Background: Nature employs processive and nonprocessive glycoside hydrolases to degrade polysaccharides.
Results: We solved the Serratia marcescens nonprocessive chitinase (ChiC2) structure and used simulation to identify dynamic hallmarks of processivity in S. marcescens chitinases.
Conclusion: Dynamic metrics complement structural insights in determining processivity.
Significance: Identification of hallmarks of processivity is a key step toward development of a general, molecular-level theory of glycoside hydrolase processivity.
Keywords: Cellulase, Chitinase, Computer Modeling, Glycoside Hydrolases, Molecular Dynamics, Cellulose, Chitin, Serratia marcescens
Abstract
Degradation of recalcitrant polysaccharides in nature is typically accomplished by mixtures of processive and nonprocessive glycoside hydrolases (GHs), which exhibit synergistic activity wherein nonprocessive enzymes provide new sites for productive attachment of processive enzymes. GH processivity is typically attributed to active site geometry, but previous work has demonstrated that processivity can be tuned by point mutations or removal of single loops. To gain additional insights into the differences between processive and nonprocessive enzymes that give rise to their synergistic activities, this study reports the crystal structure of the catalytic domain of the GH family 18 nonprocessive endochitinase, ChiC, from Serratia marcescens. This completes the structural characterization of the co-evolved chitinolytic enzymes from this bacterium and enables structural analysis of their complementary functions. The ChiC catalytic module reveals a shallow substrate-binding cleft that lacks aromatic residues vital for processivity, a calcium-binding site not previously seen in GH18 chitinases, and, importantly, a displaced catalytic acid (Glu-141), suggesting flexibility in the catalytic center. Molecular dynamics simulations of two processive chitinases (ChiA and ChiB), the ChiC catalytic module, and an endochitinase from Lactococcus lactis show that the nonprocessive enzymes have more flexible catalytic machineries and that their bound ligands are more solvated and flexible. These three features, which relate to the more dynamic on-off ligand binding processes associated with nonprocessive action, correlate to experimentally measured differences in processivity of the S. marcescens chitinases. These newly defined hallmarks thus appear to be key dynamic metrics in determining processivity in GH enzymes complementing structural insights.
Introduction
Enzymatic depolymerization of the structural polysaccharides chitin and cellulose is efficiently achieved in nature via the combined, synergistic effort of enzyme cocktails composed of glycoside hydrolases (GHs)6 and lytic polysaccharide monooxygenases (1–7). These synergistic cocktails contain both processive and nonprocessive GHs, which have evolved concomitantly to deconstruct insoluble, crystalline substrates in an efficient manner. Processive enzymes have been shown to bind individual chains in long tunnels or deep clefts and hydrolyze a series of glycosidic linkages along the same chain before dissociation (8–14). Nonprocessive enzymes, conversely, exhibit shallower clefts for binding and are thought to hydrolyze glycosidic linkages randomly in disordered regions of polymer crystals (15). Nonprocessive enzymes may make more than one cut per enzyme-substrate association primarily because their carbohydrate-binding modules (CBMs) may keep them loosely associated with the substrate and even with a single chain (16, 17). However, in contrast to processive enzymes, nonprocessive enzymes may release the single polymer chain bound in the active site after each catalytic event, although the exact mechanism of endoglucanase action remains unknown.
Developing a comprehensive thermodynamic and kinetic description of enzyme processivity is central to our collective understanding of carbon and nitrogen turnover in the biosphere (18) and is of significant importance in the design of enhanced enzyme mixtures for biomass conversion (1). Available structural data from model organisms such as S. marcescens, Hypocrea jecorina, Clostridium thermocellum, and Thermobifida fusca suggest that the degree of GH processivity is related to the shape and composition of the catalytic tunnel or cleft (2, 8–11, 13, 15, 19–24). Typically, nonprocessive enzymes exhibit open clefts with few aromatic amino acids, whereas enzymes with higher degrees of processivity possess closed tunnels or deep clefts with highly conserved aromatic residues that directly contact the ligand. Biochemical studies have further demonstrated that processive enzymes can be converted to nonprocessive enzymes via removal of specific aromatic residues lining substrate tunnels (25, 26), as well as through deletion of active site loop residues forming the tunnel of processive enzymes (27).
Despite these previous studies, a general theory describing the molecular-level hallmarks responsible for GH processivity, especially one incorporating enzyme dynamics, remains elusive. Differences in processivity are likely to result from structural and dynamic variations, beyond the presence of aromatic residues discussed above and possibly including fluctuations of the active site residues and the ligand, as well as differences in ligand solvation. These features also likely impact the ability of a given enzyme to decrystallize, hydrolyze, and processively remove chains from crystal surfaces, which requires a significant expenditure of work offset by the ligand binding free energy (28). Here, we present several new structural and dynamic hallmarks of processivity by combining structural studies and molecular dynamics (MD) simulations of a complete set of experimentally well characterized chitinolytic enzymes from one organism, which will aid in the development of a general molecular-level theory of processivity.
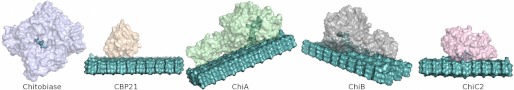
S. marcescens produces three GH family 18 chitinases, a family 20 β-hexosaminidase (chitobiase), and a lytic polysaccharide monooxygenase (Fig. 1). Structures of the two processive two-domain chitinases, ChiA and ChiB, have been solved previously (11, 13, 19, 20, 29–35). Until now, the structure of the third chitinase, ChiC (36), has remained unsolved. ChiC consists of a catalytic GH18 domain that is coupled to two C-terminal domains, putatively involved in substrate binding by a proline-rich linker (37) (the two C-terminal domains are a Fibronectin III-like module and a CBM5/12 module). Both the complete enzyme (referred to as ChiC1) and a truncated variant containing only the catalytic GH18 module (ChiC2) are observed in the culture supernatant of S. marcescens growing on chitin (38). ChiC2 is thought to be generated by proteolytic cleavage of ChiC1 (38). Several studies on the full-length enzyme (36, 39) have shown that ChiC is a nonprocessive enzyme. This is best illustrated by studies with the soluble chitin derivative chitosan, which showed that the degradation pattern obtained in a reaction with ChiC was similar to that obtained during random acid hydrolysis (39).
FIGURE 1.
The chitinolytic system from S. marcescens. The enzyme system comprises three family GH18 chitinases, processive ChiA and ChiB and nonprocessive ChiC, a family GH20 chitobiase that cleaves chitobiose and CBP21, a lytic polysaccharide monooxygenase. The structures of all enzymes, except ChiC, have been reported previously (11, 13, 19, 20, 29–35). The enzymes are shown with their substrates in light teal. An SDS-PAGE gel of the entire system has been included in supplemental Fig. S1.
