Background: The prostate-specific tumor suppressor NKX3.1 is targeted for proteasomal degradation.
Results: A proline-dependent C-terminal 21 amino acid portable degron regulates NKX3.1 ubiquitin-independent degradation in prostate cancer cells.
Conclusion: NKX3.1 proteasomal degradation is mediated by both ubiquitin-dependent and -independent pathways.
Significance: Understanding mechanisms regulating NKX3.1 turnover provides opportunities to develop tumor suppressor restoration therapies in prostate cancer.
Keywords: Prostate Cancer, Proteasome, Protein Degradation, Tumor Suppressor Gene, Ubiquitination
Abstract
Reduced expression of the homeodomain transcription factor NKX3.1 is associated with prostate cancer initiation and progression. NKX3.1 turnover requires post-translational modifications including phosphorylation and ubiquitination. Here, we demonstrate the existence of a non-canonical mechanism for NKX3.1 turnover that does not require ubiquitination. Using a structure-function approach, we have determined that the conserved, C-terminal 21-amino acid domain of NKX3.1 (C21) is required for this novel ubiquitin-independent degradation mechanism. Addition of C21 decreased half-life of enhanced green fluorescence protein (EGFP) by 5-fold, demonstrating that C21 constitutes a portable degron. Point mutational analyses of C21 revealed that a conserved proline residue (Pro-221) is central to degron activity, and mutation to alanine (P221A) increased NKX3.1 half-life >2-fold. Proteasome inhibition and in vivo ubiquitination analyses indicated that degron activity is ubiquitin-independent. Evaluating degron activity in the context of a ubiquitination-resistant, lysine-null NKX3.1 mutant (NKX3.1KO) confirmed that P221A mutation conferred additional stability to NKX3.1. Treatment of prostate cancer cell lines with a C21-based peptide specifically increased the level of NKX3.1, suggesting that treatment with degron mimetics may be a viable approach for NKX3.1 restoration.
Introduction
Protein degradation is a closely orchestrated process that plays an important role in maintaining cell homeostasis. A majority of protein degradation is mediated by the ubiquitin proteasome system, and aberrations in this system have been implicated in several diseases, including cancer (1, 2). The classical ubiquitin proteasome system involves hierarchical enzyme activities that culminate in E3-ubiquitin ligase-mediated isopeptide bond formation between ubiquitin, a 76-amino acid protein, and the ϵ-amino group of an acceptor lysine of a target protein (3, 4). Subsequently, the E3-ubiquitin ligases facilitate covalent addition of ubiquitin monomers to generate polyubiquitin chains. Polyubiquitinated proteins are recognized by the proteasome, a multisubunit proteolytic machine that degrades target proteins (5). Although ubiquitination is required for proteasomal recognition and degradation of most proteins, several studies have shown that the proteasome can function in some cases in the absence of ubiquitination. For example, proteins such as Rb, p53, and p21 the canonical tumor-suppressor as well as c-Fos and several viral oncoproteins undergo ubiquitin-independent proteasomal degradation (6–9). Understanding the complexities of protein turnover mechanism, particularly those significant in a growth regulatory context is necessary for efficient drug development based on tumor suppressor restoration.
NKX3.1 is a homeodomain transcription factor that regulates prostate organogenesis and displays hallmark features of a tumor suppressor protein (10–13). Loss of heterozygosity at 8p21-22, which encompasses the NKX3.1 locus, is associated with 50–85% of prostate cancer cases, and diminished NKX3.1 accumulation is common in prostate cancer cases (12). Moreover, NKX3.1 expression is diminished in prostate cancer precursor lesions, including prostate intraepithelial neoplasia and prostatic inflammatory atrophy, suggesting that reduced NKX3.1 expression may play a role in cancer initiation (14–16). In overt adenocarcinoma, reduced NKX3.1 level correlates with disease progression (12, 17–20). NKX3.1 is known to have anti-proliferative effects on prostate epithelial cells, and NKX3.1 restoration in xenografts in mice has been shown to induce tumor regression (17). Interestingly, diminished NKX3.1 accumulation does not strictly correlate with a reduction in mRNA, indicating that NKX3.1 post-translational regulation plays a role in supporting growth suppressive function of NKX3.1 (20, 21). Recently, restoration of tumor suppressor steady state level has emerged as a viable approach for targeted therapy several forms of cancer (22). A full understanding of NKX3.1 post-translational regulatory mechanisms would facilitate development of strategies to restore NKX3.1 in prostate cancer.
Several regions of the 234-amino acid protein NKX3.1 have been implicated in regulating steady-state level. Amino acid residues 81 to 97 comprise a Pro-Glu-Ser-Thr-rich sequence that mediates NKX3.1 turnover by polyubiquitination (23). Protein kinase CK2 phosphorylation at threonine residues within this domain protects NKX3.1 from proteasomal degradation (23). Other studies have demonstrated that amino acid residues 184 to 196 harbor phosphorylation sites that regulate ubiquitin-mediated NKX3.1 degradation in response to the inflammatory cytokine TNF-α (24). In addition, the E3 ubiquitin ligase topoisomerase 1 binding, arginine/serine-rich, E3 ubiquitin ligase has been shown to polyubiquitinate and target NKX3.1 for proteasomal degradation (25). Interactions with TOPORS and other unidentified E3 ligases require specific regions within the NKX3.1 N and C termini (24, 25).
To date, analyses of NKX3.1 turnover have focused exclusively on ubiquitin-mediated proteasomal degradation. Here, we describe a previously unidentified, ubiquitin-independent NKX3.1 degradation mechanism mediated by the conserved C-terminal 21 amino acids (see Fig. 1A). Moreover, we demonstrate the development of a successful strategy to exploit these findings to restore NKX3.1 in prostate cancer cells.
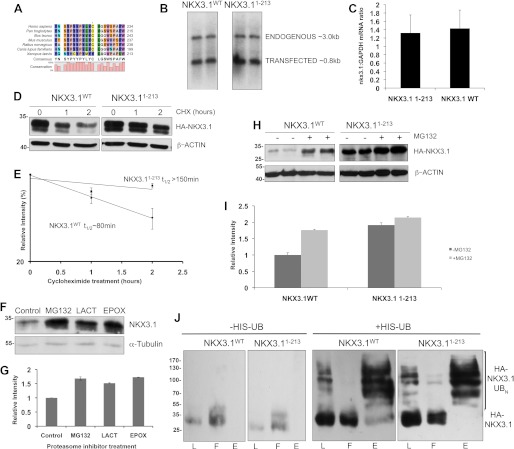
FIGURE 1.
