Background: Transforming growth factor β (TGFβ) inhibits osteocalcin (Ocn) transcription and osteoblast differentiation.
Results: The inhibition of Ocn expression and osteoblast differentiation by TGFβ is blunted upon lack of Atf4 or vimentin knockdown.
Conclusion: ATF4 and vimentin are novel downstream targets of TGFβ in osteoblasts.
Significance: Understanding mechanisms by which transcription factors are regulated is crucial for developing effective anabolic drugs for bone.
Keywords: Cell Signaling, Cytoskeleton, Gene Regulation, Gene Transcription, Osteoblasts, ATF4, Cell Signaling, Osteoblast Differentiation, TGFβ, Vimentin
Abstract
ATF4 is an osteoblast-enriched transcription factor of the leucine zipper family. We recently identified that vimentin, a leucine zipper-containing intermediate filament protein, suppresses ATF4-dependent osteocalcin (Ocn) transcription and osteoblast differentiation. Here we show that TGFβ inhibits ATF4-dependent activation of Ocn by up-regulation of vimentin expression. Osteoblasts lacking Atf4 (Atf4−/−) were less sensitive than wild-type (WT) cells to the inhibition by TGFβ on alkaline phosphatase activity, Ocn transcription and mineralization. Importantly, the anabolic effect of a monoclonal antibody neutralizing active TGFβ ligands on bone in WT mice was blunted in Atf4−/− mice. These data establish that ATF4 is required for TGFβ-related suppression of Ocn transcription and osteoblast differentiation in vitro and in vivo. Interestingly, TGFβ did not directly regulate the expression of ATF4; instead, it enhanced the expression of vimentin, a negative regulator of ATF4, at the post-transcriptional level. Accordingly, knockdown of endogenous vimentin in 2T3 osteoblasts abolished the inhibition of Ocn transcription by TGFβ, confirming an indirect mechanism by which TGFβ acts through vimentin to suppress ATF4-dependent Ocn activation. Furthermore, inhibition of PI3K/Akt/mTOR signaling, but not canonical Smad signaling, downstream of TGFβ, blocked TGFβ-induced synthesis of vimentin, and inhibited ATF4-dependent Ocn transcription in osteoblasts. Thus, our study identifies that TGFβ stimulates vimentin production via PI3K-Akt-mTOR signaling, which leads to suppression of ATF4-dependent Ocn transcription and osteoblast differentiation.
Introduction
Transforming growth factor β (TGFβ)2 regulates many biological processes including patterning during development, cell proliferation, differentiation, apoptosis, and other physiological and pathological conditions such as wound healing, fibrosis, and cancer growth and metastasis. In the mammalian skeleton, TGFβ is one of the most abundant cytokines stored in bone matrix; and once activated from its latent form, it promotes osteoblast proliferation and mesenchymal stem cell recruitment to active bone remodeling sites (1–5), while inhibits osteoblast differentiation (6–8). The canonical Smad-dependent signaling has been identified as a mediator of TGFβ in skeletal cells as well as in many other cell types. In response to ligand binding, TGFβ receptors phosphorylate Smad2 and/or Smad3, which in turn bind to Smad4 to induce translocation into the nucleus (9), where the Smad complex either binds directly to DNA or indirectly to other transcription factors to regulate gene transcription (10).
It has become clear now that multiple Smad-independent or noncanonical signaling pathways are activated in response to TGFβ ligands in various cell types. These include: MAP kinase (MAPK) pathways in intestine, lung epithelial cells, and breast cancer cells; Rho-like GTPase signaling pathways during epithelial to mesenchymal transition (EMT) in epithelial cells and primary keratinocytes; and mammalian targets of rapamycin (mTOR) through phosphatidylinositol 3-kinase (PI3K) and Akt pathways during EMT in fibroblasts (11–18). Although each of these signaling pathways has been shown to play an important role in skeletal biology, whether TGFβ activates non-canonical signaling pathways in osteoblasts is unknown.
Activating transcription factor (ATF4) is a leucine zipper-containing transcription factor belonging to the CREB family and was originally identified as an osteoblast-specific transcription factor required for osteocalcin (Ocn) transcription and osteoblast differentiation (19). Ocn mRNA is exclusively expressed in differentiated osteoblasts hence it is often used as a marker gene of mature osteoblasts (20). Together with Runx2, the first reported osteoblast-specific transcription factor, ATF4 activates Ocn transcription in vitro and in vivo through direct binding to its cognate osteoblast-specific element 1 (OSE1) of the Ocn promoter (19, 21, 22). In osteoblasts, TGFβ targets Runx2 via a canonical Smad signaling pathway to achieve its inhibition of both Runx2 and Ocn transcription, thereby suppressing osteoblast differentiation (6). However, it is possible that TGFβ targets other effectors at the transcriptional level to inhibit Ocn transcription in osteoblasts.
We have recently identified that in osteoblasts vimentin binds directly to ATF4 through its first leucine-zipper domain, which prevents ATF4 from binding to its cognate DNA OSE1 on the Ocn promoter, leading to inhibition of ATF4-dependent Ocn transcription and osteoblast differentiation (23). Vimentin is a member of the intermediate filament protein family and the most widely accepted molecular marker of mesenchymal cells. Moreover, its mRNA is often up-regulated in response to TGFβ during EMT and cancer progression (24, 25). Consistent with its inhibitory role during osteoblast differentiation, vimentin expression is down-regulated when osteoblasts progress toward a fully differentiated stage (23). Since this suggested that one component of vimentin regulation involves the regulatory control of its expression, we searched for the extracellular ligands that govern its expression. In doing so, we noticed that TGFβ stimulated vimentin mRNA in C2C12 myoblastic cells (26), a cell type of mesenchymal origin that can differentiate into chondrocytes and osteoblasts (27). Given that TGFβ and vimentin both negatively regulate Ocn transcription and osteoblast differentiation, we hypothesized that TGFβ targets vimentin and ATF4 to suppress Ocn transcription and osteoblast differentiation.
Here we present evidence that TGFβ requires endogenous ATF4 to inhibit Ocn transactivation in primary osteoblasts and osteoblastic cell lines. With the delivery of a monoclonal anti-TGFβ antibody to mice, we show that ATF4 is also required for TGFβ to increase bone mass. Employing a series of molecular and biochemical approaches, we demonstrate that TGFβ directly up-regulates vimentin production at post-transcriptional level, via PI3K-Akt-mTOR signaling, but not Smad signaling, to achieve its inhibition of Ocn transcription. Therefore, our study identifies two novel effectors, vimentin and ATF4, that act downstream of TGFβ in the regulation of osteoblast differentiation via PI3K-Akt-mTOR signaling.
EXPERIMENTAL PROCEDURES
Materials
Tissue culture media and fetal bovine serum were purchased from Invitrogen. Anti-vimentin antibodies were from Santa Cruz Biotechnology and Biovision for V9 anti-rat vimentin and anti-mouse vimentin (#3634), respectively. Antibodies for ATF4 (C20) and Sp1 (PEP2) were from Santa Cruz Biotechnology, HA tag was from Abcam (ab9110); Flag tag was from Sigma (M2); and phospho-Smad2/3 was from Cell Signaling. Recombinant human (rh) TGFβ1 is from R&D systems. All chemicals were from Sigma unless indicated otherwise.
Cell Culture
ROS17/2.8 rat osteosarcoma cells were grown in DMEM/F-12 medium containing 10% FBS. Mouse osteoblastic 2T3 and MC3T3-E1 cell lines were cultured in α-Minimal Essential Medium (αMEM) containing 10% FBS. C2C12 myoblasts were grown in Dulbecco's modified Eagle's medium (DMEM) that contains 10% FBS and myoblastic DMEM or differentiation medium that contains 2% horse serum. COS1 monkey kidney cells were cultured in DMEM containing 10% FBS. All media were supplemented with 1% penicillin-streptomycin, and cells were passaged every 3 days.
