Background: Clock genes are expressed in different peripheral organs.
Results: Rhythmic expression was lost with Ihh in the growth plate from mice defective of BMAL1 with a small body size.
Conclusion: Endochondral ossification is under the control by clock genes in chondrocytes.
Significance: Peripheral clocks are a target for treating cartilaginous diseases relevant to abnormal postnatal chondrogenesis.
Keywords: Chondrocytes, Circadian Rhythms, Clock Genes, Gene Knockout, Gene Transcription, Hedgehog
Abstract
We have previously shown transient promotion by parathyroid hormone of Period-1 (Per1) expression in cultured chondrocytes. Here we show the modulation by clock genes of chondrogenic differentiation through gene transactivation of the master regulator of chondrogenesis Indian hedgehog (IHH) in chondrocytes of the growth plate. Several clock genes were expressed with oscillatory rhythmicity in cultured chondrocytes and rib growth plate in mice, whereas chondrogenesis was markedly inhibited in stable transfectants of Per1 in chondrocytic ATDC5 cells and in rib growth plate chondrocytes from mice deficient of brain and muscle aryl hydrocarbon receptor nuclear translocator-like (BMAL1). Ihh promoter activity was regulated by different clock gene products, with clear circadian rhythmicity in expression profiles of Ihh in the growth plate. In BMAL1-null mice, a predominant decrease was seen in Ihh expression in the growth plate with a smaller body size than in wild-type mice. BMAL1 deficit led to disruption of the rhythmic expression profiles of both Per1 and Ihh in the growth plate. A clear rhythmicity was seen with Ihh expression in ATDC5 cells exposed to dexamethasone. In young mice defective of BMAL1 exclusively in chondrocytes, similar abnormalities were found in bone growth and Ihh expression. These results suggest that endochondral ossification is under the regulation of particular clock gene products expressed in chondrocytes during postnatal skeletogenesis through a mechanism relevant to the rhythmic Ihh expression.
Introduction
Circadian rhythm is seen in a variety of physiological and/or pathological processes in different organisms ranging from bacteria to mammals. The circadian rhythmicity has been thought to be generated at the cellular level by circadian core oscillators expressed in the suprachiasmatic nucleus (SCN)2 of the anterior hypothalamus, which is recognized as a region of the central clock in the mammalian brain (1, 2). However, recent studies have shown the expression of these core oscillators in various peripheral tissues including heart (3), adipose tissue (4), pancreas (5), and liver (2). Accordingly, the prevailing view is that the central SCN clock acts as a synchronizer rather than an essential master regulator of peripheral rhythmic oscillations. In peripheral tissues, oscillatory rhythmicity may be under the direct control by clock gene products locally expressed (6). In mice, the rhythmic transactivation of Period (Per) gene seems to be essential for the generation of circadian rhythms. Expression of mouse Per genes is positively regulated by other clock gene products belonging to the basic helix-loop-helix period/aryl hydrocarbon receptor nuclear translocator/single minded class, which are CLOCK and brain and muscle aryl hydrocarbon receptor nuclear translocator-like (BMAL1), respectively. In addition, mouse PER proteins constitute multimeric complexes with products of Cryptochrome (Cry) genes such as Cry1 and Cry2, which in turn negatively regulate the gene transcription mediated by the BMAL1/CLOCK complex after binding to BMAL1 C terminus. The PER/CRY repressor complex is ubiquitinylated for degradation by the proteasomal pathway, and then the BMAL1/CLOCK complex activates a new transcriptional cycle with oscillatory rhythmicity (7–13).
Bone is precisely orchestrated by different cells through a well organized and highly regulated mechanism, where mesenchymal precursor cells differentiate into skeletal elements by forming a cartilaginous model during embryogenesis toward bone formation in the vertebral column and long bone (14). Through this endochondral ossification process, the cartilaginous rudiment, which is a tightly regulated area of chondrocytic differentiation for maturation, undergoes developmental growth during skeletogenesis. Within the cartilaginous rudiment, chondrocytes differentiate, progressing through the resting, proliferating, hypertrophic, and calcifying stages, which leads to mineralization of the cartilage matrix around the central region of the rudiment in the area enriched of hypertrophic chondrocytes. Shortly after the mineralization process takes place, most hypertrophic chondrocytes undergo sustained apoptosis. Upon apoptotic death of hypertrophic chondrocytes after mineralization, osteoblasts, osteoclasts, and capillaries begin to invade the cartilage matrix to produce new bone, leading to the longitudinal bone elongation (15). These sequential differentiation steps are not only under the control of essential transcription factors such as SRY-type HMG box 9 (SOX9) and runt-related transcription factor-2 (RUNX2) (15), but also regulated by a variety of local autocrine/paracrine factors including parathyroid hormone-related peptide (PTHRP) (16), Indian hedgehog (IHH) (16), and Wnt (17).
In a recent study using implanted microtransducers to document longitudinal bone growth in an immature lamb model, 90% of bone elongation occurs during recumbency, but not during either standing or locomotion (18). Our previous studies have demonstrated transient up-regulation of the clock gene Per1 expression by parathyroid hormone (PTH) in mouse prechondrogenic cell line ATDC5 cells and in organotypic cultured mouse metatarsals isolated before vascularization (19). Although evidence is accumulating for the functional expression of clock genes in cartilage, not much attention has been paid to the role of clock gene products in mechanisms underlying circadian rhythmicity of both chondrogenic differentiation and longitudinal bone growth in vivo to date. In the present study, therefore, we have investigated the possible role of clock gene products in mechanisms underlying the regulation of cellular differentiation processes in the growth plates of BMAL1-null mice and mice in which this core molecular clock oscillator was conditionally knocked out from chondrocytes.
EXPERIMENTAL PROCEDURES
Animal Maintenance
The protocol employed here meets the guideline of the Japanese Society for Pharmacology and was approved by the Committee for Ethical Use of Experimental Animals at Kanazawa University (permit numbers: 71061 and 71066). Mice were maintained for 1 week under controlled temperature and humidity with a 12-h light/12-h dark cycle with access to standard laboratory food and water ad libitum. Total RNA was extracted from the rib growth plate at different diurnal times, where zeitgeber time (ZT) 0 is defined as the time of light-on and ZT12 is defined as the time of light-off, respectively. C57BL/6J strain mice defective of BMAL1 (Bmal1−/−)3 were originally generated by Dr. Shimba (Nihon University, Chiba, Japan) (20), and heterozygotes were intercrossed to obtain animals with three different genotypes in Kanazawa University. Bmal1+/fl mice (21) were obtained from The Jackson Laboratory (Bar Harbor, ME). α1(II)-collagen-Cre mice4 (22) were a generous gift from Dr. G. Karsenty (Department of Genetics and Development, Columbia University, New York, NY) through Professor Shu Takeda (Department of Internal Medicine, Keio University School of Medicine, Tokyo, Japan). Bmal1+/fl mice and α1(II)-collagen-Cre mice were backcrossed with C57BL/6J at least seven and six times, respectively.
Immunohistochemistry
Tibial sections prepared from 1-day-old neonatal mice were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min, washed with PBS, treated with 0.3% H2O2 in methanol for 30 min, and washed with 70% ethanol for 5 min. After washing with PBS, sections were subjected to blocking with PBS containing 1% bovine serum albumin and 0.1% Triton X-100 at room temperature for 1 h. Sections were then reacted with an antibody against BMAL1 (Santa Cruz Biotechnology, Santa Cruz, CA) or CLOCK (Santa Cruz Biotechnology) diluted at 1:200 with the same blocking buffer at room temperature overnight followed by reaction with a biotinylated anti-goat IgG antibody at room temperature for 30 min and subsequent incubation with VECTASTAIN Elite ABC reagent (Vector Laboratories, Burlingame, CA) for 1 h. Finally, immunostaining was done using 0.05% diaminobenzidine and 0.03% H2O2.
Cell Culture
For the culture of chondrocytes, cartilages were isolated from neonatal mouse ribs followed by incubation at 37 °C for 1.5 h in Dulbecco's modified Eagle's medium (DMEM) containing 0.3% collagenase (Wako, Osaka, Japan) and subsequent digestion with DMEM containing 0.3% collagenase for 6 h. Supernatants obtained by the second digestion were collected and centrifuged at 250 × g for 5 min. The resultant pellets were suspended in DMEM containing 10% fetal bovine serum (FBS). Cells were plated at a density of 1 × 104 cells/cm2 in appropriate dishes and then cultured for different periods at 37 °C under 5% CO2. After 1 day in culture, culture medium was changed to DMEM containing 10% FBS and 50 μg/ml ascorbic acid, 1 mm pyruvate, and 1 mm cysteine for subsequent culturing for different periods. Culture medium was changed every 2 days. Chondrogenic ATDC5 cells were purchased from the RIKEN Cell Bank. ATDC5 cells were cultured in Dulbecco's modified Eagle's medium and Ham's F-12 medium (DMEM/F12) (Invitrogen) containing 5% FBS. For induction of differentiation, culture medium were replaced with medium containing 10 μg/ml transferrin, 30 nm sodium selenite, and 10 μg/ml bovine insulin (Sigma).
