Background: Depletion of cellular polyamines results in cessation of cellular proliferation.
Results: Genomic and biochemical analysis demonstrates stress establishment in the polyamine-depleted cells.
Conclusion: Establishment of stress response accounts at least in part for growth arrest establishment in polyamine-depleted cells.
Significance: Our results suggest a novel view on the mechanistic basis accounting for growth arrest of polyamine-depleted cells.
Keywords: Cyclin D1, p38 MAPK, Polyamines, Stress Response, Unfolded Protein Response, PERK, eIF2α, Growth Regulation
Abstract
Polyamines are small organic polycations that are absolutely required for cell growth and proliferation; yet the basis for this requirement is mostly unknown. Here, we combined a genome-wide expression profiling with biochemical analysis to reveal the molecular basis for inhibited proliferation of polyamine-depleted cells. Transcriptional responses accompanying growth arrest establishment in polyamine-depleted cells or growth resumption following polyamine replenishment were monitored and compared. Changes in the expression of genes related to various fundamental cellular processes were established. Analysis of mirror-symmetric expression patterns around the G1-arrest point identified a set of genes representing a stress-response signature. Indeed, complementary biochemical analysis demonstrated activation of the PKR-like endoplasmic reticulum kinase arm of the unfolded protein response and of the stress-induced p38 MAPK. These changes were accompanied by induction of key growth-inhibitory factors such as p21 and Gadd45a and reduced expression of various cyclins, most profoundly cyclin D1, setting the basis for the halted proliferation. However, although the induced stress response could arrest growth, polyamine depletion also inhibited proliferation of PKR-like endoplasmic reticulum kinase and p38α-deficient cells and of cells harboring a nonphosphorylatable mutant eIF2α (S51A), suggesting that additional yet unidentified mechanisms might inhibit proliferation of polyamine-depleted cells. Despite lengthy persistence of the stress and activation of apoptotic signaling, polyamine-depleted cells remained viable, apparently due to induced expression of protective genes and development of autophagy.
Introduction
The polyamines spermidine and spermine and their diamine precursor putrescine are ubiquitous naturally occurring polycationic alkylamines that are essential for the processes of cell growth and proliferation (1). Under physiological conditions, polyamines exist as protonated cations, and therefore, they seem to execute their cellular functions through binding to acidic sites on cellular macromolecules such as proteins, nucleic acids, and phospholipids (2). It is inferred that through these interactions polyamines may affect RNA and DNA structure, transcription, translation, activity of kinases and phosphatases, and regulation of ion channels (1).
Accumulating evidence links polyamines to cellular transformation. Overexpression of ornithine decarboxylase, the first rate-limiting enzyme in the polyamine biosynthesis pathway, causes cellular transformation (3) and stimulates tumor formation in a mouse model (4). The importance of polyamines for fundamental cellular processes is further emphasized by the evolvement of cellular mechanisms that ensure tight regulation of their optimal intracellular levels (1). Polyamine metabolism is frequently dysregulated in cancer and other hyperproliferative conditions, thus making this metabolic pathway an attractive target for therapeutic intervention. Polyamine depletion results in cessation of cellular proliferation. In most cases, polyamine-depleted cells remain viable and can be growth-stimulated upon polyamine replenishment (2). However, the mechanistic basis that underlines growth arrest provoked by polyamine depletion is mostly unknown.
Here, we present a temporal profile of gene expression in NIH3T3 cells that were first growth-arrested by polyamine depletion and then growth-stimulated by polyamine replenishment. Analysis of these data with advanced bioinformatic tools revealed profound changes in the expression of genes that regulate fundamental processes such as transcription, cell cycle, DNA replication, DNA repair, and sterol biosynthesis. Significantly, polyamine depletion stimulated expression of a set of genes that are coordinately up-regulated during cellular stress response. These data combined with complementary biochemical analysis suggested stress response as a potential cause for arrested proliferation of polyamine-depleted cells. However, because proliferation of cells deficient in central components of the induced stress is still arrested by polyamine depletion, we conclude that growth arrest might be also established by additional yet unidentified mechanisms.
EXPERIMENTAL PROCEDURES
Cell Culture Conditions
NIH3T3 mouse fibroblasts and HeLa cells were grown in Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% (v/v) fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Biological Industries) at 37 °C in 5% CO2.
[3H]Thymidine Incorporation Assay
NIH3T3 cells were grown in 12-well plates and treated with difluoromethylornithine (DFMO)2 (a mechanism-based suicide inhibitor of ornithine decarboxylase) or growth-stimulated with spermidine for the indicated time periods. To determine thymidine incorporation into nascent DNA molecules, the cells were incubated with [3H]thymidine (7.5 μCi/well) for the last 4 h prior to harvesting. Cells were washed with PBS and stored at −80 °C for at least 1 h. Cells were then resuspended in 150 μl of PBS and transferred to a 96-well plate. [3H]Thymidine incorporation values were obtained using a Matrix automatic reader (Micromate 196 Harvester, Packard) and a Matrix 96 beta counter (Packard) following the manufacturer's instructions.
Polyamine Analysis
NIH3T3 cells grown in 10-cm dishes were harvested, pelleted, and resuspended in 100 μl of PBS. The cells were lysed in 3% perchloric acid, and precipitated material was removed by centrifugation (5 min at 13,000 rpm). The supernatant was collected for polyamine analysis, and the pellet was used for normalization by DNA quantification (DNA was quantified by resuspending the pellet in 400 μl of 4% diphenylamine (Sigma) in acetic acid, 400 μl of 10% perchloric acid, and 20 μl of 1:500 acetaldehyde (Sigma), and by incubation for 16 h at 30 °C followed by absorbance determination at 595 and 700 nm). For polyamine analysis, 100 μl of the perchloric acid supernatant was mixed with 200 μl of 6 mg/ml dansyl chloride (in acetone). After the addition of 10 mg of sodium carbonate, the mixture was incubated for 16 h in the dark. To neutralize residual dansyl chloride, 50 μl of 100 mg/ml l-proline solution were added and incubated for 1 h at room temperature. Dansylated derivatives were extracted into 250 μl of toluene by centrifugation. Portions of 50–100 μl were spotted on Silica 60 F254 (Merck) TLC plate, and the dansylated derivatives were resolved by thin layer chromatography using ethyl acetate/cyclohexane (1:1.5) as a solvent and visualized by UV illumination. Dansylated derivatives of known polyamines served as markers.
Flow Cytometry
For live sorting, NIH3T3 cells were stained with Hoechst 33342 dye (5 μg/ml) for 30 min in the dark at 37 °C. The cells were then washed with PBS, trypsinized, collected, resuspended in serum-free medium, and kept on ice during the following steps. The cells were sorted using FACSVantage SE (BD Biosciences) using a UV filter. The collected fractions were sedimented at 2000 rpm, and total RNA was isolated using TRI Reagent (Molecular Research Center, Inc.).
Determination of Protein Synthesis Activity
Cells were pulse-labeled for 10 min with 100 μCi/ml l-[35S]methionine. Cells were washed four times with PBS, collected by centrifugation, and lysed. Aliquots containing 30 μg of protein were loaded onto GF/C glass microfiber filters and subjected to TCA precipitation. Radioactivity was determined using the BAS-2500 Bio-imaging analyzer (Fujifilm).
Real Time PCR and Oligonucleotide Microarray Hybridization
Residual DNA was removed from the RNA preparation by digestion (DNA-freeTM kit, Ambion). cDNA was synthesized using the iScript kit (Roche Applied Science). Real time PCR analysis was performed using SYBR Green I (Kapa Biosystems) as a fluorescent dye, according to the manufacturer's guidelines. All experiments were carried out in triplicate, and results were normalized to GAPDH RNA. Real time PCR primers were selected from the Harvard Primer Bank (Primer Bank IDs are as follows: Egr1, 76559936b1; CYR61, 239937453b1; FOS, 31560587b1; BTG2, 84875511b1; Gadd34, 258613961b1; and GAPDH, 126012538b1). 100 ng of a total RNA portion was used for subsequent hybridization to Affymetrix (Santa Clara) Mouse Gene 1.0 ST array according to the manufacturer's instructions.
Gene Expression Data Analysis
Statistical analysis of microarray data were performed using the Partek® Genomics Suite software (Partek Inc., St. Louis, MO). CEL files (containing raw expression measurements) were imported to Partek GS. The data were preprocessed and normalized using the robust multichip average algorithm. The normalized data were explored by principal component analysis and hierarchical clustering to detect batch or other random effects that may appear if the replicates are carried out sequentially. Batch effects were corrected using the defaults of the software. To identify differentially expressed genes, one-way analysis of variance was applied. Contrasts were calculated to compare pairwise between given time points. False discovery rate was used to correct for multiple comparisons. Probe sets, whose normalized expression intensity was below 5.5 in all the arrays, were considered as not detected and filtered out. Gene expression data were next submitted to clustering using the Click algorithm from the EXPANDER suite (5). Before clustering, the probe sets were filtered to include only those with significant q values (less than 0.05 in at least one pairwise comparison) and a fold change of at least two between any two conditions. Expression levels of each gene were standardized so that the mean was equal to 0 and the variance was equal to 1. Heat maps were drawn using the Partek software.
Analysis of Functional Categories
Clusters of differentially expressed genes were categorized based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway using the Database for Annotation, Visualization, and Integrated Discovery (DAVID, david.abcc.ncifcrf.gov) (6). The PRIMA algorithm (7) implemented in EXPANDER was applied to identify cis-regulatory promoter elements that control the observed transcriptional modulation in each of the clusters previously obtained. Statistical tests identified transcription factors whose binding site signatures were significantly enriched in the target set (gene promoters in each cluster) relative to the background (all promoters in the genome).
