Background: TgSUB1 is a subtilisin protease that trims invasion proteins on the surface of Toxoplasma gondii.
Results: TgMIC5 suppresses TgSUB1 activity and structurally mimics a subtilisin prodomain, suggesting a mechanism for inhibition.
Conclusion: The C-terminal region of TgMIC5 is responsible for inhibition of TgSUB1.
Significance: We identify a novel subtilisin propeptide mimic.
Keywords: Cell Invasion, NMR, Parasitology, Protease Inhibitor, Protein Structure, MIC5, SUB1, Toxoplasma gondii, Microneme Proteins, Subtilisin
Abstract
Toxoplasma gondii is the model parasite of the phylum Apicomplexa, which contains obligate intracellular parasites of medical and veterinary importance. Apicomplexans invade host cells by a multistep process involving the secretion of adhesive microneme protein (MIC) complexes. The subtilisin protease TgSUB1 trims several MICs on the parasite surface to activate gliding motility and host invasion. Although a previous study showed that expression of the secretory protein TgMIC5 suppresses TgSUB1 activity, the mechanism was unknown. Here, we solve the three-dimensional structure of TgMIC5 by nuclear magnetic resonance (NMR), revealing that it mimics a subtilisin prodomain including a flexible C-terminal peptide that may insert into the subtilisin active site. We show that TgMIC5 is an almost 50-fold more potent inhibitor of TgSUB1 activity than the small molecule inhibitor N-[N-(N-acetyl-l-leucyl)-l-leucyl]-l-norleucine (ALLN). Moreover, we demonstrate that TgMIC5 is retained on the parasite plasma membrane via its physical interaction with the membrane-anchored TgSUB1.
Introduction
Toxoplasma gondii is an obligate intracellular parasite belonging to the phylum Apicomplexa and is the causative agent of toxoplasmosis. Toxoplasma is able to invade almost any nucleated cell and is prevalent among human populations, with a worldwide infection rate of up to 30%. Infection in humans occurs primarily following consumption of undercooked infected meat or contact with feces from infected domestic cats. In immunocompromised individuals, Toxoplasma reactivates from a semidormant state to cause acute diseases including blindness (1) or potentially fatal encephalitis (2, 3). Infection in pregnant women can result in a range of fetal birth defects or death (4). Due to its remarkably high infection rate, T. gondii constitutes the third most common cause of food-related death in both the United States (5) and France (6) after Salmonella and Listeria.
T. gondii actively invades host cells during infection. Host cell entry requires the sequential release of proteins from secretory organelles named micronemes and rhoptries (7). Microneme proteins (MICs)4 contribute to host cell attachment and provide a link to the parasite actomyosin motor that drives cell entry (8, 9). Many MICs contain lectin domains that bind specific carbohydrate ligands on the surface of host cells, providing a key attachment point for the parasite (10–14). Adhesive MICs are assembled into heteromeric complexes as they pass through the secretory pathway. Several proteolytic molecular checkpoints occur throughout their assembly and transit to the micronemes. Proteolytic processing of MICs also occurs after their release onto the parasite surface, and this is essential for successful invasion. TgSUB1 is a glycosylphosphatidylinositol (GPI)-anchored microneme protein that shows subtilisin-like serine protease activity. Lagal et al. (15) recently showed that TgSUB1 mediates the necessary surface processing of certain MICs for efficient host recognition and invasion, such as TgMIC2-M2AP and MIC4.
Using gene knock-out and proteome profiling Brydges et al. (16) showed that the protease activity of TgSUB1 is negatively regulated by another microneme protein, TgMIC5. TgMIC5 is expressed as a preproprotein, which is proteolytically processed to a proprotein by the signal peptidase before being further processed in a post-Golgi compartment to the mature protein (17). Although Brydges et al. did not determine the precise nature of TgSUB1 regulation by TgMIC5, they suggested two possible mechanisms. First, they proposed that TgMIC5 could inhibit TgSUB1 activity directly, possibly by binding to and occluding the active site. Second, they proposed that TgMIC5 associates with the substrates of TgSUB1 on the parasite surface, thereby protecting them from overdigestion by TgSUB1. The latter model was favored in part because TgMIC5 showed no amino acid sequence similarity to any protease inhibitor. Immunoprecipitation experiments failed to identify partners of TgMIC5, thus limiting further mechanistic insight of its role in regulating TgSUB1 proteolysis (16).
Combining atomic resolution studies with data from biochemical and parasite studies, we reveal that TgMIC5 is a subtilisin propeptide mimic and a potent inhibitor of TgSUB1. We discuss the implications of this inhibitory activity in the context of infection by T. gondii. Our work could also provide the inspiration for novel therapeutics design to target parasite proteases.
EXPERIMENTAL PROCEDURES
Parasite Culture
The RH strain of T. gondii, which is a type I strain that is highly pathogenic for all laboratory mice, was used in all experiments. Parasite tachyzoites (RH, RHΔmic5, and RHΔsub1) were grown in human foreskin fibroblast cells and harvested as described previously (18).
Cloning, Expression, and Purification of Recombinant TgMIC5 from Escherichia coli
Wild type (WT) MIC5 was expressed using the pTYB2 plasmid (New England Biolabs) in BL21 (DE3) E. coli strain (Stratagene) in Luria Bertani medium supplemented with 50 μg/ml carbenicillin. The protein expression was induced with 500 μm isopropyl β-d-thiogalactopyranoside for 3 h at 37 °C. All subsequent purification steps were performed at 4 °C, and protease inhibitors were absent. The cells were lysed using the Cell Disruptor TS5 (Constant Cell Disruption Systems) coupled to a Haake TC200 chiller (Thermo Electron Corporation). WT MIC5 fusion protein was purified by affinity chromatography using chitin resin (New England Biolabs). The resin was equilibrated with 10 bed volumes of column buffer (20 mm HEPES, pH 7.5, 500 mm NaCl, 1 mm EDTA). The bound intein tag was cleaved using a cleavage buffer containing 20 mm HEPES, pH 8.0, 500 mm NaCl, 1 mm EDTA, 50 mm dithiothreitol (DTT). The chitin resin column was flushed with 3 bed volumes of cleavage buffer and allowed to sit for 16 h. Pure WT MIC5 was recovered in the flow-through.
To facilitate the production of mutant and truncated proteins, WT MIC5 was cloned onto pET30 Xa/LIC plasmid (Novagen). The mutated proteins were generated using the QuikChangeTM Site-directed Mutagenesis kit (Agilent Technologies). All five mutated/truncated proteins were expressed using a pET30 Xa/LIC plasmid in BL21 (DE3) E. coli strain in Luria Bertani medium supplemented with 40 μg/ml kanamycin. The cultures were initially grown at 37 °C and followed by a temperature adjustment for 1 h at 18 °C before inducing with 500 μm isopropyl β-d-thiogalactopyranoside for 12 h. The mutated and truncated proteins were purified using a nickel-nitrilotriacetic acid resin (Qiagen). The resin was equilibrated with 10 bed volumes of binding buffer (20 mm Tris-HCl, pH 8, 500 mm NaCl, 5 mm imidazole). Bound proteins were washed with 10 bed volumes of binding buffer, wash buffer (20 mm Tris-HCl, pH 8, 500 mm NaCl, 20 mm imidazole) and eluted with 10 bed volumes of elution buffer (20 mm Tris-HCl, pH 8, 500 mm NaCl, 250 mm imidazole).
Size exclusion chromatography was carried out using an AKTA system (Amersham Biosciences) in combination with a preparative SuperdexTM 75 column. All samples were exchanged into identical buffer conditions (25 mm KH2PO4, pH 7.2, 150 mm NaCl). The protein fractions were pooled together, concentrated to ∼500 μm, and exchanged into an NMR buffer (25 mm KH2PO4, pH 7.2, 100 mm NaCl). 15N,13C-labeled samples of WT MIC5 were produced in minimal medium containing 0.07% 15NH4Cl and 0.2% 13C6-glucose.
