Background: Spiders extrude adhesive glues to form connection joints that mediate web construction and prey wrapping.
Results: DNA microarray analysis and mass spectrometry reveal new protein glue constituents that comprise connection joints.
Conclusion: Spider glue proteins represent a diverse group of polypeptides with distinct molecular architectures.
Significance: Learning how spider glues mediate the fusion of fibers is crucial for understanding adhesion mechanisms in biology.
Keywords: Mass Spectrometry (MS), Molecular Cell Biology, Protein Self-assembly, Spectroscopy, Structural Biology
Abstract
Adhesive spider glues are required to perform a variety of tasks, including web construction, prey capture, and locomotion. To date, little is known regarding the molecular and structural features of spider glue proteins, in particular bioadhesives that interconnect dragline or scaffolding silks during three-dimensional web construction. Here we use biochemical and structural approaches to identify and characterize two aggregate gland specific gene products, AgSF1 and AgSF2, and demonstrate that these proteins co-localize to the connection joints of both webs and wrapping silks spun from the black widow spider, Latrodectus hesperus. Protein architectures are markedly divergent between AgSF1 and AgSF2, as well as traditional spider silk fibroin family members, suggesting connection joints consist of a complex proteinaceous network. AgSF2 represents a nonglycosylated 40-kDa protein that has novel internal amino acid block repeats with the consensus sequence NVNVN embedded in a glycine-rich matrix. Analysis of the amino acid sequence of AgSF1 reveals pentameric QPGSG iterations that are similar to conserved modular elements within mammalian elastin, a rubber-like elastomeric protein that interfaces with collagen. Wet-spinning methodology using purified recombinant proteins show AgSF1 has the potential to self-assemble into fibers. X-ray fiber diffraction studies performed on these synthetic fibers reveal the presence of noncrystalline domains that resemble classical rubber networks. Collectively, these data support that the aggregate gland serves to extrude a protein mixture that contains substances that allow for the self-assembly of fiber-like structures that interface with dragline silks to mediate prey capture.
Introduction
Spider silk is a lightweight material that has a combination of high-tensile strength and extensibility. Anatomical studies of spider abdominal glands have revealed the presence of seven distinct silk and/or glue-silk producing glands, which include the major and minor ampullate, tubuliform, flagelliform, aciniform, pyriform, and aggregate gland (1). The silk producing glands have been hypothesized to serve as specialized biofactories that produce proteins spun into different silk types with distinct mechanical properties (2, 3). These structural proteins share common protein architectures, including the presence of internal block repeats, and nonrepetitive conserved N and C termini (4–6). Six of the seven silk-producing glands have been shown to produce fibers with the exception of the aggregate gland (7–13). Due to the morphology of the aggregate gland, its spinning duct, and the anatomical features of its spigots, it has been debated whether this gland synthesizes structural proteins that self-assemble into fibers or whether it exclusively secretes nonstructural adhesive proteins.
Chemical analysis of web constituents cast by orb-weavers has demonstrated that the aggregate gland manufactures a complex aqueous solution containing water-soluble compounds related to neurotransmitters, inorganic salts, small peptides, and glycoproteins (14, 15). These studies have shown that ecribellate spider glue consists of microscopic nodules made of glycoproteins (15). Protein sequences for two of the glycoproteins reveal these products are expressed from opposite strands of an identical DNA sequence (16). Thus, at least for orb-weavers, spider glue glycoproteins have been proposed to function as adhesive substances that facilitate prey capture; however, they have not been shown to have intrinsic properties for self-assembly into structures that resemble fibers. Because spider glues and cements are used in conjunction with dragline silk (scaffolding threads), we hypothesized that the expression of glue proteins could be subject to coordinate regulation or energetically linked to the synthesis of the MaSp1 and MaSp2 dragline silk proteins via inter-silk-gland communication. To test this hypothesis, we coupled DNA microarray analysis, mass spectrometry, and other biochemical approaches to investigate the presence of new materials that interface with dragline silk fibers to perform essential biological tasks. For this, dragline silk was collected by allowing the spiders to free fall and changes in gene expression profiles in the silk-producing glands were monitored.
In this study, we describe the identification and characterization of two gene products aggregate gland silk factors 1 and 2 (AgSF1 and AgSF2).4 AgSF1 and AgSF2 are expressed in high quantities in the aggregate gland and extruded as adhesive materials that function to interconnect scaffolding fibers in the three-dimensional architecture of the web as well as wrapping silk for prey immobilization. We also provide evidence that recombinant AgSF1 protein chains can self-assemble into synthetic fibers. Structural studies of these fibers reveal a material that is predominantly noncrystalline.
EXPERIMENTAL PROCEDURES
DNA Microarray Analysis
A cDNA library was prepared as previously described (17). After plating the cDNA library, over 3300 individual plaques were removed and placed into SM (100 mm NaCl, 8 mm MgSO4, 50 mm Tris-Cl) buffer to facilitate diffusion. The diffused recombinant viruses were used as templates for PCR. cDNAs were amplified using the universal forward and reverse primers with the sequences 5′-AGG GAT GTT TAA TAC CAC TAC-3′ and 5′-GAT CAG AGG TTA CAT GGC CAA G-3′, respectively. After confirming amplification of cDNAs using agarose gel electrophoresis, the samples were desalted and then resuspended in 3× SSC (0.9 m NaCl and 0.09 m sodium citrate, pH 7.2) and printed onto Corning UltraGAPSTM-coated slides using an OmniGrid Accent microarrayer (GeneMachines). Three different spiders were silked once every 24 h over a 5-day period. The quantity of dragline silk collected each day ranged from 150 to 200 cm. Control spiders were not silked but housed under identical conditions. Following silking, both groups were dissected and the tubuliform, aggregate, and major and minor ampullate glands were removed and used for RNA isolation. Total RNA was isolated using TRIzol® (Invitrogen). mRNAs were labeled using the CyScribe First-strand cDNA Labeling Kit with Cy3 and Cy5 dyes and hybridized to the printed spider cDNA chips according to the manufacturer's instructions (GE Healthcare). Hybridization was performed using a Hyb4 automated system (Digilab, Inc.). Data were obtained by scanning the slides using the GenePix® Personal 4100A Microarray scanner (Axon Instruments, Inc.). Genes that displayed changes in mRNA expression profiles were further analyzed and subject to sequencing at the core facility of the University of Florida.
DNA Constructs
For expression studies, the AgSF1 cDNA was amplified by PCR using the forward and reverse primers 5′-GAA TTC GGC TAC AAA AAA ACC GTT GGA AAA GAT GGA CAA ATT-3′ and 5′-GTC GAC CGG TTG ACC ACT TCC TGG TTG CCC GTT TCC-3′, respectively. EcoRI and SalI restriction sites were engineered on the 5′-termini of the forward and reverse primers to facilitate subcloning of the cDNAs. AgSF1 cDNA fragments were amplified using PCR with a cDNA library prepared from the silk-producing glands of the black widow spider, Latrodectus hesperus. Following amplification, two cDNAs, one ∼600 bp and another 1000 bp, were extracted and ligated into the pBAD TOPO ThioFusion expression vector (Invitrogen). The 600-bp cDNA fragment coded for two copies of the 64-amino acid repeat, which was denoted AgSF1G. The 1000-bp fragment coded for two copies of the 64-amino acid repeat and 3.5 copies of the 36-amino acid ensemble repeat, which was dubbed AgSF1G+XPGXG. Both cDNAs were subject to DNA sequencing to confirm the accuracy of the sequences (University of Florida). For structural studies, the cDNAs were excised from the pBAD TOPO ThioFusion vectors using the restriction enzymes EcoRI and SalI and ligated into pET-19b-Sumo.
The full-length AgSF2 cDNA was amplified using PCR with the forward and reverse primers: 5′-GAA TTC CGA CCT GAA CCT GGT GTC CAT CAT G-3′ and 5′-GTC GAC TTA ACC TTT GAA GAG ACC CTT TTT AAC ATC-3′, respectively. The forward primer was designed to remove the first 24 amino acid residues, which corresponded to the predicted cleavage site for signal peptidase. Following amplification, the cDNA was inserted into the pBAD-TOPO-TA expression vector according to the manufacturer's instructions.
Real-time PCR Analysis
Reverse transcription reactions were used for real-time PCR analysis using the DyNAmo SYBR Green qPCR kit as previously described (18). Real-time PCR fluorescence detection was monitored using an Opticon II instrument (MJ Research Inc.). Amplification products were monitored by SYBR Green detection and checked using dissociation curve software and agarose gel electrophoresis. Oligonucleotides used for the agsf1 expression analysis included the forward and reverse primers: 5′-GGA AGT GGT ACC CAA ACT TTG CTA GGG-3′ and 5′-TTA TGA ACT GTT GCT ATC TAG CAA ACC-3′, respectively. AgSF2 primers were the forward and reverse primers 5′-GGA GGA GGA CTT CCT AAT GTT-3′ and 5′-TAA ACC GCC TCC AAC TCC TCC-3′, respectively. Primers used for normalization were the forward and reverse β-actin oligonucleotides: 5′-CCC TGA GAG GAA GTA CTC CGT-3′ and 5′-ATC CAC ATC TGC TGG AAG GTG-3′, respectively.