Autophagy Detection
HeLa cells stably expressing EGFP-LC3 were grown on 9-mm coverslips in 24-well plates in DMEM supplemented with 10% FBS and antibiotics. Following treatment with DFMO (1 mm), cells were washed three times with PBS and fixed in 4% formaldehyde for 30 min at room temperature. To visualize puncta formation, coverslips were mounted on microscope glass slides and analyzed by fluorescent microscopy. Four-hour amino acid starvation was used to obtain positive control cells. LC3 processing was analyzed by Western blot using anti-LC3B antibody (L7543; Sigma).
Measurement of the ER Calcium Levels
NIH3T3 cells were grown in 24-well plates, treated with DFMO (1 mm) for the indicated times, and loaded with Fluo 2-HA (TEFLabs) cell-permeant calcium indicator (3 μm) for 30 min at 37 °C. The plates were placed on the stage of Zeiss-510 confocal microscope for further treatment and imaging under strictly standardized conditions. Time-lapse confocal imaging was performed at 488 nm excitation using green fluorescent and phase channels at the rate of 1 frame/s for a total time of 5 min. Basal staining was considered proportional to initial ER calcium store capacity. Release of ER-stored calcium was induced by 1 μm thapsigargin, and changes in fluorescence were monitored. Statistical significance was calculated using unpaired t test and one-way analysis of variance.
Immunoblot Analyses
Cells were lysed in RIPA buffer (50 mm Tris-HCl, pH 8, 150 mm KCl, 1.0% Nonidet P-40 (IGEPAL), 0.5% sodium deoxycholate, 0.1% SDS) supplemented with mixtures of protease and phosphatase inhibitors (Sigma). Equal amounts of protein were resolved by SDS-PAGE, electroblotted to a nitrocellulose membrane, and incubated with the indicated antibodies followed by horseradish peroxidase-conjugated anti-IgG antibodies. The antibodies used were as follows: rabbit mAb anti-p-PERK and rabbit polyclonal anti-caspase 12 (Cell Signaling Technology); rabbit polyclonal anti-PERK, rabbit polyclonal anti-eIF2α, rabbit polyclonal anti-ATF4, rabbit polyclonal anti-GADD153, rabbit polyclonal anti-GADD34, rabbit polyclonal anti-p38, mouse mAb anti-p-p38, and mouse mAb anti-Cyclin D1 (Santa Cruz Biotechnology); rabbit polyclonal anti-p-eIF2α (BIOSOURCE); rabbit polyclonal anti-p-IRE1α (Abcam); mouse mAb anti-p-ERK, rabbit polyclonal anti-ERK, rabbit polyclonal anti-LC3B, and mouse mAb anti-GAPDH (Sigma); rabbit polyclonal anti-TRIB3 (Calbiochem); rabbit polyclonal anti-BiP (Abcam); and mouse mAb anti-XIAP (BD Transduction Laboratories). Signals were developed using “EZ-ECL” (Biological Industries), and the membranes were exposed to x-ray films and developed.
RESULTS
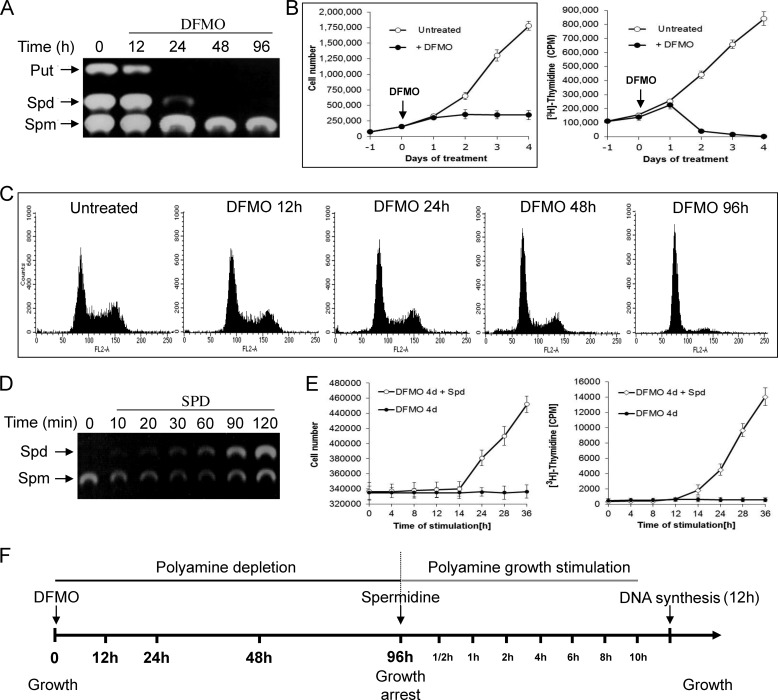
Polyamine depletion results in growth arrest that is completely reverted upon their re-addition to the growth medium of the arrested cells. We set out to characterize changes in the transcriptional program during these growth transitions. To establish a suitable experimental system, NIH3T3 mouse fibroblasts were treated with DFMO, an irreversible inhibitor of ornithine decarboxylase, the first and rate-limiting enzyme in the biosynthesis pathway of polyamines (8), and the level of polyamines and the proliferation status of the cells were determined. DFMO treatment resulted in depletion of putrescine and spermidine and a slight reduction in the level of spermine (Fig. 1A). Cell count and [3H]thymidine incorporation demonstrated establishment of growth inhibition concomitant with polyamine depletion, with complete arrest observed 4 days after addition of DFMO (Fig. 1B). Flow cytometry analysis demonstrated that DFMO-treated cells were arrested predominantly at the G0/G1 phase of the cell cycle (Fig. 1C). Rapid accumulation of intracellular spermidine was observed upon its re-addition to the growth medium of the DFMO-arrested cells, reaching normal levels within 2 h (Fig. 1D). This was accompanied by restoration of cellular proliferation noted 12 h following addition of spermidine as determined by incorporation of [3H]thymidine into DNA (Fig. 1E).
FIGURE 1.
Polyamine depletion results in reversible inhibition of cellular proliferation. A, DFMO (1 mm) was added to the medium of NIH3T3 cells, and cellular extracts were prepared at the indicated time points and subjected to polyamine analysis as described under “Experimental Procedures.” The position of polyamine markers is indicated. Put, putrescine; Spd, spermidine; Spm, spermine. B, at the indicated times DFMO-treated NIH3T3 cells were either counted or pulse-labeled with [3H]thymidine (the presented results represent triplicate experiments). Error bars indicate S.D. C, flow cytometry analysis was performed at the indicated times following the addition of DFMO. D, polyamine analysis was performed at the indicated time points following re-addition of spermidine (10 μm) to the medium of the DFMO-treated cells. E, cell count and [3H]thymidine incorporation assay were performed following re-addition of spermidine (10 mm) to the medium of the DFMO-treated cells. F, schematic presentation of the experimental frame. At the indicated time points the cells were harvested; RNA was extracted and subjected to microarray analysis.
To determine the genomic program that parallels the establishment of growth arrest and growth resumption, we characterized temporal changes in the pattern of the genome-wide transcriptional profiles manifested in NIH3T3 cells following treatment with DFMO and subsequent re-addition of polyamines to their growth medium (see Fig. 1F for the specific time points). Total RNA was extracted, amplified, and hybridized to Affymetrix MoGene 1.0 ST cDNA microarrays representing ∼29,000 genes. Because the vast majority of the DFMO-treated cells were arrested at the G0/G1 phase of the cell cycle, while growing cells were asynchronous, we have excluded from our analysis genes whose expression might be affected by the position of the cell along the cell cycle by collecting G0/G1 cells using flow cytometry. Because polyamine-stimulated cells re-entered growth synchronously, such fractionation was not required.
Transcriptional Program Following Polyamine Depletion
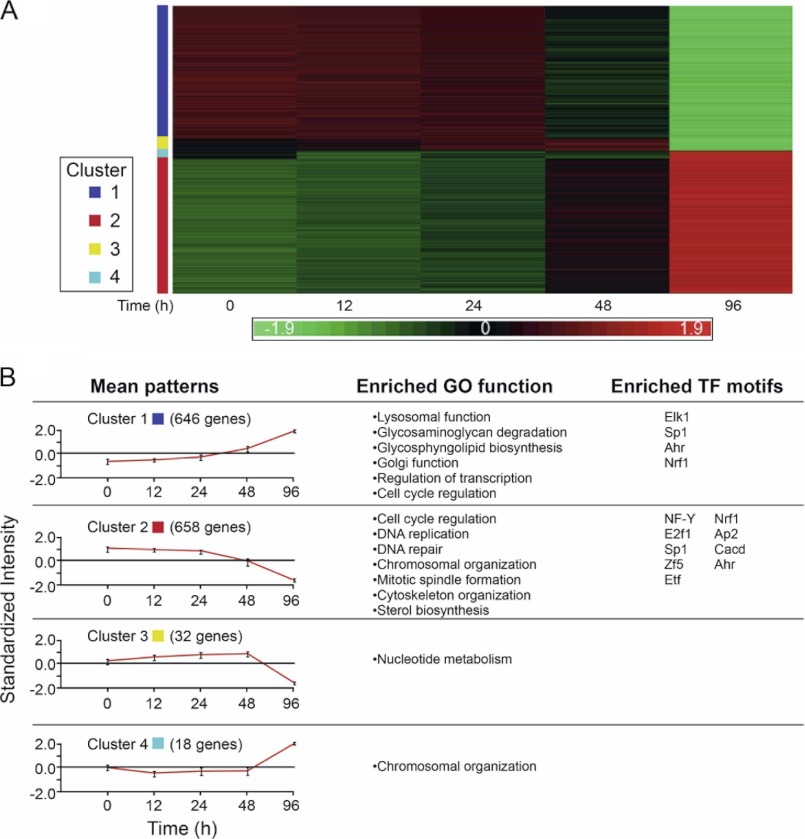
Genome-wide analysis of transcriptional responses using Affymetrix microarrays followed by analysis using the Partek genomic suite (Partek Inc.) identified 1345 genes whose expression was down- or up-regulated at least 2-fold in at least one time point during the establishment of growth arrest due to polyamine depletion (Fig. 2A and supplemental Table 1). These genes were subjected to hierarchical cluster analysis using the Click algorithm to identify prominent patterns of expression. Pattern analysis enabled their grouping into four main clusters, representing different temporal transcriptional responses to polyamine depletion (Fig. 2). The vast majority of these genes (96%) was associated with clusters 1 or 2, characterized by gradual transcriptional induction or repression initiated between 24 and 48 h following addition of DFMO to the growth medium (Fig. 2B). The other two smaller clusters (3 and 4) were characterized by delayed transcriptional response with genes down- or up-regulated as cells approached full growth arrest (96 h following addition of DFMO). Our results therefore suggest the existence of a threshold of minimal polyamine levels reached 24–48 h following DFMO addition, below which the cell becomes committed to the cell cycle exit program.