Solution Structure Determination of TgMIC5
Backbone and side chain assignments were completed using our in-house, semiautomated assignment algorithms and standard triple-resonance assignment methodology (19). Hα and Hβ assignments were obtained using HBHA (CBCACO)NH. The side chain assignments were completed using HCCH total correlation (TOCSY) spectroscopy and (H)CC(CO)NH TOCSY. Three-dimensional 1H-15N/13C NOESY-HSQC (mixing time 100 ms at 800 MHz) experiments provided the distance restraints used in the final structure calculation. The ARIA protocol (20) was used for completion of the NOE assignment and structure calculation. The frequency window tolerance for assigning NOEs was ±0.04 ppm and ±0.06 ppm for direct and indirect proton dimensions and ±0.6 ppm for both nitrogen and carbon dimensions. The ARIA parameters p, Tv, and Nv were set to default values. 144 dihedral angle restraints derived from TALOS were also implemented (21). The 10 lowest energy structures had no NOE violations >0.5 Å and dihedral angle violations >5º. Although structure calculations readily converged without the introduction of manual assignments, a systematic check of automatically assigned NOEs was carried out. The structural statistics are presented in Table 1. The heteronuclear 1H-15N NOE data were measured as described previously (22).
TABLE 1.
Structural statistics for TgMIC5 solution structure calculation
| Statistics | TgMIC5 (PDB ID code 2LU2) |
|---|---|
| Number of experimental restraints | 2,162 |
| Total NOE-derived | 2,018 |
| Ambiguous | 804 |
| Unambiguous | 1,214 |
| Intraresidue | 460 |
| Sequential | 236 |
| Medium range (|i–j|– ≤ 4) | 194 |
| Long range (|i–j|> 4) | 324 |
| Talos (φ/ψ) | 144 |
| R.m.s.d. from experimental restraintsa | |
| Distance (Å) | 0.040 ± 0.0028 |
| Dihedral angle (degrees) | 0.68 ± 0.057 |
| R.m.s.d. from idealized covalent geometry | |
| Bonds (Å) | 0.0052 ± 0.0002 |
| Improper angles (degrees) | 2.06 ± 0.095 |
| Angles (degrees) | 0.68 ± 0.012 |
| Energies (kcal mol−1) | |
| ENOE | 161.1 ± 22.6 |
| Ebond | 34.1 ± 2.9 |
| Eangle | 160.3 ± 5.9 |
| Evdw | −762.4 ± 8.7 |
| Coordinate r.m.s.d. (Å) | |
| Backbone atoms in secondary structure | 0.17 ± 0.02 |
| All backbone atoms | 0.516 ± 0.16 |
| Heavy atoms in secondary structure | 0.48 ± 0.05 |
| All heavy atoms | 0.78 ± 0.12 |
| Ramachandran plot | |
| Residues in most favored regions (%) | 85.1 |
| Residues in allowed regions (%) | 13.1 |
| Residues in generously allowed regions (%) | 1.7 |
| Residues in disallowed regions (%) | 0.1 |
a R.m.s.d., root mean square deviation.
Inhibition Assay of TgSUB1 Activity
Filter-purified RH parasites were washed three times with ice-cold D1 medium (Dulbecco's modified Eagle's medium (DMEM) containing 10 mm HEPES, pH 7.0, 1% fetal bovine serum (FBS), and 2 mm l-glutamine). Parasites were resuspended at 3 × 108 ml−1 in D1, and 90 μl of parasite resuspension was pipetted into the wells of a 96-well round bottom plate containing 10 μl of inhibitors (recombinant TgMIC5 or ALLN) and incubated for 20 min at 37 °C. Parasites were pelleted twice at 4 °C by centrifugation. Supernatants were analyzed by quantitative immunoblotting as described below. Recombinant WT and mutant TgMIC5 proteins were tested at concentrations of 8.2 × 10−3 μm, 2.5 × 10−2 μm, 7.4 × 10−2 μm, 2.2 × 10−1 μm, 6.7 × 10−1 μm, 2.0 μm, 6.0 μm, and 18.0 μm. ALLN was tested at concentrations of 4.6 × 10−2 μm, 1.4 × 10−1 μm, 4.1 × 10−1 μm, 1.2 μm, 3.7 μm, 11.1 μm, 33.3 μm, and 100.0 μm. Intensities of the TgMIC450 band were quantified with LI-COR image studio software and plotted against the corresponding concentrations of inhibitors using Prism 5 software to determine the half-maximal inhibitory concentrations (IC50). Due to the low ratio of TgMIC450 to TgMIC470, the optimal laser power for detecting TgMIC450 oversaturated the TgMIC470 band, as indicated by the white region in its middle. Lysozyme was included as a negative control because its size and globular nature are similar to TgMIC5.
SDS-PAGE and Immunoblotting
Parasite excreted-secreted antigen (ESA) fractions or cell lysates were mixed with 5× SDS-PAGE loading buffer containing 2% β-mercaptoethanol (final concentration) and boiled for 5 min before resolving on 10% or 12.5% minigels. For qualitative blots, proteins were semidry-transferred to polyvinylidene fluoride (PVDF) membrane (Millipore), blocked with phosphate-buffered saline (PBS) containing 1% skim milk, 0.05% Tween 20, and 0.05% Triton X-114, and incubated with primary antibodies. Goat anti-mouse or anti-rabbit IgGs conjugated with horseradish peroxidase (HRP) were used as secondary antibodies. Immunoblots were developed with SuperSignal West Pico chemiluminescence reagent (Pierce) and exposed to x-ray film. For quantitative blots, proteins were semidry-transferred to PVDF-FL membrane (Millipore). Blots were treated with blocking buffer (LI-COR Biosciences) and sequentially probed with rabbit polyclonal anti-TgMIC4 (23) and goat anti-rabbit IgG conjugated with IRDye 800CW (LI-COR Biosciences). Blots were air-dried and imaged by LI-COR Odyssey CLx instrument.
Immunolocalization Assays
To assess the surface distribution of proteins during invasion, tachyzoites were filter-purified, washed, and resuspended in “Endo” buffer (44.7 mm K2SO4, 10 mm MgSO4, 106 mm sucrose, 5 mm glucose, 20 mm Tris-H2SO4, 3.5 mg/ml bovine serum albumin (BSA), pH 8.2, a potassium-rich buffer that arrests parasite motility) (24) at 3 × 107 ml−1 at room temperature. Two hundred microliters of parasite resuspension was added to overnight-grown human foreskin fibroblast cells in 8-well chamber slides. The parasites were allowed to settle for 15 min at room temperature before the medium was replaced with prewarmed invasion medium (DMEM with 10 mm HEPES, pH 7.0, 3% FBS, and 2 mm l-glutamine) containing 1% ethanol. Parasites were allowed to invade for 2 min at 37 °C prior to fixation with 4% paraformaldehyde for 20 min. Parasites were stained with mouse monoclonal (mAb) B3.90 anti-TgAMA1 (25), rabbit polyclonal anti-TgMIC5 (17), or rabbit anti-Plasmodium falciparum SUB1 (PfSUB1). To assess the subcellular distribution TgMIC5 and TgSUB1 within parasites, these proteins were stained with the aforementioned antibodies along with TgAMA1 in overnight replicated parasites permeabilized with 0.1% Triton X-100. Mowiol was used as the mounting medium. Images were captured digitally with an AxioCAM MRm camera equipped Zeiss Axiovert Observer Z1 inverted fluorescence microscope at room temperature with a 100×, 1.3 NA oil objective lens and processed using Zeiss Axiovision 4.3 software. Adobe Photoshop and Illustrator CS5 programs were used to assemble the final images.
Analysis of Surface Protein Abundance by Flow Cytometry
RH, RHΔmic5, and RHΔsub1 parasites were harvested and resuspended in the invasion medium at 3 × 107 ml−1 at room temperature. Parasites were induced with invasion medium containing 1% ethanol for 2 min to stimulate release of ESAs, followed by the addition of an equal volume of 8% paraformaldehyde and the incubation at room temperature for 20 min. Fixed parasites were pelleted and blocked with 10% FBS in PBS for 10 min and immunostained with rabbit anti-PfSUB1 and rat anti-TgMIC5. Goat anti-rabbit IgG antibody conjugated with fluorescein isothiocyanate (FITC) and goat anti-rat IgG antibody conjugated with phycoerythrin were used as secondary antibodies. Intensities of FITC and phycoerythrin dyes were measured using a BD FACSCanto flow cytometer and recorded using BD FACSDiva software. Quantification was performed and plotted using FlowJo software version 9.1.