Development of Polyclonal Antibodies
Anti-AgF1 polyclonal antibodies were generated using the synthetic peptide GYKKTVGKDGQIVC (EZBiolab Inc., Carmel, IN). The anti-AgSF2 polyclonal antiserum was generated against two synthetic peptides with the sequences IQSRDPGVHHVRYC and SRGDTVYTYRTDYTC (Sigma Genosys Custom Peptide Antisera Services). Both peptides reside in the N terminus of AgSF2. To facilitate conjugation to the carrier protein keyhole limpet hemocyanin, cysteine residues were added to the C termini. For AgSF2 antibody production, both peptides were used for co-immunizations in the rabbits.
Protein Expression and Purification
Recombinant proteins were produced using 1 liter of LB. For induction experiments, each clone was grown overnight until saturation. Saturated cultures were then combined with 4 liters of fresh LB, supplemented with 0.02% arabinose or 1 mm isopropyl 1-thio-β-d-galactopyranoside for induction purposes, and grown an additional 4 h. Following induction, the bacterial cells were pelleted at 6,000 × g and then lysed in 13.3 ml of FastBreakTM Cell Lysis Reagent (Promega) per 1.0 liter of culture. To promote further lysis, the solution was sonicated at maximum setting for 1 min using a VirSonic 60 instrument (VirTis). After sonication, samples were clarified by spinning for 5 min at 8,000 × g. Supernatants were combined with 1 ml of nickel-nitrilotriacetic acid-agarose resin (50% slurry; Qiagen) and gently rocked at room temperature for 1 h to facilitate binding of the fusion proteins via their His6 tag. The resin was washed using a buffer (50 mm NaH2PO4, 300 mm NaCl) supplemented with 20 mm imidazole, followed by elution of the fusion proteins with 20 ml of same buffer containing 250 mm imidazole. Protein samples were dialyzed against water for 2–3 days and then subject to freeze-drying using a FreeZone12Plus instrument (Labconco). Typical yields for truncated AgSF1 recombinant proteins after purification using 10 liters of culture were ∼50–100 mg, respectively.
Collection and Solubilization of Silk Fibers
Spiders were housed and silk-producing glands were removed as previously described (19). For Western blot studies, silk samples were dissolved in 8 m urea, incubated at 55 °C for 2 h, and then heated at 95 °C for 5 min before SDS-PAGE analysis. Whole cell lysates were generated by solubilizing the silk-producing glands in 8 m urea and treated in a similar fashion as the fibers, with the exception that the samples were briefly sonicated prior to electrophoresis.
Mass Spectrometric Analysis
In-solution tryptic digests of the solubilized wrapping silk as well as other fiber types were performed as previously described (9, 10). The MS map and peptide MS/MS spectra were acquired with a MALDI/TOF/TOF mass spectrometer (4700 Proteomics Analyzer, Applied Biosystems, Foster City, CA) operated in reflector mode or MS/MS mode, respectively. De novo peptide sequences were obtained by manual interpretation of the high energy CAD spectra.
Scanning Electron Microscopy
Connecting junctions and wrapping silk fibers were imaged using a Hitachi S-2600 SE instrument operated with an accelerator voltage of 3 kV. Wrapping silk was collected as previously described (10). Prior to imaging, the samples were coated to a thickness of ∼14–20 nm with gold alloy using a Pelco SC-7 autosputter coater with an FTM-2 film thickness.
Atomic Force Microscopy
Spider silk junctions were dissected and mounted on a custom frame that stabilized the primary dragline fibers while preserving the junctions intact and in the view of interest. An atomic force microscope was used for examining the surface topography (Asylum Research Cypher AFM). We used silicon cantilevers with force constants of 1.9–2.3 newton/m. Images were recorded with typical scan rates of 1.0–2.0 Hz. An optical view was first used to localize the junction, and the scanning probe was operated in AC mode, enabling us to obtain both morphological data from the z-deflection, as well as time-dependent material behavior from the phase-contrast data.
X-ray Fiber Diffraction (XRFD)
X-ray fiber diffraction was performed on the BioCARS 14BM-C beamline at the Advanced Photon Source at Argonne National Laboratory, Argonne, IL. The wavelength of the x-ray beam was 0.9857 Å with a flux of 6 × 1011 photons/s and the beam size on the sample was 150 × 200 μm. Data were recorded using an ADSC Quantum-315 detector. Single fibers spun from AgSF1G+XPGXG (as spun and 3.5x post-spun fibers with average thickness ∼100 m and ∼40 μm, respectively) were held taut in a metal frame, with the fiber axis normal to the x-ray beam. The sample to detector distance was 300 mm. Data collection times were 120 s for one image. Background measurements were performed with the sample displaced from the beam and the images were recorded under the same conditions as with the samples in the beam. Multiple images were taken to get better statistics and improve on the signal/background ratio. CeO2 powder was used for instrument calibration. The two-dimensional XRFD patterns were analyzed using the software package FIT2D.5 Radial and azimuthal one-dimensional profiles were obtained from the deconvolution of two-dimensional XRFD images using FIT2D. Radial profiles are intensity as a function of radius integrated azimuthally for the whole two-dimensional pattern, whereas azimuthal profiles are integrated intensity as a function of all azimuthal angles over a thin annular ring centered at the peak maximum of the desired diffraction ring (21–23). Microcal Origin was used for the analysis of the deconvoluted one-dimensional x-ray data.
RESULTS
DNA Microarray Analysis Reveals Novel Gene Products That Are Linked to Dragline Silk Production
To identify genes that encode proteins that have functional ties with dragline silk constituents, we generated spider DNA chips for microarray analysis. Using a cDNA library prepared from the silk-producing glands of the black widow spider, along with a universal set of primers that flanked the region of cDNA insertion, we amplified over 3,300 cDNAs from individual plaques carrying recombinant viral chromosomes. Spider cDNAs were printed onto glass slides as unknown features for DNA microarray analysis. Two distinct treatments were used to collect total RNA from spiders for the DNA microarray analysis. One group of spiders, the “free fall-silked group,” was used to collect dragline silk. The other group, which represented the nonsilked or control group, was maintained under identical conditions except no dragline silk was removed from these individuals. Because the major ampullate gland is known to produce MaSp1 and MaSp2, we hypothesized that dragline silking would influence the expression pattern of fibroins and other silk components in different silk-producing glands because coordinate regulation between the glands has been proposed (1). This strategy proved successful, as ∼44 of the printed genes were observed to be down-regulated by as much as 5-fold following the silking procedure (Table 1). These cDNAs were selected for sequencing and their identities determined using the computer algorithm BLASTn to search the sequences against the NCBInr database (Table 1). Several gene products that have been demonstrated by MS/MS studies to be constituents of silks, which include the tubuliform fibroins TuSp1 (7), ECP-1 (18), and ECP-2 (24), as well as gene products previously implicated in the silk production pathway or spiral glue droplets, such as a peroxidase, arginine kinase (25), and aggregate spider glue factor (16), respectively, were identified during our DNA microarray analysis.
TABLE 1.
