Background: SARA promotes an epithelial cell phenotype, whereas its down-regulation is permissive for EMT.
Results: PI3K inhibition decreases SARA protein expression, likely through alterations in Rab5-containing endosomes.
Conclusion: PI3K signaling supports an epithelial phenotype.
Significance: PI3K has complex effects in fibrogenesis. Our data suggest an antifibrotic action of PI3K that involves maintaining SARA expression.
Keywords: EMT, Endosomes, PI 3-Kinase (PI3K), SMAD Transcription Factor, Transforming Growth Factor Beta (TGFbeta), Rab5, SARA
Abstract
SARA has been shown to be a regulator of epithelial cell phenotype, with reduced expression during TGF-β1-mediated epithelial-to-mesenchymal transition. Examination of the pathways that might play a role in regulating SARA expression identified phosphatidylinositol 3-kinase (PI3K) pathway inhibition as sufficient to reduce SARA expression. The mechanism of PI3K inhibition-mediated SARA down-regulation differs from that induced by TGF-β1 in that, unlike TGF-β1, PI3K-dependent depletion of SARA was apparent within 6 h and did not occur at the mRNA or promoter level but was blocked by inhibition of proteasome-mediated degradation. This effect was independent of Akt activity because neither reducing nor enhancing Akt activity modulated the expression of SARA. Therefore, this is likely a direct effect of p85α action, and co-immunoprecipitation of SARA and p85α confirmed that these proteins interact. Both SARA and PI3K have been shown to be associated with endosomes, and either LY294002 or p85α knockdown enlarged SARA-containing endocytic vesicles. Inhibition of clathrin-mediated endocytosis blocked SARA down-regulation, and a localization-deficient mutant SARA was protected against down-regulation. As inhibiting PI3K can activate the endosomal fusion-regulatory small GTPase Rab5, we expressed GTPase-deficient Rab5 and observed endosomal enlargement and reduced SARA protein expression, similar to that seen with PI3K inhibition. Importantly, either interference with PI3K via LY294002 or p85α knockdown, or constitutive activity of the Rab5 pathway, enhanced the expression of smooth muscle α-actin. Together, these data suggest that although TGF-β1 can induce epithelial-to-mesenchymal transition through reduction in SARA expression, SARA is also basally regulated by its interaction with PI3K.
Introduction
The expression and activation of TGF-β is a universal feature in almost every type of chronic kidney disease in both animal models and in humans (1). Exogenous expression of TGF-β1 results in renal fibrosis, and inhibition of the TGF-β1 signaling pathway suppresses fibrogenesis and prevents loss of kidney function (1). In part, the role of TGF-β1 as a principal mediator of fibrosis is attributable to its effects as an inducer of EMT. However, the signals that regulate cell phenotype under non-stimulated conditions are less understood.
We recently showed that the Smad anchor for receptor activation protein (SARA)2 is down-regulated in TGF-β1-mediated EMT in human renal proximal tubule cells. We also found that depletion of SARA was sufficient to induce phenotypic changes in these cells independently of TGF-β1 (2). Therefore, SARA appears to be an important contributor to maintenance of epithelial cell phenotype. We (3) and others (4, 5) have shown that SARA localizes to endosomes containing the early endosomal protein EEA1 and Rab5 via interaction of the SARA FYVE domain with membrane phospholipid PtdIns-3-P and that disruption of SARA function, expression, or endosomal localization impairs TGF-β1-induced Smad2 signaling (4–6). However, the mechanism of regulation of SARA expression is not known.
Phosphatidylinositol 3-kinase (PI3K) is a ubiquitous lipid kinase involved in receptor signal transduction by tyrosine kinase receptors. The large and complex PI3K family includes three classes with multiple subunits and isoforms (7, 8). The class IA PI3Ks are composed of a p85 regulatory subunit and a p110 catalytic subunit that phosphorylates phosphoinositol 4-phosphate and phosphoinositol 4,5-phosphate at their D3 position (7, 8). The p85 isoforms are a group of related proteins encoded by the Pik3r1 (p85α, p55α, p50α), Pik3r2 (p85β), and Pik3r3 (p55γ) genes. The most abundant and well characterized regulatory subunit, p85α, contains the domains within the smaller isoforms but also has an N-terminal SH3 domain and a GTPase-activating protein (GAP) domain (also called a BCR homology domain), as well as two proline-rich regions, to provide added binding functions and regulatory capacities (9). In quiescent cells, the p85-p110 complex is cytosolic, and p110-encoded PI3K activity is repressed. Activation of receptor tyrosine kinases leads to p85α SH2-domain binding-mediated recruitment of the p85-p110 complex to the plasma membrane and relieves repression of p110 catalytic activity to generate the lipid messenger PtdIns-3,4,5-P3 that relocalizes and activates a number of downstream signaling proteins, including phosphoinositide-dependent kinase-1 and Akt (also called protein kinase B (PKB)) (10). Downstream of PDK1 and Akt activation a number of pathways may be affected leading to increased cell growth, cell cycle progression, cell migration, and cell survival through the activation of numerous target proteins (11, 12).
However, p85α interacts with a number of proteins and has additional functions in addition to the regulation of p110-PI3K activity (9). One binding partner of p85α, Rab5, is the most studied member of the large and diverse family of Rab GTPases and a key regulator of endocytosis. Rab5 is involved in clathrin-coated vesicle formation, homotypic fusion between early endosomes, endosomal cargo recruitment, and endosomal motility (13). Similar to other GTPases, Rab5 is active in its GTP-bound form. It is activated by Rab-specific guanine nucleotide-exchange factor that promotes exchange of GDP for GTP and is inactivated through specific GAPs that hydrolyze the GTP to GDP.
In the present study, we demonstrate that, in human renal proximal tubule epithelial cells (HKC), endogenous SARA is basally regulated by PI3K activity. Interfering with PI3K function, either through chemical inhibition or through shRNA-mediated knockdown of p85α, results in a loss of SARA through protein degradation. We find that SARA and p85α interact and that endocytosis and appropriate endosomal localization of SARA are required for the p85α-mediated reduction in SARA expression. As reported previously, PI3K inhibition caused an alteration in endosomal structure that can be mimicked by expression of a GTPase-defective, constitutively active mutant Rab5, Rab5Q79L, which also causes a reduction of SARA at the protein level. Importantly, reduction of SARA expression by either PI3K inhibition or by Rab5Q79L expression, results in de novo expression of the EMT marker, smooth muscle α-actin (αSMA).
EXPERIMENTAL PROCEDURES
Reagents and Materials
All kinase inhibitors, including LY294002, were purchased from EMD Biosciences (San Diego, CA). MG132 was purchased from Sigma-Aldrich. Active recombinant human TGF-β1, purchased from R&D Systems (Minneapolis, MN), was maintained as a stock solution of 4 μg/ml in 4 mm HCl and used at a final concentration of 2 ng/ml. Antibody to SARA was from Proteintech (Chicago, IL); EEA1 was from Santa Cruz Biotechnology (Santa Cruz, CA); αSMA was from DAKO (Carpinteria, CA); β-actin was from Sigma-Aldrich; and phospho-Akt and p85α were from Cell Signaling (Danvers, MA).