Northern Blot Hybridization
Total RNA from indicated sources was isolated using TRIzol (Invitrogen) according to the manufacturer's protocols. Total RNA (5 μg) was resolved in 1% agarose gel and transferred onto nylon membranes. After crosslinking with UV light, the membranes were hybridized in 6× SSC buffer at 60 °C overnight with the following probes: partial cDNA of mouse vimentin from 792 to 1218, mouse Atf4 covering 287 nucleotides of 5′-untranslated region and 180 nucleotides of coding region, full-length mouse Ocn gene 2, and mouse Gapdh cDNAs.
Establishment of Permanent Reporter Cells
ROS17/2.8 cells were seeded at a density of 5 × 105 cells/well in 6-well culture dishes, and reporter construct (1 μg) of p6×OSE1-Luc, p6×mOSE1-Luc, or negative control p3xAP1-Luc was cotransfected with pcDNA3.1(+) (20 ng) at 50:1 ratio using Lipofectamine (Invitrogen) 18 h later. Cells were then allowed to grow to confluence. Neomycin (G418)-resistant colonies were then selected with G418 (300 μg/ml)-containing culture medium and pooled for experimental use.
Transient DNA Transfection and Luciferase Assay
COS1 cells were seeded at a density of 2.5 × 105/well in 6-well culture dishes for 20 h and then transfected with 2 μg/well of Flag-ATF4 or HA-vimentin expression plasmid. For reporter assays, cells were plated in 24-wells at a density of 2.5 × 104/well. p6×OSE1-Luc (0.25 μg/well) together with 0.05 μg/well of β-galactosidase (β-gal) were transfected into COS1 cells using Lipofectamine (Invitrogen). Cells were lysed 24 h post DNA transfection, and the luciferase and β-gal activity was measured using cell lysate. Fold activation or inhibition of luciferase activity was calculated by normalization of luciferase activity with β-gal activity. For vimentin knockdown, 2T3 osteoblastic cells were plated in 6-well culture dishes at 2.5 × 105/well and then transiently cotransfected with psiRNA (1 μg/well) empty vector or psiRNA-Vim with indicated reporter constructs (1 μg/well).
Primary Calvarial Osteoblast Isolation and Osteoblast Differentiation Assay
Calvariae were collected from neonatal (P3) pups, pressed on Science Brand Kimwipes to remove blood and surrounding tissues, cleaned with 1× PBS, and then subjected to series of collagenase digestions as described previously (28). Cells released from the first 2 digestions were discarded to enrich the numbers of osteoblastic cells. Cells released from the third digestion were plated in αMEM containing 10% FBS until confluence, which was defined as passage 0.
For differentiation assays, primary cells from passage 1 or osteoblastic cell lines were grown in 24-well plates in αMEM supplemented with 5 mm β-glycerophosphate and 100 μg/ml ascorbic acid for either 2 or 12 days for alkaline phosphatase staining or von Kossa staining, respectively, as previously described (29).
2G7 Treatment
Ten pairs of 10-week-old WT and Atf4−/− littermates were treated with anti-TGFβ monoclonal antibody 2G7 or control IgG at 10 mg/kg. The intraperitoneal injections of each antibody were administered 3 times per week for 4 weeks (30).
Micro-computed Tomography (μCT) Analysis
Mouse femurs were collected and fixed overnight in 4% PFA (pH 7.4) and then 70% ethanol. Trabecular bone of the metaphysis was evaluated using an ex vivo μCT imaging system (Scanco μCT40; Scanco Medical, Bassersdorf, Switzerland). Each femur was fit into a specimen tube and aligned with the scanning axis. Tomographic images were acquired at 55 kV and 145 mA with an isotropic voxel size of 12 μm, and at an integration time of 250 ms with 500 projections collected per 180 °C rotation. For 100 slices proximal to the growth plate, contours were fit to the inner layer of the cortical bone to define the total tissue volume (TV). Applying a Gaussian noise filter with a variance of 0.8 and support of 1 as well as a threshold of 300 mg of hydroxyapatite (HA) per cm3 to distinguish mineralized from non-mineralized tissue for each femur scan, we quantified the volume of trabecular bone tissue (BV) per TV (BV/TV) as well as volumetric density (in mgHA/cm3) of the bone-mineralized tissue (BMD) (31).
Western Blot Analysis
Total cell lysates from indicated cell types were isolated with radioimmunoprecipitation assay (RIPA) buffer containing 50 mm Tris-Cl, pH 7.6, 150 mm NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, and protease inhibitors (Complete ULTRA Protease Inhibitor Mixture Tablets, Roche Applied Science). Nuclear extracts were isolated using high-salt nuclear extraction buffer on hypotonic buffer-swelled cells as described (32). Total cellular proteins or nuclear extracts (50 μg per lane) were loaded onto 10% SDS-PAGE and Western blots were performed following standard protocols. Antibody concentrations were 0.5–1 μg/ml.
TGFβ and Inhibitor Treatment
ROS17/2.8 cells, primary rat or mouse calvarial osteoblasts, in 60-mm plates at 90% confluent, were starved in serum-free media overnight. rhTGFβ1 (0.2∼2 ng/ml) or vehicle (4 mm HCl containing 1 mg/ml BSA) was then added to the cell culture medium containing 1% FBS, and cells were incubated for an additional 5–6 h. For inhibitor treatment, MG115 (Piptide Institute Inc. Japan), SB505124 (Sigma), cycloheximide (Sigma), wortmannin (EMD4Biosciences), or rapamycin (EMD4 Biosciences) at indicated concentrations (0.1 nm to 5 nm) were pre-incubated with cells for 30 min prior to adding TGFβ.
Statistics
Data are presented as mean ± S.D. Statistical analyses were performed using Student's t test. All in vitro experiments were repeated at least three times.
