Background: The somatic cell reprogramming factors do not always induce pluripotency.
Results: The optimal ratio of the reprogramming factors is Oct3/4-high, Sox2-low, Klf4-high, and c-Myc-high.
Conclusion: Among the various reprogramming transcription factor combinations, high Oct3/4 and low Sox2 produced the most efficient results.
Significance: The overall gene expression profiles between the high and low efficiency conditions provide novel insights for somatic cell reprogramming.
Keywords: Epigenetics, G Protein-coupled Receptors (GPCR), Induced Pluripotent Stem (iPS) Cell, Microarray, Reprogramming
Abstract
Somatic cell reprogramming is achieved by four reprogramming transcription factors (RTFs), Oct3/4, Sox2, Klf4, and c-Myc. However, in addition to the induction of pluripotent cells, these RTFs also generate pseudo-pluripotent cells, which do not show Nanog promoter activity. Therefore, it should be possible to fine-tune the RTFs to produce only fully pluripotent cells. For this study, a tagging system was developed to sort induced pluripotent stem (iPS) cells according to the expression levels of each of the four RTFs. Using this system, the most effective ratio (Oct3/4-high, Sox2-low, Klf4-high, c-Myc-high) of the RTFs was 88 times more efficient at producing iPS cells than the worst effective ratio (Oct3/4-low, Sox2-high, Klf4-low, c-Myc-low). Among the various RTF combinations, Oct3/4-high and Sox2-low produced the most efficient results. To investigate the molecular basis, microarray analysis was performed on iPS cells generated under high (Oct3/4-high and Sox2-low) and low (Oct3/4-low and Sox2-high) efficiency reprogramming conditions. Pathway analysis revealed that the G protein-coupled receptor (GPCR) pathway was up-regulated significantly under the high efficiency condition and treatment with the chemokine, C-C motif ligand 2, a member of the GPCR family, enhanced somatic cell reprogramming 12.3 times. Furthermore, data from the analysis of the signature gene expression profiles of mouse embryonic fibroblasts at 2 days after RTF infection revealed that the genetic modifier, Whsc1l1 (variant 1), also improved the efficiency of somatic cell reprogramming. Finally, comparison of the overall gene expression profiles between the high and low efficiency conditions will provide novel insights into mechanisms underlying somatic cell reprogramming.
Introduction
In 2006, Yamanaka and colleagues (1) showed that somatic cells in mice could be reprogrammed to the pluripotent state in the presence of four reprogramming transcription factors (RTFs),3 Oct3/4, Sox2, Klf4, and c-Myc. The following year, induced pluripotent stem (iPS) cell technology was established in human cells (2), and since then, the number of potential applications for iPS technology in regenerative medicine has been growing rapidly. The technology, using transplanted iPS cells, has been used successfully in mouse and rat models of sickle cell anemia and Parkinson disease (3, 4). However, there are several problems that need resolving before iPS cells can be used safely in clinical applications. For example, although iPS cells can differentiate into any cell type in the body, it is necessary to exclude any undifferentiated cells before iPS cell-derived cells are transplanted, as the presence of undifferentiated cells may lead to tumor formation (5).
To use iPS cells in clinical applications, it is important to understand the mechanism that induces pluripotency. It is clear that the process of iPS cell generation involves certain steps (6). These can be broadly summarized as follows. Upon introduction of the four RTFs, fibroblasts down-regulate THY-1 expression; next, the genes used as markers for pluripotency are activated, including alkaline phosphatase and stage-specific embryonic antigen-1 (SSEA-1); finally, the retroviruses used for RTF introduction are silenced, whereas endogenous gene expression of pluripotency-associated molecules, such as Oct3/4 and Nanog, are activated. At this time, reactivation of an X chromosome is also seen.
On the other hand, the more detailed mechanisms underlying the induction of pluripotency are largely unknown. There are some clues, such as the involvement of cell-cell contact during the generation of iPS cells, observed during time-lapse analysis, and it is also suggested that a certain probabilistic action has been influenced during iPS cell generation (6, 7). In addition, although it is clear that the demethylation of DNA and changes in histone modifications occur in the regulatory regions of pluripotency-associated genes, such as Oct3/4 and Nanog, it is not known when these events take place (8). Furthermore, it was reported recently that the four RTFs mediated the induction of other cell types, in addition to iPS cells, including epiblast stem cells and cardiomyocytes (9, 10). Therefore, understanding the mechanism initiated in response to the introduction of the four RTFs is important, not only for the efficient induction of iPS cells but also for controlling other cell fates.
In this study, we focused on the ratio of the four RTFs. To analyze the different ratios for each factor, tagged vectors were generated and used to sort the transfected RTFs on the basis of their expression levels by FACS analysis. Using this sorting method, the efficiency of iPS cell generation was compared with the expression level of each of the four RTFs, and the optimal ratio of the four factors was identified as follows: Oct3/4-high, Sox2-low, Klf4-high, and c-Myc-high. Under these conditions, iPS cell generation efficiency was 88 times greater than the worst effective ratio (Oct3/4-low, Sox2-high, Klf4-low, and c-Myc-low). Finally, the molecular signature for sorting the high efficiency reprogramming conditions from low efficiency conditions was identified by comparing the gene expression profiles of mouse embryonic fibroblasts (MEFs) at 2 days after the RTFs infection.
EXPERIMENTAL PROCEDURES
Mice
The Nanog-GFP-IRES-puro transgenic mouse strain (RBRC02290) has been described previously (8, 11). C57BL/6 mice were purchased from Japan SLC (Shizuoka, Japan). Animal care was performed in accordance with the guidelines established by Keio University for animal and recombinant DNA experimentation. Nanog-GFP MEFs were generated by crossing the transgenic mice with C57BL/6 mice.