Genes that decrease after dragline silking spiders for 5 days
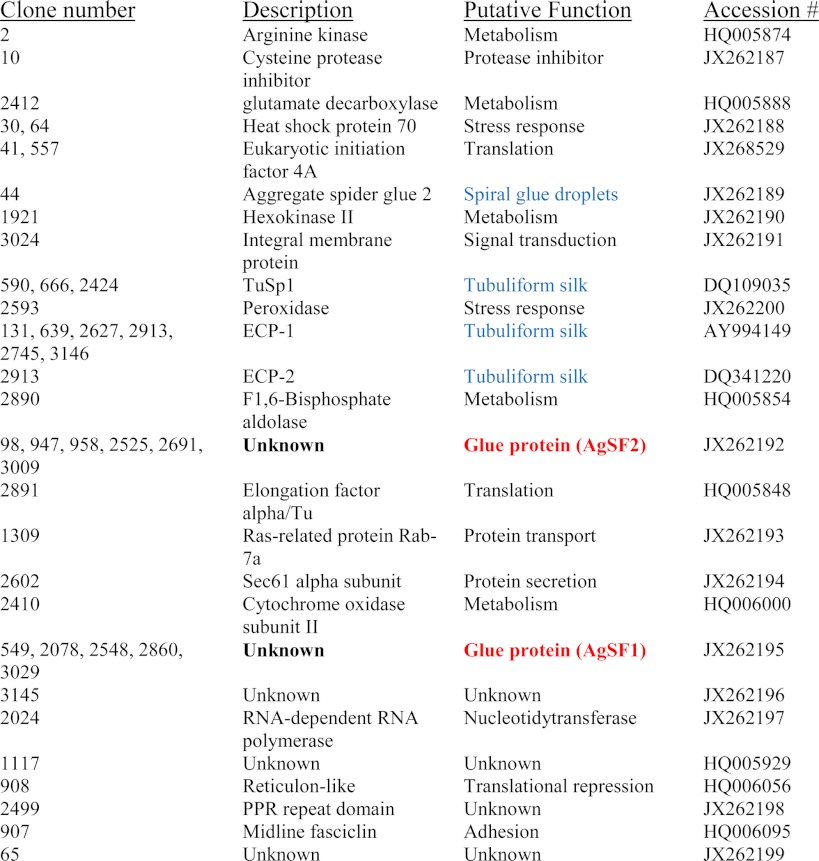
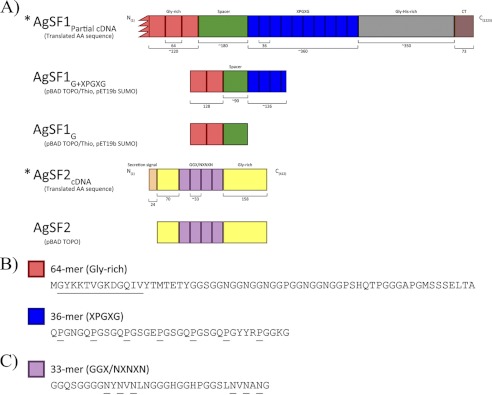
Two cDNAs were found multiple times on the DNA microarray; these corresponded to clones 549, 2078, 2548, 2860, and 3019 as well as clones 98, 947, 958, 2525, 2691, and 3009. Because these clones were identified five independent times and their sequences showed no significant similarities to sequences deposited in the nrNCBI protein database, we decided to further characterize their molecular properties and function. To obtain full-length cDNAs, we screened a cDNA library prepared from silk-producing glands using conventional nucleic acid-nucleic acid hybridization assays (data not shown). From this screen, we isolated longer cDNAs that contained regions identical to clones 549, 2078, 2548, 2860, and 3019. The longest cDNA that was retrieved represented a partial sequence that was ∼4 kb (GenBank accession JX262195); it contained a long opening reading frame encoding a putative protein that contained 1213 amino acids with a predicted molecular mass of ∼112 kDa and pI = 6.89. Protein-protein BLAST searches of the NCBInr database with the translated cDNA sequence revealed the strongest similarity to a predicted protein from the monoarch butterfly (GenBank accession EHJ72326.1), with 44% identities over 475 residues. Inspection of the spider protein C terminus revealed the presence of a glycine- and histidine-rich region (Fig. 1A). Two different internal block repeats were present within the translated cDNA. These blocks consisted of 36- or 64-amino acid segments (Fig. 1, A and B). The 36-amino acid segment was iterated 11 times. Manual inspection of each 36-amino acid block repeat revealed 5 tandem copies of X1PGX2G (X1 = Gln, Glu, or Arg; X2 = Ser or Asn), coupled to single copies of the QPGYY and RPGGKG submotifs. A striking feature was the regular spacing profile on the proline residues, which showed a conserved spacing pattern of one in every 5 residues (Fig. 1B). The X1PGX2G tandem repeats resemble pentameric modules reported in elastin (26). The residues found in the highest level included glycine, serine, proline, and glutamine at 38.5, 11.8, 11.5, and 7.2%, respectively. The second block repeat, which comprised 64 amino acids, was iterated 3 times with a fourth segment being truncated on the N terminus; this most likely reflects the partial nature of our AgSF1 cDNA on the 5′ end of the clone. This block repeat did not have sequence similarities to other repeats or submotifs in other spider silk family members. Analysis of the 64-amino acid residues revealed 32.4% glycine, 11.8% serine, and 7.4% asparagine (Fig. 1).
FIGURE 1.
Schematic of the AgSF1 and AgSF2 protein architectures and their internal block repeat modules. A, protein constructs used for heterologous protein expression. For AgSF1, the red denotes a 64-residue block repeat that is glycine-rich. Blue represents 36-amino acid block repeats that contain the X1PGX2G iterations. The green segment has been defined as a spacer region that separates the two distinct block repeats. AgSF1G or AgSF1G+XPGXG contain the glycine-rich alone or glycine-rich and XPGXG-containing block repeats, respectively. For AgSF2, the purple represents a GGX-rich and NVNXN region, whereas yellow denotes glycine-rich segments. B, representative sequence for the two different internal block repeat modules within the translated AgSF1 cDNA. Underlined proline (P) residues indicate the 5-residue spacing pattern and the GYKKTVGKDGQIV sequence shows the region used for the anti-AgSF1 polyclonal antibody production. C, representative AgSF2 internal block repeat module.
Full-length cDNAs that corresponded to clones 98, 947, 2525, 2691, and 3009 were also obtained via the library screen (GenBank accession JX262192). Analysis of the translated cDNA revealed a long ORF encoding a predicted protein with a molecular mass of ∼40.7 kDa and pI 6.61. Theoretical amino acid composition profiling demonstrated glycine (Gly) and asparagine (Asn) were the most abundant amino acids, comprising 28.4 and 14.5%, respectively. Using the program REPRO, we identified a 33-amino acid block repeat that was repeated 4 times within the internal region of the translated sequence (Fig. 1, A and C) (27). Within each block repeat two copies of the sequence NXNXN (X = Val, Asp, Leu, Phe, or Met) were found interrupted by GGX iterations, where X represents a small subset of amino acids (X = Ala, Gly, Leu, or Ser). Occasional histidine residues were embedded within the GGX iterations (Fig. 1C). GGX motifs have been reported in major ampullate fibroin sequences to adopt helical structures (28). In addition, 2 cysteine residues were present within the C terminus of translated cDNA. Protein BLAST searches against the nrNCBI database with the predicted full-length sequence revealed no significant matches to other proteins. However, consistent with proteins that are secreted, the algorithm SignalP predicted the presence of an N-terminal secretion signal.
AgSF1 and AgSF2 Display Aggregate-Gland Restricted Patterns of Expression
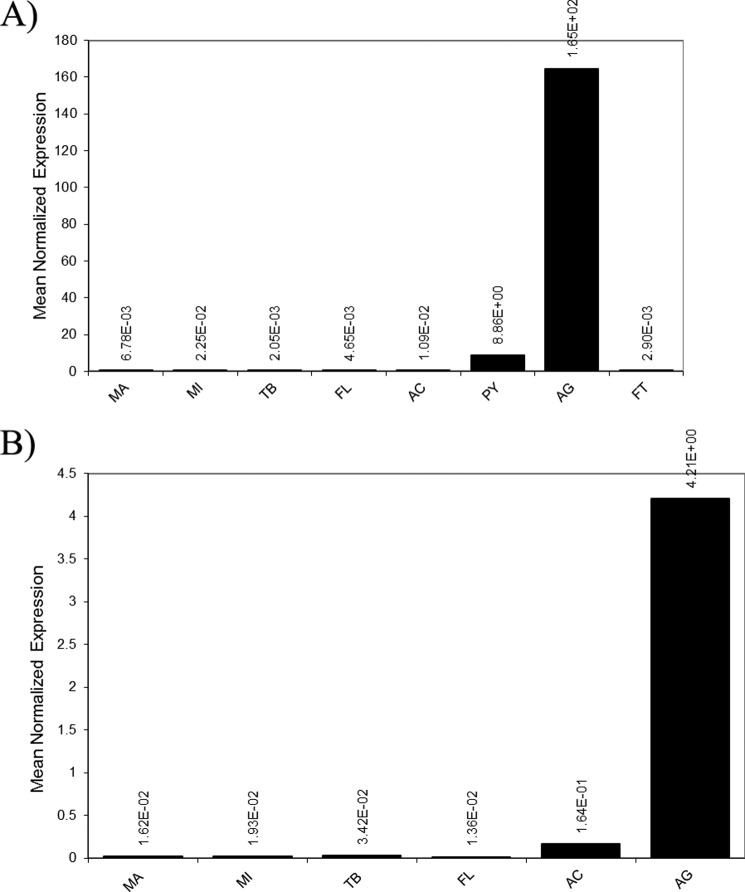
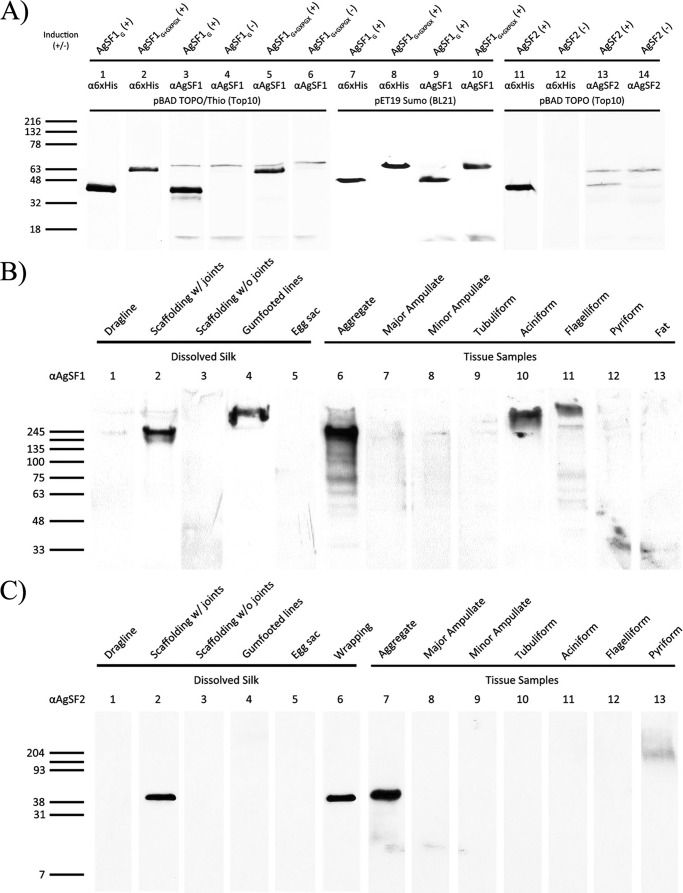
Expression patterns for these two genes were determined using quantitative real-time PCR and Western blot analyses. For quantitative RT-PCR, total RNA was isolated from the seven different silk-producing glands. Quantitative RT-PCR studies demonstrated that the aggregate gland expressed the highest quantities of mRNAs for both genes (Fig. 2, A and B). Very low levels of AgSF1 mRNA were detected in the pyriform gland; however, these levels are likely insignificant given that the aggregate gland expresses ∼19-fold more mRNA. Based upon their restricted patterns of expression, we named the two gene products AgSF1 and AgSF2. To correlate mRNA levels with endogenous protein amounts, peptides corresponding to translated regions of the AgSFs were synthesized and used to generate anti-AgSF1 and anti-AgSF2 polyclonal antibodies for Western blot analysis using lysates from the silk-producing glands. For expression studies, bacterially produced AgSF1 His-Thio-tagged or His-Sumo-tagged proteins engineered with either two copies of the 64-amino acid repeat alone (AgSF1G), or two copies of the 64-amino acid module plus 3.5 copies of the 36-amino acid repeat (AgSF1G+XPGXG) were readily detected by Western blot analysis using the anti-AgSF1 antibodies (Fig. 3A, lanes 3, 5, 9, and 10). The same recombinant proteins were detected using a monoclonal anti-His antibody (Fig. 3A, lanes 1–2 and 7–8). Bacterially expressed His-Thio-tagged full-length AgSF2 was also detected using an anti-His monoclonal antibody and anti-AgSF2 polyclonal antiserum from arabinose-induced culture extracts (Fig. 3A, lanes 11 and 13). No immunoreactive bands were detected from lysates under nonarabinose conditions (Fig. 3A, lanes 4, 6, 12, and 14). In total, these studies demonstrate that the AgSF1 and AgSF2 antisera recognize the recombinant proteins in a highly specific manner.