RESULTS
The Solution Structure of TgMIC5
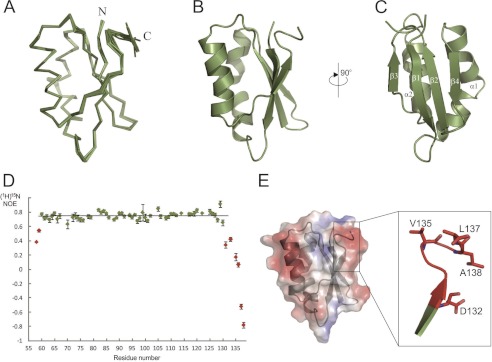
Sequence analysis revealed that TgMIC5 bears no significant sequence similarity (E value > 0.001) with any structurally characterized protein. To provide insight to the mechanism of TgMIC5 regulation of TgSUB1 processing, we determined the solution structure of recombinant TgMIC5 using heteronuclear multidimensional NMR spectroscopy. After completion of backbone assignments it became apparent that the relaxation properties of the first 54 amino acids were characteristic of high dynamics and displayed no evidence for stable secondary structure. After their removal, the NMR spectra of the globular domain remained unchanged. The NMR side chain resonance assignments and full structure calculations were continued using this construct. The structure revealed an open-sandwich antiparallel-α/antiparallel-β fold, comprising two α-helices, α1 and α2, and a four-stranded β-sheet, β1–β4 (Fig. 1, A–C). The heteronuclear 15N NOE showed that NOE values for the C-terminal six residues are significantly lower than for the main globular domain (Fig. 1D). Regions that undergo internal motion faster than overall tumbling showed a decreased NOE intensity relative to the average. This indicates that the C-terminal peptide is flexible (Fig. 1E).
FIGURE 1.
Solution structure of TgMIC5. A, ensemble of 10 lowest energy structures. Individual structures are shown as Cα traces. B and C, representations of the average structure following refinement in water. D, 1H-15N heteronuculear NOE data for TgMIC5. The distribution of 15N heteronuclear NOE across the sequence of TgMIC5 is shown. Lower NOE values indicate faster internal motions relative to the rest of the protein (22). E, electrostatic surface of a representative TgMIC5. Inset shows a sketch of the C-terminal flexible region with hydrophobic residues highlighted.
TgMIC5 Is Structurally Similar to Protease Prodomains
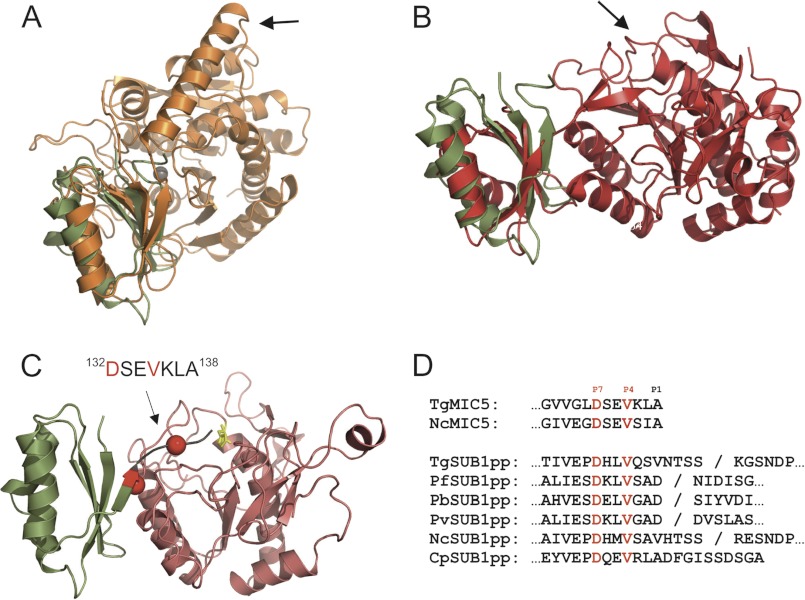
A search for protein structure similarities using programs DALI (26) and PDBeFold (27) revealed a similarity to the prodomains of zinc carboxypeptidases (Z-score = 5.8 for PDB ID code 2BOA; Fig. 2A) (28–30). Prodomains can serve as intramolecular chaperones as well as transient inhibitors to prevent premature enzymatic activity. For these metalloproteinases, the prodomains are N-terminal extensions that function as a gatekeeper by preventing substrate access to the catalytic site. The globular part of the prodomain contacts the entrance to the active site. Activation occurs through proteolysis of the connecting helical linker, which makes further interactions with the enzymatic domain and contains the cleavage site for activation. TgMIC5 does not possess these features, but is terminated with a sequence that displays significant flexibility in the solution structure (Fig. 1D). This region also possesses three hydrophobic residues (DSEVKLA; Fig. 1E) together with four charged groups (Asp, Glu, Lys, and C terminus) that could contribute to a protein-protein interaction site. Further manual analysis of similar structures revealed that the structure of TgMIC5 is also shared by prodomains of the subtilisin-like family of proteases (Z-score = 3.4 for PDB ID code 3CNQ; Fig. 2B) (31, 32). In this case the C-terminal junction point lies directly within the active site and inhibits activity. Subtilisin prodomains can be used in trans as soluble inhibitors of their cognate enzymes, as well as inhibit those from other family members (33). The C-terminal region of subtilisin prodomains is essential for enzymatic inhibition. Furthermore, a small hydrophobic surface is formed between the β-sheet surface of the prodomains and two helices from the catalytic domain (Fig. 2, A and B). The similarity with subtilisin-like prodomains raises the question of whether TgMIC5 acts in a fashion similar to that of an inhibitor of TgSUB1. The catalytic domain of TgSUB1 shows sequence identities of ∼30% to the closest nonapicomplexan subtilisins, which enables a homology model of this domain to be generated reliably. The model of TgSUB1 was calculated with SWISSmodel (34) using the crystal structure of proteinase K from Refs. 35, 36 (Fig. 2C), and TgMIC5 docked using the orientation and location expected from prodomain interactions. A striking feature of this model is that the flexible C terminus (EVLKA) could extend into the active site of the enzyme (Fig. 2C).
FIGURE 2.
Model for the interaction between of TgSUB1 and TgMIC5. A and B, structural similarity between TgMIC5 and human procarboxypeptidase A4 (Z-score = 5.8 for PDB ID code 2BOA) (28, 30) (A), and subtilisin (Z-score = 3.4 for PDB ID code 3CNQ) (31, 32) (B). Arrows indicate the cleavage sites for activation. C, homology model of TgSUB1 docked onto the solution structure of TgMIC5. Potential interaction motif is indicated with conserved side chains as red balls. The catalytic histidine is shown in yellow sticks. D, alignment of the TgMIC5 C-terminal sequence with the corresponding region of apicomplexan subtilisin propeptides. The flexible C-terminal peptide of TgMIC5 is underlined. Fully conserved residues are indicated in emboldened red and by asterisks. A forward slash (/) indicates the propeptide cleavage site, which was empirically determined for TgSUB1 (45) and PfSUB1 (46) and implied by homology for Plasmodium berghei SUB1 (PbSUB1), Plasmodium vivax SUB1 (PvSUB1) N. caninum SUB1 (NcSUB1), and Cryptosporidium parvum SUB1 (CpSUB1). GenBank accession numbers: TgMIC5, CAA70921; putative NcMIC5, NCLIV_068520, CBZ56428; TgSUB1, AAK94670; PfSUB1, CAA05627; PbSUB1, XP_679126; PvSUB1, ACT76164; NcSUB1, AAF04257; CpSUB1, CAD98301.
We reasoned that if the mechanism of action of TgMIC5 is similar to that of a subtilisin propeptide then the flexible C-terminal peptide should contain amino acids that are conserved in the corresponding region of subtilisin propeptides. Alignment of TgMIC5 with the propeptides of TgSUB1 and SUB1 sequences from other apicomplexans revealed strict conservation of Asp132 and Val135, suggesting that these residues are important for the mechanism of action (Fig. 2D). Based on this alignment and the assignment of residues relative to the Plasmodium SUB1 propeptide cleavage site, Val135 occupies the P4 position and Asp132 resides in the P7 position.