Cell Culture
The renal tubular epithelial cell line HKC was obtained from Dr. L. Racusen (14) and cultured in Dulbecco's modified Eagle's medium/F-12 supplemented with 10% fetal bovine serum, penicillin/streptomycin, amphotericin B, HEPES buffer, and glutamine.
Quantitative PCR
Total RNA was harvested from HKC using the RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer's directions. Following RNA quantification with the Quant-it RiboGreen assay (Invitrogen), RNAs were reverse-transcribed to cDNAs with the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed using the iQ SYBR Green Supermix (Bio-Rad) with the iCycler iQ real-time PCR detection system (Bio-Rad). Real time data were collected for 40 cycles of; 95 °C for 10 s and 55.3 °C for 45 s, followed by melt-curve analysis to verify the single peaks of amplicons. Primers were designed using software provided by Integrated DNA Technologies (Coralville, IA) and custom synthesized by the company. Primers used were as follows: αSMA, 5-AGCAGGCCAAGGGGCTATATAA-3 (forward) and 5′-CGTAGCTGTCTTTTTGTCCCATT-3 (reverse); SARA, 5′-GGTGAGGTGGCTCCAGTATG-3′ (forward) and 5′-CTCTGCAGTGATGCCTCCTT-3′ (reverse); p85α, 5′-CCCTATGCTTTTCAGATTCTCAG-3′ (forward) and 5′-AGGTTTTGGTGGTTTAGGAGG-3′ (reverse); human β2-microglobulin, 5′-TGTCTGGGTTTCATCCATCCGACA-3′ (forward) and 5′-TCACACGGCAGGCATACTCATCTT-3′ (reverse). Relative expression of the gene of interest was estimated by correction with the expression of β2-microglobulin, using the ΔΔCt method.
Immunoprecipitation, Western Blot Analysis, and Immunocytochemistry
Kinase inhibitor or TGF-β1 treatments were done in serum-free medium. Whole cell lysates were prepared by lysis in RIPA buffer, and immunoprecipitations were performed as described previously (15). Western blots and densitometric analysis were performed as described previously (15).
For immunocytochemistry, cells on coverslips in serum-free medium were treated with vehicle or LY294002 or Akt inhibitor IV for 16–24 h before paraformaldehyde fixation followed by permeabilization with Triton X-100. After blocking nonspecific sites with BSA, cells were incubated with SARA or EEA1 antibodies for 2 h at room temperature, washed with PBS, then incubated with 2.5 μg/ml Alexa Fluor 594-conjugated secondary antibodies for 30 min. Coverslips were mounted with Aqua-Poly/Mount (Polysciences, Warrington, PA) and viewed under a Zeiss Axiovert 200 m confocal microscope with a Zeiss plan-apochromat 100×/1.4 oil objective, and images were acquired using LSM 510 SP1 software (version 4.2). Imaging work was performed at the Northwestern University Cell Imaging Facility generously supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. Digital images were converted to TIFF files, and figures were prepared using Adobe Photoshop.
Transient Transfection and Luciferase Assay
The αSMA promoter-luciferase reporter construct was a generous gift from Dr. Robert Schwartz (Baylor College of Medicine) (16). The WT-SARA and ΔFYVE-SARA were kindly provided by Dr. Jeffrey Wrana (6). The FHRE-Luc reporter construct (17) was purchased from Addgene (Cambridge, MA; Addgene plasmid 1789). AktK179M was purchased from Upstate Biotechnology (Lake Placid, NY). The putative SARA promoter was PCR cloned from human genomic DNA using Advantage GC genomic LA polymerase from Clontech (Mountain View, CA) into pGL3-basic luciferase vector purchased from Promega (Madison, WI). Transfection was performed with the FuGENE HP transfection reagent (Roche Applied Science), and luciferase and β-galactosidase activities were measured as described previously (15).
Stable Knockdowns
Lentiviral shRNAmir p85α (PIK3R1 clone ID V2LHS_33679) and non-silencing GIPZ shRNAmir control were purchased from Open Biosystems (Huntsville, AL). To generate stable knockdown cell lines, pGIPZ clones, which are supplied as bacterial cultures in Escherichia coli, were first subjected to CaPO4 transfection for lentiviral packaging in HEK293 FT cells (Invitrogen) using psPAX2 and pMD2.G according to Open Biosystems protocol. HKC cells were incubated with viral supernatants, and after 48 h, puromycin was added in a concentration determined previously to result in 100% cell death of non-infected HKC to select infected cells. Once cells reached confluence, they were frozen or used for experiments up to passage 4–6 in the continual presence of puromycin.
Potassium Depletion
HKCs were switched to serum-free media for 24 h prior to treatment. Media were then switched to hypotonic media (50% Dulbecco's modified Eagle's medium, 50% H20). After 10 min in hypotonic buffer, the cells were either switched to isotonic media without potassium (10 mm Tris-HCl, pH 7.5, 150 mm NaCl) or isotonic media containing 10 mm KCl. Cells were depleted for 30 min prior to 6-h LY294002 treatment.
Data Analysis
Western blots were scanned using an Epson V700 photo scanner, and densitometric analysis was performed using NIH ImageJ software. Graphs represent means of at least three experiments, corrected to β-actin, and expressed as fold induction over untreated controls. For mRNA analysis, triplicate measurements of at least three independent experiments were corrected to β2-macroglobulin and expressed as fold induction over untreated controls. For promoter assays, graphs represent means of each independent measurement corrected to β-galactosidase and expressed relative to untreated controls. For all graphs, error bars represent S.D. Either one-way analysis of variance with Fishers post hoc tests, or two-tailed Student's t tests were preformed, as appropriate, using StatView (Abacus Concepts, Inc.) to evaluate differences between groups. Values of p < 0.05 were considered significant.
RESULTS
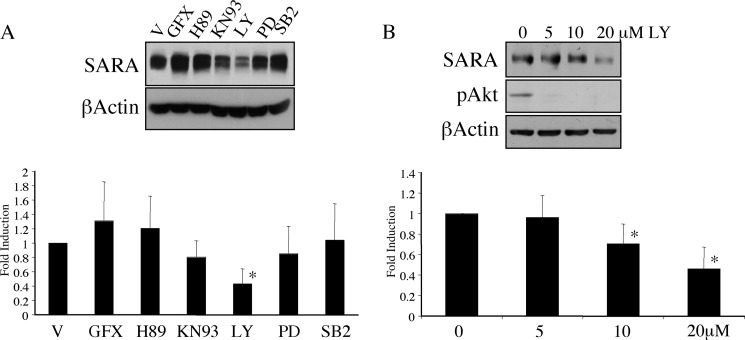
To examine the signaling pathways that might play a role in the regulation of SARA expression, we treated HKC with a panel of kinase inhibitors. Among a number of inhibitors, only the PI3K inhibitor LY294002 altered SARA expression (Fig. 1A). This reduction in SARA expression was independent of activation of TGF-β1 signaling. Wortmannin, an alternative inhibitor of PI3K, also reduced the expression of SARA (data not shown). However, although a dose of 5 μm LY294002 was sufficient to reduce basal levels of phosphorylated Akt, a dose between 10 and 20 μm was required for the reduction in SARA expression (Fig. 1B).
FIGURE 1.