Plasmids
Retroviral plasmids for iPS cell induction have been described previously (11). The following 2A sequence was used: 5′-aaaattgtcgctcctgtcaaacaaactcttaactttgatttactcaaactggctggggatgtagaaagcaatccaggtcca-3′ (12). The surface tagging antigens were obtained from pMXs-IRES-rat CD2, pMX-IRES-human CD8, and pMACS-human LNGFR (Miltenyi Biotech). Human CD25 was cloned by PCR with the following primers: 5′-GCCACCATGGATTCATACCTGCTGATG-3′ and 5′-GTCGACCTAGATTGTTCTTCTACTCTT-3′. The constructs, pMXs-IRES-rat CD2 and pMX-IRES-human CD8, were donated by Dr. Masato Kubo and Dr. Takashi Saito, respectively (13, 14). For the epigenetic modifiers, Setdb2, Smyd3, and Whsc1l1 variants 1 and 2 were cloned by PCR, inserted into the pGEM-T-easy plasmid (Promega) and converted to pMXs via the BamHI and XhoI sites. The PCR primers used were as follows: Setdb2, forward, 5′-GGATCCGCCACCATGGAAGAAAAAAATGGTGATGCA-3′; Setdb2, reverse, 5′-CTCGAGTTATATTAATTTTTTCCGACACTT-3′; Smyd3, forward, 5′-GGATCCGCCACCATGGAGGCACTGAAGGTGGAAAAG-3′; Smyd3, reverse, 5′-CTCGAGTTAGGAGGCTCGTATGTTGGCATC-3′; Whscl1l variant 1, forward, 5′-GGATCCGCCACCATGGATTTCTCTTTCTCTTTCATG-3′; Whscl1l variant 1, reverse, 5′-CTCGAGTCAGTCCACAGTTTCCTCTTTCGC-3′; and Whsc1l1 variant 2, forward, 5′-GGATCCGCCACCATGGATTTCTCTTTCTCTTTCATG-3′; Whsc1l1 variant 2, reverse, 5′-GTCGACTCACTCCTTTACTTCTTCTCCACT-3′.
Reprogramming of MEFs Using Tagged Vectors
Oct3/4–2A-hCD8, Sox2–2A-rCD2, and Klf4–2A-hCD271 with, or without, c-Myc-2A-hCD25 were introduced into MEFs by retroviruses according to the previously described method for iPS cell induction (15). Two days after infection, MEFs were collected by incubation in 0.05% trypsin EDTA for 5 min. After washing, the cells were incubated with an anti-FcγR antibody (2.4G2) (eBioscience) at 4 °C for 30 min, and then incubated with a fluorescein isothiocyanate-conjugated anti-rat CD2 monoclonal antibody (OX-34; BioLegend), a phycoerythrin-conjugated anti-human CD271 monoclonal antibody (C40–1457; BD Biosciences), and an allophycocyanin (APC)-conjugated anti-human CD8 monoclonal antibody (RPA-T8; BioLegend) for 30 min at 4 °C. For the four factor reprogramming, a phycoerythrin-Cy7-conjugated anti-human CD25 monoclonal antibody (M-A251) was also added. After washing, samples were sorted using a FACSVantage SE cytometer (BD Biosciences). Sorted cells were cultured on STO cells at a density of 30,000 cells (without c-Myc) or 4,000 cells (with c-Myc) per well in six-well plates. The numbers of Nanog-GFP positive (Nanog-GFP+) colonies were counted on days 17 or 21. Data are presented as the each dot. The median numbers are also presented as a bar. Statistical significance for difference of the medians was determined by exact Wilcoxon test using the R exactRankTests package.
Analysis of Chemokines for Reprogramming
MEFs carrying the four introduced reprogramming factors were reseeded on STO feeders 4 days after infection at a density of 4,500 cells/well in six-well plates. At that time, 100 ng/ml of each chemokine was added every 2 days to the culture until day 17. The medium was changed every second day. On day 7 after infection puromycin was added to the culture. Colony numbers were counted at day 23.
Statistical Analysis of Reprogramming Efficiency According to the Ratio of Reprogramming Factors
The Mann-Whitney U test was performed to compare differences in distribution for the number of positive colonies under the different reprogramming conditions.
Microarray Data Analysis
Expression profiles of MEFs at 2 days after the RTF infection were analyzed using the whole mouse genome 44K3D-Gene Mouse Oligo chip 24K (Agilent Technologies, Santa Clara, CA). Fluorescence intensities were detected using the Scan-Array Life Scanner (PerkinElmer Life Science) and photomultiplier tube levels were adjusted to achieve 0.1–0.5% pixel saturation. Each TIFF image was analyzed with GenePix Pro software version 6.0 (Molecular Devices, Sunnyvale, CA). The data were filtered to remove low-confidence measurements and normalized globally per array such that the median signal intensity was set at 50.
All 43,379 probes were collapsed into 21,609 genes with Entrez gene identifier (ID) by taking the maximum intensity among probe sets corresponding to the same gene ID. The standard Student's t test was performed for each comparison and the false discovery rate was estimated using the Benjamini-Hochberg procedure to obtain differentially expressed genes as a signature. In this study, a false discovery rate <5% was used as a threshold. To characterize the molecular backgrounds of the signature genes, enrichment analysis for canonical pathways and Gene Ontology biological processes (c2-cp and c5-bp gene sets in MSigDB version 3.0 (16)) was performed using the GO Term Finder (17).