A C-terminal deletion increases NKX3.1 half-life. A, sequence alignment depicting the conserved C21 sequence. B, Northern blot using NKX3.1 radiolabeled probe detecting both endogenous and transfected NKX3.1 in LNCaP lysates. C, quantitative PCR analyses in PC3 cells expressing HA-tagged NKX3.1WT or NKX3.11–213. NKX3.1 mRNA level was normalized to GAPDH mRNA levels demonstrating similar levels of NKX3.1 transcripts. D, Western blot analyses using anti-HA and anti-β-actin antibodies, of cycloheximide (CHX) chase analyses performed with PC3 cells expressing HA-tagged NKX3.1WT or NKX3.11–213. E, quantification of Western blot in C showing longer t½ of NKX3.11–213. F, Western blot analyses of HA-NKX3.1 transfected PC3 cells with a panel of different proteasome inhibitors. Lysates were prepared following MG132 (10 μm), lactacystin (LACT, 50 μm), epoximicin (EPOX, 10 μm), or dimethyl sulfoxide (Control) treatment for 12 h. Antibodies used were anti-HA and anti-α-tubulin antibodies. G, quantification of F using ImageJ software. Relative intensities indicated in arbitrary units. H, Western blot analyses of proteasome inhibition in PC3 cells expressing HA-tagged NKX3.1WT or NKX3.11–213. I, quantification of protein abundance. J, C21 deletion does not abolish ubiquitination. In vivo ubiquitination analyses of PC3 cells co-expressing His-tagged ubiquitin (His-UB) and HA-tagged NKX3.1WT or NKX3.11–213. Following proteasome inhibition, His-ubiquitinated proteins in the cell lysates were enriched by nickel resin chromatography and analyzed by Western blot using rat anti-HA antibodies. L, lysate; F, flowthrough; E, eluate.
EXPERIMENTAL PROCEDURES
Cell Culture
LNCaP cells were cultured in RPMI 1640–10% fetal bovine serum with 5% CO2 in air at 37 °C in a humidified chamber.
NKX3.1 Expression Construct Generation
The hemagglutinin (HA)-tagged NKX3.1 (pcDNA3-HA-NKX3.1) and His-tagged ubiquitin expression vectors have been described (23). Generation of C21 amino acid deletion mutant has been described previously (26). NKX3.1P218A, NKX3.1P231A, NKX3.1P221A, NKX3.1Y222A, and NKX3.1L223A mutants were generated using a QuikChange site-directed mutagenesis kit (Stratagene, San Diego, CA) according to the manufacturer's instructions. The oligonucleotides for site-directed mutagenesis used are described in the supplemental data.
The enhanced green fluorescence protein-expressing vector has been described previously (27). Codon-optimized oligonucleotides containing wild-type or mutant degron sequences were ligated in reading frame using EcoRI and KpnI at the C-terminal multiple cloning site of pEGFP-C14 vector (Clontech). The oligonucleotides used are listed in the supplemental data.
To generate the NKX3.1KO mutant, a codon-optimized gene was generated by simplified gene synthesis in which the 10 homeodomain lysines were mutated by using multiple primers listed in supplemental data (28). The remaining four lysines were mutated by QuikChange site-directed mutagenesis using primers described in supplemental data.
Proteasome Inhibition and Cycloheximide Chase Assay
PC3 cells were grown in six-well plates and transfected with 2 mg/well of NKX3.1 or EGFP expression constructs. 24 h post-transfection, cells were treated with MG132 (10 μm), lactacystin (50 μm), and epoximicin (10 μm) for 12 h (proteasome inhibition) or 10 μm cycloheximide (cycloheximide chase) for different time periods. Cell lysates were prepared using 300 mm NaCl, 100 mm Tris-HCl, pH 6.8, 2% Nonidet 40, and proteinase inhibitors and analyzed by Western blot.
Prostate Cancer Cell Transfection and Peptide Treatment
Plasmids were transiently transfected into LNCaP and PC3 cells using Lipofectamine Plus reagent (Invitrogen) or LipoD 293 transfection reagent (Signagen) according to the manufacturer's instructions; 24 h after transfection, pharmacologic agents were applied as described under “Results.”
HIV-TAT containing peptides were generated by Biopolymer Laboratories (Baltimore, MD). The peptides were solubilized in 10% glacial acetic acid and diluted to working concentrations in PBS.
Western Blot Analysis
Immunoblot analysis of prostate cancer cells transfected with HA-NKX3.1 constructs has been described previously (23). Mouse anti-GFP antibody (1:5000, Covance) was used for Western blot analysis of cells transfected with EGFP constructs.
In Vivo Ubiquitination Assay
In vivo ubiquitination assay has been described previously (23). In vivo ubiquitination assay on peptide-treated cells was done at 8 h after peptide and 20 μm MG132 treatment, lysates and eluates were analyzed by SDS-PAGE as described, and Western blot was done using rabbit anti-NKX3.1 antibodies.
Indirect Immunofluorescence Staining
Immunofluorescent microscopic analysis was done as described in Ref. 25 using rat-anti-hemagglutinin primary antibody (1:200, Roche Applied Science) and Texas Red-labeled anti-rat secondary (1:100, Vector).
Fluorescence Microscopy of Peptide-treated Cells
LNCaP cells grown in six-well plates for 24 h were treated with FITC labeled TAT-C21WT peptide or equivalent amount of FITC solution for 4 h. Mounting medium with DAPI (Vector) was used. Fluorescent images of the cells were captured using a Zeiss fluorescence microscope after replacing the medium with PBS.
RNA Preparation
LNCaP or PC3 cells were washed in sterile PBS and lysed in buffer RLT from the Qiagen RNeasy kit. The RNA extraction was done according to the manufacturer's instructions. RNA was eluted in 50 μl of RNase-free water and treated with DNase.
Northern Blot Analysis
Northern blot analysis was performed according to the manufacturer's instructions using the Ambion NorthernMax-gly kit. Radiolabeled probes against NKX3.1 and β-actin were prepared using the Ambion STRIP-EZ Probe synthesis kit.
Real-time PCR Analysis
cDNA was prepared using Fermentas Maxima cDNA synthesis kit. Real-time PCR analysis was performed in a Bio-Rad iCycler (Bio-Rad) using the setting described in Ref. 29. Primers used were as follows using Fermentas SYBR Green Master Mix: NKX3.1, 5′-CTTCCCCAAACCCCTAAGC-3′ and 5′-TCCTCTCCAACTCGATCACC-3′; GAPDH, 5′-GAAGGTGAAGGTCGGAGT-3′ and 5′-GAAGATGGTGATGGGATTTC-3′.
Relative expression of VEGF-C was calculated by the comparative threshold cycle (Ct) method using GAPDH Ct value as the reference. Average and S.D. of triplicate experiments was determined.
[3H]Thymidine Incorporation Assay
LNCaP cells were grown in a 96-well multiplate and treated with 10 μm peptide or vehicle every 8 h for 48 h. 1 mCi [3H]thymidine was added to each well after 24 h. 24 h following radionucleotide treatment, samples were harvested with cell harvester. Filter mats were sealed in plastic bags with 4 ml of Betaplate scintillation fluid (PerkinElmer Life Sciences) and [3H]thymidine incorporation was measured using a liquid scintillation counter (PerkinElmer Life Sciences).
Image Quantification and Statistical Analysis
ImageJ software was used for all image analysis (6). Signal intensity of bands detected by Western or Northern blot was normalized to signal intensity of the housekeeping proteins β-actin or α-tubulin. Relative intensities were plotted on a graph generated using Microsoft Excel. S.E. was calculated in all cases. Images shown are representative of at least duplicate experiments.