FIGURE 2.
AgSF1 and AgSF2 mRNA are expressed in elevated amounts in the aggregate gland. Real time quantitative PCR was used to determine the expression pattern of AgSF1 and AgSF2 in a variety of different silk producing glands: A, AgSF1; B, AgSF2. Total RNA was isolated from the major ampullate gland (MA), minor ampullate gland (MI), tubuliform (TB), flagelliform (FL), aciniform (AC), and the aggregate gland (AG). For AgSF1 studies, the pyriform (PY) and fat (FT) were also included. Equivalent amounts of total RNA were reverse-transcribed using SuperScript® III and aliquots used for real time quantitative PCR. Samples were analyzed in triplicates and normalized internally using the black widow actin mRNA. Data are representative of experimental results obtained from at least two independent trials.
FIGURE 3.
AgSF1 and AgSF2 proteins are expressed in the aggregate gland and localize to the connection joints of fibers. Western blot analysis of recombinant proteins expressed in Escherichia coli, extracts generated from silk-producing glands, or spun fibers. Plus or minus designations represent induced or noninduced conditions, respectively. Fat tissue was also included as a negative control. Denatured proteins were size fractionated using a 4–20% Tris-HCl precast polyacrylamide gel. A, bacterially expressed AgSF1 or AgSF2 recombinant proteins were detected using an anti-His6 antibody (α6xHis), anti-AgSF1 (αAgSF1), or anti-AgSF2 (αAgSF2) polyclonal antisera. B, solubilized proteins from different fiber types and silk-producing glands were probed with the anti-AgSF1 polyclonal antiserum. C, solubilized proteins from different fiber types and silk-producing glands were probed with the anti-AgSF2 polyclonal antiserum. Molecular masses for the protein ladder are shown on the left in kDa.
To examine the expression pattern of AgSF1 and AgSF2 in tissues, we tested whole cell lysates collected from the seven different silk-producing glands. Western blot analysis using an anti-AgSF1 antibody revealed dominant immunoreactive species in the aggregate, aciniform, and flagelliform glands (Fig. 3B, lanes 6, 10, and 11). The molecular masses of these species were ∼245 kDa or larger. Using the anti-AgSF2 antibody, a single immunoreactive band was detected in the aggregate gland extract (Fig. 3C, lane 7). The experimental molecular mass of AgSF2 closely correlated to the predicted mass, which was ∼40 kDa. No other immunoreactive species were detected in the major and minor ampullate glands, tubuliform, aciniform, or flagelliform glands (Fig. 3C, lanes 8–12). A minor immunoreactive, slower migrating species was detected in the pyriform glandular extract (Fig. 3C, lane 13).
AgSF1 and AgSF2 Are Constituents of Spider Glues
To investigate whether AgSF1 and AgSF2 were spun or deposited on fibers, we collected different fiber types from black widow spiders for biochemical studies. Dragline silk, egg sacs, wrapping silk, gumfooted lines, scaffolding fibers, and connection joints were obtained and solubilized in urea or guanidinium hydrochloride. Dissolved protein mixtures were analyzed using Western blot analysis for the presence of AgSFs using the anti-AgSF1 and anti-AgSF2 antisera. Because connection joints are difficult to individually remove from the web, we collected scaffolding fibers containing connection joints (scaffolding fibers plus connection joints) and scaffolding fibers alone. Immunoreactive AgSF1 proteins were detected in scaffolding fibers containing connection joints and gumfooted lines, but were absent in dragline, egg sacs, and scaffolding fibers without connection joints (Fig. 3B, lanes 1–5). The immunoreactive protein detected in the scaffolding fibers plus connection joints displayed a migration profile similar to the species detected in the aggregate gland lysate (Fig. 3B, compare lanes 2 and 6). The molecular mass of the immunoreactive species detected in the gumfooted lines was considerably larger, exceeding 300-kDa (Fig. 3B, lane 4). Similar fiber localization profiles were observed for AgSF2, which revealed the presence of an immunoreactive species in scaffolding fibers with connection joints, but not in the scaffolding fibers alone (Fig. 3C, lanes 2 and 3). AgSF2 was also detected in wrapping silk (Fig. 3C, lane 6). However, it was not detected in dragline silk, gumfooted lines, or egg sacs (Fig. 3C, lanes 1, 4, and 5). Taken together, the mRNA expression results and Western blot data support that AgSF1 and AgSF2 are expressed in the aggregate gland and extruded into the connection joint structures.
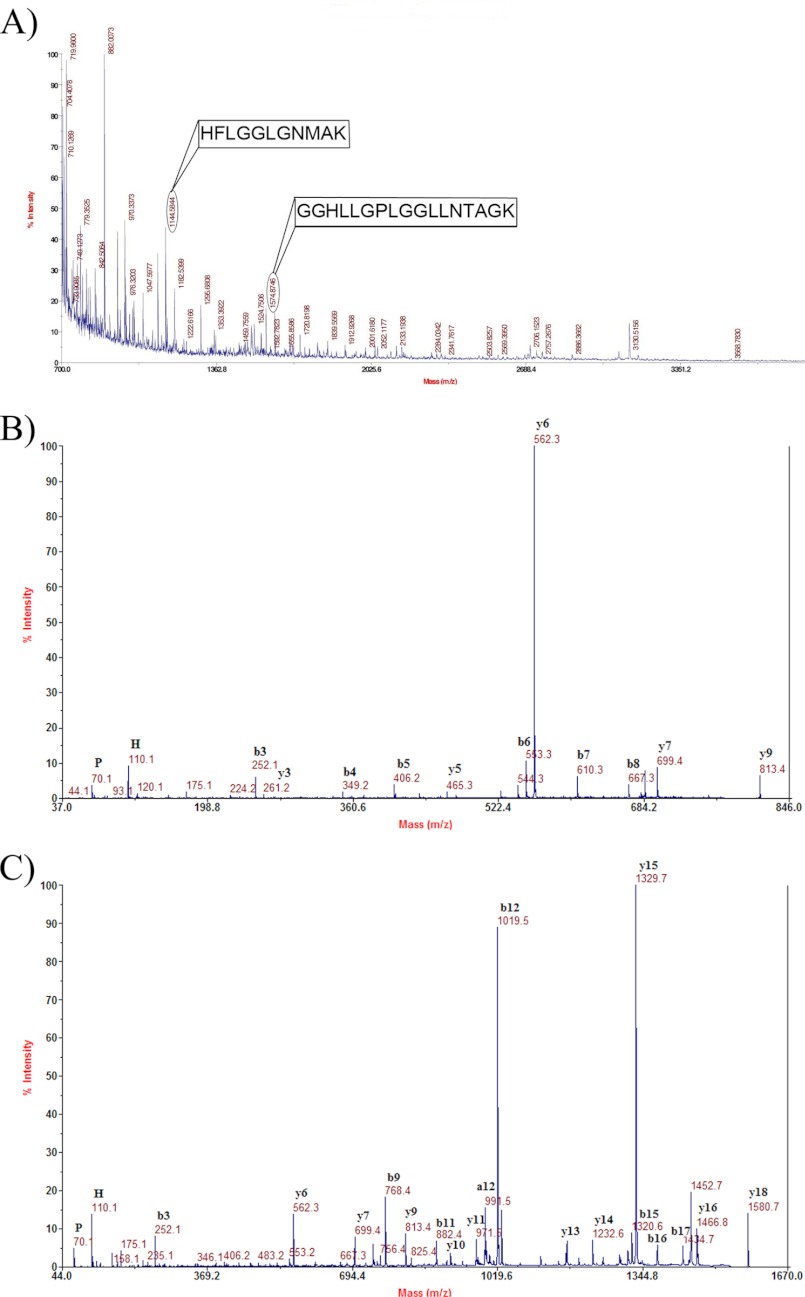
In addition to using Western blot analyses to investigate the presence of AgSF1 and AgSF2 in the different fiber types, we solubilized dragline, egg case, wrapping, gumfooted, and pyriform silks with LiBr and digested the samples with trypsin, followed by MS and MS/MS analyses. Peptide ion masses corresponding to theoretical tryptic digest products from AgSF1 and AgSF2 were not detected in dragline, egg case, gumfooted, or pyriform silks (data not shown). However, solubilized wrapping silk digested with trypsin contained two peptides that matched peptide masses located within the translated AgSF2 sequence after theoretical digestion with trypsin (Fig. 4A). These experimental precursor peptide ions displayed masses of m/z 1144.5 and 1574.8. Further analysis of these peptide ions by MS/MS revealed sequences HFLGGLGNMAK and GGHLLGPLGGLLNTAGK for ions m/z 1144.5 and 1574.8, respectively (Fig. 4A). These peptides displayed 100% matches to translated regions of the AgSF2 cDNA, with the exception that, in some cases, the de novo sequences showed Leu instead of Ile. It is sometimes difficult to distinguish these two isobaric amino acid residues by MS/MS. Two-dimensional gel electrophoresis using crude aggregate gland lysate, followed by MS/MS analysis of in-gel tryptic digestion products, did not corroborate the presence of post-translational modifications to AgSF2 (data not shown).