RESULTS
ATF4 Is Required for TGFβ to Inhibit Osteoblast Differentiation
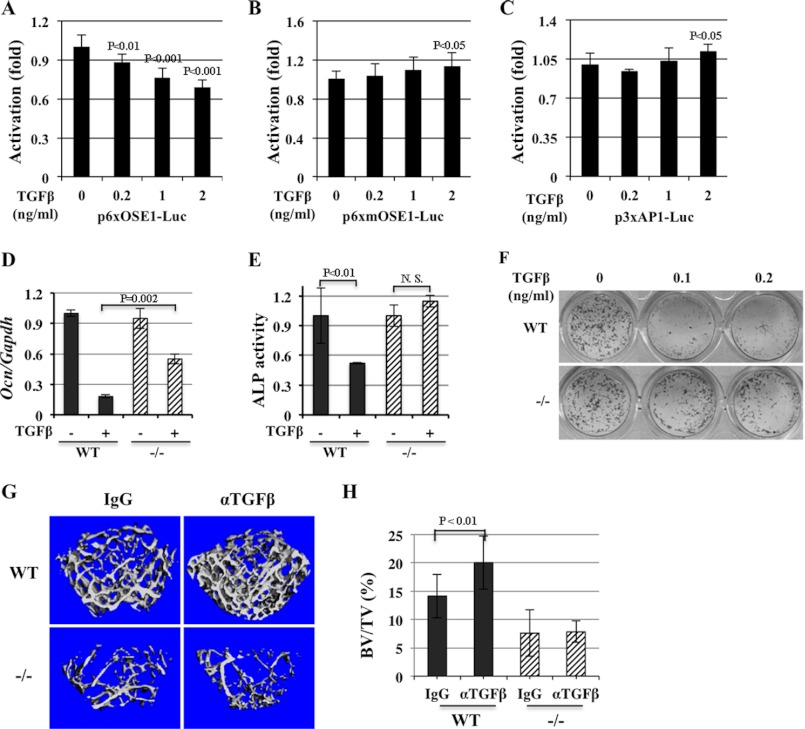
To investigate whether ATF4 was downstream of TGFβ, we first established stable ROS17/2.8 cell lines carrying a reporter luciferase gene driven by 6 repeats of wild type (WT) ATF4 binding element OSE1, p6xOSE1-Luc. As controls, we also made two other stable cell lines carrying a luciferase gene driven by either 6 repeats of a mutant ATF4 binding element, p6xmOSE1-Luc to which ATF4 fails to bind (21), or 3 repeats of the activating protein 1 (AP1) binding element, p3xAP1-Luc. Treatment of these reporter cell lines with rhTGFβ1 at various concentrations, ranging from 0.2 to 2 ng/ml, inhibited dose-dependently luciferase activity driven by 6xOSE1, but not by 6xmOSE1 (Fig. 1, A and B), indicating that the inhibitory effect of TGFβ on ATF4-dependent Ocn promoter activity relies on ATF4 binding to its cognate DNA. Furthermore, TGFβ inhibition is specific to the Ocn promoter and not a general phenomenon of leucine zipper-containing transcription factors since TGFβ failed to reduce luciferase activity in reporter cells driven by AP1 DNA binding element repeats (Fig. 1C).
FIGURE 1.
ATF4 is required for TGFβ to inhibit osteocalcin (Ocn) transcription. A–C, TGFβ inhibits ATF4-dependent reporter activity. Luciferase activity is decreased by treatment of rhTGFβ1 at indicated concentrations in stable ROS17/2.8 reporter cells. p6xOSE1-Luc, luciferase gene driven by 6 repeats of ATF4-binding elements reporter (A); p6xmOSE1-Luc, 6 repeats of mutant ATF4-binding element driving luciferase reporter (B); p3xAP1-Luc, 3 repeats of unrelated leucine-zipper protein AP1-binding site- driven reporter (C). D, inhibition of endogenous Ocn expression by TGFβ is attenuated in Atf4−/− calvarial osteoblasts. Quantitative RT-PCR (qRT-PCR) analysis of total RNA isolated from WT and Atf4−/− calvalrial osteoblasts treated with vehicle (−) or rhTGFβ1 (+, 0.5 ng/ml) in osteogenic medium for 10 days. Note that TGFβ decreased endogenous Ocn mRNA level in WT calvarial osteoblasts, which was attenuated (48% inhibition) in Atf4−/− mutant osteoblasts. E, TGFβ reduced alkaline phosphatase (ALP) activity in WT but not in Atf4−/− osteoblasts. ALP assay of WT and Atf4−/− calvarail osteoblasts treated with vehicle (−) or rhTGFβ (+, 0.5 ng/ml) in osteogenic medium for 2 days. F, TGFβ inhibits mineralized nodule formation in WT but not in Atf4−/− osteoblasts. von Kossa staining of calvarial osteoblasts treated with rhTGFβ1 (0.5 ng/ml) in osteogenic medium for 10 days. Note that TGFβ reduced the number of mineralized nodules (black colonies) dramatically (>90%) in the WT cultures but only slightly in Atf4−/− osteoblasts (<20%). G, ATF4 is required for the anabolic effect of anti-TGFβ antibody in vivo. μCT analysis of trabecular bones of WT and Atf4−/− femurs treated with control IgG antibody or anti-TGFβ monoclonal antibody (2G7, αTGFβ) neutralizing three forms of TGFβ ligand for 4 weeks. H, quantification of data shown in G. Note that 2G7 treatment increased trabecular bone volume (BV) versus total tissue volume (BV/TV) in WT femurs by 30% but failed to rescue the low BV/TV in Atf4−/− femur. n = 6.
To determine whether ATF4 was necessary for TGFβ to suppress endogenous Ocn expression and osteoblast differentiation, we utilized the Atf4-deficient (Atf4−/−) mouse model (33). Freshly isolated primary calvarial osteoblasts (passage 1) from wild type (WT) and Atf4−/− pups were treated with rhTGFβ1 (0.5 ng/ml) under osteogenic induction media for 10 days, and total RNA was collected for qRT-PCR analyses. Our data showed that TGFβ inhibited endogenous Ocn expression by 86% in WT calvarial osteoblasts but only by 43% in Atf4−/− calvarial osteoblasts (Fig. 1D). Consistent with this observation, TGFβ reduced alkaline phosphatase (ALP) activity in WT control cells by 46% but had no such effect in Atf4−/− calvarial cells (Fig. 1E). Lastly, low doses of TGFβ (0.1 and 0.2 ng/ml) decreased the number of mineralized nodule formation by 90% in WT but only 20% in Atf4−/− calvarial osteoblasts (Fig. 1F). Collectively, these data indicate that lack of endogenous ATF4 attenuates the potency of TGFβ to inhibit endogenous Ocn expression, ALP activity and differentiation or mineralization of osteoblasts in vitro.
To address whether ATF4 was also required for TGFβ in vivo and because treatment of WT mice with 1D11 an anti-TGFβ monoclonal antibody blocking all three isoforms of TGFβ (34), increased trabecular bone volume fraction in mice (30), we treated WT and Atf4−/− mice with 2G7, an monoclonal antibody with similar reactivity to the 1D11 (35). WT control and Atf4−/− mice were injected intraperitoneally with a TGFβ monoclonal antibody 2G7 for 4 weeks. Consistent with the results reported previously using 1D11 (30), μCT analysis revealed that 2G7 treatment increased bone volume fraction in WT mice by more than 30%, however, it could not improve the low bone mass in Atf4−/− mice (Fig. 1, G and H). Thus, these results confirmed that ATF4 is an important molecule downstream of TGFβ in vivo.
TGFβ Stimulates Vimentin Post-transcriptionally in Osteoblasts
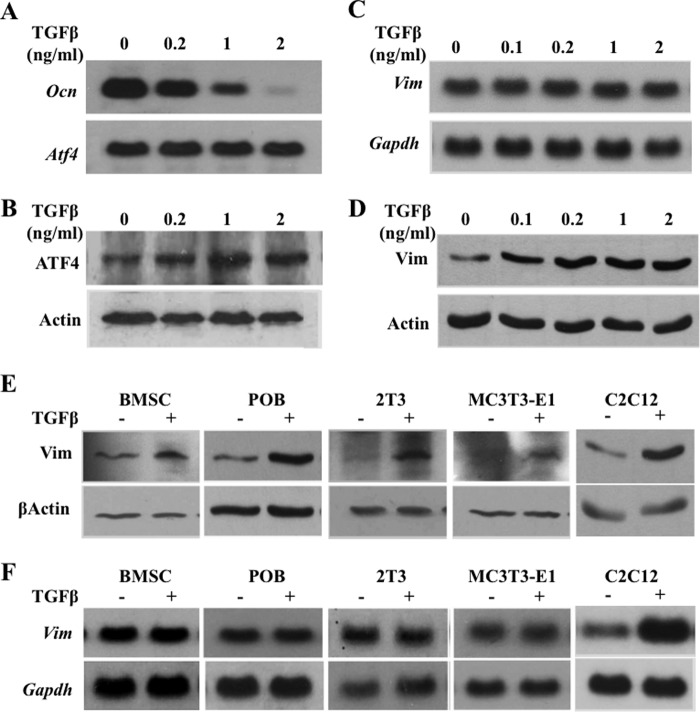
To determine whether TGFβ suppressed ATF4-dependent activation of Ocn and osteoblast differentiation (Fig. 1) via a direct effect on ATF4, we then analyzed whether TGFβ inhibited ATF4 expression. Primary calvarial osteoblasts (passage 1) were treated for 10 days with rhTGFβ1 at concentrations of 0.2, 1, or 2 ng/ml under osteogenic culture conditions, and then total RNA and protein were analyzed. As expected, endogenous Ocn expression decreased whereas ATF4 expression slightly increased, in response to TGFβ in a dose-dependent manner (Fig. 2, A and B). These results suggested that the suppression of ATF4 transcriptional activity by TGFβ is not due to a decrease in the level of ATF4 but may be via an indirect mechanism.