FIGURE 2.
Patterns of gene expression accompanying growth inhibition during polyamine depletion. A, heat map of gene expression patterns. The clusters were calculated by the Click algorithm (EXPANDER suite). Each column represents a single time point during the polyamine depletion process, and each row represents one gene across the different time points. Up-regulated genes are marked in red and down-regulated in green. The intensity values are standardized, and the scale is in standard deviation units. B, mean patterns, mean expression pattern of four clusters; GO function, gene ontology functional analysis of each cluster (DAVID), only significantly enriched functions are shown; TF motifs, significantly enriched transcription factor motifs for each cluster are calculated using the PRIMA algorithm (EXPANDER suite).
Identification of Enriched Functional Categories within Gene Clusters with Altered Expression
Gene ontology annotation and systems pathway analysis tools were used to identify biological processes enriched within each of the observed clusters (Fig. 2B and supplemental Table 2). The first major cluster composed of 658 up-regulated genes featured predominantly genes related to general lysosomal activity (V-type H+-ATPase subunits, lysosomal proteases, cathepsins, glycosidases, sulfatases, and major and minor lysosomal membrane proteins), glycosphingolipid biosynthesis (Glb1l, St3gal2, and St3gal4), Golgi-associated proteins (Golga4, Manea, Rabac1, Rab2b, Sulf2, and Nucb1), transcriptional regulation (Atf4, Atf5, Aebp1, Foxo3, Ddit3, Phf1, Mga, and Notch1), and cell cycle control (Cdk7, Cdknla, Gadd45a, Mdm2, and Rb1).
The second major cluster, containing 646 down-regulated genes, exhibited significant enrichment for genes related to major growth regulatory processes such as cell cycle regulation (Cdk2, E2F1, PCNA, Rbl1, and Uhrf1), DNA replication (Dna2, Lig1, Mcm2-7, Polal-2, Pold1-3, Pole1-2, Prim1-2, and Rfc1-5), DNA repair (Bard1, Brip1, Brca1–2, and Parp1), and chromosomal organization (different histone family members, Asf1b and Cbx5). Additional enriched pathways included mitotic spindle formation (Aurka-b and Bub1), different centromeric (CENP) complex subunits (Ndc80 and Nusap1), cytoskeleton organization (several augmin (HAUS) complex subunits and various kinesin (KIF) and tubulin family members), and sterol biosynthesis (Acat2, Cyp51, and Idi1). Because the other two clusters representing late responsive genes were relatively small (20–30 genes), the only biological processes that might be assigned to them were nucleotide metabolism (cluster 3) and chromosomal organization (cluster 4).
Transcriptional Program Following Re-addition of Polyamines
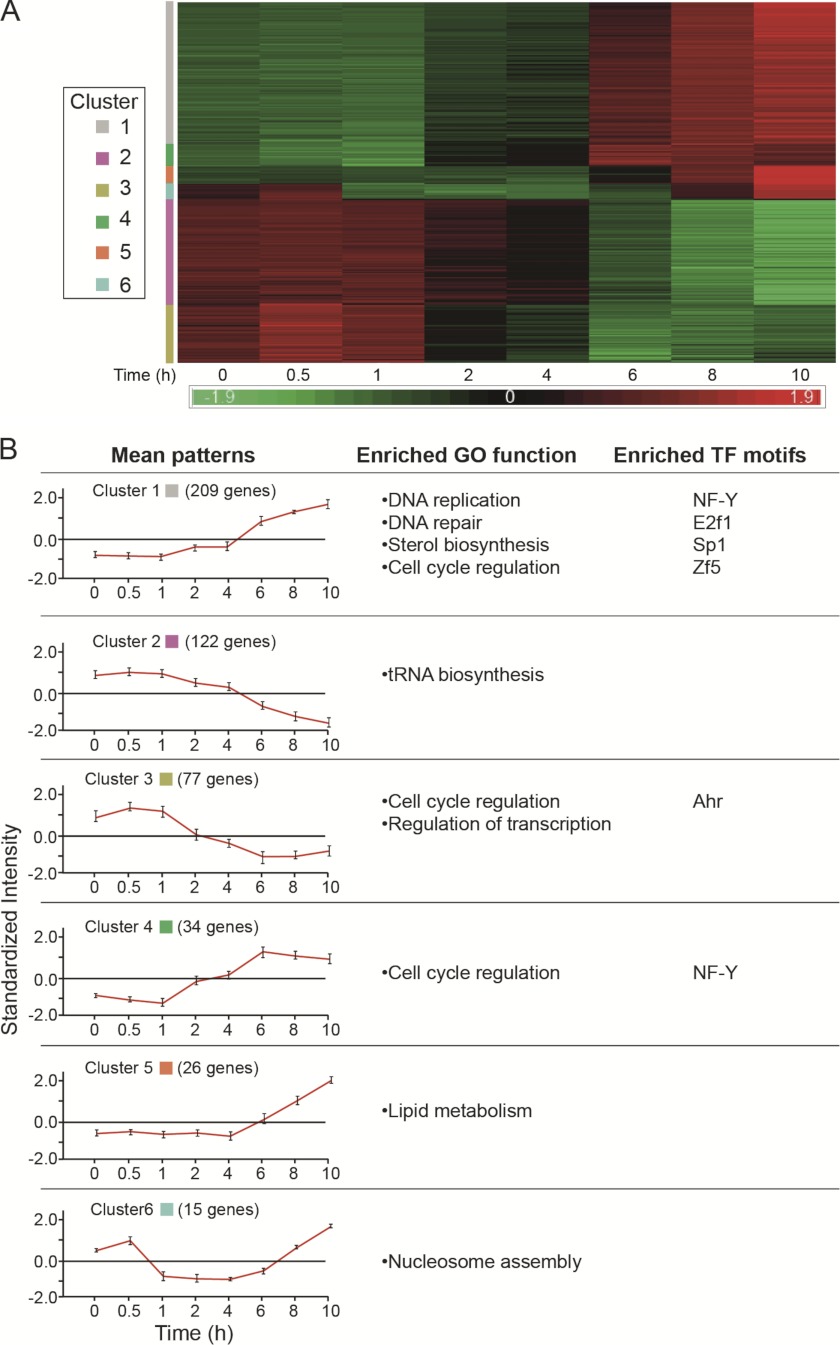
As demonstrated in Fig. 1, addition of spermidine to the growth medium of DFMO-treated (growth arrested) cells resulted in resumption of cellular proliferation. Because DNA synthesis was restored after 12 h, transcriptional responses were monitored during the first 10 h following addition of spermidine. This analysis revealed changes in the expression of 483 genes that were arranged in six clusters (Fig. 3 and supplemental Table 3). Three clusters (1, 4, and 5) contained genes whose expression was gradually up-regulated and varied in abruptness and the time lag following the addition of spermidine. Cluster 1 contained 209 genes whose expression was elevated starting at about 2 h following spermidine addition and featured enrichment in genes associated with DNA replication (Lig1, Mcm2-7, Mcm10, Pcna, Pola2, Pold2, Pole, and Prim1-2), DNA repair (Brca1, Brip1, Chaf1a-b, Exo1, and Fen1), sterol biosynthesis (Cyp51, Idi1, Mvd, and Mvk), and cell cycle regulation (Cdk2, Chek1-2, E2f1, Tfdp2, and Topbp1). Cluster 4, which contained 34 genes, was characterized by a more abrupt induction following a similar 2-h lag and included additional cell cycle-related genes such as Ccne1-2, Ccng2, Cdc25a, and E2f7. Cluster 5, which contained 26 genes with a more delayed transcriptional activation (4 h post stimulation), featured enrichment in genes involved in lipid biosynthesis (Btd, Fads2, Pcyt2, Elovl6, and Acaca) (supplemental Table 4).
FIGURE 3.
Patterns of gene expression accompanying growth resumption following polyamine re-addition. A and B are the same as in the legend to Fig. 2.