FIGURE 4.
MALDI-TOF or MALDI-tandem TOF analysis of precursor peptide ions from in-solution tryptic digestion of solubilized wrapping silk. A, MS analysis reveals the presence of precursor ion masses that are identical to products from the theoretical tryptic digestion of the translated AgSF2 cDNA. Subsequently, MS/MS analyses confirmed precursor ions with m/z 1144.5 and 1574.8 have the sequences HFLGGLGNMAK and GGHLLGPLGGLLNTAGK, respectively (boxed region). In these peptides sequences, the underlined Leu residues were determined to be the amino acid Ile (an isobaric residue) in the translated AgSF2 sequence. B, high energy collision-activated dissociation spectrum of precursor ion with m/z 813.4 (MH+, monoisotopic). The sequence of this peptide was found to be GGHPGFGGK. C, high energy collision-activated dissociation spectrum of precursor ion with m/z 1580.7 (MH+, monoisotopic). The sequence of this peptide was found to be GGHPGFGGTGGHPGFGGK. Both precursor ions with m/z 813.4 and 1580.7 after de novo sequencing were found to be present within the translated AgSF1 cDNA sequence.
In addition, after enzymatic digestion of solubilized wrapping silk, we discovered two precursor ion masses that were identical to products from the translated AgSF1 cDNA sequence following theoretical tryptic cleavage. These peptide ion masses corresponded to m/z 813.4 and 1580.7. After MS/MS analysis, these peptides were shown to be identical to regions found in the predicted sequence of AgSF1 (Fig. 4, B and C). One of the peptides, GGHPGFGGK, resulted from two tryptic cleavage events, whereas the other peptide, GGHPGFGGTGGHPGFGGK, likely resulted from a combination of acid hydrolysis (nonenzymatic) and tryptic cleavage (Fig. 4, B and C). Both aspartic acid and asparagine have been reported to represent hot spots for nonenzymatic degradation of proteins (29). Therefore, our MS/MS analyses reveal that these peptides are unique to AgSF1 and AgSF2 and are not found in the amino acid sequences of previously reported spider silk family members. Overall, these data support the assertion that the AgSF1 and AgSF2 are also extruded into wrapping silk constituents.
Connection Joints Are a Viscous Fibrous Material That Interconnects Dragline Silk Fibers
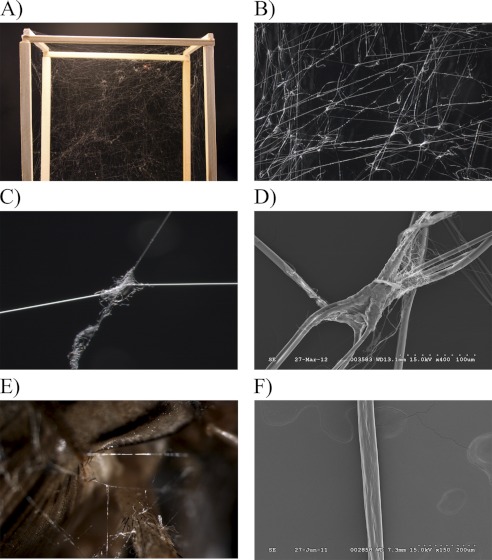
Connection joints are important structures that facilitate prey capture for orb-weavers. These structures serve to link scaffolding fibers together during the construction of three-dimensional webs (Fig. 5A). To examine the ultrastructure of connection joints, we removed web segments and analyzed these regions using microscopy. Light microscopy revealed large numbers of connection joints within the architecture of the web (Fig. 5B). These structures allow scaffolding threads to interconnect, forming a complex matrix that enhances capture of aerial insects (Fig. 5C). Scanning electron microscopy demonstrated that connection joints are composed of a fibrous material (Fig. 5D). Within this glue-like substance, small diameter fibers were observed emerging from a centralized core material; these emerging fibers contained smaller diameters relative to scaffolding threads (Fig. 5D). Connection joints were also observed in wrapping silk that was spun by spiders during immobilization of crickets (Fig. 5E).
FIGURE 5.
Black widow spider connection joints hold web- and prey-wrapping fibers together. A, digital photograph of connection joints in a web spun by a caged spider. B and C, dissection scope images of a web segment and single connecting joint at ×10 and ×115, respectively. D, S.E. of a web connecting joint at ×1500. E, dissecting scope image of a cricket immobilized with wrapping silk at ×115. F, S.E. of a synthetic fiber spun from purified recombinant AgSF1G+XPGXG protein.
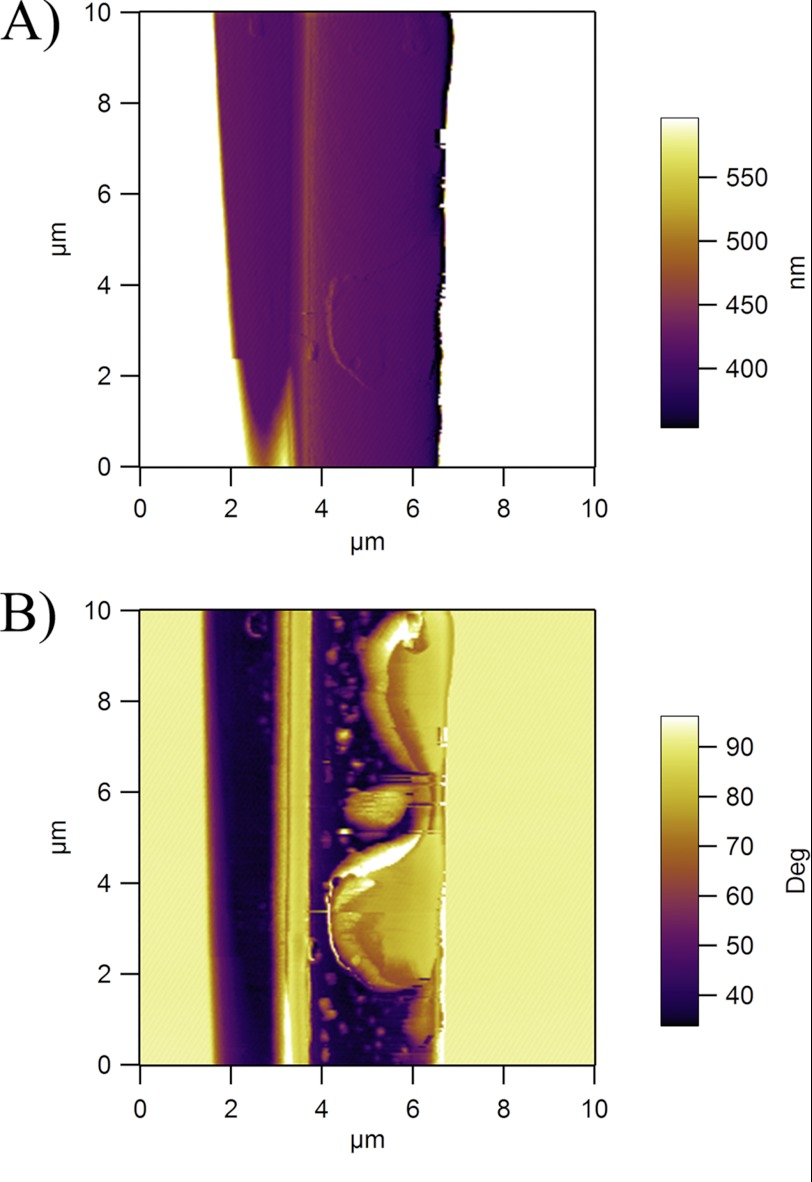
To study the biochemical properties of the connection joints, we analyzed these structures using atomic force microscopy. Within a region of the junction where a thin layer of joint material was topographically observed, we detected differences in the phase contrast at the interface between dragline and the glue (Fig. 6, A and B). Although the phase characteristics of the dragline remained fairly low, confirming the highly elastic nature of the dragline, the high values of phase shift demonstrate the highly viscous behavior of the connection joint materials, and the viscous behavior may be advantageous for dampening dynamic events (Fig. 6, A and B).