Family GH18 chitinases hydrolyze chitin through a substrate-assisted mechanism that retains the stereochemistry of the anomeric carbon. Previous studies suggest that family 18 enzymes initiate catalysis via distortion of the chitin substrate in the −1 subsite adjacent to the glycosidic bond (19, 40, 41). Substrate binding is accompanied by rotation of an aspartic acid (Asp-139 in ChiC; see below), which forms a hydrogen bond with the catalytic glutamic acid (Glu-141 in ChiC) and the N-acetyl group of the −1-bound sugar. Nucleophilic attack by the N-acetyl oxygen on the anomeric carbon leads to scission of the glycosidic bond and generates an oxazolinium ion intermediate, which is subsequently hydrolyzed to complete the reaction. Catalysis involves several additional residues (19, 42), including a conserved Tyr (Tyr-208 in ChiC) and Asp/Asn (Asn-209 in ChiC) that interact with the −1 sugar. Although this mechanism likely applies to all GH18 chitinases, processive and nonprocessive, including endo- and exo-acting, it is conceivable that active site residue flexibility and dynamics are adapted to the variations in the specific modes of actions of these enzymes.
Here, we complete the full structural complement of the S. marcescens chitinolytic mixture with the structure of the catalytic domain of the nonprocessive enzyme from S. marcescens, ChiC, at 1.68 Å resolution. This enables direct comparison of processive and nonprocessive enzymes having evolved in concert to degrade chitin. From the crystal structures, we used MD simulations to gain molecular-level insights into the structural and dynamic differences between the processive and nonprocessive enzymes in S. marcescens. MD simulations were conducted for 0.25 μs for ChiA, ChiB, ChiC2 (the abbreviation used here for the catalytic domain), and a second, representative nonprocessive chitinase from Lactococcus lactis ssp. Lactis (43), referred to here by the Protein Data Bank (PDB) identifier 3IAN. Each simulation was conducted with a chitin ligand to compare the differences in ligand solvation and flexibility as well as enzyme flexibility as a function of experimentally measured extents of processivity. Free energy calculations of the ChiC2 catalytic residue (Glu-141) conformation quantify the high flexibility of this residue, suggested by its unusual conformation in the crystal structure. The properties of ChiC2 from experimental and computational data are compared with those of ChiA and ChiB and discussed in the context of the roles of these enzymes in the chitinolytic machinery of S. marcescens. Overall, this study aids in the development of a unified, molecular-level understanding of processivity in GH enzymes by adding key dynamic and ligand binding characteristics to known static structural characteristics defining differences between processive and nonprocessive GHs.
EXPERIMENTAL PROCEDURES
For additional details, see the supplemental Materials and Methods.
Cloning, Expression, and Purification
The gene encoding ChiC was cloned, produced, and partially purified by as detailed in Ref. 44. Final purification was obtained by chitin affinity chromatography and hydrophobic interaction chromatography to isolate ChiC2.
Protein Crystallization
Crystallization of ChiC2 was obtained by sitting drop vapor diffusion crystallization through mixing 0.5 μl of 4.0 mg/ml protein solution with an equal volume of crystallization buffers from the Hampton Research PEG/Ion Screen and incubating at 25 °C. Crystals were obtained in 25 mm ammonium acetate and 20% (w/v) PEG3000.
Data Collection, Phasing, and Structure Refinement
Diffraction data were collected using an in-house Rigaku MicroMax MM-007 diffractometer. Data were scaled with CrystalClear (Rigaku/MSC Inc.), and the structure was solved by molecular replacement using AMoRe (45) with 3EBV as the template structure. 1.68 Å high-resolution data were later collected at beamline ID14-2 at the European Synchrotron Radiation Facility (ESRF) synchrotron in Grenoble, France. The new dataset was scaled using XDS (46). The structure was refined by simulated annealing energy minimization using CNS (47). Model analysis and adjustments were done with Coot (48).
MD Simulation Protocol
Crystal structures from the PDB were used to build ChiA (20), ChiB (34), and 3IAN, whereas the structure presented in this work was used to build ChiC2. For ChiC2 and 3IAN, the chitin ligand (GlcNAc7 and GlcNAc6, respectively) spanning the substrate-binding cleft was modeled from an alignment of a combination of ligands bound in structurally similar enzymes (49, 50). Ligands were docked into ChiA (GlcNAc6) and ChiB (GlcNAc5) from multiple structures as described in the supplemental Materials and Methods. In all cases, the N-acetyl group of the GlcNAc unit bound in subsite −1 was rotated to the anticipated conformation of the catalytically active complex (19). MD simulation setup, minimization, and equilibration were performed using CHARMM (51). The 250-ns simulations were performed using NAMD (52). The umbrella sampling simulations were conducted with the reaction coordinate defined as the distance between the proton on the carboxylate group of Glu-141 and the glycosidic linkage on the GlcNAc residue in the −1 site. Additional details can be found in the supplemental Materials and Methods.