To evaluate the possible circadian rhythmicity of Ihh expression in primary cultured costal chondrocytes, cells were prepared from ribs of neonatal Bmal1+/+ mice followed by culture for 14 days and subsequent exposure to 100 μm dexamethasone (Dex) for 2 h to synchronize cellular differentiation stages during culture. After medium change, total RNA was extracted for determination of Ihh levels every 4 h for 48 h after the addition of Dex by real time-based reverse transcription polymerase chain reaction (RT-PCR) analysis described below. For the analysis of cells with homologous biological activities rather than primary cells, prechondrogenic ATDC5 cells were similarly exposed to 100 μm Dex for 2 h toward subsequent determination of temporal Ihh expression profiles.
RT-PCR, Northern Blotting, and Western Blotting
RT-PCR was conducted as described previously (19, 23). Quantification of PCR products was conducted by real time-based RT-PCR using a MiniOpticonTM (Bio-Rad) with an iQ SYBR Green super mix (Bio-Rad). The relative amount of each transcript was normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in a real time-based RT-PCR. The primer sequences are summarized in Table 1. Northern blotting was conducted as described previously (24) by using digoxigenin-labeled cRNA probes. Western blotting was done by using antibodies against BMAL1, PER1 (Alpha Diagnostic, San Antonio, TX), GAPDH (Cell Signaling Technology, Danvers, MA), β-tubulin (Sigma), and V5 (Invitrogen), as described previously (25).
TABLE 1.
Primers used for RT-PCR analysis in this study
| Genes | Upstream (5′-3′) | Downstream (5′-3′) |
|---|---|---|
| Real time PCR | ||
| Gapdh | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
| Per1 | CAGGCTAACCAGGAATATTACCAGC | CACAGCCACAGAGAAGGTGTCCTGG |
| Bmal1 | TGACCCTCATGGAAGGTTAGAA | GGACATTGCATTGCATGTTGG |
| Clock | TGCTTCCTGGTAACGCGAGAAAGA | AGGAATGTGGGTTTCCAGTCCTGT |
| Sox9 | CGACTACGCTGACCATCAGA | AGACTGGTTGTTCCCAGTGC |
| Runx2 | CCGCACGACAACCGCACCAT | CGCTCCGGCCCACAAATCTC |
| Ihh | TGGACTCATTGCCTCCCAGA | CAAAGGCTCAGGAGGCTGGA |
| Col II | TGAAGACCCAGACTGCCTCAA | AGCCGCGAAGTTCTTTTCTCC |
| ColX | TTCTGCTGCTAATGTTCTTGACC | GGGATGAAGTATTGTGTCTTGGG |
| ptwrp | CATCAGCTACTGCATGACAAGG | GGTGGTTTTTGGTGTTGGGAG |
| ptcw1 | GATGGGCCTCATTGGGATC | AAAGGCCAAAGCCACGTG |
| Genotyping for BMAL1 knockout mice | ||
| Wild-type allele | CAAACCTGGTCGTCTGGAAT | GTCCTCCCCAAAAGGTGAAT |
| Mutant allele | GGGGATTTCCATCTGTGTTTAC | CTCATCTGCTTATCTGCTCTGGGG |
| Genotyping for conditional BMAL1 knockout mice | ||
| Floxed allele | ACTGGAAGTAACTTTATCAAACTG | CTGACCAACTTGCTAACAATTA |
| Δallele | CTCCTAACTTGGTTTTTGTCTGT | CTGACCAACTTGCTAACAATTA |
| Cre recombinase | CCTGGAAAATGCTTCTGTCCGTTTGCC | GAGTTGATAGCTGGCTGGCAGATG |
Establishment of ATDC5 Cells Stably Transfected with Per1
ATDC5 cells were plated at a density of 2 × 104 cells/cm2 in DMEM/F12 containing 5% FBS on culture dishes (inner diameter, 35 mm). After 24 h, cells were transfected with pcDNA3.1 containing the full-length coding region of Per1 or with empty vector (EV) using 2 μg of DNA and Lipofectamine (Invitrogen) and Plus reagent (Invitrogen). After 24 h, and every 48 h thereafter for 2 weeks, culture medium were replaced with DMEM/F12 containing 5% FBS and 500 μg/ml G418 (Invitrogen). Pools of 28 clones of ATDC5 cells resistant to G418 (ATDC5-PER1) were isolated for further studies. Pools of clones between passages 2 and 5 were used for these experiments.
Cloning, Luciferase Assay, and Electroporation
Mouse Clock, Per1, Per2, Cry1, and Cry2 and hamster Bmal1 expression plasmids were kindly donated by Dr. Reppert (University of Massachusetts Medical School, Worcester, MA). Per1-Luc reporter plasmid containing the 5′-flanking region of mouse Per1 gene (−1803/+40) was kindly provided by Dr. Paolo Sassone-Corsi (University of California, Irvine, CA). Reporter plasmids for mouse Ihh promoter were prepared as follows. Mouse Ihh promoter was at first obtained by cloning with the forward primer 5′-CTCGAG(XhoI site)-GTGACTTTCCACAAGCACCCA-3′ (−2380 to −2361) and the reverse primer 5′-AAGCTT(HindIII site)-CGGACTCAAGGGACCCGCGG-3′ (+61 to +80) with mouse tail genomic DNA. The mouse Ihh promoter fragment (−2380 to +80) was cloned into the promoterless pGL-3 basic vector (Promega, Madison, WI) to create the recombinant plasmid −2380/+80 Ihh-Luc. The deletion mutant of Ihh promoter plasmid was made from the recombinant plasmid −2380/+80 Ihh-Luc (−1792/+80 IHH-Luc and −1129/+80 Ihh-Luc) by using restriction enzyme and T4 DNA polymerase. Reporter vectors were co-transfected with a SV40-Renilla luciferase construct into ATDC5 cells or human embryonic kidney (HEK293) cells using Lipofectamine and Plus reagent. Two days after transfection, cells were lysed, and luciferase activity was determined using specific substrates in a luminometer according to the manufacturer's protocol (Promega). Transfection efficiency was normalized by determining the activity of Renilla luciferase. Transfection was also conducted into ATDC5 cells by the electroporation method using the NucleofectorTM according to the manufacturer's protocol (Lonza, Walkersville, MD) as needed.
Determination of Matrix Proteoglycan Synthesis, Alkaline Phosphatase (ALP) Activity, and Cell Proliferative Activity
For the measurement of matrix proteoglycan synthesis, cells were fixed with 10% formalin and stained with 1% Alcian blue 8GS solution (Sigma). Staining intensity was finally quantified using the computer program Scion Image. Determination of ALP activity was done as described previously (24). In brief, cells were solubilized with 0.1% Triton X-100 followed by determination of the ALP activity in lysates using p-nitrophenol phosphate as a substrate. Protein concentration was determined with a Bio-Rad protein assay kit (Bio-Rad). Cell proliferation analysis was performed by determining the reduction of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) as described previously (26).
Lentivirus (LV) Production for Infection of Primary Cultured Chondrocytes
The LV backbone vector and three helper plasmids (pRSV-REV, pMDLg/pRRE, and vesicular stomatitis virus G protein-expressing plasmid) were kindly provided by Dr. Südhof (Stanford University, Palo Alto, CA). A short hairpin sequence targeting the mouse Per1 mRNA (5′-TCGACCCCCGAATACACACTTCGAAATTCAAGAGATTTCGAAGTGTGTATTCGGTTTTTGGAAAT-3′) was inserted into the XhoI-XbaI sites downstream of the H1 promoter. Full-length cDNAs for mouse BMAL1 and mouse CLOCK were cloned into LV backbone vector. LV was produced as described previously (27). LV vector, pRSV-REV, pMDLg/pRRE, and vesicular stomatitis virus G protein-expressing plasmid were co-transfected into HEK293T cells (RIKEN, Saitama, Japan) at 10, 2.5, 5, and 3 μg of DNA per 56.7-cm2 culture area, respectively, using the calcium phosphate method. At 24 h after transfection, the HEK293T culture medium was replaced with DMEM containing 10% FBS followed by culture for 48 h and subsequent collection of culture medium. The collected culture medium was centrifuged at 500 × g for 5 min, and the supernatant containing LV particles was directly added to the culture medium for primary chondrocytes. Cultured chondrocytes were infected on day 6 for the analysis on day 14. All experimental steps were performed under the level II bio-safety conditions.
Chromatin Immunoprecipitation (ChIP) Assay
ATDC5 cells were treated with formaldehyde for cross-linking followed by sonication in lysis buffer containing several protease inhibitors (1 mm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml pepstatin). Immunoprecipitation was performed with the anti-CLOCK antibody followed by extraction of DNA with phenol/chloroform. PCR was performed using sequences found in the 5′-flanking region (−2400 to −2175, upstream 5′-GGATTGAACCCAAGGCTTTTT-3′, downstream, 5′-AGATCTTTAGAAACTGACATT-3′; −1281 to −1092, upstream 5′-GGTTTCCTTTGGTAGAGGAA-3′, downstream, 5′-CTCCGTAGGTGAGAGACTCC-3′) of mouse Ihh as primers.
In Situ Hybridization Analysis
Neonatal mouse tibiae were fixed with 10% formalin neutral buffer solution followed by decalcification with 20% EDTA and subsequent immersion in 30% sucrose. Tibiae were then dissected for frozen sections with a thickness of 10 μm in a cryostat. In situ hybridization was carried out as described previously (25) by using digoxigenin-labeled cRNA probes. Staining intensity was finally quantified using the computer program Scion Image.