RESULTS
The Four RTFs Do Not Always Induce Pluripotency in Somatic Cells
Somatic cell reprogramming is brought about by the four RTFs, Oct3/4, Sox2, Klf4, and c-Myc. Initially, these transcription factors were introduced into somatic cells by retroviral vectors; however, because these viral vectors are usually, but not always completely, inactivated toward the end of the reprogramming process, silencing of the retrovirus promoter was recognized as one of the reprogramming criteria (8). For the current study, the four RTFs were introduced into MEFs carrying green fluorescent protein (GFP) under the control of the Nanog promoter. To monitor silencing, a DsRed vector was also introduced. After induction of the four RTFs, Nanog-GFP+ and DsRed negative (DsRed−) iPS candidate cells were observed (Fig. 1a), as well as Nanog-GFP− and DsRed+ pseudo-pluripotent iPS cells (Fig. 1b). These data indicated that the RTFs did not achieve pluripotency in all somatic cells.
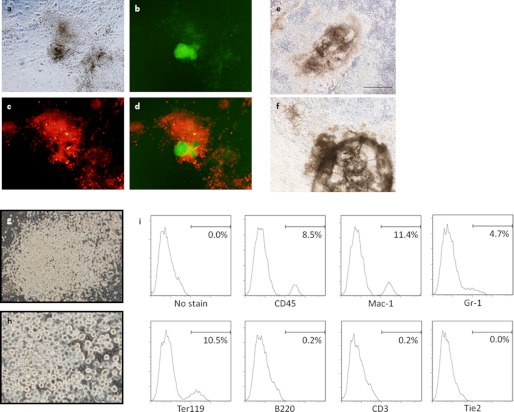
FIGURE 1.
Reprogramming factors also induce non-pluripotent cells. a–d, Nanog-GFP+, DsRed− iPS cell colony (green), and Nanog-GFP−, DsRed+ non-pluripotent pseudo cells (red); and phase-contrast (a), Nanog promoter-driven GFP expression (b), retroviral DsRed expression (c), and merged image (d). e and f, tail-tip fibroblasts-derived cardiomyocyte-like cells following four RTF infection. These cells can be seen pulsing in supplemental Movies S1 and S2. g and h, morphology of MEF-derived rounded blood-like cells following four TF infection. h, is a high magnification of g. i, flow cytometric analysis of blood-like cells. Expression levels were analyzed using the antibodies indicated.
Moreover, occasionally non-iPS cells with specific features were also seen after induction of the four RTFs; for example, Fig. 1 shows spontaneously beating cardiomyocyte-like cells generated from adult tail-tip fibroblasts (Fig. 1, e and f, and supplemental Movies S1 and S2). In addition, morphologically rounded, blood-like cells were also observed (Fig. 1, g and h). When these blood-like cells were collected by pipetting and stained for cell surface markers, they were found to be positive for the pan-hematopoietic marker, CD45 (Fig. 1i). Analysis of lineage markers revealed that these blood-like cells contained macrophages (Mac-1), granulocytes (Gr-1), and erythroid cells (Ter119) (Fig. 1i). However, B (B220) and T (CD3) lymphoid cells were not detected (Fig. 1i). The so-called “transdifferentiation” of these two lineages by the factors used in somatic cell reprogramming has also been reported by other groups (10, 18). These data indicated that the four RTFs do not only induce the pluripotent state but are also capable of producing terminally differentiated cells.
Optimal Ratio of the Four RTFs for Somatic Cell Reprogramming
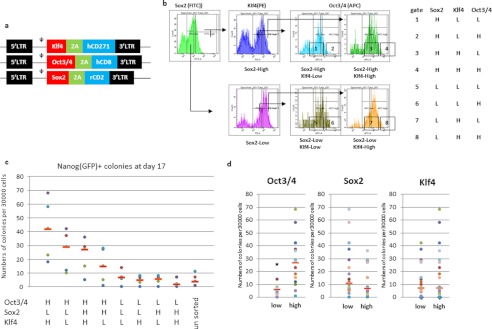
Because the reprogramming factors can also induce other cell types as well as pluripotent cells, it should be possible to fine-tune the RTFs to produce only fully pluripotent cells. Therefore, we speculated that there would be an optimal ratio of the four RTFs for efficient pluripotent cell generation. To investigate the importance of the relative expression levels of each of the RTFs in somatic cell reprogramming, Sox2, Klf4, and Oct3/4 were tagged with different rat and human cell surface antigens using a 2A sequence (Fig. 2a). After infection of MEFs with each of these constructs, flow cytometry with specific antibodies was used to sort the cells according to the expression levels of the exogenous genes (Fig. 2b). Using this strategy, the MEFs were grouped based on the ratios of the three factors, and Nanog-GFP+ colonies were counted on day 17 after infection. The effects of the expression of each of the three factors are shown in Fig. 2c, and the results indicated that the greatest numbers of Nanog-GFP+ colonies were obtained with high levels of Oct3/4. The most effective ratio of the three factors (Oct3/4-high, Sox2-low, and Klf4-high) was seven times more efficient than for the worst effective ratio (Oct3/4-low, Sox2-high, and Klf4-low).
FIGURE 2.
Somatic cell reprogramming using different ratios of Oct3/4, Sox2, and Klf4. a, retrovirus vectors with cell surface antigens. b, flow cytometric analysis of the introduced factors together with the sorting gates used. c and d, number of Nanog-GFP+ colonies after sorting on day 17 of culture. MEFs were sorted using relative gene expression levels, as indicated on the horizontal axis. Dots represent the numbers of each experiment and bar means median. The numbers on the graph (c) were recalculated based on the expression level of each factor in d. Dots represent the numbers of each experiment and the bar means median. *, p = 1.14E-06. H, high; L, low.