The C Terminus of TgMIC5 Contributes to TgSUB1 Inhibition
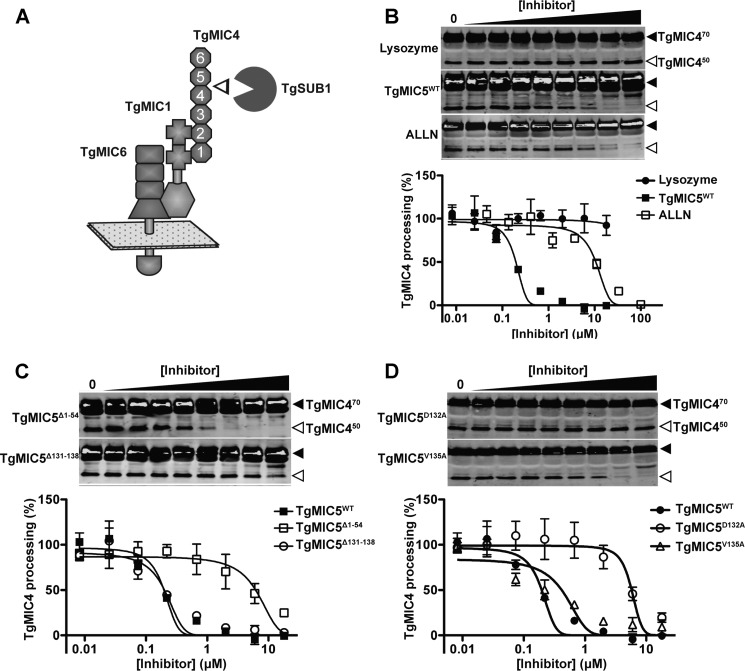
The docking model between TgMIC5 and the active site of TgSUB1 suggested a possible mechanism in which the flexible C terminus of TgMIC5 extends into the active site of TgSUB1 to inhibit its activity. To assess this we developed a novel assay for recombinant TgMIC5 inhibition of native TgSUB1. TgSUB1 is secreted from the micronemes to the parasite surface along with its substrates including TgMIC470. TgSUB1 incompletely converts TgMIC470 into TgMIC450 and TgMIC420 (Fig. 3A). All three forms of TgMIC4 are subsequently released from the surface as soluble ESAs. Based on these events we assessed the abundance of TgMIC450 in the ESA as a measure of TgSUB1 activity and inhibition thereof. Recombinant TgMIC5 was added to the parasites prior to secretion, and its inhibitory activity was compared with that of ALLN, a previously described inhibitor of TgSUB1 (15). Lysozyme, a globular protein of a size similar to TgMIC5, was included as a negative control. We found that WT TgMIC5 (TgMIC5WT) inhibited TgSUB1 activity with an IC50 of 0.22 μm whereas ALLN was less effective with an IC50 of 9.9 μm (Fig. 3B and Table 2). These findings indicate that TgMIC5 is almost 50-fold more potent than ALLN. To further test that the flexible C terminus of TgMIC5 contributes to TgSUB1 inhibition, we generated a mutant TgMIC5 protein (TgMIC5Δ131–138) lacking the last 8 amino acids. We also created an N-terminally truncated TgMIC5 mutant (TgMIC5Δ1–54) lacking the unstructured first 54 amino acids as a control. TgMIC5Δ131–138 (IC50 4.4 μm) was 20-fold less potent than TgMIC5WT whereas TgMIC5Δ1–54 showed no loss of inhibitory activity (Fig. 3C and Table 2). These findings are consistent with the model that the flexible C-terminal TgMIC5 participates in the mechanism of TgSUB1 inhibition.
FIGURE 3.
The C terminus of TgMIC5 plays a critical role in inhibition of TgSUB1. A, schematic illustrates TgMIC4 processing on the parasite surface by TgSUB1 protease. The six PAN/Apple domains of TgMIC4 are numbered from the N terminus, with cleavage by TgSUB1 occurring between domains 4 and 5 to produce an N-terminal fragment termed TgMIC450. B, TgMIC5 inhibitory activity for TgSUB1 was quantified. Recombinant TgMIC5 at the indicated concentrations was added to live tachyzoites expressing surface TgSUB1, which coverts TgMIC470 to TgMIC450 before being shed into the medium as an ESA product. ESA products were collected from the supernatant after centrifugation, separated by 12.5% SDS-PAGE and immunoblotted with rabbit anti-TgMIC4 and goat anti-rabbit IgG conjugated with IRDye 800CW. The inhibition efficacy of TgMIC5WT was compared with ALLN by infrared laser scanning (LI-COR Biosciences) to measure the intensity of TgMIC450 in the ESA. Lysosome was included as a negative control protein. C, to assess the functional region within TgMIC5 protein contributing to inhibition of TgSUB1 activity, 54 (Δ1–54) and 8 amino (Δ131–138) acids were deleted from N and C termini of WT TgMIC5, respectively. These truncation mutants were tested for inhibitory activity according to the description in B. D, to further identify the key residue(s) in the C terminus of TgMIC5 for its inhibition ability, Asp132 and Val135 were replaced with alanine to create TgMIC5D132A and TgMIC5V135A. Inhibitory activity was measured according to B. Quantitative data in B–D are from at least three independent experiments each. Error bars are S.E.
TABLE 2.
Half-maximal inhibitory concentrations (IC50) of TgSUB1 inhibitors
| Inhibitor | IC50 (μm) (mean ± S.E.) | Change relative to TgMIC5WT |
|---|---|---|
| -fold | ||
| Lysozyme | NDa | ND |
| TgMIC5WT | 0.22 ± 0.02 | 1.0 |
| ALLN | 9.9 ± 0.2 | 45 |
| TgMIC5Δ131–138 | 4.4 ± 1.2 | 20 |
| TgMIC5Δ1–54 | 0.27 ± 0.06 | 1.2 |
| TgMIC5D132A | 4.6 ± 0.9 | 21 |
| TgMIC5V135A | 0.41 ± 0.06 | 1.9 |
| TgMIC5D132A/V135A | 1.9 ± 0.2 | 8.6 |
a ND, not determined.
To test the putative role of the strictly conserved Asp132 and Val135 in protease inhibition, we generated alanine substitution mutants at each of these positions. The TgMIC5V135A mutant showed a modest 2-fold decrease in inhibitory activity compared with the WT protein (Fig. 3D and Table 2). However, the TgMIC5D132A mutant exhibited an IC50 of 4.6 μm, which is similar to that of the poorly active C-terminally truncated TgMIC5Δ131–138 mutant. This 23-fold loss of inhibitory activity suggests that TgMIC5 Asp132 is a key contributor to the mechanism of TgSUB1 inhibition. Collectively, the C terminus of TgMIC5 plays a critical role in blocking TgSUB1 activity, and Asp132 within this fragment contributes significantly to the mechanism of inhibition.
TgMIC5 Inhibition of TgSUB1 Activity Also Affects Processing of Other Secreted Micronemal Substrates
To test whether the inhibition of TgSUB1 by TgMIC5WT also affects other substrates on the parasite surface, we evaluated the proteolytic trimming of other microneme proteins including TgMIC2, TgM2AP, and TgPLP1. We also assessed TgSUB1 self-shedding from the parasite surface. The assay was performed using 0.67 μm TgMIC5, which is slightly higher than the IC90 value of TgMIC5WT (0.42 μm). TgMIC5Δ131–138 was included as a control.