RESULTS
C21 Mediates Post-translational Regulation of NKX3.1
Previous studies have shown that the conserved C-terminal 21-amino acid domain of NKX3.1 (Fig. 1A) is essential for interaction with prostate-derived Ets factor (26, 30). Interestingly, Western blot analysis demonstrated greater accumulation of the C-terminal deletion mutant (NKX3.11–213) than full-length NKX3.1. However, the underlying mechanism for C21-regulated NKX3.1 degradation remained undefined. Northern blot and quantitative PCR analyses revealed comparable levels of wild-type and NKX3.11–213 transcript, indicating that the effect occurs at the protein level (Fig. 1, B and C). Consequently, we sought to determine the effect of C21 on protein half-life by a cycloheximide chase approach. PC3 cells transfected with vectors expressing HA-tagged, full-length wild-type NKX3.1 or NKX3.11–213 were treated with cycloheximide and protein abundance was quantified by Western blot (Fig. 1, D and E). Whereas the half-life of the wild-type NKX3.1 was ∼80 min, the C21 deletion nearly doubled the half-life. Initial experiments with a panel of proteasome inhibitors demonstrated up to ∼60% NKX3.1 accumulation in prostate cancer cells expressing wild-type NKX3.1 (Fig. 1, F and G). MG132, a known proteasome inhibitor that has been used for treatment in several cancers was chosen as a standard for subsequent proteasome inhibition experiments. To determine whether C21 deletion disrupted proteasome-mediated degradation, proteasome inhibition experiments were performed. Western blot analysis following MG132 treatment on prostate cancer cells expressing NKX3.11–213 showed accumulation of both proteins; however, the increase in level of NKX3.11–213 was marginal (∼10%) in comparison with that of the wild-type NKX3.1 (∼60%, Fig. 1, H and I). Together, these experiments demonstrate that C21 deletion increased half-life by interfering with proteasome-dependent NKX3.1 degradation.
C21 Deletion Does Not Abolish NKX3.1 Ubiquitination
To explore the mechanism by which C21 deletion interfered with proteasomal degradation, we first sought to study the effect of the deletion on polyubiquitination. In several proteins, degrons mediate interaction with E3-ubiquitin ligases and mutations in these degrons prevent polyubiquitination and subsequent proteasomal degradation (31–34). In vivo ubiquitination analyses were conducted in prostate cancer cells transfected with vectors expressing HA-tagged NKX3.1WT or NKX3.11–213 and His-tagged ubiquitin. His-ubiquitinated proteins were enriched by nickel affinity chromatography, and eluates were analyzed by Western blot. High molecular weight, polyubiquitinated species were observed in case of both full-length NKX3.1 and the C21 deletion mutant (Fig. 1J), demonstrating that C21 deletion does not abolish polyubiquitination of NKX3.1. These data suggest that the C21 degron activity could operate through a direct proteasomal interaction.
C21 Constitutes a Portable Degron
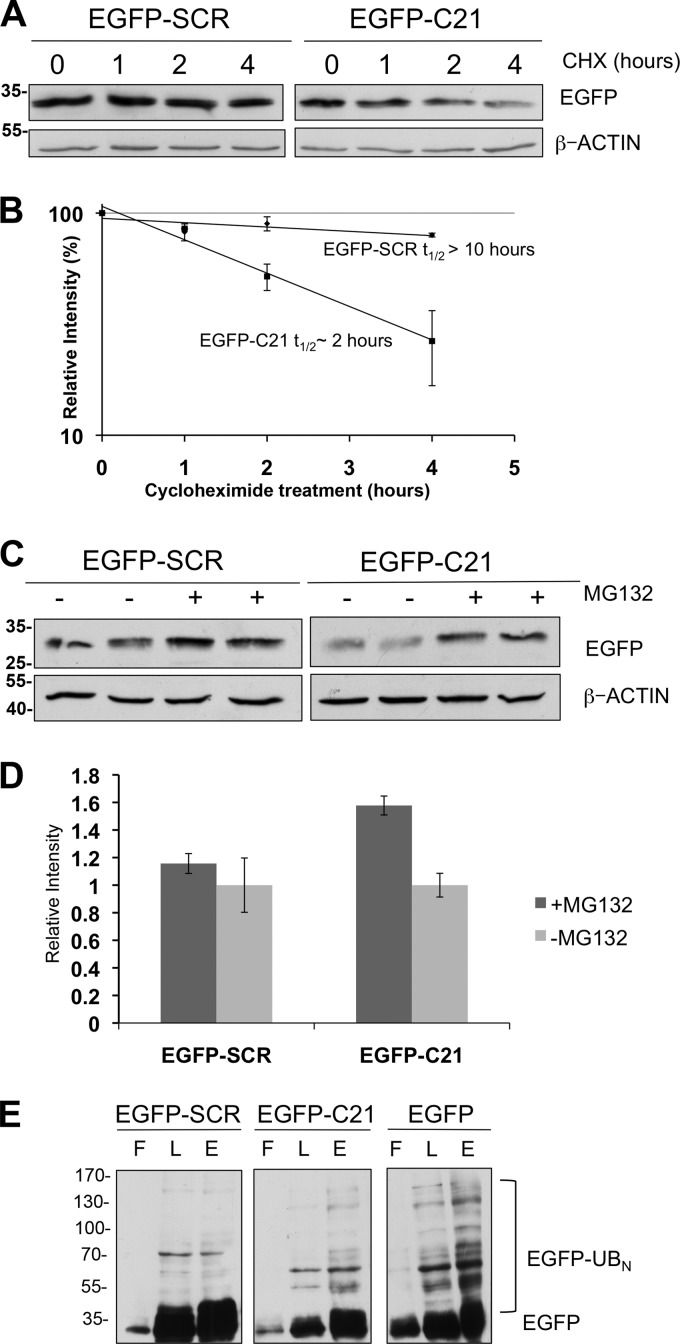
To further characterize C21 degron activity, we chose to study the degron in the context of a reporter protein. Although some degrons are effective only in their native context, others, termed portable degrons, can be transferred to heterologous proteins to which they confer instability. To determine whether C21 can function as a portable degron, a vector expressing EGFP C-terminally tagged with C21 (EGFP-C21) was transfected into LNCaP cells. Cycloheximide chase demonstrated that the half-life of EGFP-C21 was reduced 5-fold compared with native EGFP, whereas a scrambled C21 sequence had no effect on EGFP half-life (Fig. 2, A and B). Proteasome inhibition showed that addition of C21 renders EGFP susceptible to proteasomal degradation in prostate cancer cell lines (Fig. 2, C and D). Moreover, in vivo ubiquitination analyses demonstrated that C21 addition did not significantly alter EGFP polyubiquitination (Fig. 2E), thereby providing further evidence for a ubiquitin-independent degradation mechanism for C21 degron activity.
FIGURE 2.
C21 comprises a portable degron of NKX3.1. A, addition of C21 reduces half-life of EGFP. PC3 cells were transfected with vectors expressing EGFP-SCR or EGFP-C21 and treated with 10 μm cycloheximide (CHX) for different time points and lysates analyzed by Western blot with anti-EGFP and anti-β-actin antibodies. B, quantification of Western blot in A. C, addition of C21 renders EGFP susceptible to proteasomal degradation. Transfected cells were treated with 10 μm MG132 and analyzed by Western blot after 12 h. D, quantification of Western blot in C. E, In vivo ubiquitination analysis was performed as described in Fig. 1J with cells transfected EGFP and His-ubiquitin (UB) expression vectors. Cell lysates were analyzed by Western blot using mouse anti-GFP antibodies. L, lysate; F, flowthrough; E, eluate.