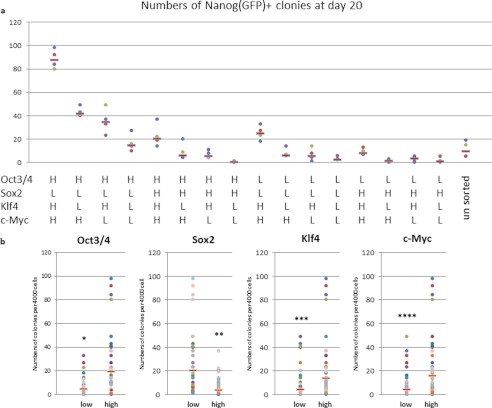
In addition to these three RTFs, the effect of c-Myc was also analyzed. A human CD25-tagged c-Myc vector was generated and used to monitor the relative expression of all four RTFs (supplemental Fig. S1). The expression levels of each of the factors were confirmed by RT-PCR (supplemental Figs. S2 and S3). The results are shown in Fig. 3. The addition of c-Myc did not affect the ratios of the other three factors. High expression of Oct3/4, Klf4, and c-Myc favored the induction of pluripotency, whereas low expression of Sox2 was better for reprogramming. Similar to induction with three RTFs, the most effective ratio of the four factors (Oct3/4-high, Sox2-low, Klf4-high, and c-Myc-high) was 50 times more efficient than for the worst effective ratio (Oct3/4-low, Sox2-high, Klf4-low, and c-Myc-high). Regardless of the efficiency, generated iPS cells showed similar gene expression patterns to ES cells and have a potential to differentiate to all three germ layers (supplemental Figs. S4 and S5).
FIGURE 3.
Somatic cell reprogramming using different ratios of Oct3/4, Sox2, Klf4, and c-Myc. a and b, number of Nanog-GFP+ colonies after sorting on day 21 of culture. MEFs were sorted using relative gene expression levels, as indicated on the horizontal axis. Dots represent the numbers of each experiment and the bar means median. The numbers on graph (a) were recalculated based on the expression level of each factor in b. Dots represent the numbers of each experiment and the bar means median. *, p = 2.69E-04; **, p = 8.96E-06; ***, p = 3.20E-03; ****, p = 8.96.98E-04. H, high; L, low.
Microarray Analysis of High (Oct3/4-high and Sox2-low) and Low (Oct3/4-low and Sox2-high) Efficiency Reprogramming Conditions
We searched for the most effective combination of the four RTFs using the relationship between Nanog-GFP+ colony numbers and the reprogramming factor ratio. Among the four factors, the Oct3/4 and Sox2 expression ratios correlated significantly with positive colony numbers. In cells with high levels of Oct3/4 and low levels of Sox2, ∼16.2 times greater numbers of positive colonies were found when all four factors were introduced (supplemental Fig. S6a). A similar result was also found when only three factors were used (supplemental Fig. S6b) even if the statistically dominant factor was only Oct3/4. To determine the molecular basis underlying these ratios and indeed, somatic cell reprogramming, microarray analysis was performed using the high (Oct3/4-high and Sox2-low) and the low (Oct3/4-low and Sox2-high) reprogramming conditions.
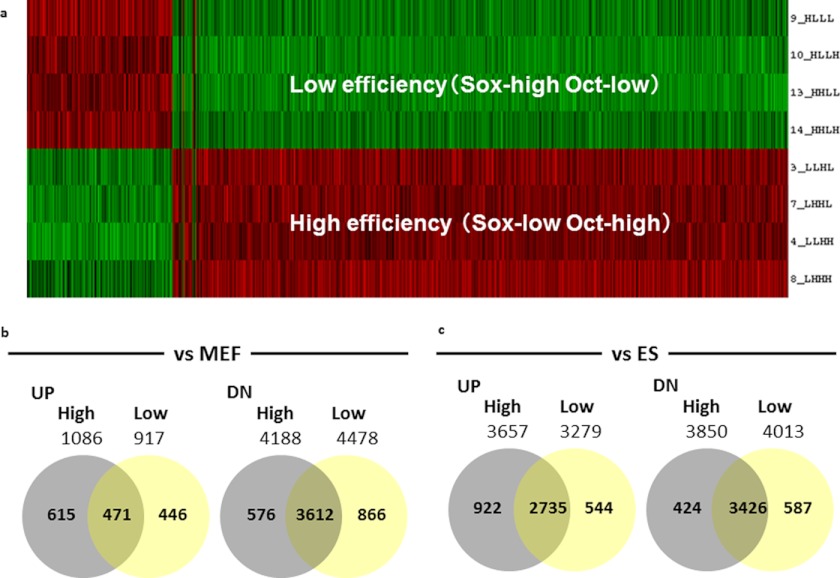
The signature genes were identified using bioinformatics calculations (Fig. 4a). First, the signature genes in MEFs at 2 days after the RTF infections were compared with those of the parental MEFs and with pluripotent embryonic stem (ES) cells. When compared with MEFs, ∼1,000 genes were up-regulated and 4,000 genes were down-regulated under both high and low efficiency conditions. Whereas about half the up-regulated genes were common to both the high and low reprogramming conditions, more than 80% of the down-regulated genes were common to both (Fig. 4b and supplemental Table S1). On the other hand, when compared with ES cells, more than 70% of the up-regulated genes and 80% of the down-regulated genes were common to both cell types under both sets of conditions (Fig. 4c and supplemental Table S2). These data indicated that the expression of many signature genes in MEFs at 2 days after the RTFs infection was altered under both high (Oct3/4-high and Sox2-low) and low (Oct3/4-low and Sox2-high) reprogramming conditions when compared with MEF and ES cells.
FIGURE 4.
Microarray analysis of the high and low efficiency conditions for reprogramming. a, array heat map of signature genes from low and high efficiency conditions. b, number of signature genes that were up- or down-regulated in the high and low efficiency conditions compared with parental MEFs. c, number of signature genes that were up- or down-regulated compared with ES cells. The names of the genes in MEFs and ES cells are listed in supplemental Tables S1 and S2, respectively.