Data Analysis
Results are all expressed as the mean ± S.E., and statistical significance was determined by two-tailed and unpaired Students' t test or one-way analysis of variance with Bonferroni's/Dunnett's post hoc test.
RESULTS
Rhythmic Expression of Clock Genes in Chondrocytes
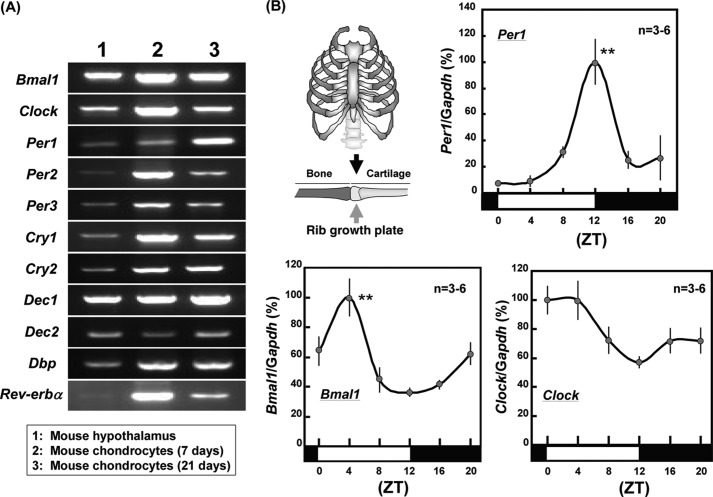
In cultured costal chondrocytes from rib cartilage of neonatal Std-ddY mice, constitutive mRNA expression was seen for all clock genes examined irrespective of culture durations of 7 or 21 days on RT-PCR analysis performed at 32 cycles (Fig. 1A). These included Bmal1, Clock, Per1, Per2, Per3, Cry1, Cry2, the basic helix-loop-helix proteins differentiated embryo chondrocyte 1 (Dec1) and Dec2, D site of albumin promoter-binding protein (Dbp), and Rev-erbα. 3-week-old male mice were bred under 12-h light and dark cycle conditions for a week followed by extraction of total RNA from rib growth plates at different times after the final light cycle initiation for real time-based RT-PCR analysis on Per1, Bmal1, and Clock. Statistically significant rhythmicity was found with expression profiles of both Per1 and Bmal1 in mouse rib growth plates with maximum expression at ZT12 for Per1 and ZT4 for Bmal1, respectively (Fig. 1B). By contrast, no significant rhythmic expression was observed in Clock in mouse rib growth plates.
FIGURE 1.
Rhythmic expression of clock genes in growth plates. A, mouse costal chondrocytes were cultured for 7 or 21 days followed by RT-PCR using primers specific for each clock gene. B, mice were bred under 12-h light and dark cycle conditions for a week followed by extraction of total RNA from rib growth plates at different times after the last light cycle initiation for real time-based RT-PCR. **, p < 0.01, significantly higher than the lowest control value for each gene.
Chondrogenic Differentiation in ATDC5 Cells Stably Transfected with Per1
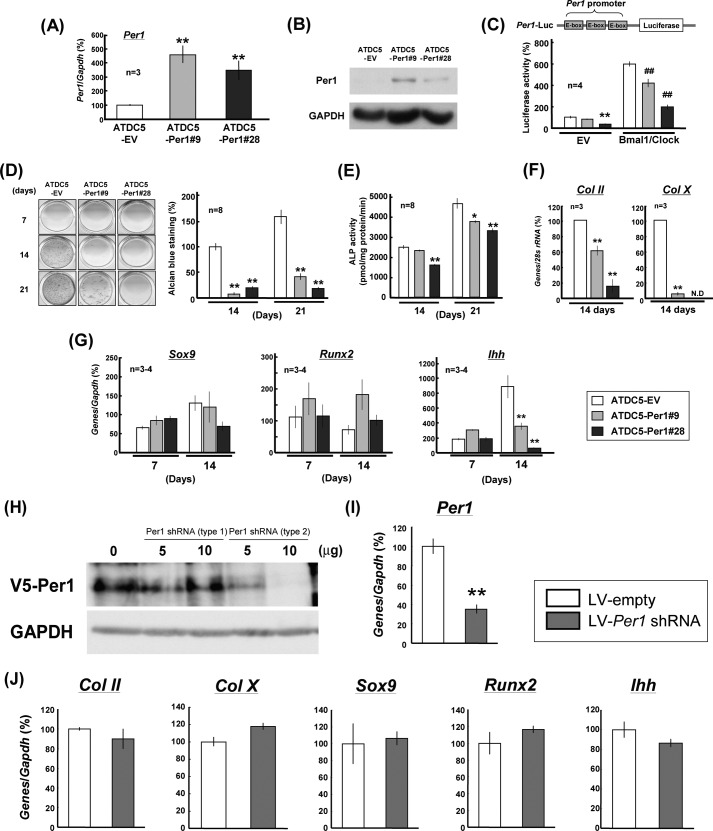
Chondrocytic ATDC5 cells were stably transfected with pcDNA3.1 containing the full-length coding region of Per1. Several different clones transfected with Per1 showed markedly elevated expression of Per1 (ATDC5-PER1) when compared with cells transfected with EV alone (ATDC5-EV). Among different clones of stable transfectants in ATDC5 cells, relative high expression was seen in both #9 and #28 clones cultured for 2 days for Per1 mRNA (Fig. 2A) and PER1 protein (Fig. 2B), respectively. As PER1 negatively regulates gene transactivation mediated by the BMAL1/CLOCK complex endowed to bind to an E-box element in the 5′-flanking region of the target genes (28), we next compared the luciferase activity of Per1-Luc reporter, containing three E-box elements in Per1 promoter, between ATDC5-EV and ATDC5-PER1 cells. Individual stable transfectants were transiently transfected with Per1-Luc in either the presence or the absence of expression vectors of BMAL1 and CLOCK followed by determination of luciferase activity 48 h after transfection. Luciferase activities were significantly lower in ATDC5-PER1#28 cells, but not in ATDC5-PER1#9 cells, in the absence of the BMAL1/CLOCK complex than in ATDC5-EV cells (Fig. 2C). Although the introduction of BMAL1/CLOCK more than sextupled the luciferase activity in ATDC5-EV cells, stable overexpression of PER1 significantly inhibited the luciferase activity elevated by the introduction of BMAL1/CLOCK irrespective of the clones examined.
FIGURE 2.
Suppression of chondrogenic differentiation in stable PER1 transfectants. A and B, ATDC5 cells were stably transfected with Per1 expression vectors or EV followed by determination of expression levels of Per1 by real time-based RT-PCR (A) and PER1 protein by Western blotting analysis (B). C, ATDC5 cells were stably transfected with Per1 expression vectors or EV followed by transient transfection with Per1 promoter, which contains three tandem copies of E-box element, linked to a luciferase construct in either the presence or the absence of Bmal1/Clock expression vector and subsequent incubation for 48 h. D, ATDC5-PER1 or ATDC5-EV cells were cultured for 7–21 days followed by fixation with 10% formalin and subsequent Alcian blue staining for acidic polysaccharide. E, stable transfectants were cultured for 14–21 days followed by determination of ALP activity. F, total RNA was extracted from stable transfectants cultured for 14 days followed by quantitative determination of mRNA expression for differentiation markers such as Col II and Col X with Northern blotting. G, total RNA was extracted from each stable transfectant cultured for 7–21 days followed by determination of Sox9, Runx2, and Ihh by real time-based RT-PCR analysis. *, p < 0.05, **, p < 0.01, significantly different from each control value obtained in ATDC5-EV cells. ##, p < 0.01, significantly different from the value obtained in ATDC5-EV cells with Bmal1/Clock. N. D, not detectable. H, HEK293T cells were transfected with V5-tagged Per1 expression vector in either the presence or the absence of two types of LV vectors for Per1 shRNA followed by determination of expression levels of V5-PER1 by Western blotting. I and J, chondrocytes cultured for 6 days were infected with LV-Per1 shRNA for 1 day followed by determination of Per1 (I) and Col II, Col X, Sox9, Runx2, and Ihh (J) by real time-based RT-PCR analysis 7 days after infection. **, p < 0.01, significantly different from each control value obtained in chondrocytes infected with LV-empty.
In ATDC5-PER1#9 and -#28 cells cultured for 14 and 21 days, significant inhibition was seen in the matrix proteoglycan synthesis evaluated by Alcian blue staining when compared with ATDC5-EV cells (Fig. 2D). The activity of ALP was markedly increased in proportion to increasing culture days from 14 to 21 days in ATDC5-EV cells, whereas ALP activity was significant lower in ATDC5-PER1#9 cells cultured for 21 days and ATDC5-PER1#28 cells cultured for 7–14 days than in ATDC5-EV cells, respectively (Fig. 2E). In stable PER1 transfectants cultured for 14 days, a marked decrease was invariably observed in mRNA expression of chondrogenic differentiation markers including type II collagen (Col II) and type X collagen (Col X) (Fig. 2F), which are both known to be highly expressed by proliferating to prehypertrophic chondrocytes and preferentially expressed by hypertrophic chondrocytes, respectively. In ATDC5-EV cells cultured from 7 to 14 days, a marked increase was seen in Ihh levels, but not in Sox9 and Runx2 levels, during the culture (Fig. 2G). In ATDC5-PER1#9 and ATDC5-PER1#28 cells, however, Ihh levels were drastically decreased during the culture for 14 days, but not for 7 days, when compared with those in ATDC5-EV cells with Sox9 and Runx2 levels being unchanged irrespective of the culture period.