RESULTS
The Overall Structure of ChiC2
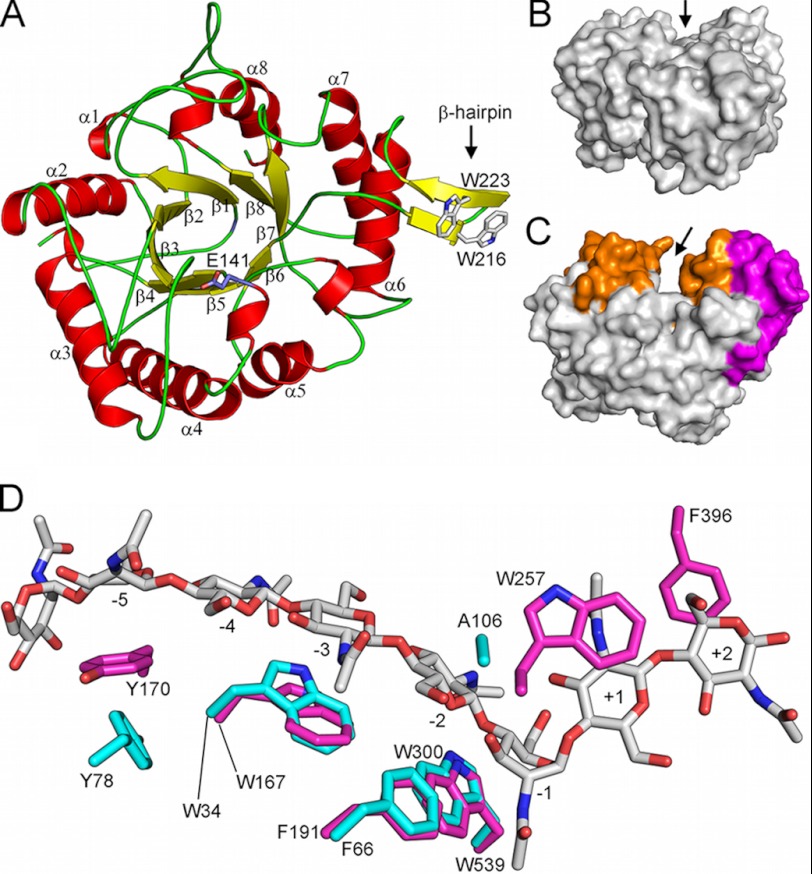
The structure of the ChiC catalytic module was refined to an R-factor of 18.6% and an Rfree factor of 21.6%, with only one residue (Gly-211, chain B) in the disallowed region of the Ramachandran plot (supplemental Table S1). As with other GH family 18 enzymes, ChiC2 consists of a (βα)8 TIM barrel fold with the catalytic glutamate of the characteristic DXXDXDXE motif positioned “on top” of the barrel in the loop connecting β-sheet 4 and α-helix 4, (Fig. 2A). The shallow substrate-binding cleft (inferred from analogies with the plant chitinase hevamine (53)) seems to be extended by a small β-hairpin subdomain (residues 213–226) with two solvent-exposed tryptophans that might aid in substrate binding (Fig. 2A). The substrate-binding cleft is shallow when compared with the processive chitinases due to the lack of several loops and the α+β subdomain that gives processive chitinases their characteristic deep substrate-binding cleft (Fig. 2, B and C). Interestingly, the structural data also revealed a hexacoordinated Ca2+ ion bound at the “bottom” of the TIM barrel and an acetate molecule bound to conserved residues in the active site, mimicking the acetate moiety of a GlcNAc positioned in the −1 subsite (see supplemental Fig. S2; the supplemental text and supplemental Figs. S3, S4, and S5 include additional information on Ca2+ binding).
FIGURE 2.
Overall structure of the catalytic module of ChiC, ChiC2. A, ChiC2 exhibits an (a/b)8 TIM barrel fold with the catalytic glutamate, Glu-141, positioned between β-strand 4 and α-helix 4 (side chain shown in stick representation). Note the β-hairpin subdomain containing two exposed tryptophans. B and C, comparison of ChiC2 (nonprocessive) with ChiA (processive, PDB ID: 1EHN (29)) shows the difference in depth of the substrate-binding clefts that are suggestive of the nonprocessive and processive properties of these enzymes, respectively. The “walls” of the ChiA substrate-binding cleft are constituted from several insertions (colored orange) in addition to a large a+b subdomain insertion (colored magenta). Arrows indicate the binding clefts. D, aromatic amino acids lining the substrate-binding cleft of ChiA (magenta sticks) and ChiC2 (cyan sticks). The substrate (GlcNAc)8 bound to the 1EHN ChiA structure is shown in gray stick representation, and subsites to which the sugars are bound are indicated. Nitrogen and oxygen atoms are colored blue and red, respectively. Note the lack of aromatic amino acids in the +1 and +2 subsites of the ChiC2 substrate-binding cleft; Tyr-275 of the +1 subsite in ChiA is replaced with an Ala in ChiC2, and Phe-396 in the +2 subsite of ChiA is part of a loop that does not exist in ChiC.
The Catalytic Center and Substrate-binding Cleft
Inspection of the amino acids in the active site shows the catalytic glutamate, Glu-141, in an unexpected position, H-bonded to the main chain amino groups of Asp-107 and Ala-108, which are part of the loop-joining β-sheet and α-helix 3 (Fig. 3A). In most other GH18 chitinases studied to date, the catalytic glutamate is almost exclusively interacting with the third Asp in the catalytic motif, Asp-139, which is important for successful catalysis (19). The unusual positioning of the catalytic glutamate has not been described previously. However, the structure of LlChi18A from L. lactis (PDB ID: 3IAN) shows a similar positioning of its catalytic glutamate, indicating that this remarkable structural detail is perhaps functionally relevant. It should be noted that both ChiC2 and LlChi18A were crystallized at neutral pH where both enzymes are catalytically active. The sequence identity between the enzymes is 50%. A more detailed analysis of the β-α3 loop (residues 102–114) reveals that this region contains a conserved sequence motif in GH18 chitinases, namely the SXGG motif (residues 102–105) that is followed by a Trp in processive chitinases (in ChiA and ChiB: SIGGW), which is vital for processivity and the ability to efficiently degrade insoluble chitin (25, 26, 36). In nonprocessive chitinases such as ChiC and LlChi18A, this Trp is substituted with an Ala (residue Ala-106; Fig. 3). A recent study on a chitinase from Bacillus cereus shows that the 102–114 loop is highly flexible and moves upon substrate binding (49).
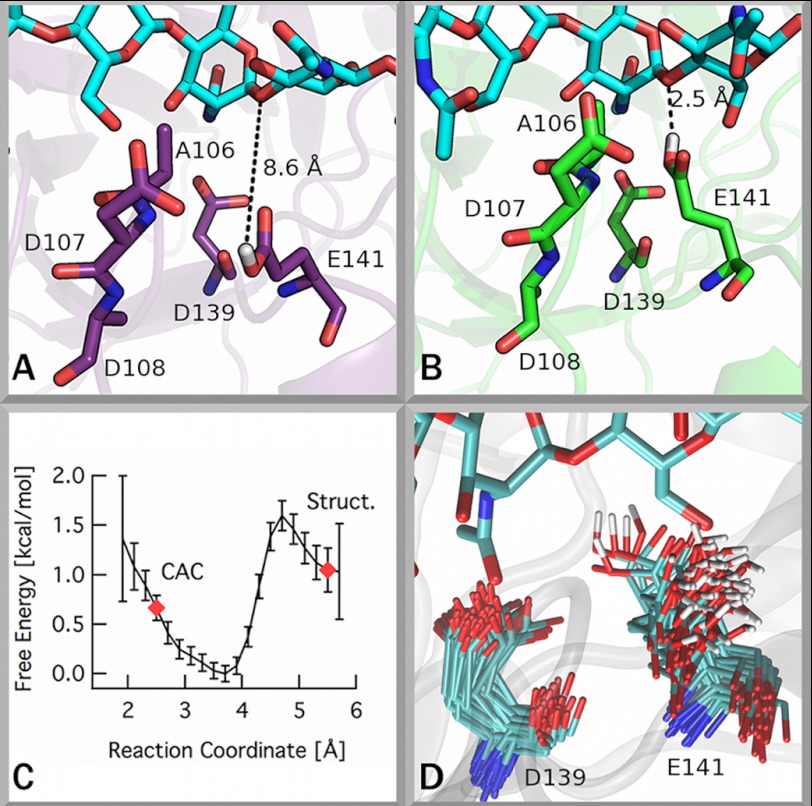
FIGURE 3.