Inhibition of PI3K reduces the level of SARA expression independently of exogenous TGF-β1 treatment. A, HKC were treated for 72 h with various inhibitors in serum-free medium. Inhibitors used were as follows: V, DMSO (vehicle); GFX, GF109203X PKC inhibitor (1 μm); H89, PKA inhibitor (50 nm); KN93, Ca2+/calmodulin-dependent protein kinase II inhibitor (10 μm); LY, LY294002 PI3K inhibitor (20 μm); PD, PD98059 ERK inhibitor (10 μm); SB2, SB203580 p38 inhibitor (10 μm). SARA expression was only significantly affected by LY294002 (*, p = 0.0354) B, serum-deprived HKC were treated with various doses of LY294002 (LY) for 72 h. β-Actin is included as a loading control. SARA was significantly reduced by 20 μm LY294002 (*, p = 0.0001).
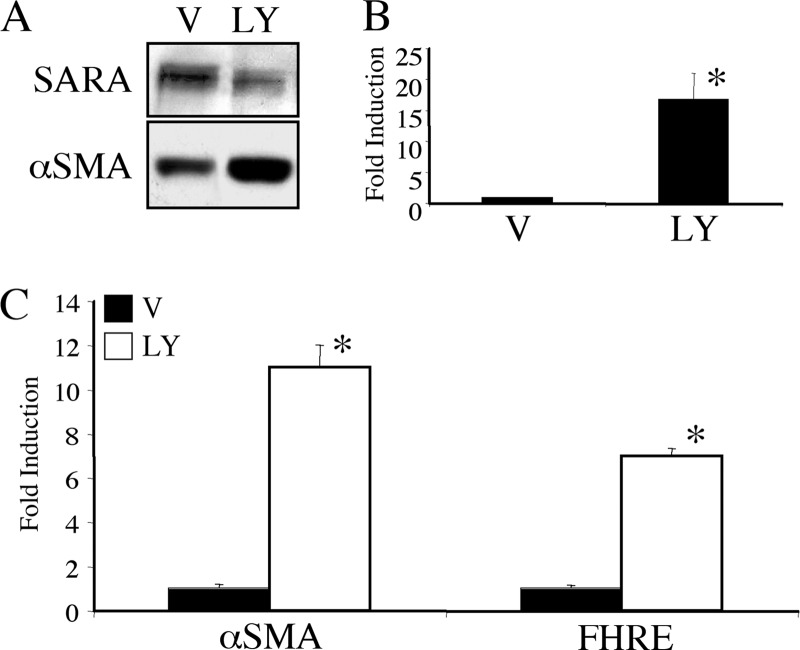
Because we had shown previously that a loss of SARA expression was sufficient to induce some of the changes associated with EMT in these cells, in particular the induction of αSMA, we investigated whether treatment with LY294002 to reduce SARA expression also caused an increase in αSMA levels. As shown in Fig. 2, inhibition of PI3K with 20 μm LY294002 increased αSMA at the protein (Fig. 2A) and mRNA (Fig. 2B) level, independent of treatment with TGF-β1. Inhibition of PI3K also induces the activity of the αSMA promoter (Fig. 2C). In this figure, the forkhead-responsive element (FHRE) was used as a control to confirm that the PI3K pathway has been inhibited. Reduced PI3K activity removes the inhibitory effect of Akt on the FOXO3a/FKHRL1 forkhead transcription factor and thereby allows enhanced binding of FOXO3a/FKHRL1 to the FHRE (17).
FIGURE 2.
PI3K inhibition results in enhanced αSMA expression. HKC were treated with 20 μm LY294002 (LY) or DMSO vehicle control (V) for 72 h and assayed for either protein (A) or mRNA with quantitative RT-PCR. B, LY294002 significantly induced αSMA RNA (*, p = 0.006). C, alternatively, HKC were transfected with either an αSMA or FHRE luciferase reporter construct along with β-galactosidase to control for transfection efficiency. 3-h post-transfection, the cells were treated with DMSO vehicle (black bars) or 20 μm LY294002 (white bars) for 48 h followed by lysis and analysis by luciferase and β-galactosidase assays. LY294002 significantly induced increases in both αSMA (*, p = 0.0001) and FHRE (*, p = 0.0001).
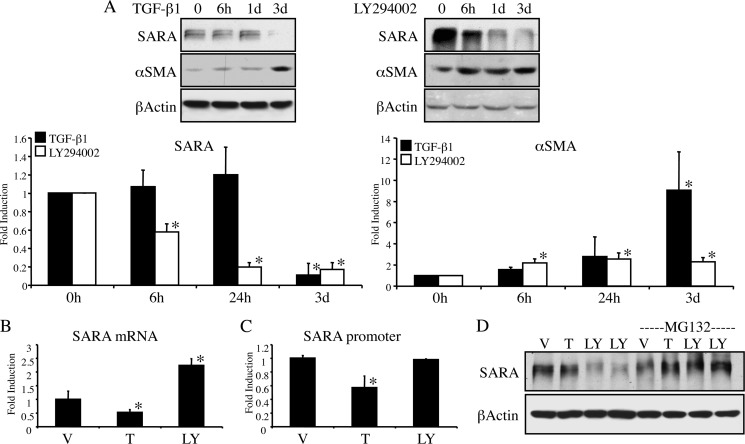
We next examined whether the LY294002-induced loss of SARA occurred via a similar mechanism to that we previously reported for TGF-β1. TGF-β1 reduced the expression of SARA between 1 and 3 days of treatment, corresponding with the timing of other phenotypic changes that have been associated with EMT (2). In contrast, cells treated with LY294002 began showing reduced SARA expression and increased αSMA after as few as 6 h of treatment (Fig. 3A). Furthermore, although TGF-β1 reduces the mRNA levels of SARA, LY294002 causes no mRNA reduction but rather appears to increase the SARA message (Fig. 3B). We cloned the putative SARA promoter into a luciferase reporter vector (pGL3) and analyzed its responsiveness to either TGF-β1 or LY294002. As shown in Fig. 3C, SARA promoter expression was reduced by TGF-β1 but was not affected by PI3K inhibition. Together, our data suggest that, unlike TGF-β1, the mechanism of LY294002-mediated decreased SARA expression may be at the level of protein degradation. To determine whether the ubiquitin proteasome-mediated degradation pathway was involved in regulation of SARA expression by PI3K inhibition, we treated the cells with the proteasome inhibitor, MG132. As shown in Fig. 3D, MG132 blocks the LY294002-mediated decline in SARA protein expression. As anticipated, at this time point of 24 h, TGF-β1 does not affect the expression of SARA.
FIGURE 3.
The mechanism of down-regulation of SARA expression differs between PI3K inhibition and TGF-β1. A, HKC were treated with either 2 ng/ml TGF-β1 (left panels) or 20 μm LY294002 (right panels) for the indicated time periods prior to lysis for Western blot. β-Actin is included as a control for loading. As demonstrated in graphs of triplicate experiments, LY294002 reduces SARA (left graph) and increases αSMA (right graph) at all treatment times compared with control (*, for SARA: 6 h, p = 0.0001; 24 h, p = 0.0001; 3 days, p = 0.0001. For αSMA, 6 h, p = 0.002; 24 h, p = 0.0005; 3 days, p = 0.0012). However, TGF-β1 only causes significant changes in SARA and αSMA at 3 days (3d). (*, for SARA, 3 days, p < 0.0001; and for αSMA, p = <0.0001). B, HKC treated for 48 h with either DMSO vehicle (V), 2 ng/ml TGF-β1 (T), or 20 μm LY294002 (LY) were assayed for mRNA expression of SARA via quantitative RT-PCR. TGF-β1 reduced SARA mRNA expression (*, p = 0.0438), but LY294002 enhanced it (*, p = 0.0001). C, HKC were transfected with the putative SARA promoter/luciferase reporter construct for 3 h prior to treatment as in B with TGF-β1 or LY294002 for an additional 48 h. TGF-β1 reduces SARA promoter activity (*, p = 0.002), LY294002 does not (p = 0.8164). D, HKC were pretreated with 10 μm MG-132 for 1 h prior to 24 h treatment with either vehicle (V), TGF-β1 (T), or LY294002 (LY). β-Actin is included as a control for loading.