FIGURE 2.
TGFβ inhibits Ocn expression and stimulates vimentin expression post-transcriptionally. A, Northern blot analysis of total RNA from primary calvarial osteoblasts showing that TGFβ inhibits endogenous Ocn expression dose-dependently. B, Western blot analysis of total protein from primary calvarial osteoblasts. C, Northern blot analysis with indicated cDNA probes showing that TGFβ does not affect endogenous vimemtin mRNA level in calvarial osteoblasts. D, Western blot analysis showing that TGFβ stimulates vimenitn protein in calvarial osteoblasts. E, Western blot analyses showing that rhTGFβ1 (0.2 ng/ml) stimulates vimentin protein expression in the indicated primary osteoblasts and osteoblastic cell lines. F, Northern blot analyses showing that rhTGFβ1 (0.2 ng/ml) does not affect endogenous vimentin mRNA level in the indicated primary osteoblasts and osteoblastic cell lines.
One of the candidates was vimentin, because we recently found that it acted as a suppressor of ATF4 in osteoblasts (23) and others have shown that its expression was stimulated by TGFβ (26). To address whether vimentin was the mediator linking TGFβ signaling and ATF4, we first treated ROS 17/2.8 cells with TGFβ and found it could induce vimentin protein expression dose-dependently. Surprisingly, however, TGFβ did not increase vimentin mRNA expression at all of the concentrations tested (Fig. 2, C and D). These data indicated that in osteoblasts TGFβ regulates vimentin expression at the protein level and not at the mRNA level, which was different from what had been observed in C2C12 cells previously (26). To further verify our findings, we also treated a panel of osteoblasts along with C2C12 cells (as a positive control) with low dose rhTGFβ1 (0.2 ng/ml), which significantly suppressed WT primary osteoblast differentiation in vitro (Fig. 1F). The results indicated that TGFβ increased vimentin protein level but not its mRNA level in all tested cells, including mouse primary bone marrow stromal cells (BMSC), calvarial osteoblasts (POB), the 2T3 mouse preosteoblastic cells, and the MC3T3-E1 mouse committed osteoblasts. However and consistent with previous report (26), TGFβ in the C2C12 cells increased both protein and mRNA vimentin levels (Fig. 2, E and F). Thus, these data demonstrated that TGFβ up-regulates vimentin expression at the post-transcriptional level in osteoblasts.
Vimentin Is Required for TGFβ to Inhibit Ocn Expression
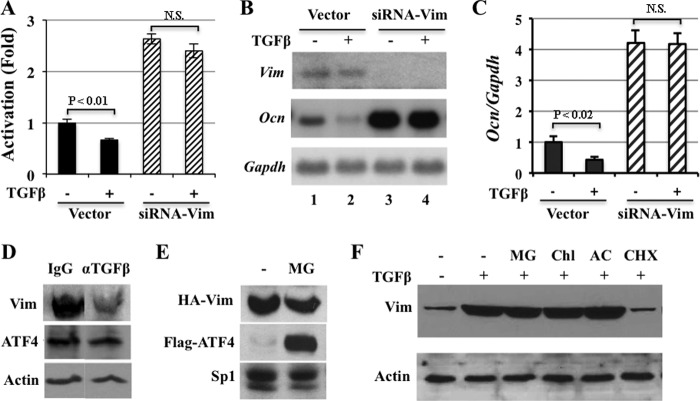
To understand whether vimentin played an essential role in mediating the inhibitory effect of TGFβ on ATF4 transcriptional activity and Ocn transcription, we knocked down endogenous vimentin expression in 2T3 mouse osteoblastic cells by transfection with siRNA-Vim (23) and the p6xOSE1-Luc as a readout for ATF4 transcriptional activity, and then tested their response to TGFβ by reporter assays. 2T3 mouse cells instead of primary osteoblasts were used to ensure the transfection efficiency. We also transfected 2T3 cells with siRNA empty vector to serve as negative control cells. Fig. 3A (lanes 1 and 2), rhTGFβ1 (0.5 ng/ml) decreased luciferase activity by 36%, which was expected and consistent with what we observed previously in ROS17/2.8 reporter cells (Fig. 1A). Luciferase activity increased 2.7-fold in 2T3 cells transfected with siRNA-Vim, due to the knockdown of endogenous vimentin that removed its suppression of ATF4 transcriptional activity (23). When TGFβ was added to these cells, ATF4 reporter activity remained 2.5-fold higher in cells containing siRNA-Vim than in cells containing siRNA control vector (Fig. 3A, lanes 3 and 4). The knockdown of endogenous vimentin was confirmed by the absence of vimentin mRNA in siRNA-Vim transfected cells, which could not be rescued by rhTGFβ1. Consistent with our previous finding (23) and transfection results shown in Fig. 3A, a low level of endogenous Ocn expression in 2T3 cells transfected with siRNA control vector was inhibited by rhTGFβ1 (0.5 ng/ml) treatment; whereas this inhibition was blunted in cells transfected with siRNA-Vim (Fig. 3B). qRT-PCR data indicated that TGFβ1 inhibited endogenous Ocn expression by 60% in cells transfected with empty siRNA vector. As expected, vimentin knockdown in 2T3 preosteoblasts by siRNA-Vim increased endogenous Ocn expression by 4-fold, which was not affected by TGFβ1 (Fig. 3C). Therefore, these data validated that endogenous vimentin is required for TGFβ to suppress ATF4-dependent Ocn transcription in vitro.
FIGURE 3.
Vimentin is required for the suppression of Ocn expression by TGFβ in osteoblasts. A, transient DNA transfection in 2T3 preosteoblastic cells demonstrating that rhTGFβ (0.5 ng/ml) decreased luciferase activity (lanes 1 and 2). Note that luciferase activity increased when siRNA-Vim was co-transfected (lanes 3 and 4). N.S., not statistically significant. B, Northern blot analysis of RNA from 2T3 osteoblastic cells transfected with siRNA vector or siRNA-Vim using indicated cDNA probes. Note that knockdown of endogenous vimentin by siRNA-Vim (top panel) blunted the inhibition of Ocn transcription by TGFβ (middle panel). C, qRT-PCR results confirming that vimentin knockdown by siRNA-Vim abolished the suppression of endogenous Ocn expression by rhTGFβ (0.5 ng/ml). D, Western blot analysis of long bone total protein extracts demonstrating that neutralizing the activity of TGFβ in vivo by anti-TGFβ decreases vimentin protein abundance. 10-week-old mice were treated with control antibody (IgG) or anti-TGFβ monoclonal antibody (αTGFβ) neutralizing three forms of TGFβ ligand for 4 weeks. E, proteasomal degradation inhibitor MG115 (MG) stabilizes ATF4 but not vimentin. Western blot analysis of nuclear extracts of COS1 cells transfected with expression plasmids of HA-Vim or Flag-ATF4 using anti-HA or anti-Flag antibodies. Sp1, loading control of nuclear extracts. F, protein synthesis is required for TGFβ to induce vimentin expression. ROS17/2.8 cells were pretreated with indicated inhibitors prior to the treatment of rhTGFβ1 (0.2 ng/ml). Note that TGFβ-induced vimentin is not affected by proteasomal inhibitor, MG115 (MG), or lysosomal inhibitors, chloroquine (ChlQ, 100 mm) and ammonium chloride (AC, 50 mm), but is diminished by protein translation inhibitor cycloheximide (CHX).