The cluster of down-regulated genes (cluster 2) included 122 genes, whose transcriptional repression started 2 h following spermidine stimulation. The only enriched process identified in this cluster represented tRNA metabolism (Cars, Mars, Rars2, and Yars). However, manual analysis based on the literature revealed additional genes associated with transcriptional regulation (Atf5, Atf6, Chd2, Churc1, and Nr1d1), cell cycle (Cdk10 and Zmat3), and fatty acid metabolism (Acad11, Acot2, Cyb5r1, and Plcxd2) that were not included in the above clusters. The remaining two clusters (clusters 3 and 6) exhibited transient modes of transcriptional activation by the stimulating spermidine. All the genes included in cluster 6 represented different core histones, whose expression was decreased during the first 6 h and raised again toward initiation of DNA synthesis (supplemental Table 4).
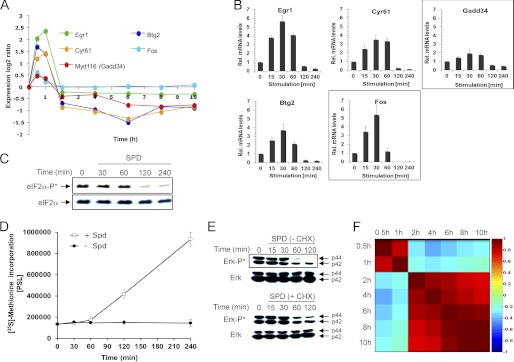
Cluster 3, which contained 77 genes, was characterized by transient up-regulated expression within the 1st h of growth stimulation, a behavior that is reminiscent of immediate-early genes, whose stimulation does not require de novo protein synthesis (9, 10). Indeed, cluster 3 contained typical immediate-early genes such as Btg2, Cyr61, Egr1, Egr2, Fos, Ier2, Ier3, Jun, and Klf6 (Fig. 4, A and B, and supplemental Table 3). Another gene in this group that might be of particular interest is Gadd34. This gene encodes a positive regulator of protein phosphatase 1α (PP1C) that dephosphorylates eIF2α (11) and therefore may potentially relieve inhibition of global protein synthesis, which we have recently demonstrated to be established in polyamine-depleted cells (12). Interestingly, eIF2α became dephosphorylated 2 h after addition of spermidine to the arrested cells (Fig. 4C), concomitant with the resumption of protein synthesis activity (Fig. 4D).
FIGURE 4.
Delayed early genes expressed following release from the translational block attenuate polyamine-dependent induction of immediate early genes. A, expression pattern of the indicated genes as determined from the arrays representing DFMO-treated (growth-arrested) cells at various time points following their stimulation with spermidine (10 μm). B, real time PCR verification of the expression pattern of genes depicted in A. C, cellular extracts were prepared at the indicated times following the addition of spermidine to the medium of DFMO-treated cells, and the phosphorylation status of eIF2α was determined by Western blot analysis. D, DFMO-treated cells were pulse-labeled with [35S]methionine at the indicated times following the addition of spermidine (10 μm) to their growth medium. E, cellular extracts were prepared at the indicated times following the addition of spermidine in the presence and absence of cycloheximide (CHX) to the medium of DFMO-treated cells, and the phosphorylation status of ERK was determined by Western blot analysis. F, heat map of Pearson correlations between the different time points. Each point was calculated as the ratio between the indicated time and the control (time 0), in log2. 1 indicates complete positive correlation (dark red); −1 indicates complete negative correlation (dark blue); 0 indicates no correlation between the time points. Only differentially expressed genes in at least one time point during the experiment were used for the analysis.
Various studies demonstrate that the expression of immediate early genes and the upstream signaling are attenuated by a negative transcriptional feedback, mediated by delayed-early genes (9, 10). Mitogenic stimulation is frequently mediated by transient activation of the ERK MAPKs (9). Interestingly, ERK, which gets phosphorylated due to polyamine depletion (13), becomes dephosphorylated upon spermidine stimulation with kinetics resembling the expression pattern of the immediate-early genes (Fig. 4E). In the presence of the translational inhibitor cycloheximide, ERK dephosphorylation was delayed, suggesting that ERK signaling and the transcription of the immediate early genes are attenuated by the products of downstream expressed delayed early genes. Indeed, as recently described in the case of growth factor-mediated growth stimulation (9), our analysis demonstrated induction of the dual specificity phosphatases Dusp5 and -6 and of the RNA-binding protein Zfp36. Their products may attenuate early signaling and expression of immediate-early genes following growth stimulation by polyamines (supplemental Table 5). The expression of these delayed early genes started 2 h following the addition of spermidine (Fig. 4F), being well correlated with the onset of protein synthesis activity.
Promoter Analysis of Genes Responding to Polyamine Depletion or Stimulation
To further identify the possible regulators that might orchestrate the observed transcriptional responses to polyamine depletion, we applied the promoter analysis algorithm PRIMA implemented in the Expander suite (14, 15). Analysis of the up-regulated genes (cluster 1) identified nine enriched binding site motifs corresponding to ELK-1, SP1, AHR, and NRF1 (supplemental Table 6). Enrichment was most significant for ELK-1 (p < 5.39·10−10), which is a member of the Ets family of transcription factors and a target of the Ras-Raf-MAPK signaling cascade that includes ERK and p38 (16).
Promoter region analysis of the down-regulated genes (cluster 2) identified enrichment for nine transcription factor binding site signatures. The highest enrichment was for NF-Y (p < 3.17·10−32), which acts as heterotrimeric complex involved in stress response and in control of proliferation-associated genes (17, 18). Other enriched transcription factor binding sites included E2F1, SP1, ETF, and AP2, all of which are well established growth control mediators (19–22). Interestingly, these four transcription factors are also targets of the MAPK pathway (23, 24), being in agreement with reports suggesting a role for MAPKs in the establishment of growth arrest induced by DFMO (13, 25). Additionally, our microarray analysis demonstrated significant reduction in E2f1 and Etf mRNA levels, suggesting their putative role in promoting growth arrest following polyamine depletion.
Promoter analysis of genes, whose expression was affected by re-addition of spermidine, identified significant over-representation of binding sites for NF-Y, E2F1, SP1, and ZF5 in cluster 1, the Ahr-binding site in cluster 3, and an additional binding site for NF-Y in cluster 4 (supplemental Table 7). Interestingly, the same transcription factor signatures identified in cluster 1 were also significantly associated with cluster 2 of genes down-regulated due to polyamine depletion, further suggesting that these four transcription factors respond to changes in polyamine concentrations.
Comparison between Expression Patterns of Growth Arrest and Growth Stimulation
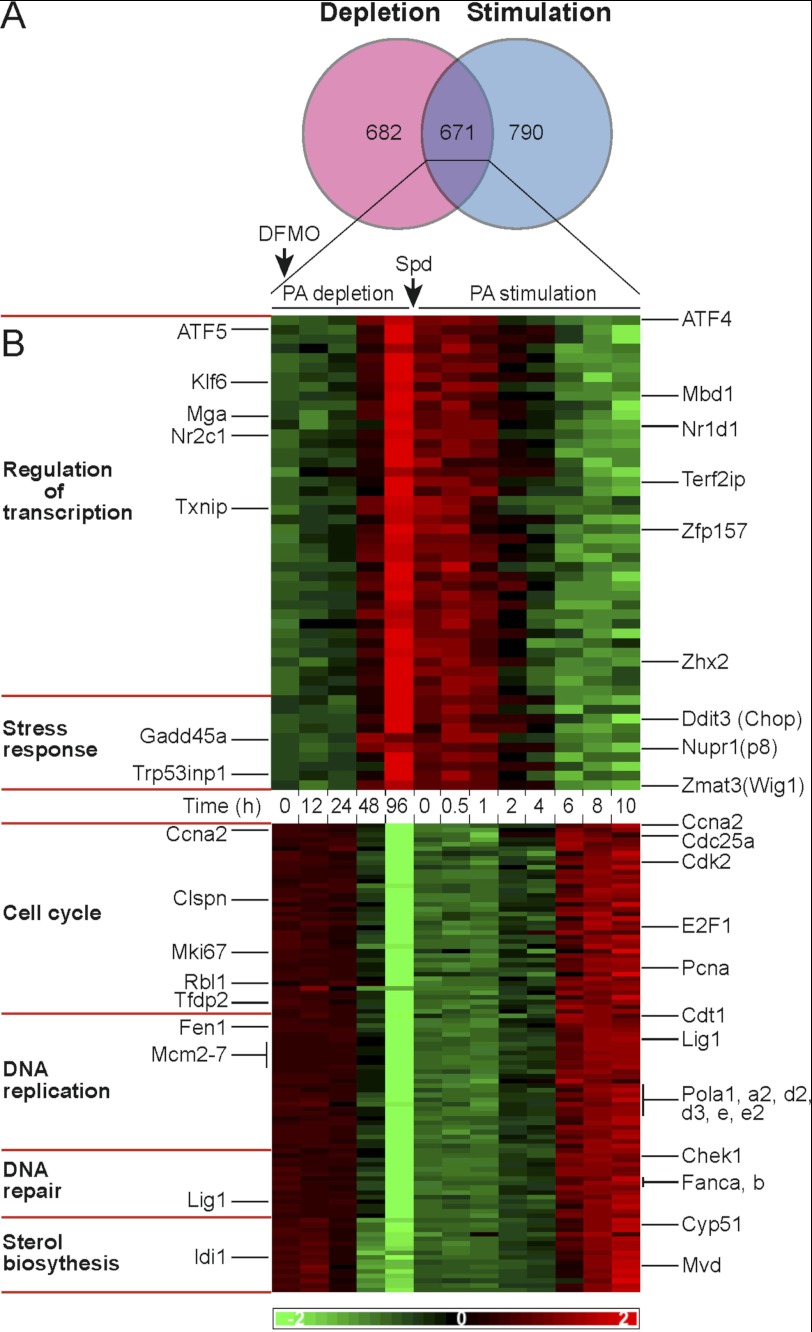
To analyze possible correlation of transcriptional responses between the states of growth arrest due to polyamine depletion and growth resumption by re-addition of polyamines, the differentially expressed genes in either experiment were compared (Fig. 5A). Genes identified in both experiments were divided based on symmetrical or asymmetrical expression patterns as cells traversed the growth arrest point, and the two groups were subjected to functional annotation analysis. Because of the limited time window of stimulation (10 h), we decided to be more permissive and take genes with differential expression of at least 1.5-fold (fold change >1.5) in at least one time point during this period (supplemental Table 8).