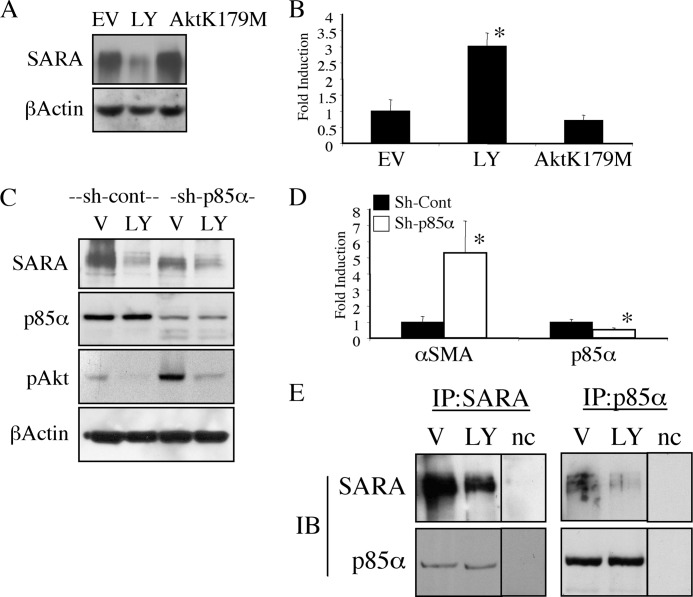
To determine whether the decrease in SARA expression by LY294002 depends on downstream PI3K-mediated signaling, we transfected HKC with a kinase-negative mutant Akt (AktK179M)). This construct did not reduce the protein expression of SARA (Fig. 4A). Furthermore, although we observed a TGF-β1-independent induction of the αSMA promoter by inhibiting PI3K in cells treated with LY294002, cells transfected with the dominant negative Akt construct did not show similar induction (Fig. 4B). We confirmed that the effect of LY294002 on the expression of SARA was in fact due to PI3K inhibition, rather than an off-target effect of the inhibitor, by creating an HKC line that stably expresses an shRNA for the regulatory p85α subunit of PI3K (Fig. 4C). Similar to chemical PI3K inhibition, knockdown of p85α reduced SARA expression (Fig. 4C, top panel). Of note, the basal phosphorylation of downstream Akt was much greater in the p85α knockdown cells (Fig. 4C). This may be due to the fact that depletion of the regulatory subunit removes the inhibition on the p110 catalytic subunit (10). Therefore, although LY294002 abolished Akt activity and p85α knockdown increased it, either treatment decreased SARA expression. This further suggests an effect on SARA that is specific to PI3K itself and is not a function of downstream Akt signaling. To determine whether p85α knockdown could affect αSMA expression, we examined αSMA mRNA and found that similar to chemical inhibition of the PI3K pathway, knockdown of p85α (Fig. 4D, left panel) increased αSMA mRNA (Fig. 4D, right panel). To determine whether this could be due to endogenous protein interaction, we immunoprecipitated either SARA, blotting for associated p85α (Fig. 4E, left panels), or p85α, blotting for SARA (Fig. 4E, right panels). Indeed, we observed interaction between p85α and SARA. Although LY294002 did not affect the level of p85α expression, it did reduce the interaction detected between p85α and SARA, which is presumably due to the reduction in overall SARA expression.
FIGURE 4.
The PI3K-mediated decrease in SARA is independent of Akt activity and may involve direct SARA interaction with p85α. A, HKC were transfected with either empty vector (EV) or AktK179M for 3 h prior to treatment with either DMSO vehicle (V), or 20 μm LY294002 (LY) for an additional 16 h and then lysed for Western blot. β-Actin is included as a control for loading. B, HKC were transfected with empty vector or AktK179M along with β-galactosidase and the αSMA promoter and treated as in A followed by lysis and luciferase β-galactosidase assays to control for transfection efficiency. LY294002 induced αSMA promoter (*, p = 0.0003), and AktK179M did not (p = 0.3168). C, HKC stably expressing either control (sh-cont) or (sh-p85α) were serum-deprived and treated for 16 h with vehicle (V, DMSO) or 20 μm LY294002. β-Actin is shown to control for even loading. D, control (black bars) or sh-p85α (white bars) expressing cells were assayed for mRNA expression of either αSMA or p85α via quantitative RT-PCR. The sh-p85α cells had significantly increased αSMA mRNA (*, p = 0.02) and decreased p85α mRNA (*, p = 0.008). E, serum-deprived HKC were treated with either vehicle (V) or 20 μm LY294002 (LY) for 16 h and lysed for immunoprecipitation and immunoblotting (IB). Negative control (nc) is a lysate-free IP lane from the same experiment run on a different region of the same gels.