The catalytic center of ChiC. The catalytic glutamate of ChiC exhibits a conformation inconsistent with the putative catalytically active conformation (CAC) (19). A and B, illustrations of these conformations are given showing the conformation found in the ChiC2 crystal structure (A) and showing the putative catalytically active conformation from an MD simulation (B). C, the free energy difference of the catalytic glutamate between the solvent-exposed conformation and the catalytically active complex. A reaction coordinate value of ∼2.5 Å corresponds to the catalytically active complex (19), and a value near 5.5 Å corresponds to the Glu-141 conformation observed in the crystal structure (4AXN). Error bars were obtained by bootstrapping in WHAM. D, superposition of frames from MD simulation of ChiC2 with a GlcNAc7 ligand bound (see supplemental text for details) illustrating the significant flexibility of Glu-141.
To examine the flexibility of Glu-141 quantitatively, the free energy differences in the likely conformations of this residue were calculated. Ultimately, the free energy difference for Glu-141 to adopt a conformation corresponding to the catalytically active complex (Fig. 3B) from its position in the crystal structure (Fig. 3A) is, within error (−0.33 ± 0.80 kcal/mol), zero as shown in Fig. 3C. The inflections and small changes in the free energy landscape along the reaction coordinate correspond to a water molecule interacting with both Glu-141 and the glycosidic linkage of the −1 GlcNAc residue at ∼3.5 Å apart.
MD simulations of ChiC2 (with and without a GlcNAc7 ligand) were used to evaluate three structural features of the catalytic center: the position of Glu-141 relative to its catalytic partner Asp-139 (Fig. 3 and supplemental Figs. S6 and S7), the root mean square deviation of the β-α3 loop adjacent to Glu-141 (supplemental Fig. S8), and the hydrogen bonding of Glu-141 with both Asp-107 and Ala-108 (supplemental Fig. S9). Three different conformational states of the Glu-141/Asp-139 arrangement (supplemental Fig. S7) were observed over the course of the simulation with ligand present, including the formation of the proposed catalytically active complex (19). The flexibility of the β-α3 loop and the occurrence of hydrogen bonding with loop residues Asp-107 and Ala-108 appear to be related to the conformation of Glu-141. Details regarding this analysis can be found in the supplemental text.
Structural and Computational Comparison of Processive and Nonprocessive Chitinases
The remarkable and previously undiscovered flexibility in the active site of ChiC2 prompted us to conduct an in-depth comparative study of processive and nonprocessive chitinases to assess the differences in structural characteristics contributing to ligand binding, and ultimately, processivity. To this end, we performed MD simulations of ChiA, ChiB, ChiC2, and the nonprocessive L. lactis chitinase, 3IAN, which is structurally similar to ChiC2. Quantitative measurements of the ratio of degradation products (dimer/trimer) on α- and β-chitin for ChiA, ChiB, and ChiC (36) allow for direct comparison of dynamic properties to known degrees of processivity. ChiB is the most processive of the three enzymes with a ratio of 11.4 on α-chitin and 12.6 on β-chitin. ChiA is less processive with ratios of 5.9 on α-chitin and 7.3 on β-chitin. ChiC is classified as a nonprocessive enzyme with product ratios of 3.0 and 4.1 on α- and β-chitin, respectively.
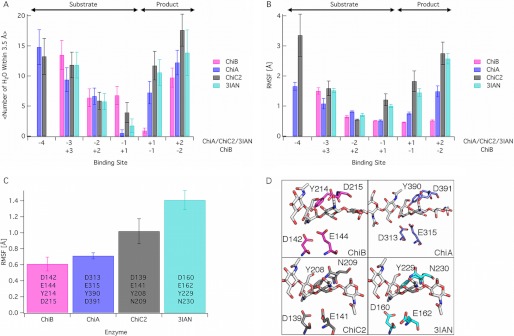
Differences between the enzymes were prominent in three structural and dynamic features of the simulations. Given the obvious differences in shape of the active sites, we hypothesized that the cleft-shaped active site region of a nonprocessive chitinase might exhibit greater solvation than that of the tunnel-shaped processive chitinase. We note that we refer to product and substrate sites, as shown in Fig. 4, in the nonprocessive chitinases that are equivalent to the product and substrate subsites in the appropriate processive chitinase (i.e. ChiC and 3IAN exhibit equivalent subsites to ChiA). The data (Fig. 4A) show higher solvation in the nonprocessive enzymes, relative to ChiA and ChiB, but only in the product subsites. There is surprisingly little difference in solvation in the substrate-binding sites. There is, however, a marked difference in the solvation between the −1/+1 sites in ChiA and ChiB in that water molecules tend to remain on the product side in the ChiA active site and on the substrate side of the hydrolysis reaction in the ChiB active site. This is likely related to the end specificity of ChiA and ChiB as reducing end- and nonreducing end-specific enzymes, respectively.
FIGURE 4.
MD simulations of two processive chitinases, ChiA and ChiB, and two nonprocessive chitinases, ChiC2 and 3IAN. All simulations were done on enzymes in which an oligomer of GlcNAc (GlcNAc5 in ChiB, GlcNAc6 in ChiA and 3IAN, and GlcNAc7 in ChiC2) had been docked to the −4 to +2 subsites of the reducing end enzymes and the +3 to −2 subsites of ChiB (see supplemental text for additional details). In panels A and B, only subsites appearing in all four enzymes are shown. The product and substrate subsite definitions are as shown above panel A. The panels show the average solvation of the ligand by binding site (A), the root mean square fluctuations (RMSF) of the ligand by the binding site (B), and the root mean square fluctuations of the tetrad of catalytic residues, listed for each enzyme (C). Error bars in panel A represent one standard deviation. Error bars in panels B and C were obtained with block averaging. D, the catalytic tetrad of each enzyme examined here with simulation with the residues labeled. These residues represent the set for analysis in panel C. The enzyme structures used for the simulation input are described in detail in the supplemental text.
Ligand fluctuations, as determined by measuring the root mean square fluctuations by binding site over the 250-ns simulations, are shown in Fig. 4B. In the nonprocessive enzymes, there is markedly increased fluctuation, which, as in the case of ligand solvation, appears primarily in the product subsites. There also appears to be significant stabilization of the ligand in the first substrate side subsite of the processive chitinases relative to the nonprocessive chitinases, which coincides with the observed flexibility of the catalytic center, including the unusual experimentally observed position of the catalytic glutamate.