To confirm the involvement of PER1 in chondrogenesis in vitro, an attempt was made to determine expression profiles of different chondrogenic marker genes in primary cultured rib chondrocytes infected with short hairpin RNA (shRNA) for PER1. To examine the validity of the two different types of PER1 shRNA LV vectors, LV-Per1 shRNA (type 1) and LV-Per1 shRNA (type 2), HEK293T cells were transfected with V5-tagged Per1 expression vector in either the presence or the absence of two types of LV vectors for Per1 shRNA followed by determination of expression levels of V5-PER1 by Western blotting. A significant decrease was seen in V5-PER1 expression in cells transfected with LV-Per1 shRNA (type 2), but not in those with LV-Per1 shRNA (type 1) (Fig. 2H). Chondrocytes cultured for 6 days were thus infected with LV-Per1 shRNA (type 2) for 1 day followed by culture in DMEM/F12 containing 5% FBS for an additional 7 days and subsequent determination of mRNA expression by real time-based RT-PCR. Per1 mRNA was significantly decreased in chondrocytes infected with LV-Per1 shRNA when compared with cells infected with LV-empty (Fig. 2I), whereas infection of LV-Per1 shRNA failed to significantly affect mRNA expression of Col II, Col X, Sox9, Runx2, and Ihh in primary chondrocytes (Fig. 2J).
Possible Involvement of Clock Genes in Ihh Transactivation
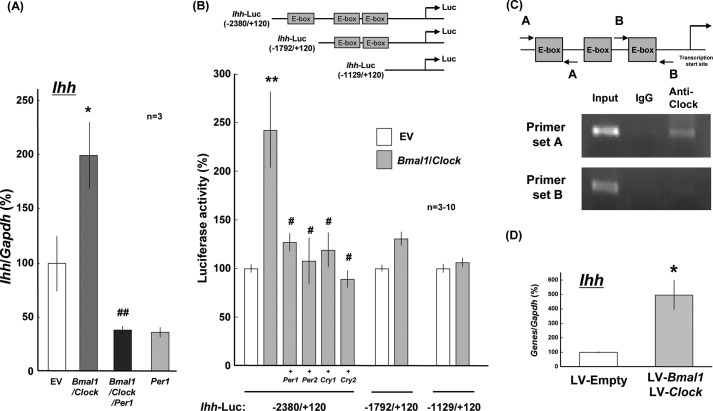
BMAL1 is shown to orchestrate a heterodimer with CLOCK for DNA binding toward the transactivation of target genes, such as Per and Cry, whose products are endowed to inhibit gene transactivation mediated by the BMAL1/CLOCK heterodimer (7, 8, 10, 13). Chondrogenic ATDC5 cells were electroporated with expression vectors of Bmal1 and Clock in either the presence or the absence of the expression vector of Per1 using NucleofectorTM prior to plating on dishes followed by determination of different cellular differentiation markers 10 days later. Real time-based RT-PCR analysis clearly confirmed a significant increase in Ihh expression in cells transfected with Bmal1/Clock, whereas further co-introduction of Per1 significantly prevented the increase by Bmal1/Clock with a concomitant inhibition by Per1 alone (Fig. 3A).
FIGURE 3.
Transactivation by clock proteins of Ihh. A, ATDC5 cells were transiently transfected with expression vectors of Bmal1 and Clock in either the presence or the absence of an expression vector of Per1 followed by determination of Ihh levels 10 days after transfection with real time-based RT-PCR. B, HEK293 cells were transiently transfected with a reporter plasmid linked to intact and mutated Ihh promoters with different lengths including −2380 to +120, −1792 to +120, and −1129 to +120 bp in either the presence or the absence of expression vectors of Per1, Per2, Cry1, and Cry2 followed by further culture for an additional 48 h and subsequent determination of luciferase activity. *, p < 0.05, **, p < 0.01, significantly different from each control value obtained in cells transfected with EV. #, p < 0.05, ##, p < 0.01, significantly different from the value obtained in cells transfected with Bmal1/Clock. C, DNA-protein complex was prepared from ATDC5 cells and treated with the anti-CLOCK antibody for ChIP assay. Typical pictures are shown in the figure, whereas similar results were invariably obtained in at least three independent determinations. D, costal chondrocytes cultured for 6 days were infected with both LV-Bmal1 and LV-Clock for 1 day followed by determination of Ihh by real time-based RT-PCR analysis 7 days after infection. *, p < 0.05, **, p < 0.01, significantly different from each control value obtained in chondrocytes infected with LV-empty.
Promoter regions of Ihh gene were subjected to Database of Transcriptional Start Sites (DBTSS) and TRANSFAC database searches. A database analysis revealed that 2.5 kb of the 5′-flanking region of the mouse Ihh gene contains three putative E-box (CACGTG) elements that are a recognition site of the BMAL1/CLOCK heterodimer. To simplify the importance of Ihh upstream elements for Ihh expression, we next employed a clonal cell line without any chondrocytic properties for determination of the luciferase reporter activity. In HEK293 cells transiently transfected with a luciferase reporter plasmid linked to the Ihh promoter region of −2388 to +120 bp in either the presence or the absence of different expression vectors, introduction of Bmal1/Clock led to a significant increase in the luciferase activity in a manner sensitive to the prevention by co-introduction of Per1, Per2, Cry1, and Cry2 (Fig. 3B, left columns). For identification of the promoter element responsible for the transactivation, deletion mutants were made from the 5′-flanking region of the mouse Ihh gene for the preparation of luciferase reporter plasmid constructs with different lengths. In HEK293 cells transfected with Ihh-Luc (−1792/+120) and Ihh-Luc (−1129/+120), no significant alterations were found in luciferase activity after the introduction of Bmal1/Clock (Fig. 3B, right columns). To determine which E-box elements are responsible for the association with BMAL1/CLOCK complex in the 5′-flanking region of mouse Ihh gene, we next performed ChIP assays. In immunoprecipitates by the anti-CLOCK antibody from lysates of ATDC5 cells cultured for 3 days, an amplified PCR product was detected with primers for the 5′-flanking region of −2400 to −2175 bp upstream, but not that of −1281 to −1092 bp upstream, in the Ihh gene (Fig. 3C).
An attempt was next made to evaluate the effect of overexpression of BMAL1/CLOCK by the LV-mediated overexpression method in primary chondrocytes. Chondrocytes cultured for 6 days were infected with both LV-Bmal1 and LV-Clock for 1 day followed by further culture in DMEM/F12 containing 5% FBS for an additional 7 days and subsequent determination of mRNA levels by real time-based RT-PCR. Infection with both LV-Bmal1 and LV-Clock led to a significant increase in Ihh mRNA expression in comparison with that in cells with LV-empty (Fig. 3D).
Skeletogenesis in Bmal1−/− Mice
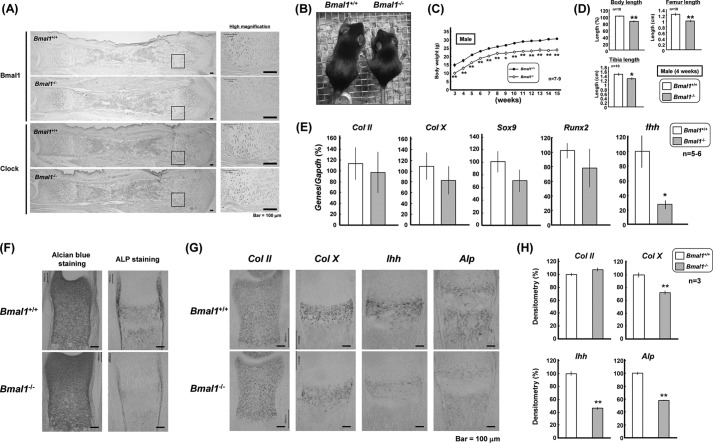
We next examined the skeletal growth phenotypes in BMAL1-null mice of C57BL6/J strain. In wild-type (Bmal1+/+) tibial sections, immunoreactive BMAL1 and CLOCK were highly expressed by prehypertrophic to hypertrophic chondrocytes with modest expression in proliferating chondrocytes (Fig. 4A). In tibial sections from Bmal1−/− mice, no marked immunoreactivity was detected for BMAL1 in any chondrocytic layers. Immunoreactive CLOCK was similarly detected in tibial sections from Bmal1−/− and Bmal1+/+ mice. In 4-week-old Bmal1−/− mice, the body size was apparently smaller than that in Bmal1+/+ mice (Fig. 4B) as reported previously (20, 29). Body weight was invariably lower throughout the growth period during 3–15 weeks after birth in male Bmal1−/− mice (Fig. 4C). In 4-week-old male Bmal1−/− mice, marked decreases were invariably seen in the body length measured from nose to anus and the longitudinal lengths of both tibia and femur when compared with those in Bmal1+/+ mice (Fig. 4D). In rib growth plates isolated from Bmal1−/− mice at 3–4 weeks old, a predominant decrease was found in Ihh levels, but not in Col II, Col X, Sox9, and Runx2 levels, when compared with those from Bmal1+/+ mice (Fig. 4E). In tibial sections of 1-day-old Bmal1−/− mice, a marked deterioration was seen in ALP staining, but not in Alcian blue staining (Fig. 4F). In situ hybridization analysis clearly revealed markedly decreased expression of Col X, Ihh, and Alp, but not Col II, in the prehypertrophic chondrocytic layer on tibial sections from 1-day-old Bmal1−/− mice (Fig. 4G). Quantification of these in situ hybridization data confirmed a significant decrease in antisense cRNA signals detected in the prehypertrophic chondrocytic layer on tibial sections of Bmal1−/− mice (Fig. 4H).