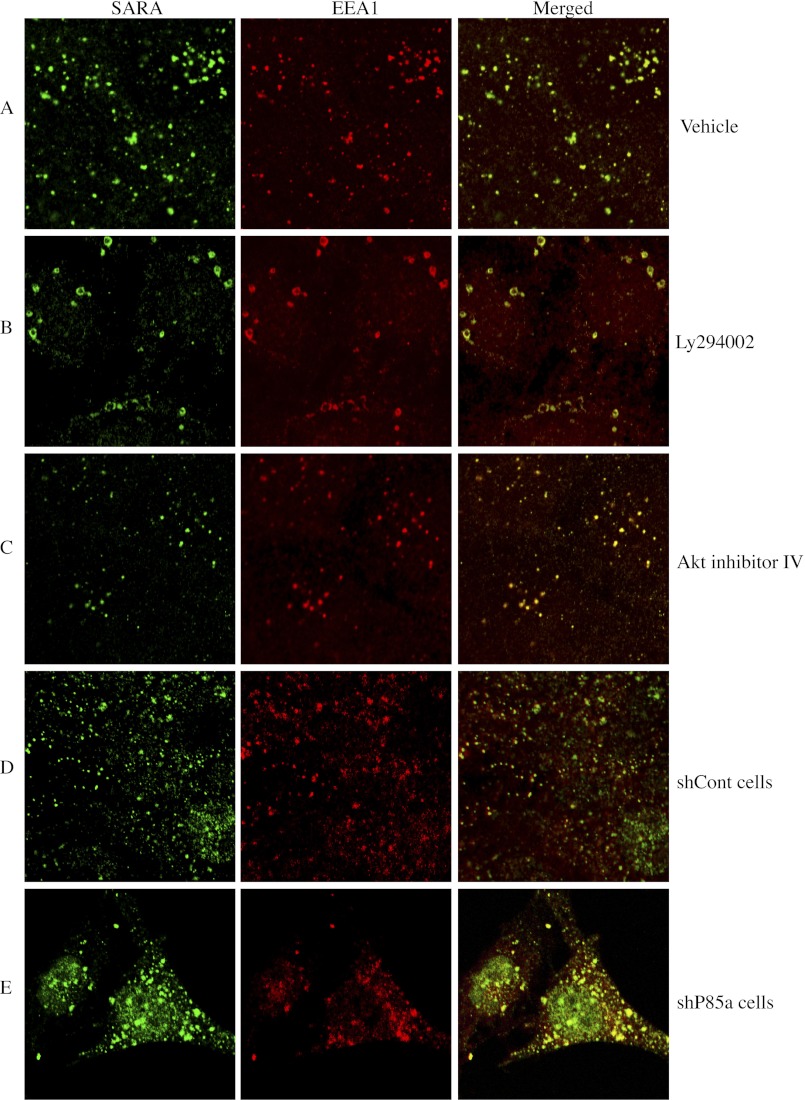
SARA localizes to early endosomal subcellular compartments (4) (5), and inhibition of PI3K may impact endocytosis (9). We therefore assessed whether inhibiting PI3K resulted in mislocalization of SARA away from endosomes leading to its degradation. However, rather than showing mislocalization of SARA, immunohistochemistry in LY294002-treated cells demonstrated that SARA remained associated with the endosomes. The endosomes appear fewer in number but also greatly enlarged compared with the small, punctate structures observed in the vehicle-treated cells (Fig. 5, A and B). Although less pronounced, the p85α shRNA-expressing cells showed similar endosomal morphology to that of LY294002-treated cells when compared with the cells expressing the control shRNA (Fig. 5, D and E). Furthermore, either treatment with LY294002 or stable expression of p85α shRNA leads to alterations in phenotype consistent with EMT-like changes (supplemental Fig. 1). In contrast, the endosomal staining of SARA in cells treated with an Akt-specific inhibitor that does not block upstream PI3K activity (Akt inhibitor IV) was not altered compared with control cells (Fig. 5, A and C).
FIGURE 5.
PI3K inhibition causes endosomal alterations. Serum-deprived HKC were treated as follows. A, vehicle control (DMSO) for 24 h; B, 20 μm LY294002 for 24 h; C, 5 μm Akt inhibitor IV for 24 h; D, sh-control (shCont) cells; E, sh-p85α cells. All cells are developed by immunocytochemistry for SARA (shown in green) and EEA1 (shown in red).
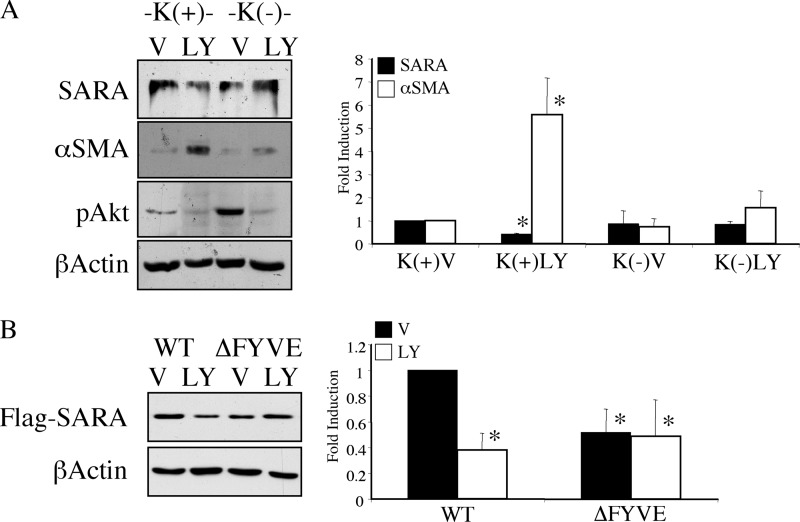
To examine the role of endocytosis in the PI3K inhibition-mediated decrease in SARA expression, we blocked clathrin-mediated endocytosis using potassium depletion (3) and examined the effect of LY294002 on SARA expression. As shown in Fig. 6A, although cells in potassium-containing control conditions (K+) had reduced SARA expression and showed an increase in αSMA in response to PI3K inhibition, inhibition of endocytosis by potassium depletion (K−) prevented both a reduction in SARA and the induction of αSMA stimulated by LY294002. To determine whether disrupting normal endosomal localization of SARA affects the ability of PI3K inhibition to result in its decreased expression, we compared the ability of LY294002 to down-regulate WT SARA versus SARA with a mutation in the FYVE domain (SARA-ΔFYVE), which is necessary for its endosomal localization. As shown in Fig. 6B, SARA-ΔFYVE appeared to have greater protection against depletion by LY294002 when compared with WT SARA. This suggests that, without appropriate endosomal localization, SARA is partially protected against depletion by PI3K inhibition.
FIGURE 6.
Endosomal localization enhances SARA down-regulation by PI3K inhibition. A, serum-deprived HKC were preincubated in isotonic media either containing potassium (K+) or without (K−) for 30 min prior to addition of either vehicle control (V) or 20 μm LY294002 (LY) for 6 h. Significant variation from control values are only seen under K+ conditions, where LY294002 reduces SARA expression (*, p = 0.0106) and enhances αSMA (*, p = 0.0001). B, HKC were transfected with either FLAG-tagged WT-SARA or FLAG-tagged ΔFYVE-SARA for 3 h followed by treatment with either control vehicle (V) or 20 μm LY294002 (LY) for an additional 16 h. β-Actin is shown as a loading control. All levels of SARA are significantly lower than untreated WT SARA expression (*, for WT+LY294002, p < 0.0001; ΔFYVE+vehicle, p = 0.0006; ΔFYVE+LY294002, p = 0.0004). However, there is no further decrease by LY294002 in ΔFYVE-expressing cells (p = 0.3290).