Proline 221 Is Required for C21 Degron Function
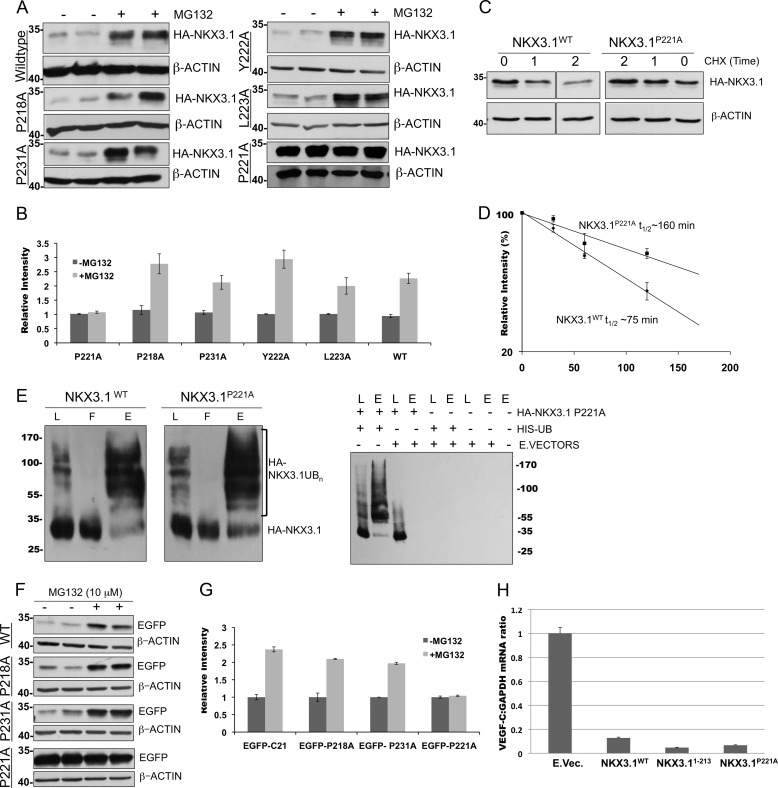
C21 is a highly conserved region of low sequence complexity. Inspection of this region for potential regulatory regions revealed a proline-containing motif (PYL, Fig. 1A). A similar C-terminal tripeptide motif (PTL) has been implicated in regulating c-Fos turnover (35). NKX3.1 point mutants at Pro-221, Tyr-222, and Leu-223 were generated along with additional proline mutants Pro-218 and Pro-231. Proteasome inhibition demonstrated that the P221A mutation abolished degron activity (Fig. 3, A and B), whereas P218A, P231A, Y222A, or L223A demonstrated a 2-fold increase in protein abundance following proteasome inhibition. Cycloheximide chase analysis demonstrated that P221A abrogates C21 degron activity generating a dramatically more stable NKX3.1 isoform (t½ ∼ 160 min, Fig. 3, C and D). Together, these findings show that Pro-221 is crucial for degron function. In vivo ubiquitination studies demonstrated that NKX3.1P221A ubiquitination was indistinguishable from that of NKX3.1WT (Fig. 3E), suggesting that the role of Pro-221 in mediating degron activity is ubiquitin-independent. Proteasome inhibition studies in the context of EGFP-C21 corroborated with our observations, where P221A mutation (EGFP-C21P221A) rescued EGFP-C21 from proteasomal degradation (Fig. 3, F and G), whereas EGFP-C21P218A and EGFP-C21P231A resembled EGFP-C21WT. These studies reveal that P221A was crucial for C21 portable degron function. Immunofluorescence analyses of PC3 cells demonstrated identical nuclear localization of transfected NKX3.1 isoforms, indicating that altered degron activity was not a consequence of protein mislocalization (data not shown).
FIGURE 3.
Pro-221 is required for C21 degron function. Proteasome inhibition in PC3 cells expressing HA-tagged NKX3.1WT, P218A, P221A, P231A, Y222A, or L223A mutants. A, Western blot analyses were performed using anti-HA and anti-β-actin antibodies to detect ∼34-kDa HA-NKX3.1 and ∼42-kDa β-actin. B, quantification of relative protein abundance. Cycloheximide (CHX) chase analyses of PC3 cells expressing HA-tagged NKX3.1WT or NKX3.1P221A. C, Western blot analyses of cell lysates was performed using antibodies as in A. D, quantification by semi-log plot of Western blot data in C. E, in vivo ubiquitination assay was performed as described in Fig. 1J with PC3 cells co-expressing His-tagged ubiquitin (His-UB) and NKX3.1WT or NKX3.1P221A and analyzed by Western blot using anti-HA antibodies. Polyubiquitinated higher molecular weight species were identified. F, proteasome inhibition in PC3 cells expressing EGFP-C21 with alanine mutations at Pro-218, Pro-221, and Pro-231. Lysates were analyzed by Western blot using anti-EGFP and β-actin antibodies to detect ∼34-kDa EGFP and ∼42-kDa β-actin. Individual mutations are indicated on the right. G, quantification of Western blot described in F. H, HA-NKX3.1P221A is a functional transcriptional repressor. Quantitative PCR analyses of mRNA obtained from PC3 cells transfected with NKX3.1WT, NKX3.11–213, or NKX3.1P221A expression vectors (E.Vec.) to demonstrate equivalent transcriptional repression of a responder gene VEGF-C. L, lysate; F, flowthrough; E, eluate.
To determine whether NKX3.1P221A was a functional transcriptional regulator, we quantified expression of VEGF-C, a known NKX3.1 target gene (29). Expression analyses in PC3 cells transfected with NKX3.1WT or NKX3.1P221A showed a 3-fold decrease in VEGF-C mRNA level in comparison with an empty vector control (Fig. 3H). These data demonstrate that NKX3.1P221A can function as a transcriptional repressor in a manner similar to NKX3.1WT in prostate cancer cell lines.