FIGURE 4.
Phenotypes in Bmal1−/− mice. A, tibiae were isolated from Bmal1+/+ or Bmal1−/− mice at 1 day of age followed by immunohistochemical analysis using an antibody against BMAL1 or CLOCK. Typical micrographic pictures are shown in the figure, whereas similar results were invariably obtained in at least three independent determinations. Scale bars, 100 μm. B, general appearance of 4-week-old male Bmal1+/+ and Bmal1−/− mice. C, body weight curves of male Bmal1+/+ and Bmal1−/− mice. D, body and bone lengths in 4-week-old male Bmal1+/+ and Bmal1−/− mice. E, rib growth plates were isolated from Bmal1+/+ and Bmal1−/− mice followed by determination of expression levels of Col II, Col X, Sox9, Runx2, and Ihh by real time-based RT-PCR. F and G, tibiae were isolated from Bmal1+/+ and Bmal1−/− mice at 1 day after birth followed by sectioning for Alcian blue and ALP staining (F) or in situ hybridization using cRNA probes (G) for Col II, Col X, Ihh, and Alp. H, quantitative data are shown with mRNA expression in the panel. *, p < 0.05, **, p < 0.01, significantly different from each control value obtained in Bmal1+/+ mice. Scale bars, 100 μm.
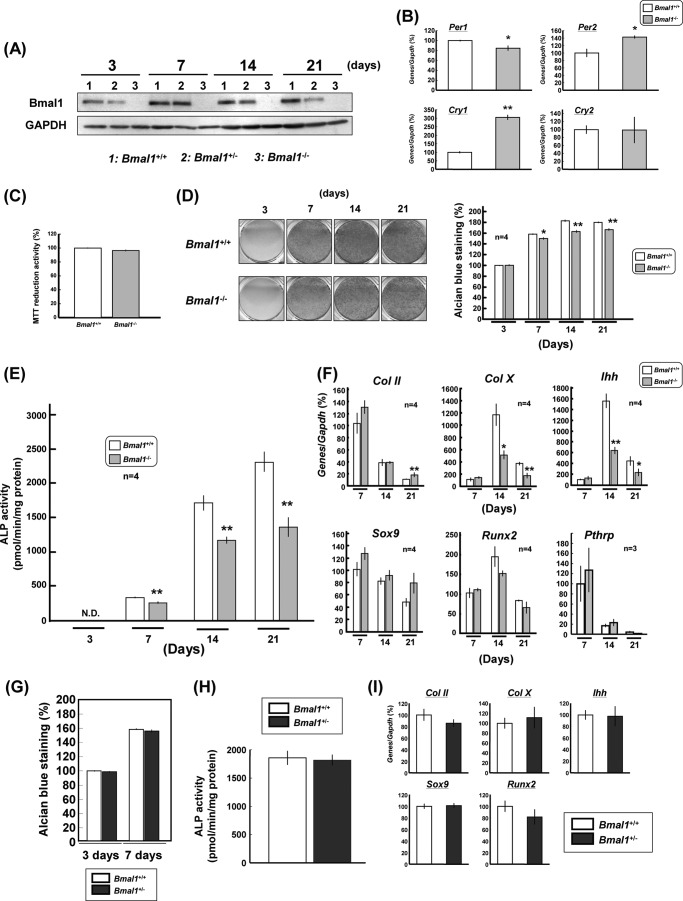
Chondrogenic Differentiation in Bmal1−/− Chondrocytes
Costal chondrocytes were prepared from ribs of neonatal Bmal1−/− or Bmal1+/+ mice as usual for the culture for 3–21 days. Western blotting analysis clearly confirmed the complete absence of BMAL1 from Bmal1−/− chondrocytes with a decreased level in Bmal1+/− chondrocytes irrespective of the culture period from 3 to 21 days (Fig. 5A). In Bmal1−/− chondrocytes cultured for 14 days, moreover, a slight but statistically significant decrease was seen in Per1 expression with concomitant significant increases in both Per2 and Cry1 levels, but not Cry2 (Fig. 5B). However, BMAL1 deficiency did not significantly affect cellular proliferation as determined by MTT reduction in chondrocytes cultured for 3 days (Fig. 5C). In chondrocytes from Bmal1+/+ mice, a marked increase was seen in the matrix proteoglycan synthesis evaluated by Alcian blue staining in proportion to the duration of culture from 3 to 14 days (Fig. 5D). In costal chondrocytes from Bmal1−/− mice, the temporal increase was markedly suppressed for Alcian blue staining throughout the culture period from 7 to 21 days. Quantitative densitometry confirmed a slight but statistically significant attenuation of Alcian blue staining in Bmal1−/− chondrocytes cultured for 7–21 days (Fig. 5D). Although ALP activity was markedly increased in proportion to the culture period from 3 to 21 days in cultured chondrocytes from Bmal1+/+ mice, knock-out of BMAL1 led to a significant inhibition of the temporal increase in ALP activity in chondrocytes cultured for 7–21 days (Fig. 5E). No significant difference was found in Sox9, Runx2, and Pthrp levels between Bmal1+/+ and Bmal1−/− chondrocytes cultured for 7–21 days, whereas a marked decrease was induced in both Col X and Ihh levels in Bmal1−/− chondrocytes cultured for 14–21 days when compared with those in Bmal1+/+ chondrocytes, with a significantly increased Col II level in cells cultured for 21 days only (Fig. 5F). However, no significant alterations were seen in Alcian blue staining (Fig. 5G), ALP activity (Fig. 5H), and Col II, Col X, Ihh, Sox9, and Runx2 expression (Fig. 5I) between Bmal1+/− and Bmal1+/+ chondrocytes cultured for 14 days.
FIGURE 5.
Suppression of chondrogenic differentiation in Bmal1−/− mice. A, costal chondrocytes were prepared from the ribs of Bmal1+/+, Bmal1+/−, and Bmal1−/− mice followed by culture for several days and subsequent determination of BMAL1 expression by Western blotting. B, costal chondrocytes were prepared from homozygous Bmal1−/− mice followed by culture for 14 days and subsequent determination of expression levels of Per1, Per2, Cry1, and Cry2. C–F, costal chondrocytes were also cultured for 3–21 days for subsequent determination of MTT reduction (C), Alcian blue staining (D), ALP activity (E), and expression levels (F) of Col II, Col X, Ihh, Sox9, Runx2, and Pthrp. **, p < 0.05, **, p < 0.01, significantly different from each control value obtained in chondrocytes isolated from Bmal1+/+ mice. N. D, not detectable. G–I, costal chondrocytes were prepared from heterozygous Bmal1+/− mice followed by culture for 3–21 days and subsequent determination of Alcian blue staining (G), ALP activity (H), and expression levels (I) of Col II, Col X, Ihh, Sox9, and Runx2.
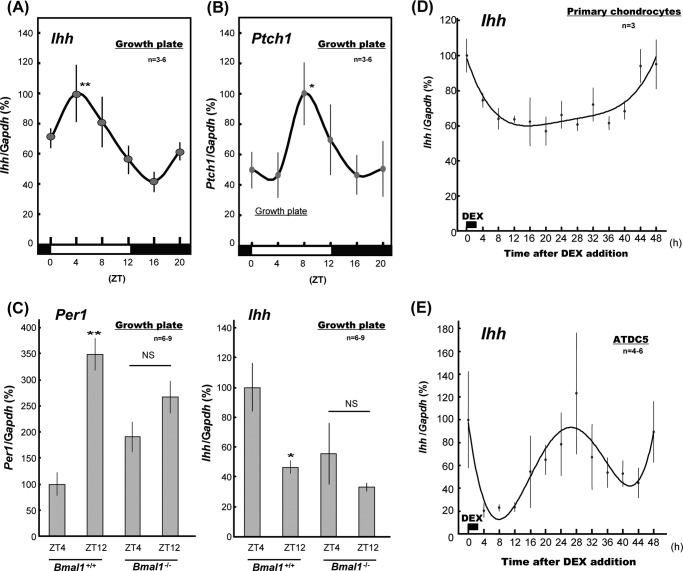
Circadian Rhythmicity of Ihh Expression in Growth Plate
A clear 24-h rhythmicity was found in Ihh levels in the rib growth plate isolated every 4 h from young (3-week-old) male ddY mice throughout a single 24-h period with maximum expression at ZT4 (Fig. 6A), in line with the aforementioned expression profiles of Bmal1. Similar rhythmicity was also seen for mRNA levels of the IHH target gene Patched homolog-1 (Ptch1) in the rib growth plate with a peak at ZT8 (Fig. 6B). However, IHH was not detected in the rib growth plate isolated every 4 h from young mice by Western blotting analysis using the anti-IHH antibody (data not shown). In rib growth plates isolated from Bmal1+/+ mice at ZT12, Per1 levels were significantly higher with significantly lower Ihh levels than in those at ZT4 (Fig. 6C). In rib growth plates isolated at ZT4 and ZT12 from Bmal1−/− mice, by contrast, no significant rhythmic alternations were observed in expression profiles of both Per1 and Ihh.