The flexibility of the catalytic machinery was further assessed by analyzing the root mean square fluctuations for a tetrad of catalytic residues over the entire simulation trajectory (Fig. 4, C and D). It is apparent that the catalytic centers of the nonprocessive chitinases are prone to higher degrees of fluctuation in comparison with processive chitinases. It also appears that the most processive of the set of enzymes, ChiB, has the lowest level of catalytic residue fluctuation, whereas the second processive enzyme, ChiA, lies between ChiA and the nonprocessive enzymes in the magnitude of catalytic residue fluctuations.
DISCUSSION
There exist ample combined structural and functional data for processive family 18 chitinases with deep substrate-binding clefts, in particular ChiA and ChiB from S. marcescens (13, 19, 20, 22, 24–26, 29–32, 34–36, 42, 54, 55). Published structures on (putatively) endo-acting and (putatively) nonprocessive family 18 chitinases are limited to those of the hevamine (53) and two bacterial enzymes (49, 56). However, for these enzymes, structural data are not accompanied by detailed insight in the actual mode of action of the enzymes. Nakamura et al. (56) only describe a structure of a bacterial enzyme, whereas Hsieh et al. (49) provide a more detailed study of the mechanism and some properties of a B. cereus chitinase, without addressing processivity. The authors of the latter study also suggest that the enzyme appears, structurally, to be an endochitinase, but mechanistically an exochitinase, although this is based on product analysis of (GlcNAc)2, which is not necessarily indicative of exo-activity (57). In addition to these publications, structural consortia have recently deposited crystal structures of two putatively nonprocessive chitinases from Streptomyces coelicolor (PDB ID: 3EBV) and L. lactis (PDB ID: 3IAN).
The lack of available and clear information regarding nonprocessive family 18 chitinases highlights the importance of the addition of the ChiC2 structure to the database, completing the entire suite of the S. marcescens chitinases. This suite of enzymes, all employing the same catalytic mechanism, provides a model system by which characteristics contributing to processivity may be studied. Findings related to processivity from the S. marcescens model system have implications for other similar enzymatic systems, including cellulases used in the production of biofuels from cellulosic materials (25, 58).
It is generally thought that deep substrate-binding clefts and grooves, which often fold over the substrate upon binding (8), are important for keeping processive enzymes attached to the substrate once bound (59–61). Observation of the shallowness of the substrate-binding cleft, combined with the flat surrounding surface in ChiC2, suggests that this enzyme is more adapted to binding a flat surface (crystalline chitin) than soluble single chains. Additionally, the small β-sheet subdomain exhibiting two tryptophan residues observed adjacent to the cleft in ChiC2 possibly serves as an attachment point to the chitin surface (Fig. 2A). In addition to the steric constraints of the tunnel, carbohydrate-π interactions of aromatic residues lining the substrate-binding clefts are important for processivity and, more generally, may make large contributions to the ligand binding free energy (21, 22, 25, 26, 28, 62–65). Notably, ChiC2 has fewer such aromatic residues than ChiA and ChiB, primarily affecting the product subsites (Fig. 2D).
Previous studies suggest that a strict, structural delineation (i.e. deep tunnels or grooves versus open clefts) between processive and nonprocessive enzymes may not be straightforward (23, 25–27, 66). For example, deletion of even a single active site loop responsible for tunnel formation in processive GH family 6 enzymes, which exhibit similarity to known exoglucanases, results in a nonprocessive enzyme with an open substrate-binding groove (27). Even more surprisingly, the deletion of a single aromatic residue directly preceding the active site in S. marcescens ChiB results in the conversion of a processive enzyme to a nonprocessive enzyme (25). Other structural factors, as well as protein dynamics, may impact processivity. Considering the general importance of enzyme dynamics for enzyme function and the delicate balance between substrate and product binding, rebinding, and product expulsion that determines the mode of action of polysaccharide-depolymerizing enzymes, delineating processivity directly from static, structural features or from sequence data alone is not likely to provide a definitive measure of the degree of processivity. This notion, combined with the previously undescribed active site flexibility observed in ChiC2 (and 3IAN), prompted us to conduct MD simulations of the complete S. marcescens GH18 enzyme suite to gain additional insights. These analyses revealed three potential hallmarks of processivity.
Flexibility of the catalytic machinery seems to be a key difference between processive and nonprocessive GH18 chitinases. Free energy calculations quantitatively confirm the flexibility of the catalytic residue, Glu-141, in ChiC2 and indicate no preference for the catalytically active complex conformation over solvent-exposed conformations. Coupling the free energy result with observation of the three different conformational states of Glu-141 (supplemental Fig. S7) and the relative flexibility of active site loops in concert with the conformation changes over the course of a 250-ns MD simulation (Figs. 3D and 4C and supplemental Figs. S6 and S7), it is apparent that the active site of ChiC2 has the ability to rapidly change conformation. Although rotation of the partner Asp in the catalytic mechanism of GH18 enzymes is essential in catalysis, flexibility of the catalytic glutamate does not appear to be inherently required by the mechanism and is thus likely to relate to the nonprocessive endo-mode of action of ChiC2.
An important aspect of chitinase processivity/nonprocessivity is the mechanism of product displacement. Although the processive chitinases have a defined product site designed to efficiently dissociate chitobiose after successful catalysis, nonprocessive enzymes dissociate from carbohydrate moieties on both sides of the catalytic center. Our data suggest that this ability of ChiC2 manifests as a more dynamic binding of the ligand, which is more flexible and more solvated when bound. These dynamic features seem compatible with the more dynamic on-off substrate interactions that are expected for nonprocessive enzymes. Somewhat unexpectedly, the data clearly show that the differences between the two enzyme types almost exclusively concern the product subsites. This contrasts with previous studies on the contribution of aromatic residues to processivity in ChiA and ChiB, which revealed essential structural features on both sides of the catalytic center (25, 26). It is conceivable that this reflects the fact that the catalytically crucial but energetically unfavorable binding and distortion of the GlcNAc sugar binding to the −1 subsite requires a certain binding energy and rigidity in the other substrate subsites that is conserved among all family GH18 chitinases. The latter is also suggested by the fact that the few aromatic residues that are conserved in the binding cleft of ChiC2 are in the substrate subsites (Fig. 2D).