FIGURE 6.
Atomic force microscopy demonstrates connection joints consist of viscous biomaterials. A, the amplitude image confirms the presence of connection joint material as a thin film over the dragline fiber at the junction point. B, the phase-contrast image reveals the highly viscous nature of the biomaterial as indicated by the dramatic increase of the phase angles on the thin film.
Structural Analysis of Synthetic Fibers Spun from Recombinant AgSF1G+XPGXG Proteins Reveals Threads with Noncrystalline Morphology
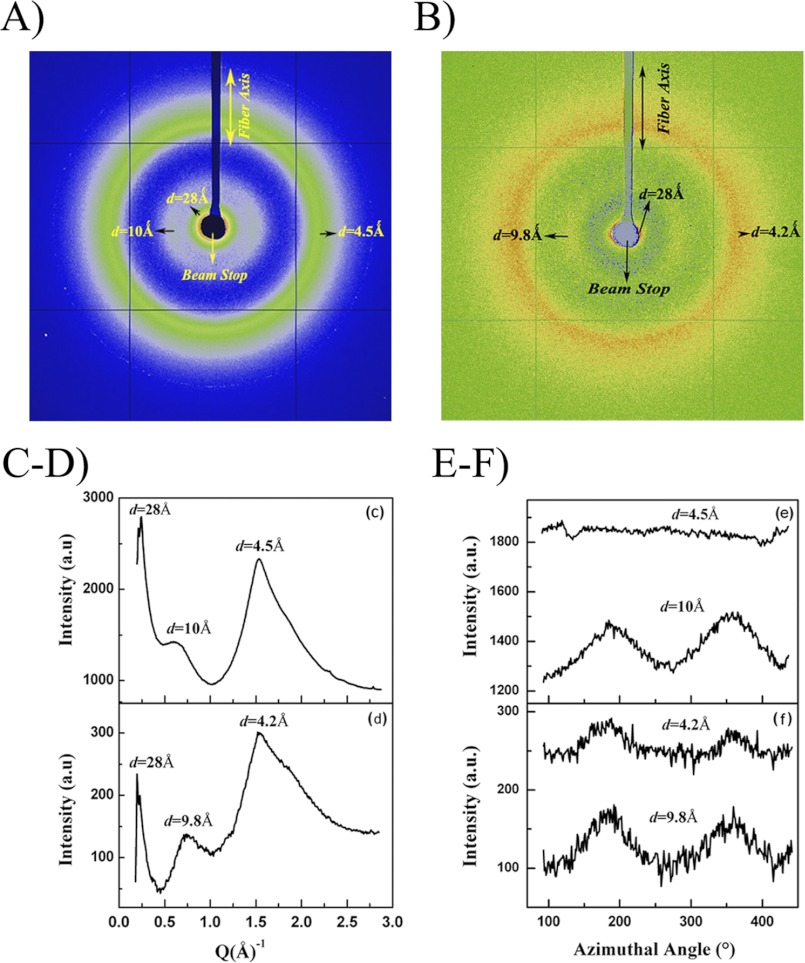
Because the primary sequence of AgSF1 contains internal block repeats that resemble elastin, we investigated whether synthetic fibers could be spun from purified recombinant AgSF1G and AgSF1G+XPGXG. Using a wet-spinning methodology (30), both recombinant proteins were capable of self-assembly into fibers. AgSF1G spun fibers were observed to be brittle and were not suited for further characterization. However, both “as-spun” and post-spun fibers were collected for AgSF1G+XPGXG spun fibers. For post-spin drawn AgSF1G+XPGXG threads, fibers were elongated to 3.5 times their original length; these were referred to as ×3.5 fibers (Fig. 5F). The XRFD two-dimensional patterns of as-spun and post-spin drawn fibers are shown in Fig. 7, A and B, respectively. Broad diffuse diffraction rings at d spacings ∼4.5 and 10 Å were observed in the two-dimensional patterns of both samples. The absence of sharp reflections indicates that the synthetic fibers were predominantly noncrystalline. The diffracted intensity in the as-spun fiber was stronger than the post-drawn fiber, mainly due to the higher thickness of the as-spun fiber. Fig. 7, C and D, show the one-dimensional radial intensity profile integrated azimuthally for the whole diffraction pattern for the as-spun and 3.5× samples, respectively. Although the width of the broad peak at d ∼ 4.5 Å is comparable in both as spun and 3.5× fibers, the smaller peak at d ∼ 10 Å becomes more defined in the 3.5× fiber. Fig. 7E shows the one-dimensional azimuthal profile of intensity as a function of angle measured from the radial position of the diffraction ring at d ∼ 4.5 and 10 Å for the as-spun fiber and Fig. 7F is the corresponding plot for the 3.5 × fiber. Although there is no axial orientation of the protein chains at d ∼ 4.5 Å in the as-spun fiber (flat profile), it is evident that the post-spin drawing aligns the protein chains with respect to the fiber axis (Fig. 7F). The azimuthal width of the peak at d ∼ 10 Å decreases from the as-spun (FWHM 74.9°) to the 3.5× fiber (FWHM 56.7°), which also suggests a higher degree of orientation of the protein chains in the 3.5 × fiber (21, 31). Taken together, the decrease in the widths of the azimuthal peaks combined with the shift of radial peaks to lower Q values in the 3.5 × fiber confirm that the post-spin draw has increased local structural order and a greater axial alignment of the protein chains. However, the post-spin draw did not result in the formation of nano-crystallites in these fibers. We also observed a strong diffraction intensity in the small angle x-ray scattering region of ∼28–34 Å in the XRFD pattern (Fig. 7, C and D), which is shadowed by the beam stop in the XRFD experiment. Diffracted intensity in the small angle x-ray scattering region in fibers generally indicate the presence of structures larger in size than unit-cell of crystallites, viz. fibrils, their size and shape, or voids between them (32).
FIGURE 7.
Synthetic fibers spun from purified recombinant AgSF1 contain amorphous domains. A, XRFD two-dimensional pattern of a single as-spun AgSF1 fiber. B, XRFD two-dimensional pattern of the AgSF1 synthetic fiber after 3.5 × post-spin draw treatment. C and D, one-dimensional radial intensity profile of the azimuthally integrated, whole two-dimensional pattern from the as-spun and 3.5 × post-spun fibers, respectively. E and F, one-dimensional azimuthal intensity profile of the radially integrated diffraction rings at d ∼ 4.5 Å and 10 Å for the as-spun and 3.5 × post-spun fibers, respectively.
DISCUSSION
After dragline silking female black widow spiders for 5 days, the DNA microarray analysis revealed gene expression profiles that could be separated into 2 categories. One class comprised genes that showed no significant alterations in their mRNA levels, which included masp1 and masp2, whereas the second class involved genes that displayed decreased mRNA quantities. The largest changes in gene activity ranged from 3.5 to 5-fold repression. Genes that were down-regulated the most included agsf1, agsf2, tusp1, ecp-1, and ecp-2. These levels were all ∼5-fold lower relative to nonsilked spiders. The presence of a collection of genes that can be down-regulated after silking implies that the different silk-producing glands are capable of shunting resources to the major ampullate gland to maintain the synthesis of dragline silk proteins, MaSp1 and MaSp2. Consistent with this assertion is that following 1 day of dragline silking, we observed elevated levels of MaSp1 and MaSp2 mRNA (data not shown); however, after 5 days of silking, whereas some gene expression profiles for other genes were declining in different silk-producing glands, MaSp1 and MaSp2 transcripts were maintained at similar levels relative to nonsilked spiders. The reshuffling of resources to the major ampullate gland is also supported by the decreased expression of tusp1, as well as the ecps, which are all genes highly expressed in females during egg case laying season and whose resources for synthesis are logically interconnected and balanced with dragline silk production. Thus, the down-regulation of egg case silk components after dragline silk removal demonstrates that our approach can be applied to identify genes that are actively expressed at high levels in other silk-producing glands, but are sensitive to the energetic costs of dragline silk synthesis.
In orb-weavers, the aggregate gland has been shown to extrude adhesive substances that include small molecular weight glycoproteins aggregate spider glue 1 and 2 (ASG1 and ASG2) (16). These glycoproteins have been reported to function as central constituents of spider viscid glue; however, whether these proteins have fiber forming properties has not been established. Sequence analysis of the ASG1 and ASG2 protein sequences showed no similarities to the connection joint proteins AgSF1 or AgSF2. Intriguingly, our DNA microarray studies did uncover one cDNA (clone 44) that contained a 65-amino acid region that showed 54% identities to a portion of the ASG2 sequence. Thus, the translated clone 44 was ASG2-like. This segment, somewhat surprisingly, did not correspond to the repetitive domain within ASG2, a region reported to share similarities with elastin and flagelliform spider silk protein repetitive sequence domains (16). Presently, it is unclear whether the product of clone 44 localizes to a specific fiber type or which silk gland expresses this gene. Attempts to use either MS or MS/MS analyses of tryptic peptides obtained from dissolved attachment discs, wrapping silk, egg case fibers, gumfooted lines, and dragline silk did not reveal peptide masses or amino acid sequences that matched theoretical digestion of the translated cDNA from clone 44. Further biochemical analysis will be required to determine the expression pattern of this gene and whether its product is extruded to coat fibers.