FIGURE 6.
Rhythmic expression of Ihh in rib growth plate. A and B, mice were bred under 12-h light and dark cycle conditions for a week followed by extraction of total RNA from the rib growth plates different times after the last light cycle initiation and subsequent determination of Ihh (A) and Ptch1 (B) levels with real time-based RT-PCR. **, p < 0.01, significantly higher than the lowest control value. C, rib growth plates were isolated from Bmal1+/+ and Bmal1−/− mice at ZT4 and ZT12 followed by determination of Per1 and Ihh levels by real time-based RT-PCR. D, costal chondrocytes were prepared from the ribs of neonatal Bmal1+/+ mice followed by culture for 14 days and subsequent exposure to 100 μm Dex for 2 h. Cells were then subjected to determination of Ihh levels every 4 h for 48 h after Dex real time-based RT-PCR. E, prechondrogenic ATDC5 cells were exposed to 100 μm Dex for 2 h followed by determination of Ihh expression every 4 h. *, p < 0.05, **, p < 0.01, significantly different from the value obtained in samples isolated at ZT4. NS, not significant.
We next attempted to demonstrate the circadian rhythmicity of Ihh expression in primary cultured costal chondrocytes prepared from ribs of Bmal1+/+ mice. Cells were cultured for 14 days followed by exposure to 100 μm Dex for 2 h and subsequent determination of Ihh levels every 4 h for 48 h by real time-based RT-PCR analysis. Contrary to the rib growth plate in vivo, however, no clear rhythmicity was found in Ihh expression by cultured costal chondrocytes within 48 h after the addition of Dex in vitro (Fig. 6D). To further evaluate the in vitro rhythmicity in chondrocytic cells with synchronized cellular biological activities during culture, prechondrogenic ATDC5 cells were next exposed to 100 μm Dex for 2 h followed by determination of Ihh expression every 4 h as well. Prior exposure to Dex led to the oscillatory expression of Ihh in ATDC5 cells within 48 h (Fig. 6E), in contrast to primary cultured chondrocytes enriched with diverse cell populations with different cellular differentiation stages.
Phenotypes of Chondrocyte-specific BMAL1-deficient Mice
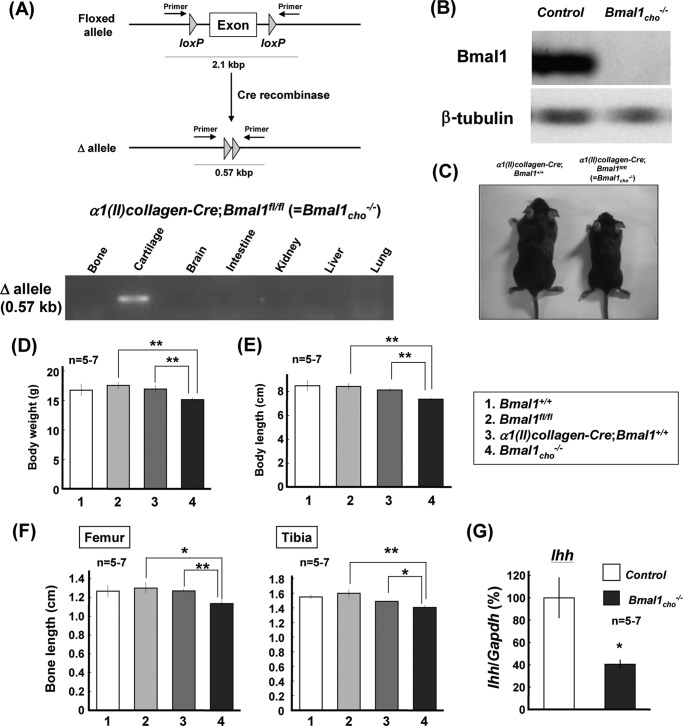
To further confirm the aforementioned regulation by BMAL1 expressed by chondrocytes of endochondral ossification during skeletogenesis, we generated mice deficient of BMAL1 from chondrocytes (α1(II)-collagen-Cre;Bmal1fl/fl = Bmal1cho−/−) by crossing α1(II)-collagen-Cre mice (α1(II)-collagen-Cre;Bmal1+/+) with mice harboring a floxed allele of Bmal1 (Bmal1fl/fl). Genotyping for Δallele confirmed the validity of α1(II)collagen-Cre-driven deletion of Bmal1 allele in cartilage (Fig. 7A), whereas BMAL1 was completely absent from cultured costal chondrocytes of Bmal1cho−/− mice for 7 days (Fig. 7B). Male Bmal1cho−/− mice indeed showed a smaller body size at 4 weeks after birth (Fig. 7C). Significant decreases were also seen in the body weight (Fig. 7D), body length (Fig. 7E), and bone lengths of femur and tibia (Fig. 7F) in 4-week-old male Bmal1cho−/− mice when compared with Bmal1fl/fl and α1(II)-collagen-Cre;Bmal1+/+ mice. In rib growth plates from these Bmal1cho−/− mice, a significant decrease was found in Ihh levels when compared with those from control mice defined as the random mixture of Bmal1+/+, Bmal1fl/fl, and α1(II)-collagen-Cre;Bmal1+/+ mice (Fig. 7G).
FIGURE 7.
Phenotypes in Bmal1cho−/− mice. A, several tissues including bone, cartilage, brain, intestine, kidney, liver, and lung were isolated from Bmal1cho−/− mice followed by extracting DNA and subsequent PCR genotyping for Δallele. B, costal chondrocytes were prepared from the ribs followed by culture for 7 days for determination of BMAL1 expression by Western blotting. C, general appearance of 4-week-old male α1(II)-collagen-Cre;Bmal1+/+ and Bmal1cho−/− mice. D–F, body weight (D), body length (E), and bone length (F) in 4-week-old male Bmal1+/+, Bmal1fl/fl, α1(II)-collagen-Cre;Bmal1+/+ and Bmal1cho−/− mice. *, p < 0.05, **, p < 0.01, significantly different from each control value obtained in Bmal1fl/fl or α1(II)-collagen-Cre;Bmal1+/+ mice. G, rib growth plates were isolated from Bmal1cho−/− and control mice grouped by the random mixture of Bmal1+/+, Bmal1fl/fl, and α1(II)-collagen-Cre;Bmal1+/+ followed by determination of expression levels of Ihh by real time-based RT-PCR. *, p < 0.05, **, p < 0.01, significantly different from each control value obtained in control mice.
DISCUSSION
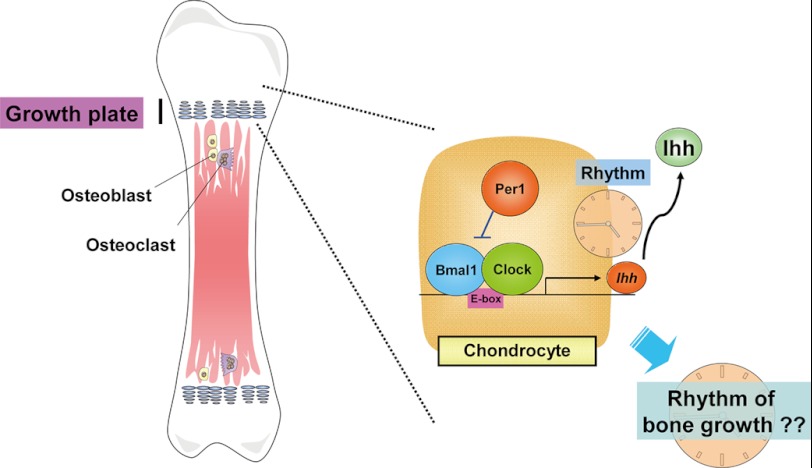
The essential importance of the present findings is that clear circadian rhythmicity was seen in expression profiles of the master regulator gene of chondrogenesis Ihh in the mouse rib growth plate in association with rhythmic expression profiles of clock genes such as Per1 and Bmal1. In rib growth plates isolated from mice defective of BMAL1, moreover, rhythmic expression was entirely lost for both Per1 and Ihh. These findings give rise to an idea that chondrogenic differentiation is under the control by clock genes constitutively expressed by chondrocytes through a mechanism related to direct regulation of the transactivation of Ihh as illustrated in Fig. 8. Period proteins are known to be synthesized through gene transactivation mediated by the positive BMAL1/CLOCK complex at upstream E-box elements, but known to inhibit the gene transactivation as a negative feedback loop together with CRY proteins after the accumulation in the cytoplasm for subsequent translocation into the nucleus. Both PER and CRY repressor members are polyubiquitinylated for degradation by the proteasome pathway, which in turn releases the inhibition by the negative PER/CRY complex of gene transactivation mediated by the positive BMAL1/CLOCK complex. This positive and negative feedback loop occurs at 24-h cycle rhythmicity (7–13). PER1 would interfere with rhythmic promotion by the BMAL1/CLOCK complex of the transcription of Ihh at the upstream E-box element in growth plate chondrocytes toward disturbance of chondrogenesis. Accordingly, rhythmic expression of Ihh could lead to oscillated rhythmic bone elongation through endochondral ossification during skeletogenesis. Although several independent laboratories including ours have already demonstrated the functional expression of clock genes by cells outside the CNS, this is the first direct demonstration of rhythmic expression of different clock genes, such as Per1 and Bmal1, by chondrocytes from the rib growth plate of mice under postnatal development. To our knowledge, rhythmic expression of Ihh was for the first time demonstrated in the rib growth plate in a manner dependent on BMAL1 in vivo in this study as well.