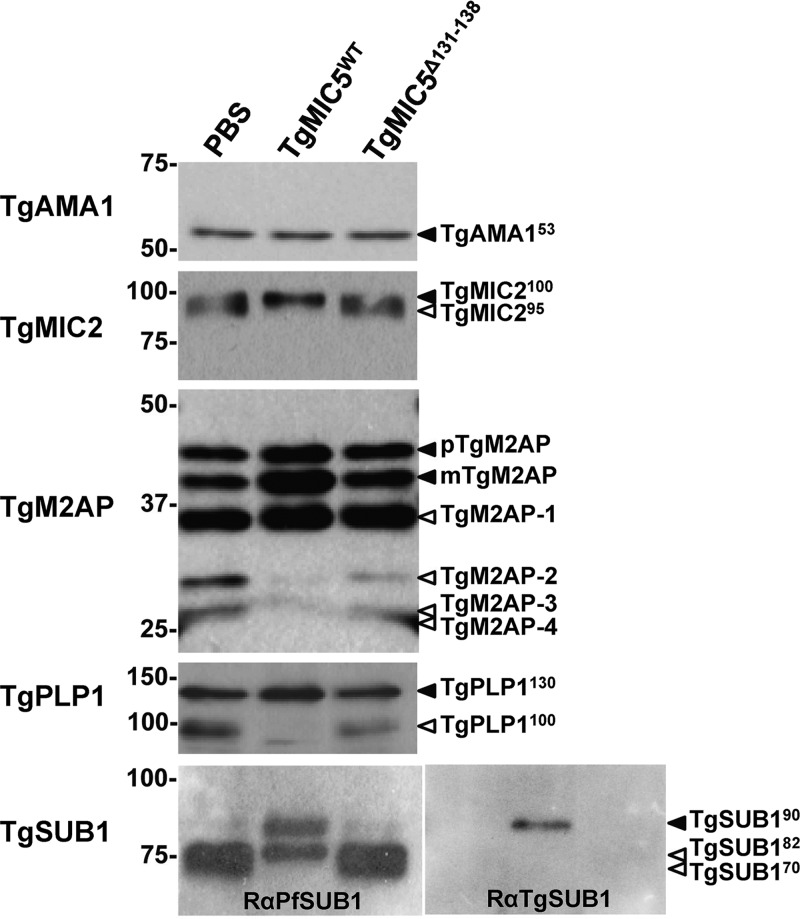
The abundance and migration of secreted TgAMA1 were not affected by TgMIC5WT or TgMIC5Δ131–138 (Fig. 4), consistent with it not being a substrate of TgSUB1 (15). TgMIC2 is shed from the parasite surface as a mixture of species including a 100-kDa form with an intact N terminus and a 95-kDa form resulting from N-terminal trimming by TgSUB1. TgMIC5WT inhibited trimming to the 95-kDa species whereas the C-terminally truncated TgMIC5Δ131–138 mutant failed to disrupt trimming (Fig. 4). TgM2AP forms a complex with TgMIC2 and is processed by TgSUB1 into four species termed TgM2AP-1, -2, -3, and -4 (37). TgM2AP-1 is thought to be processed by TgSUB1 prior to or coincident with nascent secretion from the micronemes based on its resistance to inhibition by ALLN (15). Consistent with this, TgMIC5WT strongly inhibited the production of TgM2AP-2, -3, and -4 in the ESA and resulted in the accumulation of TgM2AP-1 and the other unprocessed species proTgM2AP (pTgM2AP) mature M2AP (mM2AP). In contrast, the C-terminally truncated TgMIC5Δ131–138 showed minimal inhibitory activity, slightly affecting the production of TgM2AP-2, -3, and -4. Similarly, mature TgPLP1 migrating at 130 kDa was not cleaved by TgSUB1 into a 100-kDa species (15) upon addition of TgMIC5WT but was processed in the presence of the C-terminally truncated TgMIC5 mutant.
FIGURE 4.
TgMIC5 inhibition of TgSUB1 activity also impairs processing of other secreted micronemal substrates. PBS, recombinant TgMIC5WT, or recombinant TgMIC5Δ131–138 was incubated with live tachyzoites expressing surface TgSUB1. ESA products were collected and immunoblotted for the indicated antigens. TgAMA1 was included as the negative control and an indicator of loading consistency.
TgSUB1 is stored in the micronemes as a 90-kDa GPI-anchored species (TgSUB190), which is secreted onto the surface where it encounters its substrates. TgSUB190 is released from the surface into the ESA as 82- and 70-kDa species (TgSUB182 and TgSUB170). Binder et al. generated an rabbit anti-peptide antibody (RαTgSUB1) against a C-terminal antigenic peptide immediately adjacent to its GPI anchor (38). This antibody only recognizes TgSUB190 and the larger precursor forms whereas a rabbit polyclonal antibody against the catalytic domain of PfSUB1 (RαPfSUB1) recognizes all of the species based on the antigenic similarity between TgSUB1 and PfSUB1. When the ESA fraction was incubated with PBS or C-terminally truncated TgMIC5Δ131–138, TgSUB190 was quantitatively processed to a mixture of TgSUB182 and TgSUB170, which migrated near one another (Fig. 4). Exogenously added TgMIC5WT inhibited processing to TgSUB170, resulting in exclusive detection of the TgSUB190 and TgSUB182 (Fig. 4). RαTgSUB1 recognized TgSUB190, indicating this species in the ESA has an intact C-terminal epitope recognized by this antibody. Altogether, our findings indicate that TgMIC5 inhibits TgSUB1 processing of micronemal substrates and itself.
TgMIC5 Physically Interacts with TgSUB1
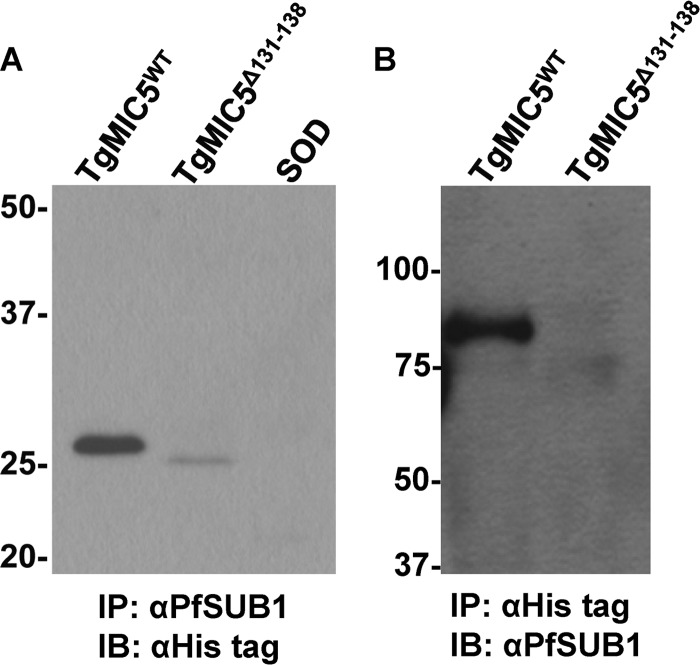
The docking model predicted that the C terminus of TgMIC5 interacts with the active site of TgSUB1 to act as the inhibitor. We tried to immunoprecipitate the endogenous TgMIC5-TgSUB1 complex from RH parasite ESA and cell lysate, but these efforts were unsuccessful, indicating a fragile interaction between TgMIC5 and TgSUB1. To compensate for a weak interaction, we incubated 0.67 μm recombinant N-terminally His6-tagged TgMIC5WT with ESA containing TgSUB1. As controls, the N-terminally His6-tagged TgMIC5Δ131–138 and superoxide dismutase were also incubated with ESA at the same concentration. When we used a polyclonal anti-PfSUB1 antibody to immunoprecipitate TgSUB1 and probed it with a monoclonal anti-His antibody, TgMIC5WT was detected more readily than the C-terminally truncated mutant TgMIC5Δ131–138 (Fig. 5A). Superoxide dismutase did not interact with TgSUB1, as expected. However, when we attempted to perform the reciprocal experiment, we failed to detect the TgSUB1 on the blot under the conditions used. Interestingly, when recombinant TgMIC5WT and TgMIC5131–138 were incubated with live parasites during secretion, TgMIC5WT interacted with TgSUB190 in the ESA. In contrast, TgSUB190 was not detected in association with TgMIC5131–138, and instead an apparent weak association of this truncation mutant with TgSUB182 and TgSUB170 was detected (Fig. 5B). These results are consistent with TgMIC5 interacting physically with TgSUB1 to regulate its activity, and the findings suggest that the TgMIC5 C-terminal domain contributes to the interaction.
FIGURE 5.
TgMIC5 physically interacts with TgSUB1 protein. A, recombinant TgMIC5WT, recombinant TgMIC5Δ131–138, or an irrelevant protein (superoxide dismutase) carrying His6 tags at their N termini was incubated with the ESA from RH parasites to test for a molecular interaction between TgMIC5 and TgSUB1. A polyclonal antibody against PfSUB1 (αPfSUB1) was used to immunoprecipitate (IP) TgSUB1, and a monoclonal antibody recognizing the His6 tag (αHis tag) was used to detect recombinant protein associated with TgSUB1 by immunoblotting (IB). B, recombinant TgMIC5WT or recombinant TgMIC5Δ131–138 was incubated with RH parasites before collecting ESA for immunoprecipitation with αHis tag and detection with αPfSUB1.