Although this study focuses on identifying universal molecular-level hallmarks of processivity in glycoside hydrolases, primary observations here relate only to the catalytic domains of these enzymes. The possibility of additional features beyond the catalytic domain contributing to processivity in these multimodular enzymes remains. In the S. marcescens chitinases, for example, ChiC exhibits two CBMs connected to the C terminus via a short proteolytically susceptible linker. It is well known that the extra domains add to enzyme efficiency, in particular toward insoluble substrates (67), and it has been shown that this is also the case for ChiC (37).7 This is often ascribed to proximity effects (68). It seems unlikely that these additional domains affect processivity of ChiC because the full-length enzyme is not processive (39) and because recent work on the two-domain chitinases ChiA and ChiB has shown that processivity is governed by arrangements of aromatic residues close to the catalytic center (25, 26). Although additional heuristics of processivity beyond the catalytic domain may exist, we have identified three interrelated noteworthy metrics, flexibility of the catalytic center, solvation of the bound substrate, and flexibility of the bound substrate, that are linked to processivity in GHs.
CONCLUSION
This study reports the structure of the S. marcescens nonprocessive chitinase, ChiC2, which completes the GH family 18 suite of chitinase catalytic domains from S. marcescens. From this model system, we applied simulation to gain insights into the structural and dynamic differences between processive and nonprocessive enzymes. The ChiC2 structure draws attention to the unusual flexibility of the catalytic residue, Glu-141, a feature currently only observed in one other chitinase, which like ChiC is a nonprocessive endochitinase (43). Our simulations demonstrate that characteristic dynamic features of the catalytic domain, primarily near the site of hydrolysis, correlate with processivity. The most visible and frequently acknowledged hallmark of processivity is the geometry of the active site (i.e. cleft, for nonprocessive enzymes, and tunnel, for processive enzymes), but previous studies have demonstrated that the degree of processivity in GH enzymes is likely more complex. Here, we have identified three additional hallmarks of GH processivity, namely, flexibility of the active site region, in particular the catalytic residues, ligand solvation proximal to the product subsites, and ligand fluctuations in the product subsites. These key features may be universal among processive and nonprocessive enzymes from other GH families.
Acknowledgments
Computer time for this research was provided by the National Renewable Energy Laboratory Computational Sciences Center supported by the United States Office of Energy Efficiency and Renewable Energy under Contract Number DE-AC36-08GO28308 and by the Texas Advanced Computing Center Ranger cluster and the National Institute for Computational Sciences Kraken cluster under the National Science Foundation XSEDE Grant MCB090159. We are grateful to the beamline staff at the ESRF, Grenoble, France for providing assistance in using beamline ID14-2.

This article contains supplemental Materials and Methods and Results, Table S1, and Figs. S1–S9.
The atomic coordinates and structure factors (code 4AXN) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
G. Vaaje-Kolstad and M. Sørlie, unpublished observations.
- GH
- glycoside hydrolase
- CBM
- carbohydrate-binding module
- MD
- molecular dynamics.
REFERENCES
- 1. Himmel M. E., Ding S. Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., Foust T. D. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807 [DOI] [PubMed] [Google Scholar]
- 2. Vaaje-Kolstad G., Westereng B., Horn S. J., Liu Z., Zhai H., Sørlie M., Eijsink V. G. (2010) An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222 [DOI] [PubMed] [Google Scholar]
- 3. Mba Medie F., Davies G. J., Drancourt M., Henrissat B. (2012) Genome analyses highlight the different biological roles of cellulases. Nat. Rev. Microbiol. 10, 227–234 [DOI] [PubMed] [Google Scholar]
- 4. Bayer E. A., Belaich J. P., Shoham Y., Lamed R. (2004) The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58, 521–554 [DOI] [PubMed] [Google Scholar]
- 5. Demain A. L., Newcomb M., Wu J. H. (2005) Cellulase, Clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69, 124–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fontes C. M., Gilbert H. J. (2010) Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 79, 655–681 [DOI] [PubMed] [Google Scholar]
- 7. Horn S. J., Vaaje-Kolstad G., Westereng B., Eijsink V. G. (2012) Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 5, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rouvinen J., Bergfors T., Teeri T., Knowles J. K., Jones T. A. (1990) Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science 249, 380–386 [DOI] [PubMed] [Google Scholar]
- 9. Divne C., Ståhlberg J., Reinikainen T., Ruohonen L., Pettersson G., Knowles J. K., Teeri T. T., Jones T. A. (1994) The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science 265, 524–528 [DOI] [PubMed] [Google Scholar]
- 10. Divne C., Ståhlberg J., Teeri T. T., Jones T. A. (1998) High-resolution crystal structures reveal how a cellulose chain is bound in the 50 Å long tunnel of cellobiohydrolase I from Trichoderma reesei. J. Mol. Biol. 275, 309–325 [DOI] [PubMed] [Google Scholar]
- 11. van Aalten D. M., Synstad B., Brurberg M. B., Hough E., Riise B. W., Eijsink V. G., Wierenga R. K. (2000) Structure of a two-domain chitotriosidase from Serratia marcescens at 1.9 Å resolution. Proc. Natl. Acad. Sci. U.S.A. 97, 5842–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guimarães B. G., Souchon H., Lytle B. L., David Wu J. H., Alzari P. M. (2002) The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum cellulosome. J. Mol. Biol. 320, 587–596 [DOI] [PubMed] [Google Scholar]
- 13. Aronson N. N., Jr., Halloran B. A., Alexyev M. F., Amable L., Madura J. D., Pasupulati L., Worth C., Van Roey P. (2003) Family 18 chitinase-oligosaccharide substrate interaction: subsite preference and anomer selectivity of Serratia marcescens chitinase A. Biochem. J. 376, 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Igarashi K., Uchihashi T., Koivula A., Wada M., Kimura S., Okamoto T., Penttilä M., Ando T., Samejima M. (2011) Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science 333, 1279–1282 [DOI] [PubMed] [Google Scholar]