Molecular Signature for Sorting the High (Oct3/4-high and Sox2-low) Low (Oct3/4-low and Sox2-high) Efficiency Reprogramming Conditions
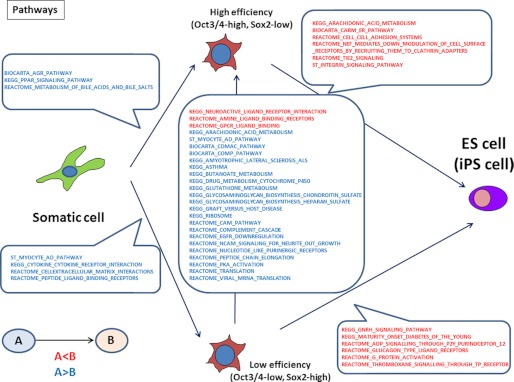
To determine the difference between the high (Oct3/4-high and Sox2-low) and low (Oct3/4-low and Sox2-high) efficiency conditions, the microarray data for these two conditions were compared. GO analysis showed that under the high efficiency condition, positive regulation of MAP kinase activity was down-regulated (supplemental Fig. S7) in iPS cells, which is significant because it is known that inhibition of the MAP kinase pathway is important for pluripotency (19). Furthermore, pathway analysis of the microarray data revealed that certain pathways were up-regulated preferentially under the high condition, compared with the low condition (Fig. 5). Under the high efficiency condition, we focused on enrichment of the GPCR pathways, and in particular, the chemokine members of the GPCR superfamily. To analyze the involvement of chemokines during somatic cell reprogramming, the effect of several chemokines on the generation of iPS cells was examined. Of these, the addition of CCL2 achieved a 12.3 times greater reprogramming efficiency than in untreated cells (Fig. 6a). These results suggested that the microarray data contained clues for the optimization of pluripotency induction.
FIGURE 5.
Pathway analysis of microarray data. Microarray data of MEFs at 2 days after the RTF infection under high (Oct3/4-high, Sox2-low) and low (Oct3/4-low, Sox2-high) efficiency conditions were compared with MEFs and ES cells, and the up- and down-regulated pathways between each cell type are shown. Up-regulated pathways are shown in red and down-regulated pathways are shown in blue.
FIGURE 6.
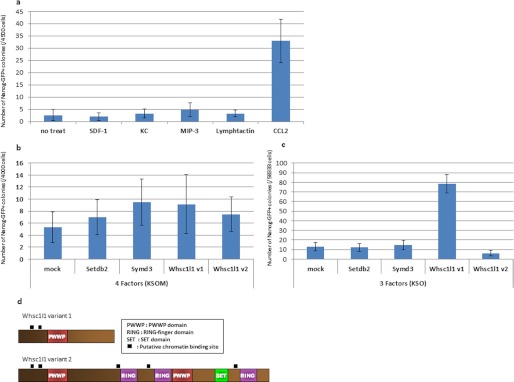
The effect of chemokines and epigenetic modifiers on somatic cell reprogramming. a, MEFs were infected with the four RTFs and the chemokines indicated were added from days 4 to 17 of the culture. The numbers of Nanog-GFP+ colonies on day 23 of culture are indicated. b and c, MEFs were infected with the epigenetic factors indicated, together with four (b) or three (c) of the RTFs. The numbers of Nanog-GFP+ colonies at 17 days after infection are shown.
To understand the mechanism further, transcription factors and epigenetic modifiers were analyzed as these factors direct cell fate and alter the regulation of multiple genes. Although under the low efficiency condition only nine TFs were up-regulated, 60 TFs were up-regulated under the high efficiency condition (supplemental Fig. S8a and Table 1). Furthermore, when the epigenetic modifiers were investigated, only one gene was up-regulated under the low efficiency condition and four under the high condition (supplemental Fig. S8b and Table 2). These data indicated that more transcription factors and epigenetic modifiers appear to be up-regulated under the high condition.
TABLE 1.