TgMIC5 Is Associated with the Parasite Surface via Its Interaction with TgSUB1
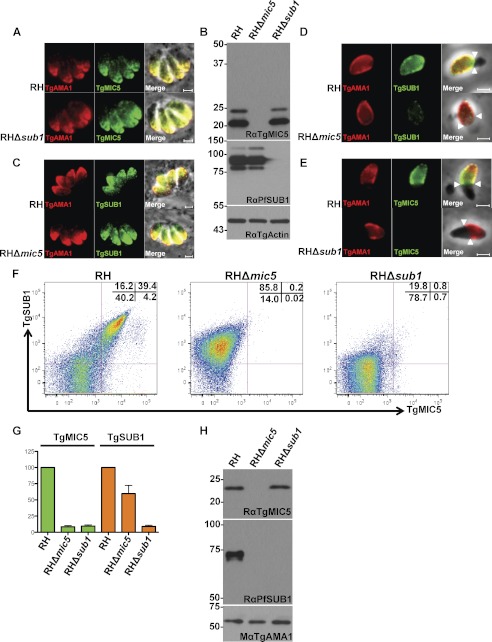
TgMIC5 resides on the parasite surface during cell invasion despite not possessing a membrane anchor (16, 17), raising the question of how TgMIC5 is retained on the parasite surface. Our finding that TgMIC5 interacts with and inhibits TgSUB1 suggests a possible mechanism of surface retention via TgSUB1. To test this, we first determined whether the expression and intracellular trafficking of each protein are independent of the other based on analysis of RHΔsub1 and RHΔmic5 mutants (Fig. 6, A–C). TgMIC5 expression and localization were normal in RHΔsub1 as assessed by immunofluorescence co-localization with TgAMA1, a marker for micronemes (Fig. 6A), and immunoblotting (Fig. 6B). Similarly, TgSUB1 expression and localization in the micronemes were normal in RHΔmic5 parasites (Fig. 6, B and C). To assess their interdependence for surface expression during cell invasion, we stained invading RHΔmic5 and RHΔsub1 parasites for TgSUB1 or TgMIC5, respectively, and TgAMA1. TgSUB1 expression on the surface of RHΔmic5 parasites was moderately reduced (Fig. 6D), suggesting that TgMIC5 influences the surface abundance of TgSUB1. More strikingly, RHΔsub1 parasites were completely negative for surface TgMIC5 (Fig. 6E), indicating that TgSUB1 is required for TgMIC5 retention on the parasite surface. We further quantified the abundance of TgMIC5 or TgSUB1 proteins on the surface of fixed RHΔsub1 and RHΔmic5 mutants by flow cytometry after inducing microneme secretion by treatment with 1% ethanol (39). Consistent with the invasion findings, the intensity of TgSUB1 on the surface of RHΔmic5 parasites was ∼40% lower than that of RH parasites, and no TgMIC5 was detected on the surface of RHΔsub1 parasites (Fig. 6, F and G).
FIGURE 6.
TgMIC5 is associated with the parasite surface via its interaction with TgSUB1. A, TgMIC5 traffics normally to the micronemes in RHΔsub1 mutant. Overnight replicated parasites were fixed and stained with rabbit anti-TgMIC5 and mouse monoclonal antibody B3.90 anti-TgAMA1, which were detected with species-specific secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 594. Scale bars, 2 μm. B, immunoblots of parasite lysates probed with the indicated antibodies show normal expression of TgMIC5 in RHΔsub1 mutant parasites and diminished expression of TgSUB1 in RHΔmic5 parasites. TgActin was detected as a loading control. C, TgSUB1 traffics normally to the micronemes in RHΔmic5 parasites. Parasites were stained with rabbit anti-PfSUB1 and monoclonal B3.90 anti-TgAMA1. Scale bars, 2 μm. D, TgSUB1 is displayed on the surface of RHΔmic5 parasites during invasion albeit with diminished intensity. Scale bars, 2 μm. E, TgMIC5 requires TgSUB1 expression for retention on the parasite surface during cell invasion. Parasites were pulse-invaded into human foreskin fibroblast cells as described under “Experimental Procedures,” fixed, and stained with antibodies to the indicated proteins. F, abundance of TgMIC5 and TgSUB1 on the parasite surface was measured by flow cytometry. Microneme secretion was stimulated with ethanol, and parasites were fixed and stained with rabbit anti-PfSUB1 and rat anti-TgMIC5, which were detected with species-specific secondary antibodies conjugated with FITC or phycoerythrin. G, flow cytometry relative intensity analysis showed the absence of TgMIC5 protein on the RHΔsub1 mutant surface, whereas the amount of TgSUB1 on the RHΔmic5 mutant surface is ∼60% of that on the surface of RH parasites. The intensity of staining for RH parasites was set to 100%. H, immunoblots of ESA show WT and mutant parasites detected with the indicated antibodies.
We also assessed the abundance of secreted TgMIC5 or TgSUB1 in the ESA derived from RHΔsub1 or RHΔmic5 parasites. Similar amounts of TgMIC5 were released to the ESA between RH and RHΔsub1 parasites (Fig. 6H), whereas TgSUB1 was not detectable in the ESA of RHΔmic5 (Fig. 6H). Because the lack of TgSUB1 in the ESA is more severe than the reduction of TgSUB1 on the parasite surface, TgSUB1 appears to be unstable and is possibly degraded upon its release into the ESA in the absence of TgMIC5. Altogether, our results suggest that the expression of TgSUB1 is partially dependent on TgMIC5 and that whereas TgMIC5 expression and localization are independent of TgSUB1, its surface localization is dependent entirely on the presence of TgSUB1.
DISCUSSION
TgSUB1 proteolytically processes several Toxoplasma proteins that are secreted from the micronemes on the parasite surface. This processing has been termed “trimming” because in most cases it results in the clipping of small peptides from the termini of MIC proteins (40). Although the role of surface trimming is not fully understood, the findings of two studies suggest that trimming enhances the adhesive function of T. gondii MIC proteins. Barragan et al. showed that an N-terminally trimmed species of TgMIC2 binds more readily to intercellular adhesion molecule-1 (ICAM-1) than the untrimmed species (41). T. gondii uses ICAM-1 as a host surface receptor for paracellular (between cell) transmigration across epithelial cell layers during infection. More recently, Lagal et al. (15) reported that the failure of TgSUB1 knock-out parasites to process MIC proteins including TgMIC2, TgM2AP, and TgMIC4 corresponded with diminished parasite attachment and invasion of host cells. TgSUB1 knock-out parasites also displayed aberrant gliding motility and attenuated virulence in mice. A distinct role in immune modulation has also been recently proposed for TgSUB1 processing of TgMIC4, which generates a galactose-binding product (TgMIC420) from the C terminus of TgMIC4 (42). Earlier work revealed a function for TgMIC5 in regulating TgSUB1 activity (16), but the molecular mechanism of regulation remained unclear. Our current findings strongly suggest that TgMIC5 suppresses TgSUB1 activity directly by mimicking the inhibitory mechanism of a subtilisin propeptide.
The NMR solution structure of amino acid residues 54–132 from TgMIC5 revealed a globular domain resembling the structure of propeptides from certain metalloproteinases and subtilisins. Typically, these fold into a compact structure with a four-stranded β-sheet and two α-helices. Although prodomains tend to be unstructured in isolation and their stability is often linked to the folding of the catalytic domain, the prodomain from PfSUB1 has been shown to be autonomously folded and interacts tightly with the mature enzyme (43). TgMIC5 is also able to fold spontaneously, and its structural similarity to subtilisin prodomains suggests that it likely interacts with the mature enzyme in a similar fashion. We confirmed the presence of a direct interaction between TgMIC5 and TgSUB1 (Fig. 5) and derived a homology model for the complex based on known prodomain-subtilisin structures (Fig. 2C). Intriguingly, our model places the flexible C-terminal peptide (amino acids 132–138) of TgMIC5 within reach of the active site, and its extension positions the C terminus directly over the catalytic triad (Fig. 2C). In this scenario, the C-terminal region of TgMIC5 would block substrate access to the TgSUB1 active site groove. NCLIV_068520 from Neospora caninum is the only likely orthologue of TgMIC5, possessing 55% sequence identity (Fig. 2D). Comparison of the C-terminal sequences of TgMIC5, NCLIV_068520 and known prodomains of apicomplexan subtilisins reveals two conserved residues, namely a valine at P4 and aspartate at P7. Although the P4 (Val) residue has been shown to be crucial for substrate recognition of PfSUB1 (44), it is not essential for the inhibition properties of its propeptide (43). However, removal of the C-terminal 11 residues from the PfSUB1 prodomain does reduce potency. The P4 position is also valine in TgMIC5 (Fig. 2D), and consistent with these results, replacement with alanine is not substantially deleterious to its inhibitory properties. We have, however, identified a conserved aspartate residue at P7 and showed that a point mutation at this position significantly reduced the potency of TgSUB1 inhibition. This residue lies at the proposed interface of the TgMIC5-TgSUB1 complex (Fig. 2C). Furthermore, our NMR dynamics measurements indicate that Asp132 forms a hinge point at the end of the globular domain, which would allow for the correct positioning of the flexible C terminus within the active site groove.
