteins, e.g. RovA(T4P), or did not lead to a significant increase in protein stability at 37 °C, e.g. RovA(Q102E/V103M/D104E) and RovA(N41H/R42Q) (supplemental Fig. S7). We conclude that only a few specific amino acid substitutions are sufficient to transform a thermotolerant protein into an intrinsic thermosensor and vice versa.
FIGURE 7.
Thermo-dependent stability of the RovA(G116A/S127I/G128K) and SlyA(A116G/I127S/K128G) proteins. The stability of RovA(G116A/S127I/G128K) produced in Y. pseudotuberculosis YPIII (A) and SlyA(A116G/I127S/K128G) synthesized in S. enterica serovar Typhimurium SL1344 (B) was investigated at 37 °C. RovA (A) and SlyA (B) are indicated by the arrows. A higher molecular weight protein (c) that reacted with the polyclonal antisera was used as a loading control.
Conclusion
The thermal upshift encountered upon host entry is the most crucial signal for many pathogens to induce and adjust their virulence functions. Frequently, thermo-induced structural changes in the DNA (e.g. supercoiling and bending) and mRNAs (thermoswitches) are used to control virulence gene expression, and the molecular mechanisms underlying these control systems have been elucidated in several pathogens. Recently, also regulatory proteins that function as “molecular thermometers” have been identified (11, 23–25). However, the mechanism of the primary thermosensing event and the structural determinants defining the thermosensitive properties of these regulators are largely unknown. In this study, we have shown that the overall structure of the thermosensitive master regulator RovA of Yersinia resembles the thermotolerant homologous protein SlyA of Salmonella in the free and DNA-bound forms. However, it contains a small number of distinct single amino acid substitutions leading to major changes in the biochemical properties of the regulator. Here, we have provided evidence that RovA thermosensing depends mainly on a flexible loop situated between two α-helical structures involved in dimer formation. Intrinsic thermal sensing is supported by additional residues, including G1y-128, which introduces more conformational flexibility in the adjacent helix α6. It is very likely that these determinants promote partial and reversible unfolding of the intervening helices upon a thermal upshift without disruption of the dimer. This conformational change is transmitted to the flexible DNA-binding domain of RovA, contacting mainly the DNA backbone. A low-affinity DNA-binding mode appears to enable the immediate release of RovA from its operator sites and allows rapid degradation of nonfunctional protein.
We further conclude that even minor changes that cause only negligible alterations of the overall structure of the SlyA/RovA regulators can provoke significant differences in the biochemical properties that affect their ability to promote transcription and to reprogram virulence-associated processes. We posit that similarly small evolutionary changes in master regulators could generate significant phenotypic changes. A clear advantage is that it allows pathogens to respond to external pressures (e.g. temperature), which allows them to adapt rapidly and long-term to changing reservoirs, different hosts, or host environments. The fact that thermo-induced conformational changes in RovA are reversible makes this regulator also a useful tool for the development of inducible expression systems for biotechnological applications.
Acknowledgments
We thank Martin Fenner for helpful discussion and Boris Grujic and Dr. Jörn Krauβe for construction of pBG1 and help with the structure determination. We also thank Carolin Schaper for technical assistance.
Footnotes
This work was supported by Grant IRTG1273 from the German Research Foundation (to C. M.) and the Fonds der Chemischen Industrie (to P. D. and D. W. H.).

This article contains supplemental “Experimental Procedures,” Figs. S1–S7, Tables S1 and S2, and additional references.
The atomic coordinates and structure factors (codes 4aih, 4aij, and 4aik) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
REFERENCES
- 1. Konkel M. E., Tilly K. (2000) Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2, 157–166 [DOI] [PubMed] [Google Scholar]
- 2. Klinkert B., Narberhaus F. (2009) Microbial thermosensors. Cell. Mol. Life Sci. 66, 2661–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schumann W. (2007) Thermosensors in eubacteria: role and evolution. J. Biosci. 32, 549–557 [DOI] [PubMed] [Google Scholar]
- 4. Perera I. C., Grove A. (2010) Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol. Cell. Biol. 2, 243–254 [DOI] [PubMed] [Google Scholar]
- 5. Ellison D. W., Miller V. L. (2006) Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9, 153–159 [DOI] [PubMed] [Google Scholar]
- 6. Buchmeier N., Bossie S., Chen C. Y., Fang F. C., Guiney D. G., Libby S. J. (1997) SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65, 3725–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. von Rhein C., Bauer S., López Sanjurjo E. J., Benz R., Goebel W., Ludwig A. (2009) ClyA cytolysin from Salmonella: distribution within the genus, regulation of expression by SlyA, and pore-forming characteristics. Int. J. Med. Microbiol. 299, 21–35 [DOI] [PubMed] [Google Scholar]