To determine whether vimentin protein level also fluctuated in vivo in response to TGFβ administration, we analyzed vimentin content in the long bones of 2G7 or control antibody-treated mice. Western blot showed that 2G7 treatment decreased the abundance of endogenous vimentin in long bone lysates compared with IgG control antibody treatment while ATF4 abundance remained constant in both control and 2G7 treated long bone samples (Fig. 3D). Thus, these results strongly suggested that TGFβ suppresses Ocn transcription and osteoblast differentiation via up-regulation of vimentin expression.
An increased protein level can be attributed to increased synthesis and/or decreased degradation. To determine which of these two possibilities contributed to the TGFβ-induced accumulation of vimentin protein in osteoblasts, we tested the effects of cycloheximide, a protein synthesis inhibitor, MG115, a proteasome inhibitor, on vimentin expression. We included Flag-ATF4 as a positive control, because it accumulates in many non-osteoblastic cell lines upon MG115 treatment (36). Analysis of nuclear extracts of COS1 cells transfected with HA-vimentin or Flag-ATF4 expression vector showed that treatment of MG115 (10 μg/ml) did not alter the abundance of HA-vimentin but did, as expected, increase Flag-ATF4 considerably (Fig. 3E). To address whether endogenous vimentin in osteoblasts behaved similarly to HA-vimentin in COS1 cells, we co-treated ROS17/2.8 cells with rhTGFβ1 (0.5 ng/ml), MG115 (10 μg/ml), or two lysosomal inhibitors chloroquine and ammonium chloride. As can be seen in Fig. 3F, the TGFβ-induced increase in vimentin expression was not affected by MG115, chloroquine (100 μm), or ammonium chloride (50 mm), but strongly decreased to basal level by cycloheximide (15 μg/ml). Collectively, these data indicated that protein neosynthesis is required for TGFβ to stimulate vimentin expression in osteoblasts.
PI3K-Akt-mTOR Signaling Is Required for TGFβ to Induce Vimentin Synthesis
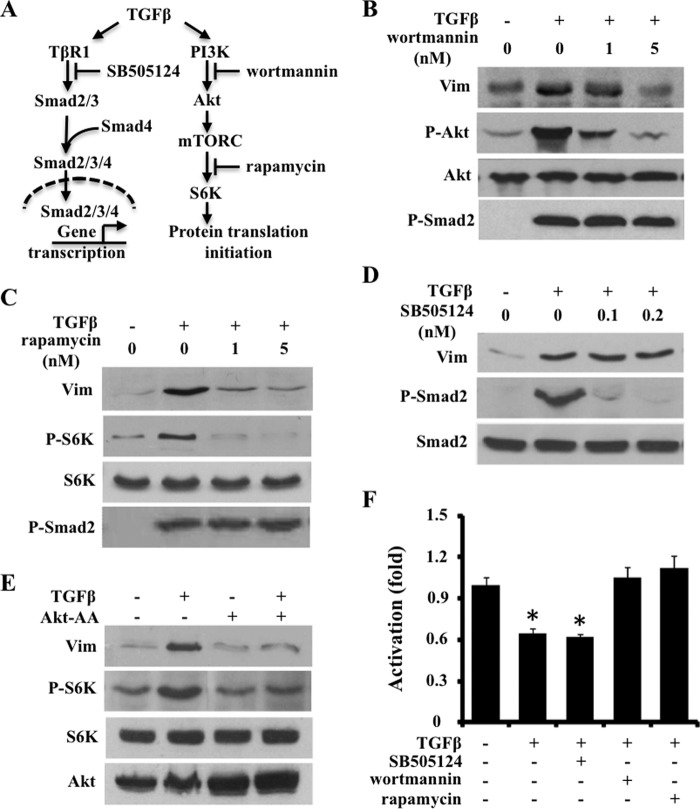
Because TGFβ has been shown to stimulate protein translation initiation via the PI3K-Akt-mTOR signaling pathway (Fig. 4A, adapted from Ref. 14), we then examined whether the PI3K-Akt-mTOR and/or the canonical Smad-signaling is responsible for TGFβ to induce vimentin in osteoblasts (Figs. 2 and 3). Wortmannin, an inhibitor of Akt phosphorylation by PI3K; rapamycin (1 and 5 nm), an inhibitor of S6K phosphorylation by mTOR (14, 37–39); and SB505124, an inhibitor of Smad2/3 phosphorylation downstream of the type I TGFβ receptor (40), were used to treat ROS17/2.8 cells. Western blot demonstrated that both wortmannin and rapamycin (1 and 5 nm) drastically reduced vimentin protein expression induced by TGFβ, which correlated with a strong decrease in Akt and S6K phosphorylation. SB505124 (0.1 and 0.2 nm) effectively decreased TGFβ-induced Smad2 phosphorylation but not TGFβ-induced vimentin expression (Fig. 2, B–D). As expected, wortmannin and rapamycin (1 and 5 nm) did not inhibit the TGFβ-induced Smad2 phosphorylation (Fig. 4, B--E), demonstrating that these inhibitors do not have an off-target effect at these concentrations. Moreover, overexpression of a dominant negative form of Akt, Akt-AA (38), blocked the increase in vimentin expression induced by TGFβ, which also correlated with an inhibition of S6K phosphorylation induced by TGFβ (Fig. 4E). Collectively, these data indicated that activation of PI3K-Akt-mTOR signaling, but not the canonical Smad signaling, is required for the stimulation of vimentin protein expression by TGFβ.
FIGURE 4.
TGFβ stimulates vimentin protein synthesis via PI3K-mTOR-Akt signaling but not Smad signaling. A, schematic presentation of inhibitors that block canonical and noncanonical signaling pathways downstream of TGFβ. B and C, Western blot of ROS17/2.8 cells indicating that wortmannin (B) and rapamycin (C) dose-dependently blunted rhTGFβ1 (0.2 ng/ml) induced vimentin and phosphorylation of downstream targets Akt or S6K. D, Western blot analysis of ROS17/2.8 cells showing that SB505124 did not inhibit rhTGFβ1 (0.2 ng/ml) to stimulate vimentin expression. Note that SB505124 effectively inhibited Smad2 phosphorylation. E, Western blot analysis showing that overexpression of dominant negative form of Akt (Akt-AA) inhibited rhTGFβ1 (0.2 ng/ml)-induced vimentin protein level in ROS17/2.8. F, ATF4-dependent activation of Ocn transcription requires PI3K-Akt-mTOR signaling. Luciferase activity in ROS17/2.8 reporter cells containing p6xOSE1-Luc was effectively (30%) inhibited by rhTGFβ (1 ng/ml) and SB505124 (0.4 μm), which was blunted by wortmannin (5 nm) or rapamycin (5 nm). *, p < 0.01.