Proline 221 Mediates Ubiquitin-independent NKX3.1 Degradation
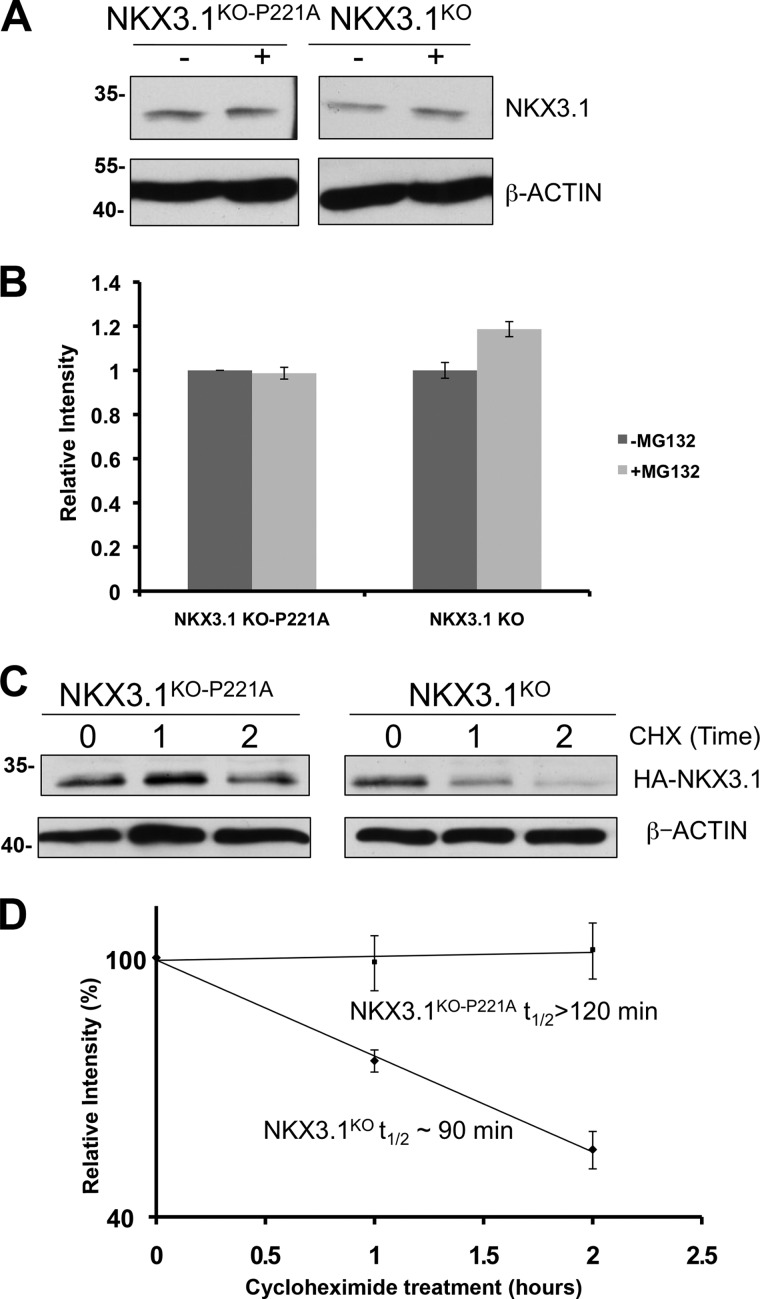
To definitively demonstrate the existence of a proteasome-dependent, ubiquitin-independent NKX3.1 degradation pathway, we generated a lysine-null NKX3.1 mutant (NKX3.1KO) that did not undergo in vivo ubiquitination (data not shown). Proteasome inhibition studies in PC3 cells confirmed that NKX3.1KO remained susceptible to proteasomal degradation, albeit to a reduced degree compared with NKX3.1WT. To determine whether Pro-221 mediates proteasome-dependent ubiquitin-independent degradation of NKX3.1, we studied the effect of P221A in the context of NKX3.1KO. Interestingly, P221A mutation in NKX3.1KO (NKX3.1KO-P221A) abolished susceptibility to proteasomal degradation (Fig. 4, A and B). Moreover, half-life studies using cycloheximide showed that P221A mutation increased NKX3.1KO stability by at least 3-fold (Fig. 4, C and D). Together, these data clearly demonstrate that Pro-221 mediates C21 ubiquitin-independent degron activity.
FIGURE 4.
P221A mutation increases stability of NKX3.1 lysine-null mutant. A, proteasome inhibition of PC3 cells expressing lysine null (NKX3.1KO) and P221A lysine-null mutant (NKX3.1KO-P221A). Lysates were analyzed by Western blot using anti-HA antibody and anti-β-actin to detect ∼34-kDa HA-NKX3.1 and ∼42-kDa β-actin. B, quantification of Western blot described in A. C, P221A mutation increases half-life of lysine-null NKX3.1. Western blot analysis of PC3 cells expressing HA-NKX3.1KO and HA-NKX3.1KO-P221A and treated with 10 μm cycloheximide (CHX) for the time indicated. Lysates were analyzed by Western blot using antibodies as in A. D, quantification of Western blot of described in C.
C21 Degron-based Peptide Increases Endogenous NKX3.1 Steady-state Level
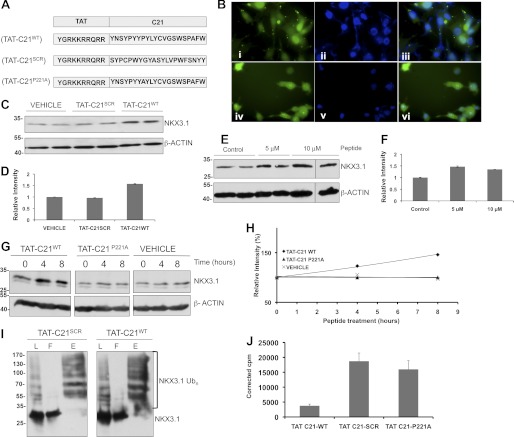
Peptidomimetics targeting the protein degradation machinery have been extensively used in cancer therapy (36–38). Based on our findings reported here, we envisioned a peptide-based strategy to increase NKX3.1 stability in prostate cancer cells. To enable cellular uptake of the degron peptide, we appended an 11-amino acid protein transduction domain derived from the HIV transactivator of transcription protein (TAT), to the N terminus of the C21 sequence (Fig. 5A) (39, 40). Fluorescent microscopic analysis of LNCaP cells treated with FITC-labeled TAT-degron peptide (TAT-C21WT) showed efficient uptake and nuclear localization (Fig. 5B). The TAT-C21WT peptide treatment increased endogenous NKX3.1 steady-state level, whereas a scrambled TAT-C21 peptide did not (Fig. 5, C and D). Northern blot analyses did not reveal changes in NKX3.1 mRNA level upon peptide treatment, indicating that the effect was post-transcriptional (data not shown). To provide further evidence of a role for Pro-221 in mediating C21 degron activity, we generated a P221A mutant peptide (TAT-C21P221A). Treatment of cells with TAT-C21P221A failed to increase the level of endogenous NKX3.1 (Fig. 5E), consistent with our data demonstrating the importance of Pro-221 in the context of native NKX3.1. In vivo ubiquitination analyses of NKX3.1 in the presence of TAT-C21WT peptide did not alter the pattern of polyubiquitination (Fig. 5F). To assess the effect of NKX3.1 restoration following peptide treatment on prostate cancer cell proliferation, we performed [3H]thymidine uptake experiments. TAT-C21WT treatment alone resulted in low [3H]thymidine incorporation, unlike TAT-C21P221A or TAT-C21SCR treatment (Fig. 5G), demonstrating that TAT-C21WT treatment and subsequent NKX3.1 steady-state level increase reduces prostate cancer cell proliferation.
FIGURE 5.