Transcription factors up-regulated under high and low efficiency conditions
| Symbol | Description |
|---|---|
| High efficiency condition | |
| POU5F1 | POU class 5 homeobox 1 |
| HOXC4 | Homeobox C4 |
| IRX4 | Iroquois homeobox 4 |
| NEUROG1 | Neurogenin 1 |
| BARHL1 | BarH-like homeobox 1 |
| FOXN1 | Forkhead box N1 |
| KLF17 | Kruppel-like factor 17 |
| NR5A1 | Nuclear receptor subfamily 5, group A, member 1 |
| ZNF43 | Zinc finger protein 43 |
| POU4F1 | POU class 4 homeobox 1 |
| RFX4 | Regulatory factor X, 4 (influences HLA class II expression) |
| ESRRG | Estrogen-related receptor gamma |
| FOXH1 | Forkhead box H1 |
| SOX15 | SRY (sex determining region Y)-box 15 |
| LHX1 | LIM homeobox 1 |
| TOPORS | Topoisomerase I binding, arginine/serine-rich |
| HNF4A | Hepatocyte nuclear factor 4, α |
| NKX61 | NK6 homeobox 1 |
| PROP1 | PROP paired-like homeobox 1 |
| CAMTA1 | Calmodulin binding transcription activator 1 |
| ARID5B | AT-rich interactive domain 5B (MRF1-like) |
| SOX17 | SRY (sex determining region Y)-box 17 |
| FOXQ1 | forkhead box Q1 |
| MAF | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) |
| TCF2 | HNF1 homeobox B |
| FEV | FEV (ETS oncogene family) |
| HES2 | Hairy and enhancer of split 2 (Drosophila) |
| PITX3 | Paired-like homeodomain 3 |
| HOXA3 | Homeobox A3 |
| HNF4G | Hepatocyte nuclear factor 4, γ |
| TCF7L2 | Transcription factor 7-like2 (T-cell specific, HMG-box) |
| TP73 | Tumor protein p73 |
| NR3C2 | Nuclear receptor subfamily 3, group C, member 2 |
| HSF1 | Heat shock transcription factor 1 |
| GLI1 | GLI family zinc finger 1 |
| SOX1 | SRY (sex determining region Y)-box 1 |
| ZNF124 | Zinc finger protein 124 |
| CDK2 | Cyclin-dependent kinase 2 |
| FOXE3 | Forkhead box E3 |
| RBPJ | Recombination signal-binding protein for immunoglobulin κJ region |
| CREBBP | CREB-binding protein |
| HOXB9 | Homeobox B9 |
| FOXL2 | Forkhead box L2 |
| FOXF2 | Forkhead box F2 |
| NCX | T-cell leukemia homeobox 2 |
| TFDP2 | Transcription factor Dp-2 (E2F dimerization partner 2) |
| ATBF1 | Zinc finger homeobox 3 |
| NR1I3 | Nuclear receptor subfamily 1, group I, member 3 |
| SOX12 | SRY (sex determining region Y)-box 12 |
| LMO3 | LIM domain only3 (rhombotin-like 2) |
| ABL1 | c-abl oncogene 1, receptor tyrosine kinase |
| GTF2IRD1 | GTF2I repeat domain containing 1 |
| IRF1 | Interferon regulatory factor 1 |
| NFIA | Nuclear factor I/A |
| SS18L1 | Synovial sarcoma translocation gene on chromosome 18-like 1 |
| NFATC2 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| STAT5B | Signal transducer and activator of transcription 5B |
| FOXO4 | Forkhead box O4 |
| HOXB6 | Homeobox B6 |
| RUNX2 | Runt-related transcription factor 2 |
| Low efficiency condition | |
| ID3 | Inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| XPA | Xeroderma pigmentosum, complementation group A |
| LEF1 | Lymphoid enhancer-binding factor 1 |
| KLF2 | Kruppel-like factor 2 (lung) |
| HEY1 | Hairy/enhancer of split related with YRPW motif 1 |
| PRDM1 | PR domain containing 1, with ZNF domain |
| ELOF1 | Elongation factor 1 homolog (Saccharomyces cerevisiae) |
| SREBF1 | Sterol regulatory element binding transcription factor 1 |
| TBX2 | T-box 2 |
TABLE 2.
Epigenetic modifiers upregulated under high and low efficiency conditions
| Symbol | Description |
|---|---|
| High efficiency condition | |
| SETDB2 | SET domain, bifurcated 2 |
| WHSC1L1 | Wolf-Hirschhorn syndrome candidate 1-like 1 |
| CREBBP | CREB-binding protein |
| SMYD3 | SET and MYND domain containing 3 |
| Low efficiency condition | |
| PRDM1 | PR domain containing 1, with ZNF domain |
To assess the function of these epigenetic modifiers for somatic cell reprogramming, retrovirus vectors were prepared for Setdb2, Smyd3, and Whsc1l1 variants 1 and 2, epigenetic modifiers that were up-regulated under the high condition. These factors were introduced into MEFs together with three or four of the RTFs, and Nanog-GFP+ colonies were counted on day 17 after infection (Fig. 6, b and c). When introduced with the three RTFs, Whsc1l1 variant 1 produced many more colonies than the control; however, variant 2 had no significant effect (Fig. 6c).
DISCUSSION
From investigations into the mechanisms governing somatic cell reprogramming that underlies iPS cell technology, several groups have reported that specific combinations of individual transcription factors can induce the generation of particular cell types (20, 21). In contrast to the induction of pluripotent stem cells, the technology for the generation of lineage-restricted cells is known as transdifferentiation or direct reprogramming. A particular combination of specific transcription factors, which are critical for the development and/or maintenance of the lineage-restricted cells, is used for transdifferentiation. On the other hand, it is reported that pluripotency inducible factors also mediate transdifferentiation (9, 10, 18). Therefore, it is both interesting and feasible to analyze the fine-tuning of the RTFs required for pluripotency. In the present study, the relative ratio of the four RTFs was examined and the results demonstrated that there is, indeed, an optimal ratio (Oct3/4-high, Sox2-low, Klf4-high, and c-Myc-high) of these factors for iPS cell generation and, moreover, that the ratio, Oct3/4-high and Sox2-low, is critical.
It was reported previously that high expression of Oct3/4 improves reprogramming efficiencies and that modified Oct3/4 with greater transcriptional activity further enhances the reprogramming efficiency (22, 23). Furthermore, control of Oct3/4 expression is essential for maintaining ES cells in the undifferentiated state, and both the overexpression and down-regulation of Oct3/4 can induce ES cell differentiation, suggesting that tightly controlled regulation of Oct3/4 expression levels controls the maintenance of pluripotency (24). In the current study, we have shown that, in the presence of other factors, high Oct3/4 expression is critical for somatic cell reprogramming, whereas low levels of Oct3/4 result in a lower induction efficiency (Figs. 2 and 3).