For the production of the anti-AgSF1 polyclonal antiserum, we used a synthetic peptide that was located within the N-terminal 64-amino block repeat. Based upon protein-protein BLAST searches of the NCBInr database, this region showed no similarities to other spidroin family members. Western blot analyses performed with this antibody revealed immunoreactive species in the aggregate, aciniform, and flagelliform glands despite the aggregate gland restricted pattern of expression of AgSF1. The molecular mass of the immunoreactive species detected in the aggregate gland co-migrated with the protein detected in the connecting joints. However, the immunoreactive species detected in the aciniform and flagelliform were substantially larger molecular weight proteins that did not co-migrate with the immunoreactive species in the aggregate gland. Taken together, these data support that the aciniform and flagelliform glands produce higher molecular weight proteins that share sequence similarity to the 64-amino acid block repeat of AgSF1. In addition, we detected one immunoreactive species within the gumfooted lines. Based upon our MS studies of solubilized gumfooted silk, however, it suggests this immunoreactive species is also distinct from AgSF1. In the future, it will be interesting to determine the identity of these other immunoreactive species and their sequence relationships to AgSF1.
The AgSF2 protein sequence has a single internal block repeat that is present in 4 copies. This block repeat is also distinct from other reported spider silk fibroin modules. Further structural studies will need to be performed to determine how this primary sequence folds into a secondary structure. Based upon our Western blot data, AgSF2 was shown to have a substantially smaller molecular mass relative to traditional fibroin family members, which have masses in the range of 250 to 350 kDa. Thus, despite the presence of internal blocks repeats, whether AgSF2 can self-assemble into fibers will require further investigation. We have not yet attempted to spin recombinant AgSF2 proteins into synthetic fibers. The observation that AgSF2 is extruded into a fast drying glue-like interface material that serves to fuse individual scaffolding fibers could imply that it functions as a novel bioadhesive during web construction to help enhance the structural integrity of the web. Although we made numerous attempts to determine whether AgSF2 is N- or O-glycosylated in vivo, our MS and MS/MS studies did not support the presence of glycans covalently attached to AgSF2 (data not shown). Because highly glycosylated proteins, such as mucins are known to have bioadhesive properties, the lack of detectable glycans on AgSF2 tryptic peptides implies it has evolved a different molecular mechanism to interact with the protein chains of major ampullate silk fibroins. AgSF2 was observed to contain glycine and asparagine content that approaches 43%. One possibility could involve the side chain atoms of asparagine, which could lend themselves to hydrogen bond with other protein chains. In addition, two cysteine residues were present within the C-terminal region of AgSF2, but whether these residues participate in interdisulfide bond formation with the conserved cysteine residue in the C terminus of MaSp1 or MaSp2 remains to be determined.
Although AgSF1 and AgSF2 are co-extruded into connection joints, their protein chain lengths and internal block repeat sequences, in addition to their N and C termini, were substantially divergent from each other and spidroin family members. For example, spidroin family members, such as PySp1 and PySp2, which are known as glue silk fibroins, contain significantly distinct repeat modules (9, 33, 34). Based upon these differences, it implies distinct roles for the AgSFs and the presence of at least two distinct glue protein families in the connection joints. Furthermore, the presence of two distinct block repeats in the AgSF1 protein chain also implies these regions perform distinct biological functions. One of the internal block repeats, the 36-amino acid segment, could be divided into the following submotifs: 1) five tandem copies of the pentameric sequence X1PGGX2; 2) one hexameric sequence RPGGKG; and 3) one pentameric sequence QPGYY. These 36-amino acid block repeats were iterated 11 times, comprising over 55 copies of the X1PGGX2 subrepeats; these pentameric sequences resemble VPGXG repeats found within elastin (35). Structurally, VPGXG iterations have been proposed to adopt a long-range β-spiral conformation with spring-like structures (36, 37). Whether iterations of X1PGGX2 sequences fold into a similar secondary structure is unclear, but the PG residues and their presence in the pentameric modules strikingly resemble the VPGXG and GPGQQ modules of elastin and the flagelliform fibroin in spiral capture silk, respectively (13, 35). Despite these similarities, there are some differences between the VPGXG and X1PGGX2 modules. One difference is the hydrophobic nature of the valine elastin relative to the hydrophilic properties of X1 (X1 = Gln, Glu, or Arg) in AgSF1. Differences in chemical properties are also present within the last residue of the pentameric sequence, where elastin contains the hydrophobic glycine residue and X2 was polar (X2 = Asn or Ser). Elastin also has been established to form intermolecular cross-linking via conserved lysine residues within its amino acid sequence. The conservation and spacing of the lysine residues within the RPGGKG motifs of AgSF1 could point to a similar function, providing an intermolecular cross-linking hub to enhance molecular alignments. However, based upon our biochemical studies, we have not been able to provide direct evidence that these residues are oxidized via chemical or enzymatic processes. The third submodule QPGYY, which contains dityrosine residues in tandem, was highly conserved throughout the 36-amino acid block repeats. Aromatic residues have been reported to be important mediators of protein assembly, and specifically tyrosine residues, have been reported to participate in intermolecular cross-linking (38–41). Thus, it raises the question whether these residues have an important role in controlling the nucleation, aggregation, polymerization, and/or intermolecular cross-linking of AgSF1 monomers during the extrusion process.
The exact mechanism that spiders use to assemble glue proteins into fibrous-like materials is highly debatable. Previous morphological studies of the spinneret silk spigots from black widow spiders show the posterior lateral spinneret contains two aggregate gland spigots with large flap-like openings, suggesting the glue-like proteinaceous material exits via a spraying process (20). Our microdissections of the aggregate glands support this report, revealing a large duct that exits the spinneret through a large flap-like spigot structure (19). Thus, unlike traditional spigots, which extrude threads, the proteins released from the aggregate gland appear to experience a different mechanism of self-assembly relative to fibroins spun by the major ampullate gland. Spraying the liquid substance through the large flap-like structure implies the secreted protein mixture is capable of undergoing assembly after extrusion. Preliminary data that removes the Sumo-tag from the recombinant AgSF1G+XPGXG, followed by gentle agitation, resulted in protein aggregation in vitro.6 Thus, the observation that AgSF1G+XPGXG monomers can spontaneously assemble is consistent with the assertion that extrusion, followed by extension of the liquid via the movement of the abdomen, can lead to fiber formation. In agreement with this proposal is the ability of AgSF1G+XPGXG recombinant proteins to spontaneously form aggregates without shear as well as form synthetic fibers with shear, and the presence of small diameter fibers emerging from centralized regions of the connection joint material. In light of our findings, these studies provide the first insight into the diverse architectures of spider glue protein sequences that function to fasten web and wrapping fibers, uncovering a rich source of new structural biomaterials.
Acknowledgments
We thank Dr. Jason Li, Asylum Research, for technical assistance on the AFM imaging and Jeff Yarger, Arizona State University, Department of Chemistry & Biochemistry, for help with the x-ray diffraction data collection. Use of the Advanced Photon Source was supported by the United States Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. DE-AC02-06CH11357. Use of the BioCARS Sector 14 was also supported by grants from the National Center for Research Resources Grant 5P41RR007707 and NIGMS Grant 8P41GM103543 from the National Institutes of Health.
This work was supported by National Science Foundation RUI Grants MCB-0950372 and DMR-1105310.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) JX262195, JX262192, and EHJ72326.1.
A. Hammersley and S. Sampath, unpublished observation.
K. Vasanthavada, X. Hu, T. Tuton-Blasingame, Y. Hsia, S. Sampath, R. Pacheco, J. Freeark, A. M. Falick, S. Tang, J. Fong, K. Kohler, C. La Mattina-Hawkins, and C. Vierra, unpublished data.
- AgSF1
- aggregate gland silk factor 1
- AgSF2
- aggregate gland silk factor 2
- MaSp1
- major ampullate spidroin 1
- TuSp1
- tubuliform spidroin 1
- BLAST
- basic local alignment search tool
- XRFD
- x-ray fiber diffraction
- ASG
- aggregate spider glue.