FIGURE 8.
Schematic representation of proposal. In the growth plate, chondrogenic differentiation would be under the oscillatory regulation by IHH expressed with circadian rhythmicity mediated by a mechanism relevant to the chondrocytic clock genes toward the diurnal and nocturnal longitudinal bone growth during skeletogenesis.
Nevertheless, longitudinal bone growth is shown to predominate during recumbence rather than locomotion in an immature lamb model (18). Longitudinal bone elongation occurs at the growth plate seen between epiphysis and metaphysis of long bones through a process referred to as endochondral ossification, in which matured chondrocytes are replaced with osteoblasts and osteoclasts toward bone orchestration after apoptotic cell death as a consequence of the progression of chondrogenic differentiation from proliferation and hypertrophy to secretion of the extracellular matrix. Therefore, the present results obtained in Bmal1−/− mice argue in favor of an idea that longitudinal bone elongation would be concurrently regulated by the rhythmic expression of clock gene products by chondrocytes. Longitudinal bone elongation is governed at the growth plate not only by cell autonomous sequential steps including transcriptional network and local autocrine/paracrine system, but also by circulating endocrine factors including growth hormone, insulin-like growth factor-1 (IGF1), glucocorticoid, thyroid hormone, and adrenaline (26, 30). Among these endocrine factors, growth hormone is a determinant of longitudinal bone elongation by way of a local direct action at the growth plate as well as by transformation into IGF1 in the liver (30) after the rhythmic secretion from the anterior pituitary in a manner highly related to the sleep cycle (31). The fact that most endocrine systems are undoubtedly under the control by the central circadian clock in SCN gives rise to speculation that the present growth retardation could be brought about by dysfunctions of the central clock due to the absence of BMAL1 from SCN neurons in Bmal1−/− mice, but not by the impairment of the putative peripheral clock located in chondrocytes. However, the present findings in Bmal1cho−/− mice clearly ruled out this possibility. In brain-rescued Bmal1−/− mice generated by mating between Bmal1−/− mice and Scg2::tTA; tetO::Bmal1 double transgenic mice, moreover, circadian rhythm is fully recovered with the wheel running task impaired in Bmal1−/− mice, but not with the growth retardation measured by body weight (29). The possibility that longitudinal bone elongation is under the oscillatory control with chondrocytic circadian rhythmicity during postnatal skeletogenesis is thus not negligible at present.
One possible interpretation for the discouraging findings on Col II expression in BMAL1-null mice is that COL II is preferentially expressed at an early differentiation stage with IHH at a later stage during chondrogenesis (15). In addition, possible artifacts and/or pitfalls would be at least in part responsible for the paradoxical data between Per1 and Ihh levels, in addition to Per1 promoter activity, in different stable PER1 transfectant clones in terms of the transfection and subsequent cloning techniques employed in this study. From this point of view, the in vivo results obtained from mice deficient of BMAL1 specifically in chondrocytes (Bmal1cho−/−) are quite noteworthy for confirmation of the pivotal role of BMAL1 expressed by chondrocytes in association with IHH in endochondral ossification during postnatal skeletogenesis. Taken together, the possibility that the chondrocytic circadian clock would play a pivotal role in the diurnal and nocturnal regulation of longitudinal bone elongation during postnatal skeletogenesis through a mechanism relevant to the rhythmic expression by chondrocytes of the autocrine factor IHH highly responsible for chondrogenesis is not ruled out so far. The differential circadian rhythmicity of Ihh expression would be due to the higher diversity and heterogeneity of cell populations in primary chondrocytes rather than cloned chondrogenic cell lines.
In our previous study, different clock genes, such as Per, Bmal1, Clock, and Cry, are constitutively expressed by chondrogenic ATDC5 cells together with direct stimulation by PTH of Per1 expression through the protein kinase A/cAMP-responsive element-binding protein signaling pathway (19). As PER1 is translocated into the nucleus to inhibit gene transactivation by the BMAL1/CLOCK heterodimer as a negative feedback loop after the accumulation in the cytoplasm (7, 8, 10, 13), PER1 transiently up-regulated by PTH would negatively regulate the transactivation mediated by the BMAL1/CLOCK complex of different chondrocytic marker genes through the E-box element located at 5′-flanking region in chondrocytes. A database analysis indeed revealed the presence of three putative E-box elements within 2.5 kb of the 5′-flanking region of Ihh gene: −2334/−2329, −1420/−1415, −1249/−1244. By contrast, IHH is locally produced by prehypertrophic to early hypertrophic chondrocytes to play a role as a master regulator of bone development, coordinating chondrocytic proliferation and differentiation (32, 33), as well as osteoblastic differentiation (34), through activation of the receptor, Patched-1. In CFK-2 chondrogenic cells, indeed, overexpression of IHH leads to up-regulation of the expression of different chondrocytic differentiation markers including ALP, COL II, and COL X (32). In IHH-null mutant mice, profound defects are found in chondrocytic proliferation and differentiation, in addition to impaired osteoblastic formation (33). Chondrocyte-specific deletion of IHH leads to complete recapitulation of all skeletal defective features observed in IHH-null mice (35). From the present findings in luciferase reporter and ChIP assays, it is conceivable that circulating PTH would induce PER1 expression through the cAMP/cAMP-responsive element-binding protein cascade in hypertrophic chondrocytes under regulation by the central clock in SCN, which in turn leads to the interference with transactivation by the BMAL1/CLOCK complex of Ihh for subsequent disturbance of chondrogenic differentiation mediated by IHH under the regulation by the peripheral clock in chondrocytes.
The present findings that PER1 shRNA failed to induce any significant changes in chondrogenic cellular differentiation processes could be accounted for by taking into consideration the fact that overflowed PER1 is highly responsible for negative regulation of the transactivation mediated by the BMAL1/CLOCK complex (7, 8, 10, 13). The fact that PER1 induced by light causes the reset of the central clock at SCN (11) would raise the possibility that PTH may entrain the peripheral clock expressed by chondrocytes in the growth plate to rhythmically synchronize and coordinate sequential chondrogenic differentiation processes toward refined longitudinal bone elongation over whole body. It is noteworthy that the pancreatic islet has a self-sustained molecular clock system endowed to coordinate insulin secretion with the sleep-wake cycle without inputs from the central clock (36). The circadian rhythmicity mediated by the chondrocytic peripheral clock expressed at the growth plate in longitudinal bone elongation during skeletogenesis in vivo, however, remains to be elucidated in future studies despite several enduring technical difficulties.
It thus appears that clock genes are functionally expressed by chondrocytes to rhythmically regulate Ihh expression in the mouse rib growth plate. The present study would not only promote our understanding of the peripheral circadian clock for longitudinal bone elongation during skeletogenesis, but also open a new window for the therapy and treatment of a variety of cartilaginous diseases relevant to abnormal development and maturation of chondrocytes in human beings.
Acknowledgments
We thank Professor Gerard Karsenty (Department of Genetics and Development, Columbia University, New York, NY) for kindly providing α1(II)-collagen-Cre mice through Professor Shu Takeda (Department of Internal Medicine, Keio University School of Medicine, Tokyo, Japan). Mouse CLOCK, PER1, PER2, CRY1, and CRY2 and hamster BMAL1 expression plasmids were kindly donated by Dr. Reppert (University of Massachusetts Medical School, Worcester, MA). PER1-Luc reporter plasmid was kindly provided by Dr. Paolo Sassone-Corsi (University of California, Irvine, CA). The LV backbone vector, pRSV-REV, pMDLg/pRRE, and vesicular stomatitis virus G protein-expressing plasmid were kindly donated by Dr. Südhof (Stanford University, Palo Alto, CA).
This work was supported in part by Grant-in-aid for Scientific Research 20790250 from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to T. T.) and by a Research Grant from Foundation of Growth Science, Japan (to T. T.).
Throughout this manuscript, the following designations were used for mouse strains: Bmal1−/−, mice defective of BMAL1; Bmal1+/+, wild-type mice; Bmal1fl/fl, mice harboring a floxed allele of BMAL1; Bmal1cho−/−, mice deficient of BMAL1 from chondrocytes.
Throughout this manuscript, the following designations were used: α1(II)-collagen-Cre mice, Cre recombinase transgenic mice under the control of a α1(II)-collagen promoter; α1(II)-collagen-Cre;Bmal1+/+ mice, Cre recombinase transgenic mice under the control of a α1(II)-collagen promoter, harboring a wild-type allele of Bmal1.