In contrast to Oct3/4, low Sox2 expression is more efficient for the acquisition of pluripotency, and it is reported that low Sox2 expression increased the reprogramming efficiency by repressing ectoderm and mesoderm marker genes (25). In the array data presented here, the ectoderm maker, CryM, showed a statistically significant decrease in expression under low Sox2 conditions (supplemental Fig. S9a and Table S3). Although another ectoderm marker, Sox13, also decreased in the presence of low Sox2, the expression of Sox21 was not linked to the level of Sox2 (supplemental Table S3). On the other hand, expression of the mesoderm marker, Myh2, did not change. However, when Klf4 expression was altered (high or low), Myh2 expression was lower in cells under low Sox2 conditions than under high Sox2 conditions (supplemental Fig. S9b and Table S3). These data indicated that although low Sox2 expression may repress ectoderm and mesoderm markers, the other RTFs are also involved in the repression of ectoderm and mesoderm marker genes. Furthermore, it has been proposed that a two-step reprogramming mechanism is necessary for the induction of pluripotency, and that Sox2 functions in the latter stages of reprogramming (26). Our data and a previous report suggest that Sox2 expression levels are low during the early phase of reprogramming (25). Thus, it is important to analyze the effects of Sox2 during the different phases of somatic cell reprogramming.
To understand the molecular basis for these events, we performed microarray analyses of the high (Oct3/4-high and Sox2-low) and low (Oct3/4-low and Sox2-high) reprogramming conditions. We observed that 50% of the up-regulated and 80% of the down-regulated genes were common to both conditions when iPS cells were compared with MEF and ES cells (Fig. 4, b and c). Because all four RTFs were introduced for this analysis, it is conceivable that many genes were commonly up- and down-regulated compared with MEF and ES cells. However, when we focused on gene expression levels between the two conditions, the GO terms showed down-regulation of cellular recognition under the low efficiency condition (supplemental Fig. S7), whereas GPCR signaling emerged as a significant pathway under the high condition (Fig. 5). As reported previously, for transdifferentiation using the four RTFs, culture conditions are important for defining cell fate (9, 10), and it is interesting that, in the current study, the high efficiency condition up-regulated the signaling pathway from cell surface molecules, whereas the low efficiency condition down-regulated cellular recognition as demonstrated by the GO terms. One could predict that the four RTFs alter the original program in the somatic cells and up-regulate cell surface molecules to produce favorable signals, including those involved in cell adhesion, required to direct different cell fates. It has been reported that cells adhered together during iPS cell generation, through the up-regulation of the cell adhesion molecule, E-cadherin (7, 27, 28). Furthermore, in the present study, we have confirmed the importance of the GPCR pathway by the addition of the chemokine, CCL2, which binds to the GPCR, CCR2. Interestingly, addition of CCL2 was effective for the high (Oct3/4-high and Sox2-low) but not low (Oct3/4-low and Sox2-high) reprogramming conditions (supplemental Fig. S10). CCL2 was recently reported to maintain pluripotency in ES cells by inducing Klf4 via the activation of STAT3 (29). In the current study, we demonstrated that CCL2 also has a function in the induction of pluripotency. In the case of iPS cell induction, Klf4 is introduced exogenously; therefore, it is important to know whether other pathways are activated during the induction of pluripotency.
When we focused on the role of transcription factors and epigenetic modifiers of the signature genes, the results showed that the high efficiency condition had more activated genes than the low condition. Thus, because epigenetic modifiers affect the expression of multiple genes, it is important to analyze the listed factors. SETDB2 and SMYD3 contain a SET domain, which has putative methyltransferase activity (30, 31), whereas WHSC1L1 is linked to Wolf-Hirschhorn syndrome (32). None of these genes have been well analyzed with respect to their roles in the induction of pluripotency. However, we found that Whsc1l1 variant 1, but not variant 2, enhances the reprogramming efficiency in the presence of Oct3/4, Sox2, and Klf4 (Fig. 6c). WHSC1L1 variant 1 is shorter and about the half the length of variant 2, and interestingly, variant 1 lacks the SET domain, which has putative histone methyltransferase activity (Fig. 6d). In future, to improve our understanding of somatic cell reprogramming, it will be important to analyze the reprogramming activity and the supporting roles played by the other genes identified as pluripotency signature genes in this study.
Acknowledgments
We thank N. Tago for cell sorting and A. Kumakubo for technical assistance.
This work was supported by PRESTO of the Japan Science and Technology Agency and Scientific Research (C), a grant from the Project for Realization of Regenerative Medicine, support for the core institutes for iPS cell research was provided by MEXT, a grant-in-aid for the Global century COE program from MEXT to Keio University, and the Keio University Medical Science Fund.

This article contains supplemental Figs. S1–S10, Tables S1–S3, and Movies S1 and S2.
- RTF
- reprogramming transcription factor
- iPS
- induced pluripotent stem
- MEF
- mouse embryonic fibroblast
- ES
- embryonic stem.