REFERENCES
- 1. Foelix R. (1996) Biology of Spiders, Oxford University Press, New York [Google Scholar]
- 2. Kovoor J. (1987) in The Ecophysiology of Spiders (Nentwig W., ed) pp. 160–186, Springer-Verlag, New York [Google Scholar]
- 3. Guerette P. A., Ginzinger D. G., Weber B. H., Gosline J. M. (1996) Silk properties determined by gland-specific expression of a spider fibroin gene family. Science 272, 112–115 [DOI] [PubMed] [Google Scholar]
- 4. Gatesy J., Hayashi C., Motriuk D., Woods J., Lewis R. (2001) Extreme diversity, conservation, and convergence of spider silk fibroin sequences. Science 291, 2603–2605 [DOI] [PubMed] [Google Scholar]
- 5. Motriuk-Smith D., Smith A., Hayashi C. Y., Lewis R. V. (2005) Analysis of the conserved N-terminal domains in major ampullate spider silk proteins. Biomacromolecules 6, 3152–3159 [DOI] [PubMed] [Google Scholar]
- 6. Sponner A., Unger E., Grosse F., Weisshart K. (2004) Conserved C-termini of Spidroins are secreted by the major ampullate glands and retained in the silk thread. Biomacromolecules 5, 840–845 [DOI] [PubMed] [Google Scholar]
- 7. Hu X., Lawrence B., Kohler K., Falick A. M., Moore A. M., McMullen E., Jones P. R., Vierra C. (2005) Araneoid egg case silk. A fibroin with novel ensemble repeat units from the black widow spider, Latrodectus hesperus. Biochemistry 44, 10020–10027 [DOI] [PubMed] [Google Scholar]
- 8. Hayashi C. Y., Blackledge T. A., Lewis R. V. (2004) Molecular and mechanical characterization of aciniform silk. Uniformity of iterated sequence modules in a novel member of the spider silk fibroin gene family. Mol. Biol. Evol. 21, 1950–1959 [DOI] [PubMed] [Google Scholar]
- 9. Blasingame E., Tuton-Blasingame T., Larkin L., Falick A. M., Zhao L., Fong J., Vaidyanathan V., Visperas A., Geurts P., Hu X., La Mattina C., Vierra C. (2009) Pyriform spidroin 1, a novel member of the silk gene family that anchors dragline silk fibers in attachment discs of the black widow spider, Latrodectus hesperus. J. Biol. Chem. 284, 29097–29108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vasanthavada K., Hu X., Falick A. M., La Mattina C., Moore A. M., Jones P. R., Yee R., Reza R., Tuton T., Vierra C. (2007) Aciniform spidroin, a constituent of egg case sacs and wrapping silk fibers from the black widow spider Latrodectus hesperus. J. Biol. Chem. 282, 35088–35097 [DOI] [PubMed] [Google Scholar]
- 11. Hinman M. B., Lewis R. V. (1992) Isolation of a clone encoding a second dragline silk fibroin. Nephila clavipes dragline silk is a two-protein fiber. J. Biol. Chem. 267, 19320–19324 [PubMed] [Google Scholar]
- 12. Colgin M. A., Lewis R. V. (1998) Spider minor ampullate silk proteins contain new repetitive sequences and highly conserved nonsilk-like “spacer regions.” Protein Sci. 7, 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hayashi C. Y., Lewis R. V. (1998) Evidence from flagelliform silk cDNA for the structural basis of elasticity and modular nature of spider silks. J. Mol. Biol. 275, 773–784 [DOI] [PubMed] [Google Scholar]
- 14. Vollrath F., Fairbrother W. J., Williams R. J. P., Tillinghast E. K., Bernstein D. T., Gallagher K. S., Townley M. A. (1990) Compounds in the droplets of the orb spider's viscid spiral. Nature 345, 526–528 [Google Scholar]
- 15. Vollrath F., Tillinghast E. K. (1991) Glycoprotein glue beneath a spider web's aqueous coat. Naturwissenschaften 78, 557–559 [Google Scholar]
- 16. Choresh O., Bayarmagnai B., Lewis R. V. (2009) Spider web glue. two proteins expressed from opposite strands of the same DNA sequence. Biomacromolecules 10, 2852–2856 [DOI] [PubMed] [Google Scholar]
- 17. Lawrence B. A., Vierra C. A., Moore A. M. (2004) Molecular and mechanical properties of major ampullate silk of the black widow spider, Latrodectus hesperus. Biomacromolecules 5, 689–695 [DOI] [PubMed] [Google Scholar]
- 18. Hu X., Kohler K., Falick A. M., Moore A. M., Jones P. R., Sparkman O. D., Vierra C. (2005) Egg case protein-1. A new class of silk proteins with fibroin-like properties from the spider Latrodectus hesperus. J. Biol. Chem. 280, 21220–21230 [DOI] [PubMed] [Google Scholar]
- 19. Jeffery F., La Mattina C., Tuton-Blasingame T., Hsia Y., Gnesa E., Zhao L., Franz A., Vierra C. (2011) Microdissection of black widow spider silk-producing glands. J. Vis. Exp. 47, 2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coddington J. (1989) Spinneret silk spigot morphology. Evidence for the monophyly of orb-weaving spiders, Cyrtophorinae, and the group Theridiidae plus Nesticidae. J. Arachnol. 17, 71–95 [Google Scholar]
- 21. Zussman E., Burman M., Yarin A. L., Khalfin R., Cohen Y. (2006) Tensile deformation of electrospun nylon-6,6 nanofibers. J. Polym. Sci. B Polym. Phys. 44, 1482–1489 [Google Scholar]
- 22. Grubb D. T., Jelinski L. W. (1997) Fiber morphology of spider silk. The effects of tensile deformation. Macromolecules 30, 2860–2867 [Google Scholar]
- 23. Riekel C., Bränden C., Craig C., Ferrero C., Heidelbach F., Müller M. (1999) Aspects of x-ray diffraction on single spider fibers. Int. J. Biol. Macromol. 24, 179–186 [DOI] [PubMed] [Google Scholar]
- 24. Hu X., Kohler K., Falick A. M., Moore A. M., Jones P. R., Vierra C. (2006) Spider egg case core fibers. Trimeric complexes assembled from TuSp1, ECP-1, and ECP-2. Biochemistry 45, 3506–3516 [DOI] [PubMed] [Google Scholar]
- 25. Pouchkina N. N., Stanchev B. S., McQueen-Mason S. J. (2003) From EST sequence to spider silk spinning. identification and molecular characterization of Nephila senegalensis major ampullate gland peroxidase NsPox. Insect. Biochem. Mol. Biol. 33, 229–238 [DOI] [PubMed] [Google Scholar]
- 26. Sandberg L. B., Leslie J. G., Leach C. T., Alvarez V. L., Torres A. R., Smith D. W. (1985) Elastin covalent structure as determined by solid phase amino acid sequencing. Pathol. Biol. 33, 266–274 [PubMed] [Google Scholar]
- 27. George R. A., Heringa J. (2000) The REPRO server. Finding protein internal sequence repeats through the Web. Trends Biochem. Sci. 25, 515–517 [DOI] [PubMed] [Google Scholar]
- 28. van Beek J. D., Hess S., Vollrath F., Meier B. H. (2002) The molecular structure of spider dragline silk. Folding and orientation of the protein backbone. Proc. Natl. Acad. Sci. U.S.A. 99, 10266–10271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geiger T., Clarke S. (1987) Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 262, 785–794 [PubMed] [Google Scholar]
- 30. Hsia Y., Gnesa E., Pacheco R., Kohler K., Jeffery F., Vierra C. (2012) Synthetic spider silk production on a laboratory scale. J. Vis. Exp. 65, 4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keum J. K., Song H. H. (2005) Thermal deformations of oriented noncrystalline poly (ethylene terephthalate) fibers in the presence of mesophase structure. Polymer 46, 939–945 [Google Scholar]
- 32. Murthy S. (2004) Recent developments in polymer characterization using x-ray diffraction. Rigaku J. 21, 15–24 [Google Scholar]
- 33. Geurts P., Zhao L., Hsia Y., Gnesa E., Tang S., Jeffery F., La Mattina C., Franz A., Larkin L., Vierra C. (2010) Synthetic spider silk fibers spun from pyriform spidroin 2, a glue silk protein discovered in orb-weaving spider attachment discs. Biomacromolecules 11, 3495–3503 [DOI] [PubMed] [Google Scholar]
- 34. Perry D. J., Bittencourt D., Siltberg-Liberles J., Rech E. L., Lewis R. V. (2010) Piriform spider silk sequences reveal unique repetitive elements. Biomacromolecules 11, 3000–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kushner A. M., Guan Z. (2011) Modular design in natural and biomimetic soft materials. Angew. Chem. Int. Ed. Engl. 50, 9026–9057 [DOI] [PubMed] [Google Scholar]
- 36. Urry D. W. (1988) Entropic elastic processes in protein mechanisms. II. Simple (passive) and coupled (active) development of elastic forces. J. Protein Chem. 7, 81–114 [DOI] [PubMed] [Google Scholar]
- 37. Urry D. W. (1988) Entropic elastic processes in protein mechanisms. I. Elastic structure due to an inverse temperature transition and elasticity due to internal chain dynamics. J. Protein Chem. 7, 1–34 [DOI] [PubMed] [Google Scholar]
- 38. Brown-Augsburger P., Tisdale C., Broekelmann T., Sloan C., Mecham R. P. (1995) Identification of an elastin cross-linking domain that joins three peptide chains. Possible role in nucleated assembly. J. Biol. Chem. 270, 17778–17783 [DOI] [PubMed] [Google Scholar]
- 39. Reches M., Gazit E. (2003) Casting metal nanowires within discrete self-assembled peptide nanotubes. Science 300, 625–627 [DOI] [PubMed] [Google Scholar]
- 40. Gazit E. (2002) A possible role for π-stacking in the self-assembly of amyloid fibrils. FASEB J. 16, 77–83 [DOI] [PubMed] [Google Scholar]
- 41. Souza J. M., Giasson B. I., Chen Q., Lee V. M., Ischiropoulos H. (2000) Dityrosine cross-linking promotes formation of stable α-synuclein polymers. Implication of nitrative and oxidative stress in the pathogenesis of neurodegenerative synucleinopathies. J. Biol. Chem. 275, 18344–18349 [DOI] [PubMed] [Google Scholar]