To further elucidate the functional relevance of this non-canonical signaling pathway in controlling vimentin expression on ATF4-dependent activation of Ocn expression, we treated ROS17/2.8 reporter cells containing p6xOSE1-Luc with various kinase inhibitors. Consistent with their effects in abolishing the effect of TGFβ on vimentin production, wortmannin and rapamycin both blunted the inhibition of luciferase activity by TGFβ, whereas SB505124 did not affect it (Fig. 4F). Taken together, these results strengthened the finding that PI3K-Akt-mTOR signaling, but not Smad signaling, acts downstream of TGFβ to mediate the inhibition of Ocn transcription.
DISCUSSION
In this study, we show that TGFβ up-regulates post-transcriptionally vimentin expression in osteoblasts through a PI3K-Akt-mTOR non-canonical TGFβ pathway to inhibit osteoblast differentiation. Thus, this study defines vimentin and ATF4 as downstream mediators of TGFβ, providing two additional modulation points of this ancient and important pathway.
ATF4 and Vimentin as Downstream Targets of TGFβ in Osteoblasts
The finding of the monoclonal TGFβ-neutralizing antibody 1D11 to promote mechanical strength in young adult mice (30) motivated us to investigate the mechanism(s) whereby anti-TGFβ treatment promotes bone mass and fracture resistance. Here, we provided evidence that the inhibition of Ocn expression, ALP activity, and mineralized nodule formation by TGFβ was attenuated by 50% in Atf4−/− osteoblasts compared with the WT osteoblasts. Moreover, the anabolic effect of 2G7 in WT mice (a 30% increase in trabecular bone volume fraction) is completely blunted in Atf4−/− mice (Fig. 1). These data allow us to conclude that ATF4 is required for transmitting TGFβ signals in osteoblasts in both cultured primary cells and in vivo. Interestingly, TGFβ did not decrease ATF4 expression but increased vimentin protein abundance in osteoblasts and bones in vivo (Figs. 2 and 3), which in turn led to the suppression of ATF4 transcriptional activity. Furthermore, silencing endogenous vimentin by siRNA-Vim attenuated the effects of TGFβ (Fig. 3). Therefore, we conclude that both ATF4 and vimentin are downstream targets of TGFβ. Further investigations are necessary to confirm the in vivo roles that vimentin play in osteoblasts.
Studies using cell and transgenic mouse models have shown that vimentin promotes cell growth in Ras-transformed cells (41) but inhibits cell differentiation (42). Loss of vimentin (Vim−/−) in mice results in failures of vascular adaptation, which leads to pathological conditions such as reduced renal mass (43), glial cellmalformation (44), impaired wound healing (45), decreased arterial resistance to sheer stress (46), and disturbed leukocyte homing to lymph nodes (47). It will be of interest to investigate whether all or part of these defects are attributable to the essential role that vimentin plays in transmitting the signals of TGFβ, given the versatile and essential roles that TGFβ plays in many physiological and pathological conditions, including the ones affected in Vim−/− mice. Furthermore, vimentin is often strongly expressed in undifferentiated mesenchymal cells. Overexpressing vimentin in mesenchymes under its own promoter inhibited cell differentiation (48, 49). Thus, it is plausible to view TGFβ as one of the local growth factors that is required to maintain the “stemness” of mesenchymal stem cells in bone through its up-regulation of vimentin in bone. Thirdly, as an intermediate filament (IF) protein, vimentin also undergoes spatial reorganization in a variety of cell types, in response to stimulation with physiological signals, including TGFβ (data not shown). It will be important to understand the molecular basis whereby vimentin, a molecule that mostly resides in cytosol, travels into the nucleus to modulate the activity of tissue-specific transcription factors.
Analogous to the signaling axis of TGFβ-vimentin-ATF4 is that of TGFβ-Smad3-Runx2, which regulates Ocn transcription and osteoblast differentiation. In the latter pathway, upon TGFβ ligand stimulation, Smad3 is first phosphorylated by TGFβ receptors and then it binds to Smad4 and translocates into the nucleus, where the complex of Smad3/4 interacts with Runx2 (6), a master regulator of osteoblast differentiation (50–53). Subsequent studies demonstrated that the Smad3/4 complex also recruits histone deacetylases (HDAC) 4 and -5 to the Runx2-binding site of Ocn promoter to suppress Ocn transcription and osteoblast differentiation (54). It is currently unknown whether the vimentin-ATF4 complex can recruit additional repressors, such as HDAC family members, to the OSE1 binding site. More importantly, the relative contribution and potential interactions of TGFβ-vimentin-ATF4 and TGFβ-Smad3-Runx2 pathways remain to be studied.
The Role of PI3K-Akt-mTOR Signaling in Osteoblasts
It has previously been reported that specific inhibition of PI3K signaling with wortmannin stimulates human mesenchymal stem cells to differentiate into osteoblasts (55). Similarly, rapamycin also promotes human embryonic stem cells to differentiate into osteoblasts (56). A positive role of these kinase inhibitors of the PI3K-Akt-mTOR pathway in the regulation of osteoblast differentiation is substantiated by the evidence that both wortmannin and rapamycin blunted the inhibitory effect of TGFβ on ATF4-dependent Ocn transcription in ROS17/2.8 cells containing the p6xOSE1-Luc reporter (Fig. 4). At odds with these results, a low concentration (0.1 nm) of rapamycin has been shown to inhibit Ocn expression and osteoblast differentiation by suppressing the expression of Runx2, in primary mouse bone marrow stromal cells and MC3T3-E1 mouse osteoblastic cells (57). It is currently unknown whether these conflicting observations are a reflection of difference in cell lines, i.e. human versus mouse cell lines, or in drug concentrations. Furthermore, it needs to be further evaluated the in vivo relevance of the newly discovered TGFβ-PI3K-Akt-mTOR-vimentin-ATF4-Ocn axis. Regardless, our current findings represent a novel paradigm involving vimentin and ATF4 as downstream effectors of TGFβ signaling in the regulation of osteoblast differentiation.
This study does not exclude factors other than TGFβ acting as physiological upstream regulators of vimentin expression in osteoblasts. Indeed, parathyroid hormone (PTH), an important bone anabolic agent when used intermittently in vivo, has been reported to suppress the de novo biosynthesis of vimentin in human osteoblastic cells (58). Interestingly, PTH has also been shown to increase the expression and activity of ATF4 in osteoblasts (59). Thus, it is possible that PTH down-regulates vimentin expression in differentiating osteoblasts, which may counteract the action of TGFβ and lead to stimulation of ATF4's transcriptional activity and osteoblast differentiation. From this perspective, our study provides the first step toward to understanding the complexity of signaling cascades that control the anabolic action of TGFβ and PTH in bone. It is important to further determine whether vimentin is a convergent point that integrates both TGFβ and PTH signals to finely tune the differentiation process of osteoblasts.
TGFβ as a potent osteotropic factor has been extensively studied (60) and our understanding of how it regulates bone development and remodeling has continued to evolve (4, 5, 30, 61–63). It is encouraging that in vivo inhibition of TGFβ activity can stimulate bone formation (30). However, evaluating the long-term effects of TGFβ neutralizing antibodies on the skeleton will be important, since TGFβ promotes osteoblast proliferation and migration (63–65), two functions that are also required for bone formation. In addition, better understanding the molecular signaling pathways downstream of TGFβ should allow one to target selective molecules in osteoblast, thereby limiting off-target effects of general inhibition of TGFβ activity and the possible development of an immune response caused by the use of an antibody-based bone anabolic strategy.
Acknowledgments
We thank Dr. Florent Elefteriou and S. Kathryn Masood for critical reading of this manuscript.
This work was supported by the Vanderbilt University career developmental funds (to X. Y.) and the Dept. of Veterans Affairs, Biomedical Laboratory Research and Development (to J. S. N.).