- 15. Kleywegt G. J., Zou J. Y., Divne C., Davies G. J., Sinning I., Ståhlberg J., Reinikainen T., Srisodsuk M., Teeri T. T., Jones T. A. (1997) The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 Å resolution, and a comparison with related enzymes. J. Mol. Biol. 272, 383–397 [DOI] [PubMed] [Google Scholar]
- 16. Murphy L., Cruys-Bagger N., Damgaard H. D., Baumann M. J., Olsen S. N., Borch K., Lassen S. F., Sweeney M., Tatsumi H., Westh P. (2012) Origin of initial burst in activity for Trichoderma reesei endo-glucanases hydrolyzing insoluble cellulose. J. Biol. Chem. 287, 1252–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nimlos M. R., Beckham G. T., Matthews J. F., Bu L., Himmel M. E., Crowley M. F. (2012) Binding preferences, surface attachment, diffusivity, and orientation of a family 1 carbohydrate-binding module on cellulose. J. Biol. Chem. 287, 20603–20612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stern R., Jedrzejas M. J. (2008) Carbohydrate polymers at the center of life's origins: the importance of molecular processivity. Chem. Rev. 108, 5061–5085 [DOI] [PubMed] [Google Scholar]
- 19. van Aalten D. M., Komander D., Synstad B., Gåseidnes S., Peter M. G., Eijsink V. G. (2001) Structural insights into the catalytic mechanism of a family 18 exo-chitinase. Proc. Natl. Acad. Sci. U.S.A. 98, 8979–8984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aronson N. N., Jr., Halloran B. A., Alexeyev M. F., Zhou X. E., Wang Y., Meehan E. J., Chen L. (2006) Mutation of a conserved tryptophan in the chitin-binding cleft of Serratia marcescens chitinase A enhances transglycosylation. Biosci. Biotechnol. Biochem. 70, 243–251 [DOI] [PubMed] [Google Scholar]
- 21. Koivula A., Kinnari T., Harjunpää V., Ruohonen L., Teleman A., Drakenberg T., Rouvinen J., Jones T. A., Teeri T. T. (1998) Tryptophan 272: an essential determinant of crystalline cellulose degradation by Trichoderma reesei cellobiohydrolase Cel6A. FEBS Lett. 429, 341–346 [DOI] [PubMed] [Google Scholar]
- 22. Watanabe T., Ariga Y., Sato U., Toratani T., Hashimoto M., Nikaidou N., Kezuka Y., Nonaka T., Sugiyama J. (2003) Aromatic residues within the substrate-binding cleft of Bacillus circulans chitinase A1 are essential for hydrolysis of crystalline chitin. Biochem. J. 376, 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Proctor M. R., Taylor E. J., Nurizzo D., Turkenburg J. P., Lloyd R. M., Vardakou M., Davies G. J., Gilbert H. J. (2005) Tailored catalysts for plant cell-wall degradation: redesigning the exo/endo preference of Cellvibrio japonicus arabinanase 43A. Proc. Natl. Acad. Sci. U.S.A. 102, 2697–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baban J., Fjeld S., Sakuda S., Eijsink V. G., Sørlie M. (2010) The roles of three Serratia marcescens chitinases in chitin conversion are reflected in different thermodynamic signatures of allosamidin binding. J. Phys. Chem. B. 114, 6144–6149 [DOI] [PubMed] [Google Scholar]
- 25. Horn S. J., Sikorski P., Cederkvist J. B., Vaaje-Kolstad G., Sørlie M., Synstad B., Vriend G., Vårum K. M., Eijsink V. G. (2006) Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc. Natl. Acad. Sci. U.S.A. 103, 18089–18094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zakariassen H., Aam B. B., Horn S. J., Vårum K. M., Sørlie M., Eijsink V. G. (2009) Aromatic residues in the catalytic center of chitinase A from Serratia marcescens affect processivity, enzyme activity, and biomass converting efficiency. J. Biol. Chem. 284, 10610–10617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davies G. J., Brzozowski A. M., Dauter M., Varrot A., Schülein M. (2000) Structure and function of Humicola insolens family 6 cellulases: structure of the endoglucanase, Cel6B, at 1.6 Å resolution. Biochem. J. 348, 201–207 [PMC free article] [PubMed] [Google Scholar]
- 28. Beckham G. T., Matthews J. F., Peters B., Bomble Y. J., Himmel M. E., Crowley M. F. (2011) Molecular-level origins of biomass recalcitrance: decrystallization free energies for four common cellulose polymorphs. J. Phys. Chem. B. 115, 4118–4127 [DOI] [PubMed] [Google Scholar]
- 29. Perrakis A., Tews I., Dauter Z., Oppenheim A. B., Chet I., Wilson K. S., Vorgias C. E. (1994) Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure 2, 1169–1180 [DOI] [PubMed] [Google Scholar]
- 30. Houston D. R., Eggleston I., Synstad B., Eijsink V. G., van Aalten D. M. (2002) The cyclic dipeptide CI-4 cyclo-(l-Arg-d-Pro) inhibits family 18 chitinases by structural mimicry of a reaction intermediate. Biochem. J. 368, 23–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Houston D. R., Shiomi K., Arai N., Omura S., Peter M. G., Turberg A., Synstad B., Eijsink V. G., van Aalten D. M. (2002) High-resolution structures of a chitinase complexed with natural product cyclopentapeptide inhibitors: mimicry of carbohydrate substrate. Proc. Natl. Acad. Sci. U.S.A. 99, 9127–9132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Houston D. R., Synstad B., Eijsink V. G., Stark M. J., Eggleston I. M., van Aalten D. M. (2004) Structure-based exploration of cyclic dipeptide chitinase inhibitors. J. Med. Chem. 47, 5713–5720 [DOI] [PubMed] [Google Scholar]
- 33. Vaaje-Kolstad G., Horn S. J., van Aalten D. M., Synstad B., Eijsink V. G. (2005) The noncatalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 280, 28492–28497 [DOI] [PubMed] [Google Scholar]
- 34. Vaaje-Kolstad G., Houston D. R., Rao F. V., Peter M. G., Synstad B., van Aalten D. M., Eijsink V. G. (2004) Structure of the D142N mutant of the family 18 chitinase ChiB from Serratia marcescens and its complex with allosamidin. Biochim. Biophys. Acta 1696, 103–111 [DOI] [PubMed] [Google Scholar]
- 35. Vaaje-Kolstad G., Vasella A., Peter M. G., Netter C., Houston D. R., Westereng B., Synstad B., Eijsink V. G., van Aalten D. M. (2004) Interactions of a family 18 chitinase with the designed inhibitor HM508 and its degradation product, chitobiono-δ-lactone. J. Biol. Chem. 279, 3612–3619 [DOI] [PubMed] [Google Scholar]
- 36. Horn S. J., Sørbotten A., Synstad B., Sikorski P., Sørlie M., Vårum K. M., Eijsink V. G. (2006) Endo/exo mechanism and processivity of family 18 chitinases produced by Serratia marcescens. FEBS J. 273, 491–503 [DOI] [PubMed] [Google Scholar]
- 37. Suzuki K., Taiyoji M., Sugawara N., Nikaidou N., Henrissat B., Watanabe T. (1999) The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem. J. 343, 587–596 [PMC free article] [PubMed] [Google Scholar]