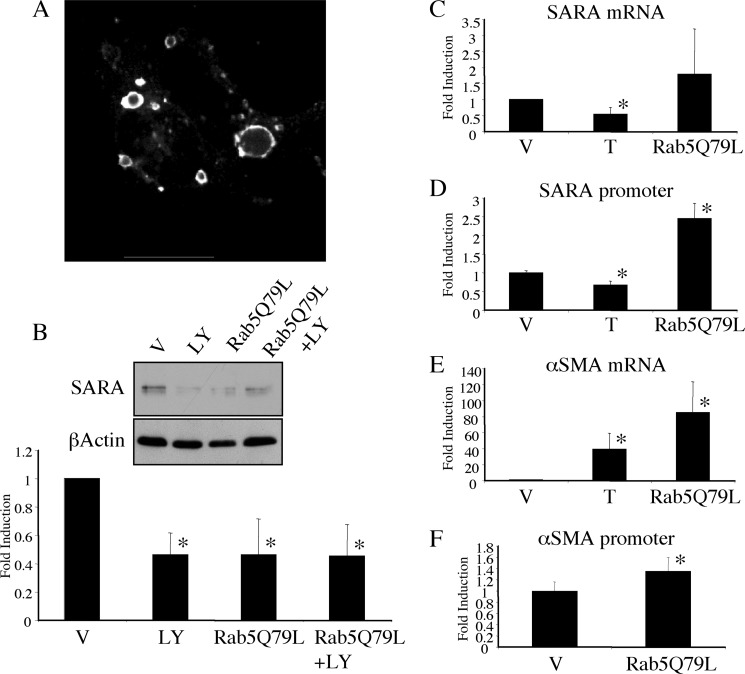
The p85α subunit of PI3K has been shown to bind directly to the early endosomal protein, Rab5, and to inactivate it through GAP activity toward Rab5 (18). Therefore, inhibition of PI3K could result in increased activity of Rab5. Previous reports have demonstrated enlarged endosomal structures in response to expression of a GTPase-deficient, constitutively active Rab5 (Rab5Q79L) (19), similar to those that we observed after PI3K inhibition. If the decrease in SARA expression by PI3K inhibition was related to enlargement of endosomes due to increased Rab5 activity, then Rab5Q79L should be able to mimic the LY294002 effect independently of PI3K signaling. To test this proposed model, we first confirmed that expression of Rab5Q79L resulted in alteration of endosomal structure. As previously reported, expression of GFP-tagged Rab5Q79L generated enlarged intracellular vesicles (Fig. 7A). These enlarged vesicular structures were not evident in cells transfected with empty GFP vector, and they co-localized with the early endosomal marker EEA1 (supplemental Fig. 2). Rab5Q79L expression also reduced SARA expression to a level similar to that observed with LY294002 (Fig. 7B). We examined whether Rab5Q79L reduced SARA expression via a mechanism similar to PI3K inhibition and found that, as with PI3K inhibition, reduced SARA expression in response to Rab5Q79L did not occur at the mRNA (Fig. 7C) or promoter (Fig. 7D) level. Rather, we saw an enhancement of SARA promoter activation by Rab5Q79L similar to what we saw with PI3K inhibition (Fig. 3B).
FIGURE 7.
Rab5Q79L depletes SARA similarly to PI3K inhibition. A, GFP immunofluorescence imaging of HKC transfected with GFP-Rab5Q79L overnight. B–F, HKC were transfected with either empty vector (EV = pEGFP-C1) or GFP-Rab5Q79L for 3 h prior to an additional 16 h treatment with either vehicle (V), 20 μm LY294002 (LY) or 2 ng/ml TGF-β1 (T). B, protein expression of SARA is shown with β-actin included as a control for loading. SARA levels are reduced in all treatments compared with vehicle treated controls (*, for LY294002, p = 0.0014; Rab5Q79L, p = 0.0014; Rab5Q79L+LY294002, p = 0.0012); however, there was no additive reduction by LY294002 and Rab5Q79L (p = 0.9548). C, SARA mRNA detected by quantitative PCR. TGF-β1 reduces SARA mRNA (*, p = 0.0438), but Rab5Q79L does not (p = 0.0919). D, SARA promoter co-transfected with either empty vector or Rab5Q79L and β-galactosidase. Luciferase values are corrected with β-galactosidase readings and represented as a fold induction over control. TGF-β1 significantly reduces the SARA promoter (*, p = 0.0442), but Rab5Q79L significantly enhances it (*, p = 0.001). E, αSMA mRNA detected by quantitative PCR. Both TGF-β1 and Rab5Q79L significantly enhance αSMA mRNA expression (*, for TGF-β1, p = 0.0092; Rab5Q79L, p = 0.0011). F, αSMA promoter co-transfected with either empty vector or Rab5Q79L and β-galactosidase. Luciferase values are corrected with β-galactosidase readings and represented as a fold induction over control. Rab5Q79L caused a modest enhancement of αSMA promoter response (*, p = 0.0089).
In agreement with our previous data showing that a loss of SARA was sufficient to alter markers of EMT (2), Rab5Q79L enhanced the expression of αSMA mRNA 88-fold over control, untreated cells and ∼2-fold over TGF-β1 treatment (Fig. 7E) Rab5Q79L also slightly but reproducibly enhanced the activity of the αSMA promoter without requiring PI3-kinase inhibition (Fig. 7F).
DISCUSSION
We report here that either PI3K inhibition or constitutive activation of Rab5 inhibits the expression of SARA, a protein important for maintaining epithelial cell phenotype. This down-regulation is associated with structural alterations in endocytic vesicles. Although a number of studies have suggested that PI3K signals through Akt to cause EMT (20, 21), our data demonstrate an effect on SARA expression that is independent of the activation status of Akt. Neither overexpression of a kinase-negative Akt mutant nor enhanced Akt activity through p85α knockdown affected the depletion of SARA or the induction of αSMA (Fig. 4). These results suggest that the effect of LY924002 on SARA is not mediated through downstream signaling and may explain why the dose of LY294002 required to decrease SARA expression is higher than that required to inhibit Akt phosphorylation (Fig. 1B). Furthermore, our co-immunoprecipitation data demonstrating an interaction between p85α and SARA (Fig. 4E) support the role of PI3K in SARA expression as being a direct effect of PI3K independent of downstream signaling. A recent report by Chamberlain and Anderson (22) demonstrated that p85α also binds directly to, and possesses GAP activity toward, Rab5. In that report, expression of a GAP-defective mutant p85α resulted in sustained levels of activated platelet-derived growth factor receptor (PDGFR) and increased downstream signaling due to decreased PDGFR degradation. Furthermore, PDGFRs that are mutated so that they cannot interact with p85α failed to be down-regulated correctly (23). Although PI3K binding does not appear required for internalization of PDGFR, it may be required to divert the PDGFR to a degradative pathway (24). Consistent with our finding that LY294002-induced SARA depletion was not mediated through an effect on Akt signaling, Chen and Wang (25) showed that the PI3K inhibitor wortmannin stimulates Rab5-mediated increases in endosome size and subsequent EGFR degradation independent of Akt activity.
A number of studies have observed that PI3K inhibition causes endosomal enlargement similar to what we found using LY294002 or shRNA-mediated p85α knockdown (25–30). However, the impact on signaling associated with endosomal enlargement varies, and it is not entirely clear what these structures represent. Contributing to the complexity of the role of PI3K in endocytosis, there are a number of different steps at which PI3K could act. In the initial stage of internalization through SH3 binding, p85α can up-regulate the activity of the large GTPase dynamin (31). As the function of dynamin is to mediate scission events to allow internalization of endocytic vesicles, this represents one way in which disruption of p85α could inhibit internalization (27). Indeed, although they are greatly enlarged, we do see fewer total endosomes in LY294002-treated cells, suggesting that the internalization step of clathrin-mediated endocytosis may also be affected. However, in other reports, the internalization rate of receptor complexes were enhanced by PI3K inhibition under conditions that resulted in endosomal enlargement (32, 33). In fact, enlarged endosomes resulting from inhibition of PI3K have been suggested to affect a number of endocytic events such as trans-Golgi network transport (34, 35), lysosomal targeting (28, 34), endosome fusion (36, 37), and endosomal recycling (32, 33, 38, 39).