TAT-C21 treatment of prostate cancer cell lines. A, diagram showing the peptides used in treatment of prostate cancer cells. B, peptide localization by immunofluorescence microscopic analyses. Cells were treated with 5 μm FITC-labeled TAT-C21WT peptide or equivalent amount of fluorescein isothiocyanate solution for 4 h. DAPI was used for nuclear staining. i–iii, LNCaP cells treated with TAT-C21WT peptide. i, FITC staining; ii, DAPI staining; iii, overlay of i and ii. iv–vi, LNCaP cells treated with FITC solution. iv, FITC staining; v, DAPI staining; vi, overlay. C, Western blot analyses using anti-NKX3.1 and anti-β-actin antibodies of LNCaP cells treated with 5 μm TAT-C21WT, TAT-C21SCR peptide, or vehicle for 8 h. D, quantification of Western blot. E, Western blot analyses of LNCaP cells treated with 5 μm and 10 μm of TAT-C21WT peptide for 8 h. F, quantification of Western blot showing maximum increase in NKX3.1 level at 5 μm. G, Western blot analyses of LNCaP cells treated with TAT-C21 peptides indicated on the top of the panel. Lysates were prepared after time indicated. H, quantification of Western blot. Increase in endogenous NKX3.1 level was observed only upon TAT-C21WT peptide treatment. I, in vivo ubiquitination assay was performed as described under “Experimental Procedures” in LNCaP cells transfected with His-ubiquitin (Ub) vectors and treated with peptides indicated. Western blot analyses using anti-NKX3.1 antibodies. L, lysate; F, flowthrough; E, eluate. Peptide treatment does not abolish endogenous NKX3.1 ubiquitination. G, [3H]thymidine incorporation assay demonstrating increased uptake by cells treated with vehicle and scrambled peptide and not by cells treated with TAT-C21WT.
DISCUSSION
NKX3.1 plays a prominent role in prostate development and functions as a prostate-specific tumor suppressor in adult animals (11, 12, 41). Previous studies have shown that despite a reduction in protein level, NKX3.1 mRNA level remains normal or nearly so, in many prostate cancer cases, underscoring the importance of post-translational modifications in regulating NKX3.1 steady-state level (20). Although phosphorylation and ubiquitination are known to regulate NKX3.1, our knowledge of post-translational mechanisms that maintain steady-state level is incomplete (23–25, 42). NKX3.1 restoration in human prostate cancer xenografts has been shown to decrease disease progression, suggesting that this may serve as a novel prostate cancer therapeutic approach (42). However, development of strategies to stabilize NKX3.1 must be predicated upon a thorough understanding of pathways regulating this protein in prostate cancer cells. Here, we have demonstrated that NKX3.1 is regulated by a ubiquitin-independent proteasomal degradation pathway and that perturbation of the pathway stabilizes NKX3.1.
The NKX3.1 C terminus is a known protein-protein interaction domain, and deletion of this region was shown to stabilize NKX3.1 (24, 26). Here, we show that C21 constitutes a portable degron. Several short-lived proteins, including transcription factors Myc and c-Fos, contain degrons that regulate turnover in a context-dependent manner (34, 35). In some proteins, degron activity regulates E3-ubiquitin ligase recruitment that subsequently promotes polyubiquitination (32, 33, 43, 44). However, our studies have demonstrated that C21 deletion did not alter polyubiquitination, suggesting that C21 degron activity is independent of E3-ubiquitin ligase recognition or interaction. Distinct pathways activate alternative degradation mechanisms for proteins such as Rb and the Fos-family members upon viral infection, oxygen depletion, or other physiological conditions (27, 45). The data presented here demonstrate the presence of additional, context-dependent NKX3.1 turnover mechanism. Tightly regulated NKX3.1 turnover is crucial for prostate homeostasis, and alternative NKX3.1 degradation mechanisms could become physiologically significant during tumor progression.
Mutational analyses of C21 indentified Pro-221 as being necessary for degron activity. Pro-221 resides in a PYL tripeptide sequence in NKX3.1 and a similar motif (PS/TL) has been implicated in cell cycle-dependent degradation of the Fos family proteins (35). However, Y222A or L223A mutations did not inhibit proteasomal degradation, emphasizing the central role for Pro-221 in NKX3.1 steady-state maintenance. Proline residues play important roles in protein structure and function maintenance (46, 47). The cyclic nature of the proline side chain affects secondary structure and protein folding (48, 49). Apart from regulating protein structure, γ-C atom hydroxylation by prolyl hydroxylases is another key regulatory mechanism (47). For example, oxygen-dependent proline hydroxylation mediates HIF-1a ubiquitination by the vHL E3-ubiquitin ligase (50). In our hands, treatment of prostate cancer cells with prolyl hydroxylase inhibitor, dimethyoxalylglycine did not appear to alter transfected NKX3.1 accumulation or in vivo ubiquitination detectably altered (data not shown), suggesting that C21 degron activity is independent of prolyl hydroxylation.
Previous studies have implicated proline residues in regulating proteasomal degradation (51). A proline-rich domain in the homeodomain protein, PRH, has been shown to mediate proteasomal interaction, in a ubiquitin-independent manner. Remarkably, the P221A mutation in the context of NKX3.1KO conferred resistance to proteasomal degradation, demonstrating that Pro-221 regulates NKX3.1 turnover even in the absence of ubiquitin acceptor sites. Moreover, regions on proteins lacking defined structures have been implicated in regulating ubiquitin-independent proteasomal degradation of several proteins where the disordered region may regulate protein unfolding and proteasome entry (52). The C21 sequence has a central helical region flanked by two disordered regions. Based on these observations, we hypothesize that the disordered C21 region mediates proteasomal interaction and regulates NKX3.1 degradation.
Tumor suppressor restoration in cancer using small molecules, peptides, or peptido-mimectics has emerged as a potential strategy for therapy. For example, Velcade (bortezomib) is a global proteasome inhibitor that has shown efficacy in treating multiple myeloma and ovarian carcinoma (53–55). In a more targeted approach, MDM2 peptidomimetics have been shown to interfere with MDM2/p53 interaction, thereby restoring p53 level in hematologic malignancies (56–60). In addition, a naturally occurring proline- and arginine-rich peptide, PR39, has been shown to restore IκB level by proteasome inhibition (61). In light of these observations, we designed a C21-based peptide, TAT-C21WT and demonstrated its ability to specifically block NKX3.1 degradation. These data not only support the central role for C21 in regulating NKX3.1 turnover but also underscore the potential of peptide-based strategies to restore NKX3.1 in prostate cancer. It will be of great interest to extend these pharmacological studies to mouse prostate cancer models that display reduced NKX3.1 to further explore the utility of TAT-C21WT peptide as a therapeutic agent.
In summary, we have described a key role for the NKX3.1 far C terminus in regulating steady-state level of this potent prostate-specific tumor suppressor. Our studies demonstrate a ubiquitin-independent, proteasome-dependent pathway of NKX3.1 turnover. These data add to the complexity of known mechanisms regulating NKX3.1 turnover but provide promising therapeutic avenues for peptide-based NKX3.1 restoration in prostate cancer cells.
This work was supported by Grant W81XWH-07-1-0232 from the Congressionally Directed Medical Research Prostate Cancer Program (to C. J. B.).

This article contains supplemental data and additional references.
- EGFP
- enhanced green fluorescence protein
- TAT
- transactivator of transcription protein.