- 38. Watanabe T., Kimura K., Sumiya T., Nikaidou N., Suzuki K., Suzuki M., Taiyoji M., Ferrer S., Regue M. (1997) Genetic analysis of the chitinase system of Serratia marcescens 2170. J. Bacteriol. 179, 7111–7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sikorski P., Sørbotten A., Horn S. J., Eijsink V. G., Vårum K. M. (2006) Serratia marcescens chitinases with tunnel-shaped substrate-binding grooves show endo activity and different degrees of processivity during enzymatic hydrolysis of chitosan. Biochemistry 45, 9566–9574 [DOI] [PubMed] [Google Scholar]
- 40. Brameld K. A., Goddard W. A. (1998) Substrate distortion to a boat conformation at subsite-1 is critical in the mechanism of family 18 chitinases. J. Am. Chem. Soc. 120, 3571–3580 [Google Scholar]
- 41. Tews I., van Scheltinga A. C., Perrakis A., Wilson K. S., Dijkstra B. W. (1997) Substrate-assisted catalysis unifies two families of chitinolytic enzymes. J. Am. Chem. Soc. 119, 7954–7959 [Google Scholar]
- 42. Synstad B., Gåseidnes S., Van Aalten D. M., Vriend G., Nielsen J. E., Eijsink V. G. (2004) Mutational and computational analysis of the role of conserved residues in the active site of a family 18 chitinase. Eur. J. Biochem. 271, 253–262 [DOI] [PubMed] [Google Scholar]
- 43. Vaaje-Kolstad G., Bunaes A. C., Mathiesen G., Eijsink V. G. (2009) The chitinolytic system of Lactococcus lactis ssp. lactis comprises a nonprocessive chitinase and a chitin-binding protein that promotes the degradation of α- and β-chitin. FEBS J. 276, 2402–2415 [DOI] [PubMed] [Google Scholar]
- 44. Synstad B., Vaaje-Kolstad G., Cederkvist F. H., Saua S. F., Horn S. J., Eijsink V. G., Sørlie M. (2008) Expression and characterization of endochitinase C from Serratia marcescens BJL200 and its purification by a one-step general chitinase purification method. Biosci. Biotechnol. Biochem. 72, 715–723 [DOI] [PubMed] [Google Scholar]
- 45. Navaza J. (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 57, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 46. Kabsch W. (2010) Integration, scaling, space-group assignment, and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 48. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 49. Hsieh Y. C., Wu Y. J., Chiang T. Y., Kuo C. Y., Shrestha K. L., Chao C. F., Huang Y. C., Chuankhayan P., Wu W. G., Li Y. K., Chen C. J. (2010) Crystal structures of Bacillus cereus NCTU2 chitinase complexes with chitooligomers reveal novel substrate binding for catalysis: a chitinase without chitin binding and insertion domains. J. Biol. Chem. 285, 31603–31615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bokma E., Rozeboom H. J., Sibbald M., Dijkstra B. W., Beintema J. J. (2002) Expression and characterization of active site mutants of hevamine, a chitinase from the rubber tree Hevea brasiliensis. Eur. J. Biochem. 269, 893–901 [DOI] [PubMed] [Google Scholar]
- 51. Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Terwisscha van Scheltinga A. C., Kalk K. H., Beintema J. J., Dijkstra B. W. (1994) Crystal Structures of Hevamine, A plant defense protein with chitinase and lysozyme activity, and its complex with an inhibitor. Structure 2, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 54. Hult E. L., Katouno F., Uchiyama T., Watanabe T., Sugiyama J. (2005) Molecular directionality in crystalline β-chitin: hydrolysis by chitinases A and B from Serratia marcescens 2170. Biochem. J. 388, 851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Horn S. J., Sorlie M., Vaaje-Kolstad G., Norberg A. L., Synstad B., Varum K. M., Eijsink V. G. (2006) Comparative studies of chitinases A, B, and C from Serratia marcescens. Biocatal. Biotransform. 24, 39–53 [Google Scholar]
- 56. Nakamura T., Mine S., Hagihara Y., Ishikawa K., Uegaki K. (2007) Structure of the catalytic domain of the hyperthermophilic chitinase from Pyrococcus furiosus. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 63, 7–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brurberg M. B., Nes I. F., Eijsink V. G. (1996) Comparative studies of chitinases A and B from Serratia marcescens. Microbiology 142, 1581–1589 [DOI] [PubMed] [Google Scholar]
- 58. Eijsink V. G., Vaaje-Kolstad G., Vårum K. M., Horn S. J. (2008) Toward new enzymes for biofuels: lessons from chitinase research. Trends Biotechnol. 26, 228–235 [DOI] [PubMed] [Google Scholar]
- 59. Meinke A., Damude H. G., Tomme P., Kwan E., Kilburn D. G., Miller R. C., Jr., Warren R. A., Gilkes N. R. (1995) Enhancement of the endo-β-1,4-glucanase activity of an exocellobiohydrolase by deletion of a surface loop. J. Biol. Chem. 270, 4383–4386 [DOI] [PubMed] [Google Scholar]
- 60. Davies G., Henrissat B. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 61. von Ossowski I., Ståhlberg J., Koivula A., Piens K., Becker D., Boer H., Harle R., Harris M., Divne C., Mahdi S., Zhao Y., Driguez H., Claeyssens M., Sinnott M. L., Teeri T. T. (2003) Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Ce17A: a comparison with Phanerochaete chrysosporium Cel7D. J. Mol. Biol. 333, 817–829 [DOI] [PubMed] [Google Scholar]
- 62. Payne C. M., Bomble Y. J., Taylor C. B., McCabe C., Himmel M. E., Crowley M. F., Beckham G. T. (2011) Multiple functions of aromatic-carbohydrate interactions in a processive cellulase examined with molecular simulation. J. Biol. Chem. 286, 41028–41035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Norberg A. L., Karlsen V., Hoell I. A., Bakke I., Eijsink V. G., Sørlie M. (2010) Determination of substrate binding energies in individual subsites of a family 18 chitinase. FEBS Lett. 584, 4581–4585 [DOI] [PubMed] [Google Scholar]
- 64. Igarashi K., Koivula A., Wada M., Kimura S., Penttilä M., Samejima M. (2009) High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J. Biol. Chem. 284, 36186–36190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vuong T. V., Wilson D. B. (2009) Processivity, synergism, and substrate specificity of Thermobifida fusca Cel6B. Appl. Environ. Microbiol. 75, 6655–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pages S., Kester H. C., Visser J., Benen J. A. (2001) Changing a single amino acid residue switches processive and nonprocessive behavior of Aspergillus niger endopolygalacturonase I and II. J. Biol. Chem. 276, 33652–33656 [DOI] [PubMed] [Google Scholar]
- 67. Watanabe T., Ito Y., Yamada T., Hashimoto M., Sekine S., Tanaka H. (1994) The roles of the C-terminal domain and Type-III domains of chitinase A1 from Bacillus circulans WL-12 in chitin degradation. J. Bacteriol. 176, 4465–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]