FIGURE 5.
Growth arrest traversing gene expression patterns, comparison of polyamine depletion to polyamine stimulation. A, Venn diagram illustrating comparison of differentially expressed genes during polyamine depletion (fold change >2) to differentially expressed genes during polyamine stimulation (fold change >1.5). B, heat map of genes with symmetrical expression patterns. Differentially expressed genes in both experiments (Venn diagram cross-section) were subjected to gene ontology annotation analysis (left panel). Selected genes in enriched pathways are illustrated in the right panel.
Comparison of the transcriptional changes demonstrated asymmetry in the expression of genes related to cell division and governing chromosomal organization, mitotic spindle formation, and cytoskeleton organization. To a large extent, this apparent asymmetry was a result of the limited time window (0–10 h) used to monitor alteration of expression during the stimulation phase, and the observed changes were mostly associated with G1/S transition. In contrast, symmetry was observed in the behavior of 671 genes, out of which about 300 genes could be assigned to cell cycle regulation, DNA replication, DNA repair, stress response, sterol biosynthesis, and transcription regulation (Fig. 5B and supplemental Table 9). Interestingly, most of the symmetrical changes in the expression of transcription regulators involved transcriptional repressors like Mbd1, Mga, Txnip, Zfp157, and Zhx2, whose expression was induced during growth repression and decreased upon growth stimulation. Among these, ZHX2 was previously demonstrated to negatively regulate transcriptional activity of NF-Y (26), whose binding motif was the most enriched within promoter regions of genes down-regulated in polyamine-depleted cells and among genes up-regulated during growth stimulation by polyamines (Figs. 2, cluster 2, and 3, cluster 1).
Symmetrical Gene Expression Reveals a Signature of a Stress Response
The above results could not trivially discriminate between genes whose altered expression triggered inhibition of proliferation from genes, whose expression was altered as a result of the arrested growth. Interestingly, one of the processes identified among the observed transcriptional changes was cellular stress response. The genes associated with stress response included Ddit3 (Chop), Gadd45a, Nupr1(p8), and ATF4, a central stress-related transcription factor regulating expression of CHOP (27). Further analysis identified several CHOP targets, such as Gadd34, Trib3, Chac1, Car6, and Odz4, thus suggesting induction of the ATF4-CHOP pathway (28, 29). In addition, during polyamine depletion we observed increased expression of genes associated with lysosomal activity, glycosphingolipid metabolism, Golgi functions, and the down-regulated expression of genes in the sterol biosynthesis pathway, changes previously associated with establishment of cellular stress responses (30–35). Further in this line, the transcription factors, ELK1, SP1, AHR, and NrF1, whose signatures were over-represented in genes up-regulated in polyamine-depleted cells, were demonstrated to mediate stress responses and regulate cellular proliferation (36–39). Interestingly, stress responses usually arrest cells at the G0/G1 phase of the cell cycle (40–43), as we show here for polyamine-depleted cells (Fig. 1). Activation of this process represents a highly conserved adaptation to several types of cellular stresses such as ER stress, amino acid starvation, oxidative stress, and hypoxia (27, 28, 44), cumulatively known as the integrated stress response (27). Thus, together with our recent demonstration that polyamine depletion leads to phosphorylation of eIF2α (12), our results suggest that induction of a stress response potentially contributes to the establishment of growth arrest in polyamine-depleted cells.
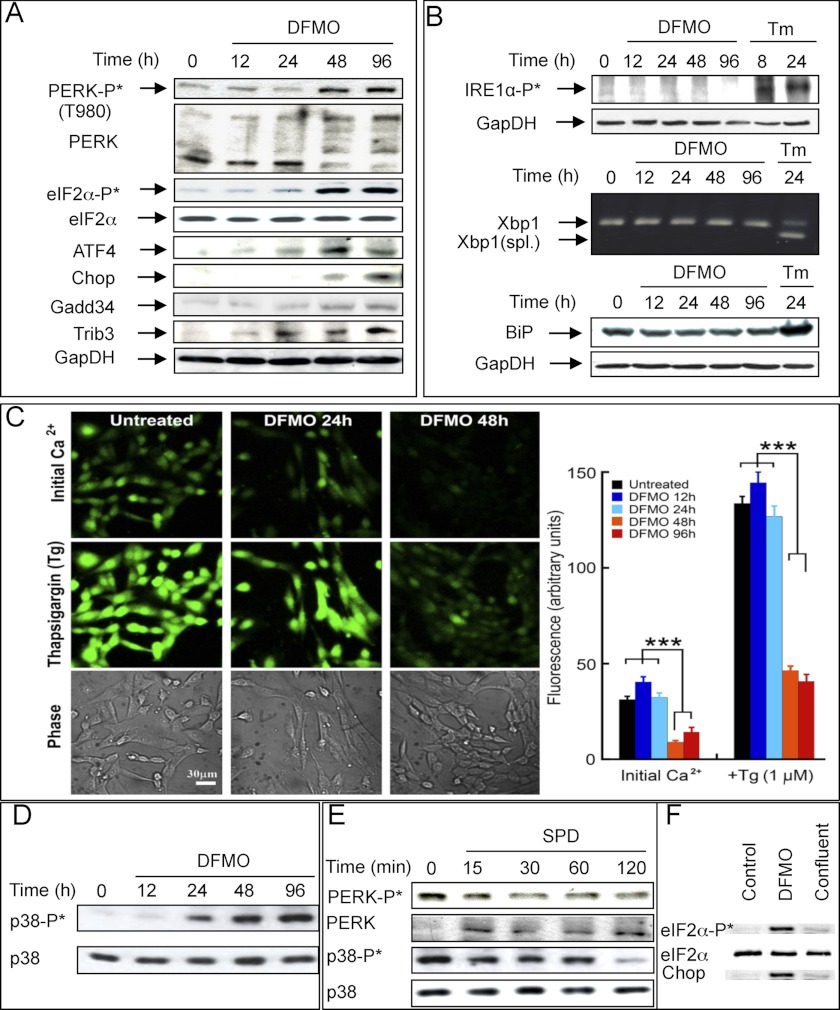
Polyamine Depletion Elicits a Noncanonical ER Stress Response
Previous studies indicated that polyamine depletion causes swelling and distortion of the rough ER, suggesting that the rough ER may be a target where polyamines affect cell proliferation (45, 46). It is therefore possible that the observed distortion reflects manifestation of ER stress, eliciting unfolded protein response (UPR) (47), a process known to activate the ATF4-CHOP pathway (27). We show here that in DFMO-treated cells phosphorylation of eIF2α was paralleled by activation of PERK, one of the four known eIF2α kinases, and by stimulated expression of ATF4 and CHOP, transcription factors that activate downstream stress-responding genes (Fig. 6A). PERK is an ER membrane resident kinase that is activated by ER stress, leading to phosphorylation of eIF2α and translational arrest thus relieving protein load in the ER (28). In response to a typical ER stress, PERK activation is paralleled by activation of another ER membrane kinase, IRE1α, and the transcription factor ATF6, representing two additional arms of the unfolded protein response (47). However, in contrast to PERK activation, we could not demonstrate IRE1α or ATF6 activation in the polyamine-depleted cells. Further in this line, XBP1 mRNA, which is spliced upon IRE1α activation (47), remained unspliced (Fig. 6B), and none of the ATF6-responsive genes (48) were induced by DFMO treatment (array results). Additionally, the level of the central ER chaperon protein GRP78 (BiP) was not elevated in the polyamine-depleted cells, in contrast to its induction during canonical UPR (42, 49), as exemplified here by tunicamycin treatment (Fig. 6B). Nevertheless, the ER seems to be involved in the established stress as evident by the reduction of Ca2+ levels in the ER of polyamine-depleted cells (Fig. 6C).
FIGURE 6.
Establishment of stress in polyamine-depleted cells. A, cellular extracts were prepared at the indicated times following the addition of DFMO, fractionated, blotted, and probed with the indicated antibodies. B, levels of phospho-IRE1α and BiP (GRP78) following DFMO or tunicamycin (Tm) treatments were determined by Western blot analysis. Xbp1 mRNA splicing was determined by polymerase chain reaction as described under “Experimental Procedures.” C, total (initial) calcium levels were measured by loading cells with Fluo2-AM indicator. ER calcium capacity at each time point was measured by inducing efflux from ER by 1 μm thapsigargin (Tg). Confocal images at ×20 magnification (left panel) and statistical analysis (right panel) are presented. Results are based on three independent experiments. D, level of phospho-p38 following treatment with DFMO; E, levels of phospho-PERK and phospho-p38 following re-addition of spermidine (SPD) to DFMO-treated cells were determined by Western blot analysis. F, levels of eIF2α, phosphorylated eIF2α and CHOP were determined in growing cells (Control), polyamine-depleted cells (DFMO), and cells that were harvested 5 days after reaching confluence (Confluent).