In our studies, it appears that retention in the endosome as occurs with endosomal enlargement likely enhances the degradation of SARA. This model is supported by the fact that inhibiting clathrin-mediated endocytosis also inhibits the ability of LY294002 to reduce SARA expression or to enhance αSMA expression (Fig. 6A). Consistent with this notion, ΔFYVE-SARA, a localization-deficient mutant (6) was relatively protected from LY294002-mediated down-regulation (Fig. 6B). It should be noted that although the effect of LY294002 was blunted in cells expressing the ΔFYVE-SARA mutant, the overall expression of this localization-deficient mutant appeared less that of WT SARA. Therefore, although SARA might be degraded in a PI3K-sensitive manner in the endosome, it may also be degraded by other means if improperly localized. However, although we found that either the activation of Rab5 or the inhibition of PI3K resulted in alteration of endosomes and decreased expression of SARA, our studies do not conclusively determine whether the effect of PI3K inhibition is through a PI3K-mediated effect on Rab5 or whether any manipulation resulting in endosomal enlargement or disruption would similarly affect SARA expression. Additionally, the induction of the αSMA promoter by Rab5Q79L was modest in comparison with that seen with LY294002 (compare Fig. 7F with Fig. 4B). Therefore, it is not clear that the Rab5-mediated effects completely mimic those of PI3K inhibition.
The interaction between SARA and PI3K may represent the first report of SARA interaction with a non-TGF-β1 signaling pathway. However, in addition to its interaction with p85α demonstrated here, SARA has been shown to interact with the TGFβ Receptor (6), and the TβR can also interact with p85α (40). Thus, it remains to be determined whether the TβR is a part of the same complex with SARA and p85α.
SARA previously was shown to associate with Rab5-positive early endosomes (5, 41), and one study by Itoh et al. (5) demonstrated a redistribution of SARA away from endosomes due to PI3K inhibition by wortmannin. However, this was only examined in the context of overexpressed SARA, and the effect of wortmannin treatment on endosome morphology was not examined (5). In our studies, PI3K inhibition did not increase the detectable presence of endogenous SARA in the cytosol. Rather, SARA appeared to be retained in the enlarged endosomes. Another study showed that overexpression of SARA caused endosomal enlargement similar to what we have shown for PI3K inhibition or expression of Rab5Q79L, which resulted in inhibition of transferrin endocytosis (41). Although these investigators were unable to detect any interaction between SARA and Rab5 in in vitro binding assays, the fact that a dominant negative mutant of Rab5 could abrogate the inhibition of transferrin recycling due to SARA overexpression suggests that SARA may act as a Rab5 effector (41). Together with our data in the present study showing an effect on SARA expression due to activation of Rab5, this result suggests that the relationship between SARA expression and Rab5 activation may be complex and that there might be mutual feedback between the two proteins.
Activation of the PI3K/Akt has been suggested to be an important feature of EMT, and inhibition of PI3K has been shown to reduce TGF-β1-mediated EMT as well as reducing fibrogenesis in various models (21). However, other reports suggest that PI3K may also have a protective effect in fibrosis. It was recently reported that hemin (heme oxygenase-1 inducer) prevented fibrosis in a rat model of renovascular hypertensive cardiomyopathy (42). In this study, hemin increased the expression of the p85α subunit of PI3K and inhibition of PI3K with LY294002 blocked the protective effects of hemin. Futhermore, a recent study by Winbanks et al. (43) examining the effect of mTOR and PI3K on regulation of fibroblasts in tubulointerstitial fibrosis found that inhibition of PI3K with LY294002 increased the proportion of αSMA-positive cells and hence myofibroblasts. Together with our data in the present study showing that SARA protein can be depleted by PI3K inhibition, these results suggest that although there are a number of reports that PI3K can play a role in inhibition of fibrosis, it may also have a deleterious role basally in altering cell phenotype independently of fibrotic stimuli.

This article contains supplemental Figs. 1 and 2.
- SARA
- Smad anchor for receptor activation protein
- GAP
- GTPase-activating protein
- αSMA
- smooth muscle α-actin
- FHRE
- forkhead-responsive element
- PDGFR
- platelet-derived growth factor receptor
- DMSO
- dimethyl sulfoxide
- EEA1
- Early Endosome Antigen 1
- HKC
- Human Poximal Tubule-Kidney.
REFERENCES
- 1. Liu Y. (2004) Epithelial to mesenchymal transition in renal fibrogenesis: pathologic significance, molecular mechanism, and therapeutic intervention. J. Am. Soc. Nephrol. 15, 1–12 [DOI] [PubMed] [Google Scholar]
- 2. Runyan C. E., Hayashida T., Hubchak S., Curley J. F., Schnaper H. W. (2009) Role of SARA (SMAD anchor for receptor activation) in maintenance of epithelial cell phenotype. J. Biol. Chem. 284, 25181–25189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Runyan C. E., Schnaper H. W., Poncelet A. C. (2005) The role of internalization in transforming growth factor β1-induced Smad2 association with Smad anchor for receptor activation (SARA) and Smad2-dependent signaling in human mesangial cells. J. Biol. Chem. 280, 8300–8308 [DOI] [PubMed] [Google Scholar]
- 4. Panopoulou E., Gillooly D. J., Wrana J. L., Zerial M., Stenmark H., Murphy C., Fotsis T. (2002) Early endosomal regulation of Smad-dependent signaling in endothelial cells. J. Biol. Chem. 277, 18046–18052 [DOI] [PubMed] [Google Scholar]
- 5. Itoh F., Divecha N., Brocks L., Oomen L., Janssen H., Calafat J., Itoh S., Dijke Pt P. (2002) The FYVE domain in Smad anchor for receptor activation (SARA) is sufficient for localization of SARA in early endosomes and regulates TGF-β/Smad signaling. Genes Cells 7, 321–331 [DOI] [PubMed] [Google Scholar]
- 6. Tsukazaki T., Chiang T. A., Davison A. F., Attisano L., Wrana J. L. (1998) SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell 95, 779–791 [DOI] [PubMed] [Google Scholar]
- 7. Fruman D. A. (2011) Regulatory subunits of class IA PI3K. Curr. Top Microbiol Immunol. 346, 225–244 [DOI] [PubMed] [Google Scholar]
- 8. Kok K., Geering B., Vanhaesebroeck B. (2009) Regulation of phosphoinositide 3-kinase expression in health and disease. Trends Biochem. Sci. 34, 115–127 [DOI] [PubMed] [Google Scholar]
- 9. Mellor P., Furber L. A., Nyarko J. N., Anderson D. H. (2012) Multiple roles for the p85α isoform in the regulation and function of PI3K signaling and receptor trafficking. Biochem. J. 441, 23–37 [DOI] [PubMed] [Google Scholar]
- 10. Miled N., Yan Y., Hon W. C., Perisic O., Zvelebil M., Inbar Y., Schneidman-Duhovny D., Wolfson H. J., Backer J. M., Williams R. L. (2007) Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science 317, 239–242 [DOI] [PubMed] [Google Scholar]
- 11. Stephens L., Anderson K., Stokoe D., Erdjument-Bromage H., Painter G. F., Holmes A. B., Gaffney P. R., Reese C. B., McCormick F., Tempst P., Coadwell J., Hawkins P. T. (1998) Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science 279, 710–714 [DOI] [PubMed] [Google Scholar]