- TGFβ
- transforming growth factor β
- EMT
- epithelial to mesenchymal transition
- ATF
- activating transcription factor
- mTOR
- mammalian targets of rapamycin
- Ocn
- osteocalcin; μCT, micro-computed tomography
- HA
- hydroxyapatite
- HDAC
- histone deacetylase
- PTH
- parathyroid hormone.
REFERENCES
- 1. Robey P. G., Young M. F., Flanders K. C., Roche N. S., Kondaiah P., Reddi A. H., Termine J. D., Sporn M. B., Roberts A. B. (1987) Osteoblasts synthesize and respond to transforming growth factor-type β (TGF-β) in vitro. J. Cell Biol. 105, 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lucas P. A. (1989) Chemotactic response of osteoblast-like cells to transforming growth factor β. Bone 10, 459–463 [DOI] [PubMed] [Google Scholar]
- 3. Pfeilschifter J., Wolf O., Naumann A., Minne H. W., Mundy G. R., Ziegler R. (1990) Chemotactic response of osteoblastlike cells to transforming growth factor β. J. Bone Miner. Res. 5, 825–830 [DOI] [PubMed] [Google Scholar]
- 4. Jian H., Shen X., Liu I., Semenov M., He X., Wang X. F. (2006) Smad3-dependent nuclear translocation of β-catenin is required for TGF-β1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev. 20, 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang Y., Wu X., Lei W., Pang L., Wan C., Shi Z., Zhao L., Nagy T. R., Peng X., Hu J., Feng X., Van Hul W., Wan M., Cao X. (2009) TGF-β1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat. Med. 15, 757–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alliston T., Choy L., Ducy P., Karsenty G., Derynck R. (2001) TGF-β-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 20, 2254–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Breen E. C., Ignotz R. A., McCabe L., Stein J. L., Stein G. S., Lian J. B. (1994) TGF β alters growth and differentiation related gene expression in proliferating osteoblasts in vitro, preventing development of the mature bone phenotype. J. Cell Physiol. 160, 323–335 [DOI] [PubMed] [Google Scholar]
- 8. Centrella M., Horowitz M. C., Wozney J. M., McCarthy T. L. (1994) Transforming growth factor-β gene family members and bone. Endocr. Rev. 15, 27–39 [DOI] [PubMed] [Google Scholar]
- 9. Abdollah S., Macías-Silva M., Tsukazaki T., Hayashi H., Attisano L., Wrana J. L. (1997) TβRI phosphorylation of Smad2 on Ser-465 and Ser-467 is required for Smad2-Smad4 complex formation and signaling. J. Biol. Chem. 272, 27678–27685 [DOI] [PubMed] [Google Scholar]
- 10. Massagué J. (1998) TGF-β signal transduction. Annu. Rev. Biochem. 67, 753–791 [DOI] [PubMed] [Google Scholar]
- 11. Bhowmick N. A., Ghiassi M., Bakin A., Aakre M., Lundquist C. A., Engel M. E., Arteaga C. L., Moses H. L. (2001) Transforming growth factor-β1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell 12, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edlund S., Landström M., Heldin C. H., Aspenström P. (2002) Transforming growth factor-β-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol. Biol. Cell 13, 902–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frey R. S., Mulder K. M. (1997) Involvement of extracellular signal-regulated kinase 2 and stress-activated protein kinase/Jun N-terminal kinase activation by transforming growth factor β in the negative growth control of breast cancer cells. Cancer Res. 57, 628–633 [PubMed] [Google Scholar]
- 14. Lamouille S., Derynck R. (2007) Cell size and invasion in TGF-β-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J. Cell Biol. 178, 437–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mulder K. M., Morris S. L. (1992) Activation of p21ras by transforming growth factor beta in epithelial cells. J. Biol. Chem. 267, 5029–5031 [PubMed] [Google Scholar]
- 16. Wilkes M. C., Mitchell H., Penheiter S. G., Doré J. J., Suzuki K., Edens M., Sharma D. K., Pagano R. E., Leof E. B. (2005) Transforming growth factor-β activation of phosphatidylinositol 3-kinase is independent of Smad2 and Smad3 and regulates fibroblast responses via p21-activated kinase-2. Cancer Res. 65, 10431–10440 [DOI] [PubMed] [Google Scholar]
- 17. Yan Z., Winawer S., Friedman E. (1994) Two different signal transduction pathways can be activated by transforming growth factor β1 in epithelial cells. J. Biol. Chem. 269, 13231–13237 [PubMed] [Google Scholar]
- 18. Zhang Y. E. (2009) Non-Smad pathways in TGF-β signaling. Cell Res. 19, 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H. C., Schinke T., Li L., Brancorsini S., Sassone-Corsi P., Townes T. M., Hanauer A., Karsenty G. (2004) ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell 117, 387–398 [DOI] [PubMed] [Google Scholar]
- 20. Hauschka P. V. (1986) Osteocalcin: the vitamin K-dependent Ca2+-binding protein of bone matrix. Haemostasis 16, 258–272 [DOI] [PubMed] [Google Scholar]
- 21. Ducy P., Karsenty G. (1995) Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell Biol. 15, 1858–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schinke T., Karsenty G. (1999) Characterization of Osf1, an osteoblast-specific transcription factor binding to a critical cis-acting element in the mouse Osteocalcin promoters. J. Biol. Chem. 274, 30182–30189 [DOI] [PubMed] [Google Scholar]
- 23. Lian N., Wang W., Li L., Elefteriou F., Yang X. (2009) Vimentin inhibits ATF4-mediated osteocalcin transcription and osteoblast differentiation. J. Biol. Chem. 284, 30518–30525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kokkinos M. I., Wafai R., Wong M. K., Newgreen D. F., Thompson E. W., Waltham M. (2007) Vimentin and epithelial-mesenchymal transition in human breast cancer–observations in vitro and in vivo. Cells Tissues Organs 185, 191–203 [DOI] [PubMed] [Google Scholar]
- 25. Steinert P. M., Roop D. R. (1988) Molecular and cellular biology of intermediate filaments. Annu. Rev. Biochem. 57, 593–625 [DOI] [PubMed] [Google Scholar]
- 26. Wu Y., Zhang X., Salmon M., Lin X., Zehner Z. E. (2007) TGFbeta1 regulation of vimentin gene expression during differentiation of the C2C12 skeletal myogenic cell line requires Smads, AP-1 and Sp1 family members. Biochim. Biophys. Acta 1773, 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katagiri T., Yamaguchi A., Komaki M., Abe E., Takahashi N., Ikeda T., Rosen V., Wozney J. M., Fujisawa-Sehara A., Suda T. (1994) Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell Biol. 127, 1755–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ecarot-Charrier B., Glorieux F. H., van der Rest M., Pereira G. (1983) Osteoblasts isolated from mouse calvaria initiate matrix mineralization in culture. J. Cell Biol. 96, 639–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang X., Ji X., Shi X., Cao X. (2000) Smad1 domains interacting with Hoxc-8 induce osteoblast differentiation. J. Biol. Chem. 275, 1065–1072 [DOI] [PubMed] [Google Scholar]
- 30. Edwards J. R., Nyman J. S., Lwin S. T., Moore M. M., Esparza J., O'Quinn E. C., Hart A. J., Biswas S., Patil C. A., Lonning S., Mahadevan-Jansen A., Mundy G. R. (2010) Inhibition of TGF-β signaling by 1D11 antibody treatment increases bone mass and quality in vivo. J. Bone Miner. Res. 25, 2419–2426 [DOI] [PubMed] [Google Scholar]