We showed using a novel live cell assay that recombinant TgMIC5 inhibits parasite-derived TgSUB1 processing of TgMIC2, TgM2AP, TgMIC4, and TgPLP1. This finding supports a model wherein TgMIC5 regulates the activity of TgSUB1, permitting an optimal level of proteolysis and precluding the degradation of off targets or the hyperprocessing of its normal substrates. Consistent with this model, TgGRA1 is less abundant in the ESA and appears to be degraded following secretion from TgMIC5-deficient parasites (16). TgGRA1 is not normally a substrate of TgSUB1 because it is secreted after parasite invasion into the parasitophorous vacuole, a site where TgSUB1 is not found. Several other unidentified proteins also seem to be degraded upon secretion from TgMIC5-deficient parasites. Additionally, whereas TgSUB1 processes TgMIC4 into TgMIC450 and TgMIC420 in WT parasites, it hyperprocesses TgMIC4 into a new species, TgMIC418, in the absence of TgMIC5 (16). That TgMIC5 parasites do not appear to have an invasion or virulence phenotype might be the result of offsetting effects: higher efficiency trimming of the major adhesin TgMIC2 promotes parasite attachment and invasion whereas hyperprocessing or degradation of other substrates diminishes their functionality.
Interestingly, our findings indicate that TgMIC5 also affects the processing and stability of TgSUB1 itself. TgSUB190 is the GPI-anchored species in the micronemes and presumably on the parasite surface. Recombinant TgMIC5 inhibited the processing of TgSUB190 to the TgSUB170 soluble form in the ESA. This implies that TgSUB1 activity is required for self-shedding from the parasite surface and the release of TgSUB170 into the ESA. TgMIC5 inhibition also resulted in the abnormal presence TgSUB190 into the ESA. It remains unclear in this situation whether the TgSUB190 ESA form is still GPI-anchored or if it is shed from the membrane by a phospholipase or another protease. If it is shed by another protease then the cleavage site must be immediately adjacent to the GPI anchor because the TgSUB190 ESA form still contains the C-terminal proximal epitope recognized by RαTgSUB1. That the abundance of the TgSUB182-soluble form in the ESA was not affected by TgMIC5 inhibition of TgSUB1 activity suggests that it is shed by another surface protease. We also found that TgSUB1 is moderately less abundant on the surface of RHΔmic5 parasites and essentially absent from the ESA of these parasites. Both of these findings can be explained by the enhanced activity of TgSUB1, resulting in increased self-shedding from the parasite surface and self-degradation in the ESA.
It was previously unclear how TgMIC5 remained associated with the parasite surface during cell invasion because an analysis of its primary structure did not suggest a transmembrane domain or lipid anchor. Consistent with the absence of intrinsic membrane association, the heterologous expression of TgMIC5 in mammalian cells resulted in its secretion into the medium as a soluble protein, with no discernable retention on the plasma membrane (16). Our findings strongly suggest that TgMIC5 protein is retained on the parasite plasma membrane via its interaction with TgSUB1. Nonetheless, despite the multiple lines of evidence linking TgMIC5 to TgSUB1, the interaction between these proteins appears to be somewhat fragile based on the difficulties of capturing the endogenous TgMIC5-TgSUB1 complex from parasite lysates and ESA. The addition of excess recombinant TgMIC5WT to RH parasite ESA compensated for the apparent low affinity between TgMIC5 and TgSUB1, resulting in the detection of an interaction complex. The modest interaction between these proteins likely favors a balanced regulation of TgSUB1, thereby permitting an appropriate level of processing to ensure efficient cell invasion without adverse proteolysis. Accordingly, our findings reveal a novel mechanism of proteolytic regulation by a propeptide mimic that fine tunes proteolysis during infection.
Acknowledgment
We thank Dr. Pete Simpson for support and useful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant AI063263 (to V. B. C.). This work was also supported by Medical Research Council Grants G0800038 (to S. M.) and an American Heart Association postdoctoral fellowship (to Z. D.).
- MIC
- microneme
- ALLN
- N-[N-(N-acetyl-l-leucyl)-l-leucyl]-l-norleucine
- ESA
- excreted-secreted antigen
- GPI
- glycosylphosphatidylinositol
- PDB
- Protein Data Bank.
REFERENCES
- 1. Rothova A. (2003) Ocular manifestations of toxoplasmosis. Curr. Opin. Ophthalmol. 14, 384–388 [DOI] [PubMed] [Google Scholar]
- 2. Richards F. O., Jr., Kovacs J. A., Luft B. J. (1995) Preventing toxoplasmic encephalitis in persons infected with human immunodeficiency virus. Clin. Infect. Dis. 21, S49-S56 [DOI] [PubMed] [Google Scholar]
- 3. Sell M., Klingebiel R., Di Iorio G., Sampaolo S. (2005) Primary cerebral toxoplasmosis: a rare case of ventriculitis and hydrocephalus in AIDS. Clin. Neuropathol. 24, 106–111 [PubMed] [Google Scholar]
- 4. Carruthers V. B. (2002) Host cell invasion by the opportunistic pathogen Toxoplasma gondii. Acta Trop. 81, 111–122 [DOI] [PubMed] [Google Scholar]
- 5. Mead P. S., Slutsker L., Dietz V., McCaig L. F., Bresee J. S., Shapiro C., Griffin P. M., Tauxe R. V. (1999) Food-related illness and death in the United States. Emerg. Infect. Dis. 5, 607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vaillant V., de Valk H., Baron E., Ancelle T., Colin P., Delmas M. C., Dufour B., Pouillot R., Le Strat Y., Weinbreck P., Jougla E., Desenclos J. C. (2005) Foodborne infections in France. Foodborne Pathog. Dis. 2, 221–232 [DOI] [PubMed] [Google Scholar]
- 7. Carruthers V. B., Boothroyd J. C. (2007) Curr. Opin. Microbiol. 10, 83–89 [DOI] [PubMed] [Google Scholar]
- 8. Sibley L. D. (2011) Invasion and intracellular survival by protozoan parasites. Immunol. Rev. 240, 72–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Soldati-Favre D. (2008) Molecular dissection of host cell invasion by the apicomplexans: the glideosome. Parasite 15, 197–205 [DOI] [PubMed] [Google Scholar]
- 10. Lai L., Bumstead J., Liu Y., Garnett J., Campanero-Rhodes M. A., Blake D. P., Palma A. S., Chai W., Ferguson D. J., Simpson P., Feizi T., Tomley F. M., Matthews S. (2011) The role of sialyl glycan recognition in host tissue tropism of the avian parasite Eimeria tenella. PloS Pathog. 7, e1002296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blumenschein T. M., Friedrich N., Childs R. A., Saouros S., Carpenter E. P., Campanero-Rhodes M. A., Simpson P., Chai W., Koutroukides T., Blackman M. J., Feizi T., Soldati-Favre D., Matthews S. (2007) Atomic resolution insight into host cell recognition by Toxoplasma gondii. EMBO J. 26, 2808–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Friedrich N., Matthews S., Soldati-Favre D. (2010) Sialic acids: key determinants for invasion by the Apicomplexa. Int. J. Parasitol. 40, 1145–1154 [DOI] [PubMed] [Google Scholar]
- 13. Friedrich N., Santos J. M., Liu Y., Palma A. S., Leon E., Saouros S., Kiso M., Blackman M. J., Matthews S., Feizi T., Soldati-Favre D. (2010) Members of a novel protein family containing microneme adhesive repeat domains act as sialic acid-binding lectins during host cell invasion by apicomplexan parasites. J. Biol. Chem. 285, 2064–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saouros S., Edwards-Jones B., Reiss M., Sawmynaden K., Cota E., Simpson P., Dowse T. J., Jäkle U., Ramboarina S., Shivarattan T., Matthews S., Soldati-Favre D. (2005) A novel galectin-like domain from Toxoplasma gondii micronemal protein 1 assists the folding, assembly, and transport of a cell adhesion complex. J. Biol. Chem. 280, 38583–38591 [DOI] [PubMed] [Google Scholar]