Further support for the establishment of stress in the polyamine-depleted cells was provided by the observed phosphorylation of the stress-associated p38 MAPK (Fig. 6D), which could also potentially explain the induction of the ELK1 target genes (16). Importantly, the induced stress was rapidly reverted upon addition of polyamines to the DFMO-treated cells as evident by rapid dephosphorylation of eIF2α (Fig. 4C), PERK, and p38 (Fig. 6E).
It can be argued that the induced stress is actually secondary to growth arrest. In this respect, it was demonstrated that CHOP is not uniformly induced due to arrested growth (50). We have extended this observation by comparing eIF2α phosphorylation and CHOP induction in cells that were growth-arrested due to polyamine depletion or by contact inhibition. We show that although eIF2α phosphorylation and CHOP were prominently induced in the DFMO-treated cells, no induction was observed in cells that reached confluence and kept growth-arrested for 5 days (Fig. 6F).
Cell Cycle Inhibition by Polyamine Depletion
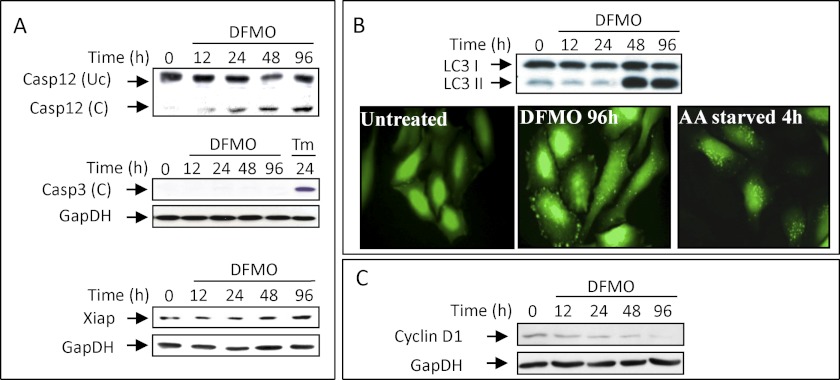
Growth arrest can be manifested as a result of reduced availability of growth-stimulating proteins such as cyclin D1, whose level was previously demonstrated to decline rapidly in stressed cells due to attenuated translation and accelerated degradation (40, 51, 52). We demonstrate here that the level of the cyclin D1 protein declines in DFMO-treated cells (Fig. 7C), most likely playing a key role in promoting the establishment of the observed G1 arrest. Additional changes that may also support growth arrest include a decline in the level of mRNAs encoding other cyclins such as cyclin A and E and other growth-promoting proteins like CDK2, E2F1, and PCNA, and increased levels of the mRNAs encoding p21 and GADD45a, proteins that mediate G1 arrest in response to variety of stress stimuli (53–55).
FIGURE 7.
Polyamine-depleted cells are growth-arrested but maintain viability. A, cellular extracts were prepared at the indicated times from DFMO or tunicamycin (Tm)-treated cells, and the level of the indicated proteins was determined by Western blot analysis. B, HeLa cells stably expressing LC3 fused to GFP were either treated with DFMO for 96 hours in complete DMEM or starved for serum and amino acids for 4 hours in Earles balanced salt solution. Punctation was visualized using fluorescent microscopy and LC3 cleavage by Western blot analysis. C, the level of cyclin D1 in extracts of DFMO treated cells was determined by Western blot analysis.
Polyamine-depleted NIH3T3 Cells Are Protected from Apoptosis
In most cases, polyamine depletion results in reversible growth arrest without provoking cell death (56, 57). We demonstrate here that polyamine depletion results in lengthy activation of the established stress (Fig. 6A), without inducing apoptosis that often accompanies prolonged stress (58, 59). This is particularly interesting, as polyamine depletion led to activation of the ER membrane resident caspase-12, but not the executioner caspase-3 (Fig. 7A). This might be a result of transcriptional repression of caspase-3 (supplemental Table 10) or increased expression of the XIAP protein (Fig. 7A), an inhibitor of caspase-3 activation (60, 61). Additional protection might be provided by induction of autophagy as encoded by key autophagy genes such as Atg12, Atg7, Atg4c, Map11c3b (LC3), and Lamp2, processing and punctation of LC3, and induction of genes that belong to the lysosomal degradation pathway in the polyamine-depleted cells (Fig. 7B).
DISCUSSION
Comprehensive transcription profiling combined with bioinformatic and complementary biochemical analysis suggest induction of a stress response as the potential cause for growth arrest establishment in polyamine-depleted cells.
Using Affymetrix DNA array hybridization and bioinformatic tools, we revealed the most prominent changes in gene expression that accompany the establishment of growth arrest due to polyamine depletion and during growth resumption following re-addition of polyamines to the medium of the arrested cells. Transcriptional reprogramming was initiated 24–48 h following addition of DFMO, being correlated with the time required for complete depletion of putrescine and spermidine. Genes related to cell cycle regulation, DNA replication, DNA repair, chromosomal organization, mitotic spindle formation, cytoskeleton organization, nucleotide metabolism, and sterol biosynthesis were down-regulated. Conversely, genes related to lysosomal functions, glycosaminoglycan degradation, glycosphingolipid biosynthesis, Golgi functions, transcription, and cell cycle regulation were up-regulated. Addition of spermidine to the growth-arrested cells resulted in robust transcriptional response driving cell cycle resumption, which was initiated by a transient up-regulation of typical immediate early genes and followed by induction of delayed early genes, whose products attenuate the expression of immediate early genes (supplemental Tables 3 and 11).
Promoter analysis of the responsive clusters identified the binding motif of ELK1, which is a target of the ERK and p38 components of the MAPK pathway as the most significant signature among the up-regulated genes. This is in agreement with previous studies reporting ERK activation following polyamine depletion (13, 25). Binding motifs for key growth regulators such as NF-Y, E2F1, SP1, and ETF were the most enriched motifs among the down-regulated genes. Interestingly, these same motifs were also found among genes up-regulated after growth stimulation by polyamines.
Most interestingly, inspection of genes in the various pathways responding to growth inhibition due to polyamine depletion and their comparison with genes responding to growth stimulation by polyamine replenishment revealed expressional changes in genes that collectively act as stress-response executioners, leading to inhibition of cellular proliferation and sometimes also to apoptosis. This included genes like Nupr1, ATF4, Cebpβ, Gadd34, and Chop and downstream Chop targets such as Trib3, Chac1, Odz4, and Car6, which were transiently up-regulated 48 h after DFMO addition (supplemental Table 12). In addition, we also observed transient up-regulated expression of genes associated with amino acid synthesis and transport, shown to be induced during stress response (28).
The notion that polyamine depletion may arrest cellular proliferation by inducing stress is further supported by our recent demonstration of eIF2α phosphorylation in polyamine-depleted cells (12). eIF2α phosphorylation functions as a central hub integrating cellular response to different types of cellular stress such as ER stress, oxidative stress, hypoxia, and amino acids starvation (27, 28, 44). We demonstrate here that PERK, an ER membrane resident kinase, which is one of the four kinases known to phosphorylate eIF2α, is activated in polyamine-depleted cells (Fig. 6A). This suggests establishment of ER stress and UPR activation in the polyamine-depleted cells. Interestingly, it was recently demonstrated that overexpression of ornithine decarboxylase resulting in increased intracellular levels of polyamines suppresses thapsigargin-induced apoptosis accompanied by reduced PERK phosphorylation and CHOP protein level (62). We could not, however, obtain evidence for parallel activation of the other two arms of the UPR initiated by IRE1 and ATF6, whose concerted action accompany establishment of a canonical ER stress. Lack of up-regulation of BiP and of other ER chaperons further argues against possible establishment of a typical ER stress due to accumulation of unfolded proteins and development of canonical UPR. Moreover, studies demonstrating morphological changes in the rough ER of polyamine-depleted cells (45, 46) and our present demonstration of reduced ER calcium levels in polyamine-depleted cells suggest that perturbation of ER functions may be involved in the establishment of growth arrest. Interestingly, the PERK arm of the UPR was not the only stress response activated in polyamine-depleted cells. We also noted parallel activation of the p38 MAPK that is activated in response to variety of stress stimuli (63). Because both are known to negatively regulate cyclin D1 (40, 63), each of them can account for the inhibited proliferation of polyamine-depleted cells. This is compatible with the ability of DFMO treatment to inhibit proliferation of the eIF2α-S51A mutant and of the p38α-deficient cells. However, because cells containing both deficiencies are not available, we cannot rule out the existence of additional causes for the establishment of growth arrest in polyamine-depleted cells.
Stressed cells are usually growth-arrested at the G1 phase of the cell cycle (40, 41, 43, 64), as we demonstrate here for polyamine-depleted cells. The G1 arrest in polyamine-depleted cells was associated with increased transcription of p21 and Gadd45a, decreased transcription of Cdk2, E2f1, Pcna, cyclins A and E, and most profoundly with decreased levels of the cyclin D1 protein, a regulator of the Cdks required for G1/S transition, whose decline was demonstrated to suffice for arresting cellular proliferation (40, 65).