REFERENCES
- 1. Hoeller D., Hecker C. M., Dikic I. (2006) Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat. Rev. Cancer 6, 776–788 [DOI] [PubMed] [Google Scholar]
- 2. Hershko A. (2005) The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 12, 1191–1197 [DOI] [PubMed] [Google Scholar]
- 3. Hershko A., Ciechanover A., Rose I. A. (1979) Resolution of the ATP-dependent proteolytic system from reticulocytes: a component that interacts with ATP. Proc. Natl. Acad. Sci. U.S.A. 76, 3107–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilkinson K. D., Urban M. K., Haas A. L. (1980) Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J. Biol. Chem. 255, 7529–7532 [PubMed] [Google Scholar]
- 5. Hershko A., Ciechanover A., Heller H., Haas A. L., Rose I. A. (1980) Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl. Acad. Sci. U.S.A. 77, 1783–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin Y., Lee H., Zeng S. X., Dai M. S., Lu H. (2003) MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 22, 6365–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sdek P., Ying H., Chang D. L., Qiu W., Zheng H., Touitou R., Allday M. J., Xiao Z. X. (2005) MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol. Cell 20, 699–708 [DOI] [PubMed] [Google Scholar]
- 8. Ying H., Xiao Z. X. (2006) Targeting retinoblastoma protein for degradation by proteasomes. Cell Cycle 5, 506–508 [DOI] [PubMed] [Google Scholar]
- 9. Hwang J., Winkler L., Kalejta R. F. (2011) Ubiquitin-independent proteasomal degradation during oncogenic viral infections. Biochim. Biophys. Acta 1816, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sciavolino P. J., Abrams E. W., Yang L., Austenberg L. P., Shen M. M., Abate-Shen C. (1997) Tissue-specific expression of murine Nkx3.1 in the male urogenital system. Dev. Dyn. 209, 127–138 [DOI] [PubMed] [Google Scholar]
- 11. He W. W., Sciavolino P. J., Wing J., Augustus M., Hudson P., Meissner P. S., Curtis R. T., Shell B. K., Bostwick D. G., Tindall D. J., Gelmann E. P., Abate-Shen C., Carter K. C. (1997) A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics 43, 69–77 [DOI] [PubMed] [Google Scholar]
- 12. Bowen C., Bubendorf L., Voeller H. J., Slack R., Willi N., Sauter G., Gasser T. C., Koivisto P., Lack E. E., Kononen J., Kallioniemi O. P., Gelmann E. P. (2000) Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 60, 6111–6115 [PubMed] [Google Scholar]
- 13. Prescott J. L., Blok L., Tindall D. J. (1998) Isolation and androgen regulation of the human homeobox cDNA, NKX3.1. Prostate 35, 71–80 [DOI] [PubMed] [Google Scholar]
- 14. De Marzo A. M., Platz E. A., Sutcliffe S., Xu J., Grönberg H., Drake C. G., Nakai Y., Isaacs W. B., Nelson W. G. (2007) Inflammation in prostate carcinogenesis. Nat. Rev. Cancer 7, 256–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abdulkadir S. A., Magee J. A., Peters T. J., Kaleem Z., Naughton C. K., Humphrey P. A., Milbrandt J. (2002) Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol. Cell. Biol. 22, 1495–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iwata T., Schultz D., Hicks J., Hubbard G. K., Mutton L. N., Lotan T. L., Bethel C., Lotz M. T., Yegnasubramanian S., Nelson W. G., Dang C. V., Xu M., Anele U., Koh C. M., Bieberich C. J., De Marzo A. M. (2010) MYC overexpression induces prostatic intraepithelial neoplasia and loss of Nkx3.1 in mouse luminal epithelial cells. PLoS One 5, e9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim M. J., Bhatia-Gaur R., Banach-Petrosky W. A., Desai N., Wang Y., Hayward S. W., Cunha G. R., Cardiff R. D., Shen M. M., Abate-Shen C. (2002) Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res. 62, 2999–3004 [PubMed] [Google Scholar]
- 18. Magee J. A., Abdulkadir S. A., Milbrandt J. (2003) Haploinsufficiency at the Nkx3.1 locus. A paradigm for stochastic, dosage-sensitive gene regulation during tumor initiation. Cancer Cell 3, 273–283 [DOI] [PubMed] [Google Scholar]
- 19. Tulchin N., Chambon M., Juan G., Dikman S., Strauchen J., Ornstein L., Billack B., Woods N. T., Monteiro A. N. (2010) BRCA1 protein and nucleolin colocalize in breast carcinoma tissue and cancer cell lines. Am. J. Pathol. 176, 1203–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bethel C. R., Faith D., Li X., Guan B., Hicks J. L., Lan F., Jenkins R. B., Bieberich C. J., De Marzo A. M. (2006) Decreased NKX3.1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia, and adenocarcinoma: association with gleason score and chromosome 8p deletion. Cancer Res. 66, 10683–10690 [DOI] [PubMed] [Google Scholar]
- 21. Bethel C. R., Bieberich C. J. (2007) Loss of Nkx3.1 expression in the transgenic adenocarcinoma of mouse prostate model. Prostate 67, 1740–1750 [DOI] [PubMed] [Google Scholar]
- 22. Patel S., Player M. R. (2008) Small-molecule inhibitors of the p53-HDM2 interaction for the treatment of cancer. Expert Opin. Investig. Drugs 17, 1865–1882 [DOI] [PubMed] [Google Scholar]
- 23. Li X., Guan B., Maghami S., Bieberich C. J. (2006) NKX3.1 is regulated by protein kinase CK2 in prostate tumor cells. Mol. Cell. Biol. 26, 3008–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Markowski M. C., Bowen C., Gelmann E. P. (2008) Inflammatory cytokines induce phosphorylation and ubiquitination of prostate suppressor protein NKX3.1. Cancer Res. 68, 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guan B., Pungaliya P., Li X., Uquillas C., Mutton L. N., Rubin E. H., Bieberich C. J. (2008) Ubiquitination by TOPORS regulates the prostate tumor suppressor NKX3.1. J. Biol. Chem. 283, 4834–4840 [DOI] [PubMed] [Google Scholar]
- 26. Chen H., Bieberich C. J. (2005) Structural and functional analysis of domains mediating interaction between NKX-3.1 and PDEF. J. Cell. Biochem. 94, 168–177 [DOI] [PubMed] [Google Scholar]
- 27. Ferrara P., Andermarcher E., Bossis G., Acquaviva C., Brockly F., Jariel-Encontre I., Piechaczyk M. (2003) The structural determinants responsible for c-Fos protein proteasomal degradation differ according to the conditions of expression. Oncogene 22, 1461–1474 [DOI] [PubMed] [Google Scholar]
- 28. Wu G., Wolf J. B., Ibrahim A. F., Vadasz S., Gunasinghe M., Freeland S. J. (2006) Simplified gene synthesis: a one-step approach to PCR-based gene construction. J. Biotechnol. 124, 496–503 [DOI] [PubMed] [Google Scholar]
- 29. Zhang H., Muders M. H., Li J., Rinaldo F., Tindall D. J., Datta K. (2008) Loss of NKX3.1 favors vascular endothelial growth factor-C expression in prostate cancer. Cancer Res. 68, 8770–8778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen H., Nandi A. K., Li X., Bieberich C. J. (2002) NKX-3.1 interacts with prostate-derived Ets factor and regulates the activity of the PSA promoter. Cancer Res. 62, 338–340 [PubMed] [Google Scholar]