- 15. Lagal V., Binder E. M., Huynh M. H., Kafsack B. F., Harris P. K., Diez R., Chen D., Cole R. N., Carruthers V. B., Kim K. (2010) Toxoplasma gondii protease TgSUB1 is required for cell surface processing of micronemal adhesive complexes and efficient adhesion of tachyzoites. Cell. Microbiol. 12, 1792–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brydges S. D., Zhou X. W., Huynh M. H., Harper J. M., Mital J., Adjogble K. D., Däubener W., Ward G. E., Carruthers V. B. (2006) Targeted deletion of MIC5 enhances trimming proteolysis of Toxoplasma invasion proteins. Eukaryot. Cell 5, 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brydges S. D., Sherman G. D., Nockemann S., Loyens A., Däubener W., Dubremetz J. F., Carruthers V. B. (2000) Molecular characterization of TgMIC5, a proteolytically processed antigen secreted from the micronemes of Toxoplasma gondii. Mol. Biochem. Parasitol. 111, 51–66 [DOI] [PubMed] [Google Scholar]
- 18. Harper J. M., Huynh M. H., Coppens I., Parussini F., Moreno S., Carruthers V. B. (2006) A cleavable propeptide influences Toxoplasma infection by facilitating the trafficking and secretion of the TgMIC2-M2AP invasion complex. Mol. Biol. Cell 17, 4551–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marchant J., Sawmynaden K., Saouros S., Simpson P., Matthews S. (2008) Complete resonance assignment of the first and second apple domains of MIC4 from Toxoplasma gondii, using a new NMRView-based assignment aid. Biomol. NMR Assign. 2, 119–121 [DOI] [PubMed] [Google Scholar]
- 20. Rieping W., Habeck M., Bardiaux B., Bernard A., Malliavin T. E., Nilges M. (2007) ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics 23, 381–382 [DOI] [PubMed] [Google Scholar]
- 21. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 23. Brecht S., Carruthers V. B., Ferguson D. J., Giddings O. K., Wang G., Jakle U., Harper J. M., Sibley L. D., Soldati D. (2001) The toxoplasma micronemal protein MIC4 is an adhesin composed of six conserved apple domains. J. Biol. Chem. 276, 4119–4127 [DOI] [PubMed] [Google Scholar]
- 24. Endo T., Tokuda H., Yagita K., Koyama T. (1987) Effects of extracellular potassium on acid release and motility initiation in Toxoplasma gondii. J. Protozool. 34, 291–295 [DOI] [PubMed] [Google Scholar]
- 25. Donahue C. G., Carruthers V. B., Gilk S. D., Ward G. E. (2000) The Toxoplasma homolog of Plasmodium apical membrane antigen-1 (AMA-1) is a microneme protein secreted in response to elevated intracellular calcium levels. Mol. Biochem. Parasitol. 111, 15–30 [DOI] [PubMed] [Google Scholar]
- 26. Holm L., Sander C. (1995) DALI: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 27. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 28. García-Castellanos R., Bonet-Figueredo R., Pallarés I., Ventura S., Avilés F. X., Vendrell J., Gomis-Rütha F. X. (2005) Detailed molecular comparison between the inhibition mode of A/B-type carboxypeptidases in the zymogen state and by the endogenous inhibitor latexin. Cell. Mol. Life Sci. 62, 1996–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marx P. F., Brondijk T. H., Plug T., Romijn R. A., Hemrika W., Meijers J. C., Huizinga E. G. (2008) Crystal structures of TAFI elucidate the inactivation mechanism of activated TAFI: a novel mechanism for enzyme autoregulation. Blood 112, 2803–2809 [DOI] [PubMed] [Google Scholar]
- 30. Vendrell J., Querol E., Avilés F. X. (2000) Metallocarboxypeptidases and their protein inhibitors: structure, function and biomedical properties. Biochim. Biophys. Acta 1477, 284–298 [DOI] [PubMed] [Google Scholar]
- 31. Nakagawa M., Ueyama M., Tsuruta H., Uno T., Kanamaru K., Mikami B., Yamagata H. (2010) Functional analysis of the cucumisin propeptide as a potent inhibitor of its mature enzyme. J. Biol. Chem. 285, 29797–29807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruan B., London V., Fisher K. E., Gallagher D. T., Bryan P. N. (2008) Engineering substrate preference in subtilisin: structural and kinetic analysis of a specificity mutant. Biochemistry 47, 6628–6636 [DOI] [PubMed] [Google Scholar]
- 33. Fugère M., Limperis P. C., Beaulieu-Audy V., Gagnon F., Lavigne P., Klarskov K., Leduc R., Day R. (2002) Inhibitory potency and specificity of subtilase-like pro-protein convertase (SPC) prodomains. J. Biol. Chem. 277, 7648–7656 [DOI] [PubMed] [Google Scholar]
- 34. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 35. Helland R., Larsen A. N., Smalås A. O., Willassen N. P. (2006) The 1.8 Å crystal structure of a proteinase K-like enzyme from a psychrotroph Serratia species. FEBS J. 273, 61–71 [DOI] [PubMed] [Google Scholar]
- 36. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou X. W., Blackman M. J., Howell S. A., Carruthers V. B. (2004) Proteomic analysis of cleavage events reveals a dynamic two-step mechanism for proteolysis of a key parasite adhesive complex. Mol. Cell. Proteomics 3, 565–576 [DOI] [PubMed] [Google Scholar]
- 38. Binder E. M., Lagal V., Kim K. (2008) The prodomain of Toxoplasma gondii GPI-anchored subtilase TgSUB1 mediates its targeting to micronemes. Traffic 9, 1485–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carruthers V. B., Moreno S. N., Sibley L. D. (1999) Ethanol and acetaldehyde elevate intracellular [Ca2+] and stimulate microneme discharge in Toxoplasma gondii. Biochem. J. 342, 379–386 [PMC free article] [PubMed] [Google Scholar]
- 40. Carruthers V. B., Blackman M. J. (2005) A new release on life: emerging concepts in proteolysis and parasite invasion. Mol. Microbiol. 55, 1617–1630 [DOI] [PubMed] [Google Scholar]
- 41. Barragan A., Brossier F., Sibley L. D. (2005) Transepithelial migration of Toxoplasma gondii involves an interaction of intercellular adhesion molecule 1 (ICAM-1) with the parasite adhesin MIC2. Cell. Microbiol. 7, 561–568 [DOI] [PubMed] [Google Scholar]
- 42. Marchant J., Cowper B., Liu Y., Lai L., Pinzan C., Marq J. B., Friedrich N., Sawmynaden K., Liew L., Chai W., Childs R. A., Saouros S., Simpson P., Roque Barreira M. C., Feizi T., Soldati-Favre D., Matthews S. (2012) Galactose recognition by the apicomplexan parasite Toxoplasma gondii. J. Biol. Chem. 287, 16720–16733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jean L., Hackett F., Martin S. R., Blackman M. J. (2003) Functional characterization of the propeptide of Plasmodium falciparum subtilisin-like protease-1. J. Biol. Chem. 278, 28572–28579 [DOI] [PubMed] [Google Scholar]
- 44. Silmon de Monerri N. C., Flynn H. R., Campos M. G., Hackett F., Koussis K., Withers-Martinez C., Skehel J. M., Blackman M. J. (2011) Global identification of multiple substrates for Plasmodium falciparum SUB1, an essential malarial processing protease. Infect. Immun. 79, 1086–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miller S. A., Binder E. M., Blackman M. J., Carruthers V. B., Kim K. (2001) A conserved subtilisin-like protein TgSUB1 in microneme organelles of Toxoplasma gondii. J. Biol. Chem. 276, 45341–45348 [DOI] [PubMed] [Google Scholar]
- 46. Sajid M., Withers-Martinez C., Blackman M. J. (2000) Maturation and specificity of Plasmodium falciparum subtilisin-like protease-1, a malaria merozoite subtilisin-like serine protease. J. Biol. Chem. 275, 631–641 [DOI] [PubMed] [Google Scholar]