Prolonged stress usually leads to apoptosis (58, 59). However, although polyamine depletion elicited prolonged stress in NIH3T3 cells as evident from the sustained phosphorylation of eIF2α, the polyamine-depleted cells remain viable. Inspection of possible apoptotic signals in these cells revealed increased expression of proapoptotic genes such as caspase-12, Trib3, Fas, Bax, Bid, and Dr5, and activation of caspase-12, changes that usually lead to activation of the downstream executioner caspase-9 and -3 (66–69). Nevertheless, polyamine-depleted cells remained viable and resumed proliferation upon re-addition of polyamines to their growth medium. This might be a result of impaired caspase-3 activation, possibly due to induction of XIAP, an inhibitor of caspase-3 that was recently demonstrated to inhibit apoptosis in polyamine-depleted intestinal epithelial cells (61). Moreover, our results demonstrate induction of autophagy in the DFMO-treated cells, which can provide additional cytoprotective effect (44, 46, 70). Lack of these cytoprotective mechanisms may underline the few cases in which polyamine depletion is cytotoxic.
Overall, our results demonstrate stress establishment in polyamine-depleted cells through activation of the PERK arm of the UPR and parallel activation of the p38 MAPK. This results in activation of downstream stress-responding genes that affect the expression of cell cycle regulators that can lead to growth arrest at the G1 phase of the cell cycle. We demonstrate here that these stress responses are not established in cells that are growth-arrested due to contact inhibition. This strongly supports the notion that these stress responses are causative in opposition to being a result of the established growth arrest. Because eIF2α-S51A mutant or p38-deficient cells are still growth-arrested by DFMO treatment, it is possible these two stresses are redundant in their ability to induce growth arrest in polyamine-depleted cells. Moreover, because cells carrying both deficiencies are not available, we cannot rule out the possibility that additional yet unidentified mechanisms can also account for arresting growth of polyamine-depleted cells.
This work was supported by grants from the Israel Academy of Science and Humanities, the M. D. Moross Institute for Cancer Research, and the Kahn Family Research Center for Systems Biology of the Human Cell at the Weizmann Institute.

This article contains supplemental Tables 1–12.
- DFMO
- difluoromethylornithine
- ER
- endoplasmic reticulum
- dansyl
- 5-dimethylaminonaphthalene-1-sulfonyl
- UPR
- unfolded protein response
- PERK
- PKR-like endoplasmic reticulum kinase.
REFERENCES
- 1. Pegg A. E. (2009) Mammalian polyamine metabolism and function. IUBMB Life 61, 880–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pegg A. E. (1988) Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res. 48, 759–774 [PubMed] [Google Scholar]
- 3. Auvinen M., Paasinen A., Andersson L. C., Hölttä E. (1992) Ornithine decarboxylase activity is critical for cell transformation. Nature 360, 355–358 [DOI] [PubMed] [Google Scholar]
- 4. Clifford A., Morgan D., Yuspa S. H., Soler A. P., Gilmour S. (1995) Role of ornithine decarboxylase in epidermal tumorigenesis. Cancer Res. 55, 1680–1686 [PubMed] [Google Scholar]
- 5. Sharan R., Maron-Katz A., Shamir R. (2003) CLICK and EXPANDER. A system for clustering and visualizing gene expression data. Bioinformatics 19, 1787–1799 [DOI] [PubMed] [Google Scholar]
- 6. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 7. Ulitsky I., Maron-Katz A., Shavit S., Sagir D., Linhart C., Elkon R., Tanay A., Sharan R., Shiloh Y., Shamir R. (2010) Expander. From expression microarrays to networks and functions. Nat. Protoc. 5, 303–322 [DOI] [PubMed] [Google Scholar]
- 8. Mamont P. S., Duchesne M. C., Grove J., Bey P. (1978) Anti-proliferative properties of dl-α-difluoromethylornithine in cultured cells. A consequence of the irreversible inhibition of ornithine decarboxylase. Biochem. Biophys. Res. Commun. 81, 58–66 [DOI] [PubMed] [Google Scholar]
- 9. Amit I., Citri A., Shay T., Lu Y., Katz M., Zhang F., Tarcic G., Siwak D., Lahad J., Jacob-Hirsch J., Amariglio N., Vaisman N., Segal E., Rechavi G., Alon U., Mills G. B., Domany E., Yarden Y. (2007) A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 39, 503–512 [DOI] [PubMed] [Google Scholar]
- 10. Lau L. F., Nathans D. (1987) Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells. Coordinate regulation with c-fos or c-myc. Proc. Natl. Acad. Sci. U.S.A. 84, 1182–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Novoa I., Zeng H., Harding H. P., Ron D. (2001) Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Landau G., Bercovich Z., Park M. H., Kahana C. (2010) The role of polyamines in supporting growth of mammalian cells is mediated through their requirement for translation initiation and elongation. J. Biol. Chem. 285, 12474–12481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Manni A., Washington S., Mauger D., Hackett D. A., Verderame M. F. (2004) Cellular mechanisms mediating the anti-invasive properties of the ornithine decarboxylase inhibitor α-difluoromethylornithine (DFMO) in human breast cancer cells. Clin. Exp. Metastasis 21, 461–467 [DOI] [PubMed] [Google Scholar]
- 14. Elkon R., Linhart C., Sharan R., Shamir R., Shiloh Y. (2003) Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 13, 773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shamir R., Maron-Katz A., Tanay A., Linhart C., Steinfeld I., Sharan R., Shiloh Y., Elkon R. (2005) EXPANDER. An integrative program suite for microarray data analysis. BMC Bioinformatics 6, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Whitmarsh A. J., Yang S. H., Su M. S., Sharrocks A. D., Davis R. J. (1997) Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol. Cell. Biol. 17, 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhattacharya A., Deng J. M., Zhang Z., Behringer R., de Crombrugghe B., Maity S. N. (2003) The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 63, 8167–8172 [PubMed] [Google Scholar]
- 18. Gurtner A., Fuschi P., Martelli F., Manni I., Artuso S., Simonte G., Ambrosino V., Antonini A., Folgiero V., Falcioni R., Sacchi A., Piaggio G. (2010) Transcription factor NF-Y induces apoptosis in cells expressing wild-type p53 through E2F1 up-regulation and p53 activation. Cancer Res. 70, 9711–9720 [DOI] [PubMed] [Google Scholar]
- 19. Grinstein E., Jundt F., Weinert I., Wernet P., Royer H. D. (2002) Sp1 as G1 cell cycle phase-specific transcription factor in epithelial cells. Oncogene 21, 1485–1492 [DOI] [PubMed] [Google Scholar]
- 20. Kageyama R., Merlino G. T., Pastan I. (1989) Nuclear factor ETF specifically stimulates transcription from promoters without a TATA box. J. Biol. Chem. 264, 15508–15514 [PubMed] [Google Scholar]
- 21. Qin X. Q., Barsoum J. (1997) Differential cell cycle effects induced by E2F1 mutants. Oncogene 14, 53–62 [DOI] [PubMed] [Google Scholar]
- 22. Zeng Y. X., Somasundaram K., el-Deiry W. S. (1997) AP2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat. Genet. 15, 78–82 [DOI] [PubMed] [Google Scholar]
- 23. Tuli R., Seghatoleslami M. R., Tuli S., Howard M. S., Danielson K. G., Tuan R. S. (2002) p38 MAPK regulation of AP-2 binding in TGF-β1-stimulated chondrogenesis of human trabecular bone-derived cells. Ann. N.Y. Acad. Sci. 961, 172–177 [DOI] [PubMed] [Google Scholar]
- 24. Zellmer S., Schmidt-Heck W., Godoy P., Weng H., Meyer C., Lehmann T., Sparna T., Schormann W., Hammad S., Kreutz C., Timmer J., von Weizsäcker F., Thürmann P. A., Merfort I., Guthke R., Dooley S., Hengstler J. G., Gebhardt R. (2010) Transcription factors ETF, E2F, and SP-1 are involved in cytokine-independent proliferation of murine hepatocytes. Hepatology 52, 2127–2136 [DOI] [PubMed] [Google Scholar]
- 25. Hu X., Washington S., Verderame M. F., Manni A. (2005) Interaction between polyamines and the mitogen-activated protein kinase pathway in the regulation of cell cycle variables in breast cancer cells. Cancer Res. 65, 11026–11033 [DOI] [PubMed] [Google Scholar]
- 26. Kawata H., Yamada K., Shou Z., Mizutani T., Yazawa T., Yoshino M., Sekiguchi T., Kajitani T., Miyamoto K. (2003) Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX family, functions as a transcriptional repressor. Biochem. J. 373, 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 28. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 29. Malhi H., Kaufman R. J. (2011) Endoplasmic reticulum stress in liver disease. J. Hepatol. 54, 795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. d'Azzo A., Tessitore A., Sano R. (2006) Gangliosides as apoptotic signals in ER stress response. Cell Death Differ. 13, 404–414 [DOI] [PubMed] [Google Scholar]