- 31. Dohmen R. J., Wu P., Varshavsky A. (1994) Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263, 1273–1276 [DOI] [PubMed] [Google Scholar]
- 32. Jin J., Shirogane T., Xu L., Nalepa G., Qin J., Elledge S. J., Harper J. W. (2003) SCFβ-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 17, 3062–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ye X., Nalepa G., Welcker M., Kessler B. M., Spooner E., Qin J., Elledge S. J., Clurman B. E., Harper J. W. (2004) Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J. Biol. Chem. 279, 50110–50119 [DOI] [PubMed] [Google Scholar]
- 34. Herbst A., Salghetti S. E., Kim S. Y., Tansey W. P. (2004) Multiple cell-type-specific elements regulate Myc protein stability. Oncogene 23, 3863–3871 [DOI] [PubMed] [Google Scholar]
- 35. Acquaviva C., Brockly F., Ferrara P., Bossis G., Salvat C., Jariel-Encontre I., Piechaczyk M. (2001) Identification of a C-terminal tripeptide motif in the control of rapid proteasomal degradation of c-Fos proto-oncoprotein during the Go-to-S phase transition. Oncogene 20, 7563–7572 [DOI] [PubMed] [Google Scholar]
- 36. Tovar C., Higgins B., Kolinsky K., Xia M., Packman K., Heimbrook D. C., Vassilev L. T. (2011) MDM2 antagonists boost antitumor effect of androgen withdrawal: implications for therapy of prostate cancer. Mol. Cancer 10, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hori T., Kondo T., Kanamori M., Tabuchi Y., Ogawa R., Zhao Q. L., Ahmed K., Yasuda T., Seki S., Suzuki K., Kimura T. (2010) Nutlin-3 enhances tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis through up-regulation of death receptor 5 (DR5) in human sarcoma HOS cells and human colon cancer HCT116 cells. Cancer Lett. 287, 98–108 [DOI] [PubMed] [Google Scholar]
- 38. Jehangir S., Dowdy S. F. (2005) Transmembrane delivery of protein and peptide drugs by TAT-mediated transduction in the treatment of cancer. Adv. Drug Del. Rev. 57, 579–596 [DOI] [PubMed] [Google Scholar]
- 39. Frankel A. D., Pabo C. O. (1988) Cellular uptake of the tat protein from human immunodeficiency virus. Cell 55, 1189–1193 [DOI] [PubMed] [Google Scholar]
- 40. Vivès E., Brodin P., Lebleu B. (1997) A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 272, 16010–16017 [DOI] [PubMed] [Google Scholar]
- 41. Bhatia-Gaur R., Donjacour A. A., Sciavolino P. J., Kim M., Desai N., Young P., Norton C. R., Gridley T., Cardiff R. D., Cunha G. R., Abate-Shen C., Shen M. M. (1999) Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 13, 966–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lei Q., Jiao J., Xin L., Chang C. J., Wang S., Gao J., Gleave M. E., Witte O. N., Liu X., Wu H. (2006) NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell 9, 367–378 [DOI] [PubMed] [Google Scholar]
- 43. Orlicky S., Tang X., Willems A., Tyers M., Sicheri F. (2003) Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112, 243–256 [DOI] [PubMed] [Google Scholar]
- 44. Sen N., Sen A., Mackow E. R. (2007) Degrons at the C terminus of the pathogenic but not the nonpathogenic hantavirus G1 tail direct proteasomal degradation. J. Virol. 81, 4323–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kalejta R. F., Shenk T. (2003) Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc. Natl. Acad. Sci. U.S.A. 100, 3263–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kay B. K., Williamson M. P., Sudol M. (2000) The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14, 231–241 [PubMed] [Google Scholar]
- 47. Kaelin W. G. (2005) Proline hydroxylation and gene expression. Annu. Rev. Biochem. 74, 115–128 [DOI] [PubMed] [Google Scholar]
- 48. Yan Y. T., Stein S. M., Ding J., Shen M. M., Abate-Shen C. (2000) A novel PF/PN motif inhibits nuclear localization and DNA binding activity of the ESX1 homeoprotein. Mol. Cell. Biol. 20, 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kanelis V., Donaldson L., Muhandiram D. R., Rotin D., Forman-Kay J. D., Kay L. E. (2000) Sequential assignment of proline-rich regions in proteins: application to modular binding domain complexes. J. Biomol. NMR 16, 253–259 [DOI] [PubMed] [Google Scholar]
- 50. Lewis M. D., Roberts B. J. (2004) Role of the C-terminal α-helical domain of the von Hippel-Lindau protein in its E3 ubiquitin ligase activity. Oncogene 23, 2315–2323 [DOI] [PubMed] [Google Scholar]
- 51. Bess K. L., Swingler T. E., Rivett A. J., Gaston K., Jayaraman P. S. (2003) The transcriptional repressor protein PRH interacts with the proteasome. Biochem. J. 374, 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Melo S. P., Barbour K. W., Berger F. G. (2011) Cooperation between an intrinsically disordered region and a helical segment is required for ubiquitin-independent degradation by the proteasome. J. Biol. Chem. 286, 36559–36567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Papandreou C. N., Logothetis C. J. (2004) Bortezomib as a potential treatment for prostate cancer. Cancer Res. 64, 5036–5043 [DOI] [PubMed] [Google Scholar]
- 54. Tahmatzopoulos A., Gudegast C., Stöckle M., Wullich B., Unteregger G., Zwergel U., Zwergel T. (2004) Proteasome inhibitors: induction of apoptosis as new therapeutic option in prostate cancer. Aktuelle Urol. 35, 491–496 [DOI] [PubMed] [Google Scholar]
- 55. Moehler T., Goldschmidt H. (2011) Therapy of relapsed and refractory multiple myeloma. Recent Results Cancer Res. 183, 239–271 [DOI] [PubMed] [Google Scholar]
- 56. Corallini F., Celeghini C. (2008) The potential role of Nutlins in the treatment of B-chronic lymphocytic leukemia (B-CLL). J. Leukoc. Biol. 84, 651. [DOI] [PubMed] [Google Scholar]
- 57. Vassilev L. T. (2004) Small-molecule antagonists of p53-MDM2 binding: research tools and potential therapeutics. Cell Cycle 3, 419–421 [PubMed] [Google Scholar]
- 58. Adams J., Kauffman M. (2004) Development of the proteasome inhibitor Velcade (Bortezomib). Cancer Invest. 22, 304–311 [DOI] [PubMed] [Google Scholar]
- 59. Kane R. C., Bross P. F., Farrell A. T., Pazdur R. (2003) Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist 8, 508–513 [DOI] [PubMed] [Google Scholar]
- 60. Mack P. C., Davies A. M., Lara P. N., Gumerlock P. H., Gandara D. R. (2003) Integration of the proteasome inhibitor PS-341 (Velcade) into the therapeutic approach to lung cancer. Lung Cancer 41, S89–96 [DOI] [PubMed] [Google Scholar]
- 61. Gaczynska M., Osmulski P. A., Gao Y., Post M. J., Simons M. (2003) Proline- and arginine-rich peptides constitute a novel class of allosteric inhibitors of proteasome activity. Biochemistry 42, 8663–8670 [DOI] [PubMed] [Google Scholar]