- 12. Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. (1997) Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 13. Zerial M., McBride H. (2001) Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2, 107–117 [DOI] [PubMed] [Google Scholar]
- 14. Racusen L. C., Monteil C., Sgrignoli A., Lucskay M., Marouillat S., Rhim J. G., Morin J. P. (1997) Cell lines with extended in vitro growth potential from human renal proximal tubule: characterization, response to inducers, and comparison with established cell lines. J. Lab. Clin. Med. 129, 318–329 [DOI] [PubMed] [Google Scholar]
- 15. Runyan C. E., Schnaper H. W., Poncelet A. C. (2004) The phosphatidylinositol 3-kinase/Akt pathway enhances Smad3-stimulated mesangial cell collagen I expression in response to transforming growth factor-β1. J. Biol. Chem. 279, 2632–2639 [DOI] [PubMed] [Google Scholar]
- 16. Min B. H., Foster D. N., Strauch A. R. (1990) The 5′-flanking region of the mouse vascular smooth muscle α-actin gene contains evolutionarily conserved sequence motifs within a functional promoter. J. Biol. Chem. 265, 16667–16675 [PubMed] [Google Scholar]
- 17. Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 18. Chamberlain M. D., Berry T. R., Pastor M. C., Anderson D. H. (2004) The p85α subunit of phosphatidylinositol 3′-kinase binds to and stimulates the GTPase activity of Rab proteins. J. Biol. Chem. 279, 48607–48614 [DOI] [PubMed] [Google Scholar]
- 19. Stenmark H., Parton R. G., Steele-Mortimer O., Lütcke A., Gruenberg J., Zerial M. (1994) Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 13, 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grille S. J., Bellacosa A., Upson J., Klein-Szanto A. J., van Roy F., Lee-Kwon W., Donowitz M., Tsichlis P. N., Larue L. (2003) The protein kinase Akt induces epithelial mesenchymal transition and promotes enhanced motility and invasiveness of squamous cell carcinoma lines. Cancer Res. 63, 2172–2178 [PubMed] [Google Scholar]
- 21. Larue L., Bellacosa A. (2005) Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3′-kinase/AKT pathways. Oncogene 24, 7443–7454 [DOI] [PubMed] [Google Scholar]
- 22. Anderson D. H., Chamberlain M. D. (2005) Assay and stimulation of the Rab5 GTPase by the p85 α subunit of phosphatidylinositol 3-kinase. Methods Enzymol. 403, 552–561 [DOI] [PubMed] [Google Scholar]
- 23. Joly M., Kazlauskas A., Fay F. S., Corvera S. (1994) Disruption of PDGF receptor trafficking by mutation of its PI-3 kinase binding sites. Science 263, 684–687 [DOI] [PubMed] [Google Scholar]
- 24. Joly M., Kazlauskas A., Corvera S. (1995) Phosphatidylinositol 3-kinase activity is required at a postendocytic step in platelet-derived growth factor receptor trafficking. J. Biol. Chem. 270, 13225–13230 [DOI] [PubMed] [Google Scholar]
- 25. Chen X., Wang Z. (2001) Regulation of epidermal growth factor receptor endocytosis by wortmannin through activation of Rab5 rather than inhibition of phosphatidylinositol 3-kinase. EMBO Rep. 2, 842–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shpetner H., Joly M., Hartley D., Corvera S. (1996) Potential sites of PI-3 kinase function in the endocytic pathway revealed by the PI-3 kinase inhibitor, wortmannin. J. Cell Biol. 132, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gillooly D. J., Melendez A. J., Hockaday A. R., Harnett M. M., Allen J. M. (1999) Endocytosis and vesicular trafficking of immune complexes and activation of phospholipase D by the human high-affinity IgG receptor requires distinct phosphoinositide 3-kinase activities. Biochem. J. 344, 605–611 [PMC free article] [PubMed] [Google Scholar]
- 28. Mousavi S. A., Brech A., Berg T., Kjeken R. (2003) Phosphoinositide 3-kinase regulates maturation of lysosomes in rat hepatocytes. Biochem. J. 372, 861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen X., Wang Z. (2001) Regulation of intracellular trafficking of the EGF receptor by Rab5 in the absence of phosphatidylinositol 3-kinase activity. EMBO Rep. 2, 68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Houle S., Marceau F. (2003) Wortmannin alters the intracellular trafficking of the bradykinin B2 receptor: role of phosphoinositide 3-kinase and Rab5. Biochem. J. 375, 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gout I., Dhand R., Hiles I. D., Fry M. J., Panayotou G., Das P., Truong O., Totty N. F., Hsuan J., Booker G. W. (1993) The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell 75, 25–36 [PubMed] [Google Scholar]
- 32. Kurashima K., Szabó E. Z., Lukacs G., Orlowski J., Grinstein S. (1998) Endosomal recycling of the Na+/H+ exchanger NHE3 isoform is regulated by the phosphatidylinositol 3-kinase pathway. J. Biol. Chem. 273, 20828–20836 [DOI] [PubMed] [Google Scholar]
- 33. Awwad H. O., Iyer V., Rosenfeld J. L., Millman E. E., Foster E., Moore R. H., Knoll B. J. (2007) Inhibitors of phosphoinositide 3-kinase cause defects in the postendocytic sorting of β2-adrenergic receptors. Exp. Cell Res. 313, 2586–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakajima Y., Pfeffer S. R. (1997) Phosphatidylinositol 3-kinase is not required for recycling of mannose 6-phosphate receptors from late endosomes to the trans-Golgi network. Mol. Biol. Cell 8, 577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davidson H. W. (1995) Wortmannin causes mistargeting of procathepsin D. Evidence for the involvement of a phosphatidylinositol 3-kinase in vesicular transport to lysosomes. J. Cell Biol. 130, 797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jones A. T., Mills I. G., Scheidig A. J., Alexandrov K., Clague M. J. (1998) Inhibition of endosome fusion by wortmannin persists in the presence of activated Rab5. Mol. Biol. Cell 9, 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mills I. G., Jones A. T., Clague M. J. (1999) Regulation of endosome fusion. Mol. Membr Biol. 16, 73–79 [DOI] [PubMed] [Google Scholar]
- 38. Ernst S., Zobiack N., Boecker K., Gerke V., Rescher U. (2004) Agonist-induced trafficking of the low-affinity formyl peptide receptor FPRL1. Cell Mol. Life Sci. 61, 1684–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao Y., Keen J. H. (2008) Gyrating clathrin: highly dynamic clathrin structures involved in rapid receptor recycling. Traffic 9, 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yi J. Y., Shin I., Arteaga C. L. (2005) Type I transforming growth factor beta receptor binds to and activates phosphatidylinositol 3-kinase. J. Biol. Chem. 280, 10870–10876 [DOI] [PubMed] [Google Scholar]
- 41. Hu Y., Chuang J. Z., Xu K., McGraw T. G., Sung C. H. (2002) SARA, a FYVE domain protein, affects Rab5-mediated endocytosis. J. Cell Sci. 115, 4755–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Worou M. E., Belmokhtar K., Bonnet P., Vourc'h P., Machet M. C., Khamis G., Eder V. (2011) Hemin decreases cardiac oxidative stress and fibrosis in a rat model of systemic hypertension via PI3K/Akt signaling. Cardiovasc Res. 91, 320–329 [DOI] [PubMed] [Google Scholar]
- 43. Winbanks C. E., Grimwood L., Gasser A., Darby I. A., Hewitson T. D., Becker G. J. (2007) Role of the phosphatidylinositol 3-kinase and mTOR pathways in the regulation of renal fibroblast function and differentiation. Int. J. Biochem. Cell Biol. 39, 206–219 [DOI] [PubMed] [Google Scholar]