- 31. Wang W., Lian N., Li L., Moss H. E., Wang W., Perrien D. S., Elefteriou F., Yang X. (2009) Atf4 regulates chondrocyte proliferation and differentiation during endochondral ossification by activating Ihh transcription. Development 136, 4143–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989) Rapid detection of octamer binding proteins with 'mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Masuoka H. C., Townes T. M. (2002) Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood 99, 736–745 [DOI] [PubMed] [Google Scholar]
- 34. Dasch J. R., Pace D. R., Waegell W., Inenaga D., Ellingsworth L. (1989) Monoclonal antibodies recognizing transforming growth factor-β. Bioactivity neutralization and transforming growth factor β2 affinity purification. J. Immunol. 142, 1536–1541 [PubMed] [Google Scholar]
- 35. Pinkas J., Teicher B. A. (2006) TGF-β in cancer and as a therapeutic target. Biochem. Pharmacol. 72, 523–529 [DOI] [PubMed] [Google Scholar]
- 36. Yang X., Karsenty G. (2004) ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J. Biol. Chem. 279, 47109–47114 [DOI] [PubMed] [Google Scholar]
- 37. Arcaro A., Wymann M. P. (1993) Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem. J. 296, 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stokoe D., Stephens L. R., Copeland T., Gaffney P. R., Reese C. B., Painter G. F., Holmes A. B., McCormick F., Hawkins P. T. (1997) Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science 277, 567–570 [DOI] [PubMed] [Google Scholar]
- 39. Wullschleger S., Loewith R., Hall M. N. (2006) TOR signaling in growth and metabolism. Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 40. DaCosta Byfield S., Major C., Laping N. J., Roberts A. B. (2004) SB-505124 is a selective inhibitor of transforming growth factor-β type I receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 65, 744–752 [DOI] [PubMed] [Google Scholar]
- 41. Olson E. N., Capetanaki Y. G. (1989) Developmental regulation of intermediate filament and actin mRNAs during myogenesis is disrupted by oncogenic ras genes. Oncogene 4, 907–913 [PubMed] [Google Scholar]
- 42. Capetanaki Y., Smith S., Heath J. P. (1989) Overexpression of the vimentin gene in transgenic mice inhibits normal lens cell differentiation. J. Cell Biol. 109, 1653–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Terzi F., Henrion D., Colucci-Guyon E., Federici P., Babinet C., Levy B. I., Briand P., Friedlander G. (1997) Reduction of renal mass is lethal in mice lacking vimentin. Role of endothelin-nitric oxide imbalance. J. Clin. Invest. 100, 1520–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Colucci-Guyon E., Giménez Y. R. M., Maurice T., Babinet C., Privat A. (1999) Cerebellar defect and impaired motor coordination in mice lacking vimentin. Glia 25, 33–43 [PubMed] [Google Scholar]
- 45. Eckes B., Colucci-Guyon E., Smola H., Nodder S., Babinet C., Krieg T., Martin P. (2000) Impaired wound healing in embryonic and adult mice lacking vimentin. J. Cell Sci. 113, 2455–2462 [DOI] [PubMed] [Google Scholar]
- 46. Henrion D., Terzi F., Matrougui K., Duriez M., Boulanger C. M., Colucci-Guyon E., Babinet C., Briand P., Friedlander G., Poitevin P., Lévy B. I. (1997) Impaired flow-induced dilation in mesenteric resistance arteries from mice lacking vimentin. J. Clin. Invest. 100, 2909–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nieminen M., Henttinen T., Merinen M., Marttila-Ichihara F., Eriksson J. E., Jalkanen S. (2006) Vimentin function in lymphocyte adhesion and transcellular migration. Nat. Cell Biol. 8, 156–162 [DOI] [PubMed] [Google Scholar]
- 48. Capetanaki Y. G., Ngai J., Lazarides E. (1984) Characterization and regulation in the expression of a gene coding for the intermediate filament protein desmin. Proc. Natl. Acad. Sci. U.S.A. 81, 6909–6913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tapscott S. J., Bennett G. S., Toyama Y., Kleinbart F., Holtzer H. (1981) Intermediate filament proteins in the developing chick spinal cord. Dev. Biol. 86, 40–54 [DOI] [PubMed] [Google Scholar]
- 50. Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 51. Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 52. Mundlos S., Otto F., Mundlos C., Mulliken J. B., Aylsworth A. S., Albright S., Lindhout D., Cole W. G., Henn W., Knoll J. H., Owen M. J., Mertelsmann R., Zabel B. U., Olsen B. R. (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89, 773–779 [DOI] [PubMed] [Google Scholar]
- 53. Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., Owen M. J. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89, 765–771 [DOI] [PubMed] [Google Scholar]
- 54. Kang J. S., Alliston T., Delston R., Derynck R. (2005) Repression of Runx2 function by TGF-β through recruitment of class II histone deacetylases by Smad3. EMBO J. 24, 2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kratchmarova I., Blagoev B., Haack-Sorensen M., Kassem M., Mann M. (2005) Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science 308, 1472–1477 [DOI] [PubMed] [Google Scholar]
- 56. Lee K. W., Yook J. Y., Son M. Y., Kim M. J., Koo D. B., Han Y. M., Cho Y. S. (2010) Rapamycin promotes the osteoblastic differentiation of human embryonic stem cells by blocking the mTOR, pathway and stimulating the BMP/Smad pathway. Stem Cells Dev. 19, 557–568 [DOI] [PubMed] [Google Scholar]
- 57. Singha U. K., Jiang Y., Yu S., Luo M., Lu Y., Zhang J., Xiao G. (2008) Rapamycin inhibits osteoblast proliferation and differentiation in MC3T3-E1 cells and primary mouse bone marrow stromal cells. J. Cell Biochem. 103, 434–446 [DOI] [PubMed] [Google Scholar]
- 58. Lomri A., Marie P. J. (1990) Changes in cytoskeletal proteins in response to parathyroid hormone and 1,25-dihydroxyvitamin D in human osteoblastic cells. Bone Miner. 10, 1–12 [DOI] [PubMed] [Google Scholar]
- 59. Yu S., Franceschi R. T., Luo M., Fan J., Jiang D., Cao H., Kwon T. G., Lai Y., Zhang J., Patrene K., Hankenson K., Roodman G. D., Xiao G. (2009) Critical role of activating transcription factor 4 in the anabolic actions of parathyroid hormone in bone. PLoS One 4, e7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mundy G. R. (1991) The effects of TGF-β on bone. Ciba Found Symp. 157, 137–143; discussion 143–151 [PubMed] [Google Scholar]
- 61. Bae S. C., Lee K. S., Zhang Y. W., Ito Y. (2001) Intimate relationship between TGF beta/BMP signaling and runt domain transcription factor, PEBP2/CBF. J. Bone Joint Surgery 83, S48–S55 [PubMed] [Google Scholar]
- 62. Maeda S., Hayashi M., Komiya S., Imamura T., Miyazono K. (2004) Endogenous TGF-β signaling suppresses maturation of osteoblastic mesenchymal cells. EMBO J. 23, 552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Qiu T., Wu X., Zhang F., Clemens T. L., Wan M., Cao X. (2010) TGF-β type II receptor phosphorylates PTH receptor to integrate bone remodeling signaling. Nat. Cell Biol. 12, 224–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Erlebacher A., Derynck R. (1996) Increased expression of TGF-β2 in osteoblasts results in an osteoporosis-like phenotype. J. Cell Biol. 132, 195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Erlebacher A., Filvaroff E. H., Ye J. Q., Derynck R. (1998) Osteoblastic responses to TGF-β during bone remodeling. Mol. Biol. Cell 9, 1903–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]