- 8. Linehan S. A., Rytkönen A., Yu X. J., Liu M., Holden D. W. (2005) SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infect. Immun. 73, 4354–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi Y., Latifi T., Cromie M. J., Groisman E. A. (2004) Transcriptional control of the antimicrobial peptide resistance ugtL gene by the Salmonella PhoP and SlyA regulatory proteins. J. Biol. Chem. 279, 38618–38625 [DOI] [PubMed] [Google Scholar]
- 10. Heroven A. K., Nagel G., Tran H. J., Parr S., Dersch P. (2004) RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol. Microbiol. 53, 871–888 [DOI] [PubMed] [Google Scholar]
- 11. Herbst K., Bujara M., Heroven A. K., Opitz W., Weichert M., Zimmermann A., Dersch P. (2009) Intrinsic thermal sensing controls proteolysis of Yersinia virulence regulator RovA. PLoS Pathog. 5, e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang F., Ke Y., Tan Y., Bi Y., Shi Q., Yang H., Qiu J., Wang X., Guo Z., Ling H., Yang R., Du Z. (2010) Cell membrane is impaired, accompanied by enhanced type III secretion system expression in Yersinia pestis deficient in RovA regulator. PLoS ONE 5, e12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cathelyn J. S., Crosby S. D., Lathem W. W., Goldman W. E., Miller V. L. (2006) RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc. Natl. Acad. Sci. U.S.A. 103, 13514–13519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Revell P. A., Miller V. L. (2000) A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol. Microbiol. 35, 677–685 [DOI] [PubMed] [Google Scholar]
- 15. Tran H. J., Heroven A. K., Winkler L., Spreter T., Beatrix B., Dersch P. (2005) Analysis of RovA, a transcriptional regulator of Yersinia pseudotuberculosis virulence that acts through antirepression and direct transcriptional activation. J. Biol. Chem. 280, 42423–42432 [DOI] [PubMed] [Google Scholar]
- 16. Cathelyn J. S., Ellison D. W., Hinchliffe S. J., Wren B. W., Miller V. L. (2007) The RovA regulons of Yersinia enterocolitica and Yersinia pestis are distinct: evidence that many RovA-regulated genes were acquired more recently than the core genome. Mol. Microbiol. 66, 189–205 [DOI] [PubMed] [Google Scholar]
- 17. Dolan K. T., Duguid E. M., He C. (2011) Crystal structures of SlyA protein, a master virulence regulator of Salmonella, in free and DNA-bound states. J. Biol. Chem. 286, 22178–22185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kong W., Weatherspoon N., Shi Y. (2008) Molecular mechanism for establishment of signal-dependent regulation in the PhoP/PhoQ system. J. Biol. Chem. 283, 16612–16621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okada N., Oi Y., Takeda-Shitaka M., Kanou K., Umeyama H., Haneda T., Miki T., Hosoya S., Danbara H. (2007) Identification of amino acid residues of Salmonella SlyA that are critical for transcriptional regulation. Microbiology 153, 548–560 [DOI] [PubMed] [Google Scholar]
- 20. Xue P., Corbett D., Goldrick M., Naylor C., Roberts I. S. (2009) Regulation of expression of the region 3 promoter of the Escherichia coli K5 capsule gene cluster involves H-NS, SlyA, and a large 5′-untranslated region. J. Bacteriol. 191, 1838–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Corbett D., Bennett H. J., Askar H., Green J., Roberts I. S. (2007) SlyA and H-NS regulate transcription of the Escherichia coli K5 capsule gene cluster, and expression of slyA in Escherichia coli is temperature-dependent, positively autoregulated, and independent of H-NS. J. Biol. Chem. 282, 33326–33335 [DOI] [PubMed] [Google Scholar]
- 22. Zhao G., Weatherspoon N., Kong W., Curtiss R., 3rd, Shi Y. (2008) A dual-signal regulatory circuit activates transcription of a set of divergent operons in Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 105, 20924–20929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hurme R., Berndt K. D., Normark S. J., Rhen M. (1997) A proteinaceous gene regulatory thermometer in Salmonella. Cell 90, 55–64 [DOI] [PubMed] [Google Scholar]
- 24. Servant P., Grandvalet C., Mazodier P. (2000) The RheA repressor is the thermosensor of the HSP18 heat shock response in Streptomyces albus. Proc. Natl. Acad. Sci. U.S.A. 97, 3538–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Elsholz A. K., Michalik S., Zühlke D., Hecker M., Gerth U. (2010) CtsR, the Gram-positive master regulator of protein quality control, feels the heat. EMBO J. 29, 3621–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uliczka F., Pisano F., Schaake J., Stolz T., Rohde M., Fruth A., Strauch E., Skurnik M., Batzilla J., Rakin A., Heesemann J., Dersch P. (2011) Unique cell adhesion and invasion properties of Yersinia enterocolitica O:3, the most frequent cause of human yersiniosis. PLoS Pathog. 7, e1002117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramachandran G. N., Sasisekharan V. (1968) Conformation of polypeptides and proteins. Adv. Protein Chem. 23, 283–438 [DOI] [PubMed] [Google Scholar]
- 29. Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr., Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. (1977) The Protein Data Bank. A computer-based archival file for macromolecular structures. Eur. J. Biochem. 80, 319–324 [DOI] [PubMed] [Google Scholar]
- 30. Heroven A. K., Dersch P. (2006) RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence, and motility of Yersinia pseudotuberculosis. Mol. Microbiol. 62, 1469–1483 [DOI] [PubMed] [Google Scholar]