- SCN
- suprachiasmatic nucleus
- ALP
- alkaline phosphatase
- BMAL1
- brain and muscle aryl hydrocarbon receptor nuclear translocator-like
- COL II
- type II collagen
- COL X
- type X collagen
- CRY
- Cryptochrome
- Dex
- dexamethasone
- DIG
- digoxigenin
- DMEM/F12
- Dulbecco's modified Eagle medium and Ham's F-12 medium
- EV
- empty vector
- IHH
- Indian hedgehog
- LV
- lentivirus
- MTT
- 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
- PBS
- phosphate-buffered saline
- PER
- period
- PTH
- parathyroid hormone
- PTHRP
- parathyroid hormone-related peptide
- RUNX2
- runt-related transcription factor-2
- SOX9
- sry-type HMG box 9
- HMG
- high mobility group
- ZT
- zeitgeber time.
REFERENCES
- 1. Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., Hogenesch J. B. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 [DOI] [PubMed] [Google Scholar]
- 2. Storch K. F., Lipan O., Leykin I., Viswanathan N., Davis F. C., Wong W. H., Weitz C. J. (2002) Extensive and divergent circadian gene expression in liver and heart. Nature 417, 78–83 [DOI] [PubMed] [Google Scholar]
- 3. Young M. E., Razeghi P., Cedars A. M., Guthrie P. H., Taegtmeyer H. (2001) Intrinsic diurnal variations in cardiac metabolism and contractile function. Circ. Res. 89, 1199–1208 [DOI] [PubMed] [Google Scholar]
- 4. Shimba S., Ishii N., Ohta Y., Ohno T., Watabe Y., Hayashi M., Wada T., Aoyagi T., Tezuka M. (2005) Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 12071–12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mühlbauer E., Wolgast S., Finckh U., Peschke D., Peschke E. (2004) Indication of circadian oscillations in the rat pancreas. FEBS Lett. 564, 91–96 [DOI] [PubMed] [Google Scholar]
- 6. Yoo S. H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H. K., Oh W. J., Yoo O. J., Menaker M., Takahashi J. S. (2004) PERIOD2::LUCIFERASE real time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., Weitz C. J. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 [DOI] [PubMed] [Google Scholar]
- 8. Griffin E. A., Jr., Staknis D., Weitz C. J. (1999) Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286, 768–771 [DOI] [PubMed] [Google Scholar]
- 9. Kiyohara Y. B., Tagao S., Tamanini F., Morita A., Sugisawa Y., Yasuda M., Yamanaka I., Ueda H. R., van der Horst G. T., Kondo T., Yagita K. (2006) The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc. Natl. Acad. Sci. U.S.A. 103, 10074–10079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X., Maywood E. S., Hastings M. H., Reppert S. M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205 [DOI] [PubMed] [Google Scholar]
- 11. Okamura H., Doi M., Fustin J. M., Yamaguchi Y., Matsuo M. (2010) Mammalian circadian clock system: molecular mechanisms for pharmaceutical and medical sciences. Adv. Drug. Deliv. Rev. 62, 876–884 [DOI] [PubMed] [Google Scholar]
- 12. Sato T. K., Yamada R. G., Ukai H., Baggs J. E., Miraglia L. J., Kobayashi T. J., Welsh D. K., Kay S. A., Ueda H. R., Hogenesch J. B. (2006) Feedback repression is required for mammalian circadian clock function. Nat. Genet. 38, 312–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamaguchi S., Mitsui S., Miyake S., Yan L., Onishi H., Yagita K., Suzuki M., Shibata S., Kobayashi M., Okamura H. (2000) The 5′ upstream region of mPer1 gene contains two promoters and is responsible for circadian oscillation. Curr. Biol. 10, 873–876 [DOI] [PubMed] [Google Scholar]
- 14. Karsenty G. (2003) The complexities of skeletal biology. Nature 423, 316–318 [DOI] [PubMed] [Google Scholar]
- 15. Kronenberg H. M. (2003) Developmental regulation of the growth plate. Nature 423, 332–336 [DOI] [PubMed] [Google Scholar]
- 16. Adams S. L., Cohen A. J., Lassová L. (2007) Integration of signaling pathways regulating chondrocyte differentiation during endochondral bone formation. J. Cell Physiol. 213, 635–641 [DOI] [PubMed] [Google Scholar]
- 17. Macsai C. E., Foster B. K., Xian C. J. (2008) Roles of Wnt signaling in bone growth, remodeling, skeletal disorders, and fracture repair. J. Cell Physiol. 215, 578–587 [DOI] [PubMed] [Google Scholar]
- 18. Noonan K. J., Farnum C. E., Leiferman E. M., Lampl M., Markel M. D., Wilsman N. J. (2004) Growing pains: are they due to increased growth during recumbency as documented in a lamb model? J. Pediatr. Orthop. 24, 726–731 [DOI] [PubMed] [Google Scholar]
- 19. Hinoi E., Ueshima T., Hojo H., Iemata M., Takarada T., Yoneda Y. (2006) Up-regulation of per mRNA expression by parathyroid hormone through a protein kinase A-CREB-dependent mechanism in chondrocytes. J. Biol. Chem. 281, 23632–23642 [DOI] [PubMed] [Google Scholar]
- 20. Shimba S., Ogawa T., Hitosugi S., Ichihashi Y., Nakadaira Y., Kobayashi M., Tezuka M., Kosuge Y., Ishige K., Ito Y., Komiyama K., Okamatsu-Ogura Y., Kimura K., Saito M. (2011) Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PloS One 6, e25231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Storch K. F., Paz C., Signorovitch J., Raviola E., Pawlyk B., Li T., Weitz C. J. (2007) Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell 130, 730–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Terpstra L., Prud'homme J., Arabian A., Takeda S., Karsenty G., Dedhar S., St-Arnaud R. (2003) Reduced chondrocyte proliferation and chondrodysplasia in mice lacking the integrin-linked kinase in chondrocytes. J. Cell Biol. 162, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takarada T., Hinoi E., Kambe Y., Sahara K., Kurokawa S., Takahata Y., Yoneda Y. (2007) Osteoblast protects osteoclast devoid of sodium-dependent vitamin C transporters from oxidative cytotoxicity of ascorbic acid. Eur. J. Pharmacol. 575, 1–11 [DOI] [PubMed] [Google Scholar]
- 24. Takarada T., Hinoi E., Takahata Y., Yoneda Y. (2008) Serine racemase suppresses chondrogenic differentiation in cartilage in a Sox9-dependent manner. J. Cell Physiol. 215, 320–328 [DOI] [PubMed] [Google Scholar]
- 25. Takarada T., Hojo H., Iemata M., Sahara K., Kodama A., Nakamura N., Hinoi E., Yoneda Y. (2009) Interference by adrenaline with chondrogenic differentiation through suppression of gene transactivation mediated by Sox9 family members. Bone 45, 568–578 [DOI] [PubMed] [Google Scholar]
- 26. Takarada T., Takahata Y., Iemata M., Hinoi E., Uno K., Hirai T., Yamamoto T., Yoneda Y. (2009) Interference with cellular differentiation by d-serine through antagonism at N-methyl-d-aspartate receptors composed of NR1 and NR3A subunits in chondrocytes. J. Cell Physiol. 220, 756–764 [DOI] [PubMed] [Google Scholar]
- 27. Pang Z. P., Cao P., Xu W., Südhof T. C. (2010) Calmodulin controls synaptic strength via presynaptic activation of calmodulin kinase II. J. Neurosci. 30, 4132–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reppert S. M., Weaver D. R. (2002) Coordination of circadian timing in mammals. Nature 418, 935–941 [DOI] [PubMed] [Google Scholar]
- 29. McDearmon E. L., Patel K. N., Ko C. H., Walisser J. A., Schook A. C., Chong J. L., Wilsbacher L. D., Song E. J., Hong H. K., Bradfield C. A., Takahashi J. S. (2006) Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science 314, 1304–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nilsson O., Marino R., De Luca F., Phillip M., Baron J. (2005) Endocrine regulation of the growth plate. Horm. Res. 64, 157–165 [DOI] [PubMed] [Google Scholar]
- 31. Hastings M., O'Neill J. S., Maywood E. S. (2007) Circadian clocks: regulators of endocrine and metabolic rhythms. J. Endocrinol. 195, 187–198 [DOI] [PubMed] [Google Scholar]
- 32. Deckelbaum R. A., Chan G., Miao D., Goltzman D., Karaplis A. C. (2002) Ihh enhances differentiation of CFK-2 chondrocytic cells and antagonizes PTHrP-mediated activation of PKA. J. Cell Sci. 115, 3015–3025 [DOI] [PubMed] [Google Scholar]
- 33. St-Jacques B., Hammerschmidt M., McMahon A. P. (1999) Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 13, 2072–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chung U. I., Schipani E., McMahon A. P., Kronenberg H. M. (2001) Indian hedgehog couples chondrogenesis to osteogenesis in endochondral bone development. J. Clin. Invest. 107, 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Razzaque M. S., Soegiarto D. W., Chang D., Long F., Lanske B. (2005) Conditional deletion of Indian hedgehog from collagen type 2α1-expressing cells results in abnormal endochondral bone formation. J. Pathol. 207, 453–461 [DOI] [PubMed] [Google Scholar]
- 36. Marcheva B., Ramsey K. M., Buhr E. D., Kobayashi Y., Su H., Ko C. H., Ivanova G., Omura C., Mo S., Vitaterna M. H., Lopez J. P., Philipson L. H., Bradfield C. A., Crosby S. D., JeBailey L., Wang X., Takahashi J. S., Bass J. (2010) Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466, 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]