REFERENCES
- 1. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 2. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 3. Hanna J., Wernig M., Markoulaki S., Sun C. W., Meissner A., Cassady J. P., Beard C., Brambrink T., Wu L. C., Townes T. M., Jaenisch R. (2007) Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 318, 1920–1923 [DOI] [PubMed] [Google Scholar]
- 4. Wernig M., Zhao J. P., Pruszak J., Hedlund E., Fu D., Soldner F., Broccoli V., Constantine-Paton M., Isacson O., Jaenisch R. (2008) Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 105, 5856–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miura K., Okada Y., Aoi T., Okada A., Takahashi K., Okita K., Nakagawa M., Koyanagi M., Tanabe K., Ohnuki M., Ogawa D., Ikeda E., Okano H., Yamanaka S. (2009) Variation in the safety of induced pluripotent stem cell lines. Nat. Biotechnol. 27, 743–745 [DOI] [PubMed] [Google Scholar]
- 6. Stadtfeld M., Maherali N., Breault D. T., Hochedlinger K. (2008) Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2, 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Araki R., Jincho Y., Hoki Y., Nakamura M., Tamura C., Ando S., Kasama Y., Abe M. (2010) Conversion of ancestral fibroblasts to induced pluripotent stem cells. Stem Cells 28, 213–220 [DOI] [PubMed] [Google Scholar]
- 8. Okita K., Ichisaka T., Yamanaka S. (2007) Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 9. Han D. W., Greber B., Wu G., Tapia N., Araúzo-Bravo M. J., Ko K., Bernemann C., Stehling M., Schöler H. R. (2011) Direct reprogramming of fibroblasts into epiblast stem cells. Nat. Cell Biol. 13, 66–71 [DOI] [PubMed] [Google Scholar]
- 10. Efe J. A., Hilcove S., Kim J., Zhou H., Ouyang K., Wang G., Chen J., Ding S. (2011) Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 13, 215–222 [DOI] [PubMed] [Google Scholar]
- 11. Nagamatsu G., Kosaka T., Kawasumi M., Kinoshita T., Takubo K., Akiyama H., Sudo T., Kobayashi T., Oya M., Suda T. (2011) A germ cell-specific gene, Prmt5, works in somatic cell reprogramming. J. Biol. Chem. 286, 10641–10648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hasegawa K., Cowan A. B., Nakatsuji N., Suemori H. (2007) Efficient multicistronic expression of a transgene in human embryonic stem cells. Stem Cells 25, 1707–1712 [DOI] [PubMed] [Google Scholar]
- 13. Komine O., Hayashi K., Natsume W., Watanabe T., Seki Y., Seki N., Yagi R., Sukzuki W., Tamauchi H., Hozumi K., Habu S., Kubo M., Satake M. (2003) The Runx1 transcription factor inhibits the differentiation of naive CD4+ T cells into the Th2 lineage by repressing GATA3 expression. J. Exp. Med. 198, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamasaki S., Ishikawa E., Sakuma M., Ogata K., Sakata-Sogawa K., Hiroshima M., Wiest D. L., Tokunaga M., Saito T. (2006) Mechanistic basis of pre-T cell receptor-mediated autonomous signaling critical for thymocyte development. Nat. Immunol. 7, 67–75 [DOI] [PubMed] [Google Scholar]
- 15. Takahashi K., Okita K., Nakagawa M., Yamanaka S. (2007) Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2, 3081–3089 [DOI] [PubMed] [Google Scholar]
- 16. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Gene set enrichment analysis. A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boyle E. I., Weng S., Gollub J., Jin H., Botstein D., Cherry J. M., Sherlock G. (2004) GO::TermFinder. Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20, 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Szabo E., Rampalli S., Risueño R. M., Schnerch A., Mitchell R., Fiebig-Comyn A., Levadoux-Martin M., Bhatia M. (2010) Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468, 521–526 [DOI] [PubMed] [Google Scholar]
- 19. Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008) The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D. A. (2008) In vivo reprogramming of adult pancreatic exocrine cells to beta cells. Nature 455, 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sekiya S., Suzuki A. (2011) Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393 [DOI] [PubMed] [Google Scholar]
- 22. Wang Y., Chen J., Hu J. L., Wei X. X., Qin D., Gao J., Zhang L., Jiang J., Li J. S., Liu J., Lai K. Y., Kuang X., Zhang J., Pei D., Xu G. L. (2011) Reprogramming of mouse and human somatic cells by high-performance engineered factors. EMBO Rep. 12, 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hirai H., Tani T., Katoku-Kikyo N., Kellner S., Karian P., Firpo M., Kikyo N. (2011) Radical acceleration of nuclear reprogramming by chromatin remodeling with the transactivation domain of MyoD. Stem Cells 29, 1349–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Niwa H., Miyazaki J., Smith A. G. (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation, or self-renewal of ES cells. Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- 25. Yamaguchi S., Hirano K., Nagata S., Tada T. (2011) Sox2 expression effects on direct reprogramming efficiency as determined by alternative somatic cell fate. Stem Cell Res. 6, 177–186 [DOI] [PubMed] [Google Scholar]
- 26. Lin Z., Perez P., Lei D., Xu J., Gao X., Bao J. (2011) Two-phase analysis of molecular pathways underlying induced pluripotent stem cell induction. Stem Cells 29, 1963–1974 [DOI] [PubMed] [Google Scholar]
- 27. Chen T., Yuan D., Wei B., Jiang J., Kang J., Ling K., Gu Y., Li J., Xiao L., Pei G. (2010) E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells 28, 1315–1325 [DOI] [PubMed] [Google Scholar]
- 28. Redmer T., Diecke S., Grigoryan T., Quiroga-Negreira A., Birchmeier W., Besser D. (2011) E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 12, 720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hasegawa Y., Takahashi N., Forrest A. R., Shin J. W., Kinoshita Y., Suzuki H., Hayashizaki Y. (2011) CC chemokine ligand 2 and leukemia inhibitory factor cooperatively promote pluripotency in mouse induced pluripotent cells. Stem Cells 29, 1196–1205 [DOI] [PubMed] [Google Scholar]
- 30. Xu P. F., Zhu K. Y., Jin Y., Chen Y., Sun X. J., Deng M., Chen S. J., Chen Z., Liu T. X. (2010) Setdb2 restricts dorsal organizer territory and regulates left-right asymmetry through suppressing fgf8 activity. Proc. Natl. Acad. Sci. U.S.A. 107, 2521–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hamamoto R., Furukawa Y., Morita M., Iimura Y., Silva F. P., Li M., Yagyu R., Nakamura Y. (2004) SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 6, 731–740 [DOI] [PubMed] [Google Scholar]
- 32. Stec I., van Ommen G. J., den Dunnen J. T. (2001) WHSC1L1, on human chromosome 8p11.2, closely resembles WHSC1 and maps to a duplicated region shared with 4p16.3. Genomics 76, 5–8 [DOI] [PubMed] [Google Scholar]