- 31. Harding H. P., Zhang Y., Khersonsky S., Marciniak S., Scheuner D., Kaufman R. J., Javitt N., Chang Y. T., Ron D. (2005) Bioactive small molecules reveal antagonism between the integrated stress response and sterol-regulated gene expression. Cell Metab. 2, 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee G. H., Kim D. S., Kim H. T., Lee J. W., Chung C. H., Ahn T., Lim J. M., Kim I. K., Chae H. J., Kim H. R. (2011) Enhanced lysosomal activity is involved in Bax inhibitor-1-induced regulation of the endoplasmic reticulum (ER) stress response and cell death against ER stress. Involvement of vacuolar H+-ATPase (V-ATPase). J. Biol. Chem. 286, 24743–24753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oku M., Tanakura S., Uemura A., Sohda M., Misumi Y., Taniguchi M., Wakabayashi S., Yoshida H. (2011) Novel cis-acting element GASE regulates transcriptional induction by the Golgi stress response. Cell Struct. Funct. 36, 1–12 [DOI] [PubMed] [Google Scholar]
- 34. Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S., Banfi S., Parenti G., Cattaneo E., Ballabio A. (2009) A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 [DOI] [PubMed] [Google Scholar]
- 35. Tessitore A., del P Martin M., Sano R., Ma Y., Mann L., Ingrassia A., Laywell E. D., Steindler D. A., Hendershot L. M., d'Azzo A. (2004) GM1-ganglioside-mediated activation of the unfolded protein response causes neuronal death in a neurodegenerative gangliosidosis. Mol. Cell 15, 753–766 [DOI] [PubMed] [Google Scholar]
- 36. Biswas M., Phan D., Watanabe M., Chan J. Y. (2011) The Fbw7 tumor suppressor regulates nuclear factor E2-related factor 1 transcription factor turnover through proteasome-mediated proteolysis. J. Biol. Chem. 286, 39282–39289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dalton T. P., Puga A., Shertzer H. G. (2002) Induction of cellular oxidative stress by aryl hydrocarbon receptor activation. Chem. Biol. Interact. 141, 77–95 [DOI] [PubMed] [Google Scholar]
- 38. Ferreiro I., Joaquin M., Islam A., Gomez-Lopez G., Barragan M., Lombardía L., Domínguez O., Pisano D. G., Lopez-Bigas N., Nebreda A. R., Posas F. (2010) Whole genome analysis of p38 SAPK-mediated gene expression upon stress. BMC Genomics 11, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gille H., Strahl T., Shaw P. E. (1995) Activation of ternary complex factor Elk-1 by stress-activated protein kinases. Curr. Biol. 5, 1191–1200 [DOI] [PubMed] [Google Scholar]
- 40. Brewer J. W., Hendershot L. M., Sherr C. J., Diehl J. A. (1999) Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc. Natl. Acad. Sci. U.S.A. 96, 8505–8510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clement A., Henrion-Caude A., Besnard V., Corroyer S. (2001) Role of cyclins in epithelial response to oxidants. Am. J. Respir. Crit. Care Med. 164, S81–S84 [DOI] [PubMed] [Google Scholar]
- 42. Igarashi K., Kashiwagi K. (2010) Characteristics of cellular polyamine transport in prokaryotes and eukaryotes. Plant Physiol. Biochem. 48, 506–512 [DOI] [PubMed] [Google Scholar]
- 43. Liu Y., László C., Liu Y., Liu W., Chen X., Evans S. C., Wu S. (2010) Regulation of G1 arrest and apoptosis in hypoxia by PERK and GCN2-mediated eIF2α phosphorylation. Neoplasia 12, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rzymski T., Milani M., Pike L., Buffa F., Mellor H. R., Winchester L., Pires I., Hammond E., Ragoussis I., Harris A. L. (2010) Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 29, 4424–4435 [DOI] [PubMed] [Google Scholar]
- 45. Hyvönen M. T., Merentie M., Uimari A., Keinänen T. A., Jänne J., Alhonen L. (2007) Mechanisms of polyamine catabolism-induced acute pancreatitis. Biochem. Soc. Trans. 35, 326–330 [DOI] [PubMed] [Google Scholar]
- 46. Parkkinen J. J., Lammi M. J., Agren U., Tammi M., Keinänen T. A., Hyvönen T., Eloranta T. O. (1997) Polyamine-dependent alterations in the structure of microfilaments, Golgi apparatus, endoplasmic reticulum, and proteoglycan synthesis in BHK cells. J. Cell Biochem. 66, 165–174 [PubMed] [Google Scholar]
- 47. Lai E., Teodoro T., Volchuk A. (2007) Endoplasmic reticulum stress. Signaling the unfolded protein response. Physiology 22, 193–201 [DOI] [PubMed] [Google Scholar]
- 48. Adachi Y., Yamamoto K., Okada T., Yoshida H., Harada A., Mori K. (2008) ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct. Funct. 33, 75–89 [DOI] [PubMed] [Google Scholar]
- 49. Lee A. S. (2005) The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods 35, 373–381 [DOI] [PubMed] [Google Scholar]
- 50. Wang X. Z., Lawson B., Brewer J. W., Zinszner H., Sanjay A., Mi L. J., Boorstein R., Kreibich G., Hendershot L. M., Ron D. (1996) Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153). Mol. Cell.Biol. 16, 4273–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fasanaro P., Magenta A., Zaccagnini G., Cicchillitti L., Fucile S., Eusebi F., Biglioli P., Capogrossi M. C., Martelli F. (2006) Cyclin D1 degradation enhances endothelial cell survival upon oxidative stress. FASEB J. 20, 1242–1244 [DOI] [PubMed] [Google Scholar]
- 52. Raven J. F., Baltzis D., Wang S., Mounir Z., Papadakis A. I., Gao H. Q., Koromilas A. E. (2008) PKR and PKR-like endoplasmic reticulum kinase induce the proteasome-dependent degradation of cyclin D1 via a mechanism requiring eukaryotic initiation factor 2α phosphorylation. J. Biol. Chem. 283, 3097–3108 [DOI] [PubMed] [Google Scholar]
- 53. Kaneto H., Kajimoto Y., Fujitani Y., Matsuoka T., Sakamoto K., Matsuhisa M., Yamasaki Y., Hori M. (1999) Oxidative stress induces p21 expression in pancreatic islet cells: possible implication in beta-cell dysfunction. Diabetologia 42, 1093–1097 [DOI] [PubMed] [Google Scholar]
- 54. Leung-Pineda V., Pan Y., Chen H., Kilberg M. S. (2004) Induction of p21 and p27 expression by amino acid deprivation of HepG2 human hepatoma cells involves mRNA stabilization. Biochem. J. 379, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liebermann D. A., Hoffman B. (2008) Gadd45 in stress signaling. J. Mol. Signal. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li L., Li J., Rao J. N., Li M., Bass B. L., Wang J. Y. (1999) Inhibition of polyamine synthesis induces p53 gene expression but not apoptosis. Am. J. Physiol. 276, C946–C954 [DOI] [PubMed] [Google Scholar]
- 57. Ray R. M., Zimmerman B. J., McCormack S. A., Patel T. B., Johnson L. R. (1999) Polyamine depletion arrests cell cycle and induces inhibitors p21(Waf1/Cip1), p27(Kip1), and p53 in IEC-6 cells. Am. J. Physiol. 276, C684–C691 [DOI] [PubMed] [Google Scholar]
- 58. Bevilacqua E., Wang X., Majumder M., Gaccioli F., Yuan C. L., Wang C., Zhu X., Jordan L. E., Scheuner D., Kaufman R. J., Koromilas A. E., Snider M. D., Holcik M., Hatzoglou M. (2010) eIF2α phosphorylation tips the balance to apoptosis during osmotic stress. J. Biol. Chem. 285, 17098–17111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Song B., Scheuner D., Ron D., Pennathur S., Kaufman R. J. (2008) Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Invest. 118, 3378–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang X., Zou T., Rao J. N., Liu L., Xiao L., Wang P. Y., Cui Y. H., Gorospe M., Wang J. Y. (2009) Stabilization of XIAP mRNA through the RNA-binding protein HuR regulated by cellular polyamines. Nucleic Acids Res. 37, 7623–7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zou T., Rao J. N., Guo X., Liu L., Zhang H. M., Strauch E. D., Bass B. L., Wang J. Y. (2004) NF-κB-mediated IAP expression induces resistance of intestinal epithelial cells to apoptosis after polyamine depletion. Am. J. Physiol. Cell Physiol. 286, C1009–C1018 [DOI] [PubMed] [Google Scholar]
- 62. Hsieh W. C., Hsu P. C., Liao Y. F., Young S. T., Wang Z. W., Lin C. L., Tsay G. J., Lee H., Hung H. C., Liu G. Y. (2010) Overexpression of ornithine decarboxylase suppresses thapsigargin-induced apoptosis. Mol. Cells 30, 311–318 [DOI] [PubMed] [Google Scholar]
- 63. Thornton T. M., Rincon M. (2009) Nonclassical p38 MAPK functions. Cell cycle checkpoints and survival. Int. J. Biol. Sci. 5, 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nishikawa Y., Yamamoto Y., Kaji K., Mitsui H. (1980) Reversible G1 arrest of a human Burkitt lymphoma cell line(Raji) induced by tunicamycin. Biochem. Biophys. Res. Commun. 97, 1296–1303 [DOI] [PubMed] [Google Scholar]
- 65. Masamha C. P., Benbrook D. M. (2009) Cyclin D1 degradation is sufficient to induce G1 cell cycle arrest despite constitutive expression of cyclin E2 in ovarian cancer cells. Cancer Res. 69, 6565–6572 [DOI] [PubMed] [Google Scholar]
- 66. Morishima N., Nakanishi K., Takenouchi H., Shibata T., Yasuhiko Y. (2002) An endoplasmic reticulum stress-specific caspase cascade in apoptosis. Cytochrome c-independent activation of caspase-9 by caspase-12. J. Biol. Chem. 277, 34287–34294 [DOI] [PubMed] [Google Scholar]
- 67. Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. (2005) TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 24, 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Strasser A., Jost P. J., Nagata S. (2009) The many roles of FAS receptor signaling in the immune system. Immunity 30, 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yamaguchi H., Wang H. G. (2004) CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J. Biol. Chem. 279, 45495–45502 [DOI] [PubMed] [Google Scholar]
- 70. Kouroku Y., Fujita E., Tanida I., Ueno T., Isoai A., Kumagai H., Ogawa S., Kaufman R. J., Kominami E., Momoi T. (2007) ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 14, 230–239 [DOI] [PubMed] [Google Scholar]