Background: Amino acid deprivation activates transcription by ATF4 action.
Results: Transcription from some ATF4-dependent genes can be rescued in Atf4-deficient cells by HDAC inhibition.
Conclusion: ATF4 functions to promote genomic histone acetylation of a sub-set of its target genes.
Significance: The mechanism of ATF4 action on this set of genes is to alter chromatin structure
Keywords: Gene Regulation, Gene Transcription, Histidine, Histone Deacetylase Inhibitors, Nutrition, ATF3, Asparagine Synthetase, Dietary Protein, Histone Demethylase, Nutrient Sensing
Abstract
Following amino acid deprivation, the amino acid response (AAR) induces transcription from specific genes through a collection of signaling mechanisms, including the GCN2-eIF2-ATF4 pathway. The present report documents that the histone demethylase JMJD3 is an activating transcription factor 4 (ATF4)-dependent target gene. The JMJD3 gene contains two AAR-induced promoter activities and chromatin immunoprecipitation (ChIP) analysis showed that the AAR leads to enhanced ATF4 recruitment to the C/EBP-ATF response element (CARE) upstream of Promoter-1. AAR-induced histone modifications across the JMJD3 gene locus occur upon ATF4 binding. Jmjd3 transcription is not induced in Atf4-knock-out cells, but the AAR-dependent activation was rescued by inhibition of histone deacetylation with trichostatin A (TSA). The TSA rescue of AAR activation in the absence of Atf4 also occurred for the Atf3 and C/EBP homology protein (Chop) genes, but not for the asparagine synthetase gene. ChIP analysis of the Jmjd3, Atf3, and Chop genes in Atf4 knock-out cells documented that activation of the AAR in the presence of TSA led to specific changes in acetylation of histone H4. The results suggest that a primary function of ATF4 is to recruit histone acetyltransferase activity to a sub-set of AAR target genes. Thus, absolute binding of ATF4 to these particular genes is not required and no ATF4 interaction with the general transcription machinery is necessary. The data are consistent with the hypothesis that ATF4 functions as a pioneer factor to alter chromatin structure and thus, enhance transcription in a gene-specific manner.
Introduction
Mammalian cells have developed mechanisms for adapting to nutritional stress, including dietary protein or amino acid (AA)2 limitation which triggers a collection of signaling processes that collectively are referred to as the amino acid response (AAR) (1, 2). Limitation of maternal dietary protein during embryogenesis has both short-term effects on fetal growth as well as long-term health effects on the resulting offspring (3–6). It is widely believed that diet-induced epigenetic changes during development causes altered gene expression during the subsequent adulthood period of the affected fetuses. These observations led to the fetal origins of adult disease (FOAD) hypothesis (7).
General control non-derepressible-2 (GCN2) is one of several independent, stress-activated kinases for which a primary substrate is the α subunit of the eukaryotic translation initiation factor eIF2 (8). GCN2 kinase is activated by binding uncharged tRNA, which is increased during AA deprivation. Once phosphorylated, p-eIF2 suppresses general protein synthesis, but paradoxically, increases translation of selected mRNA species and among them, is activating transcription factor 4 (ATF4) (9, 10). ATF4 is a member of the basic-region leucine zipper (bZIP) transcription factor family that activates transcription by homo- or heterodimerizing with other bZIP transcription factors (11). Like its functional counterpart in yeast, GCN4, ATF4 activates a large number of genes that have a wide spectrum of functions (12). ATF4 can bind as a heterodimer to a genomic sequence comprised of a half-site for the C/EBP (CCAAT-enhancer binding protein) family and a half-site for the ATF family. The phrase “C/EBP-ATF composite site” was coined by Wolfgang et al. during a regulatory study of the C/EBP homology protein (CHOP) gene (13). For this same site, Fawcett et al. later showed that in response to arsenite-induced stress ATF4 initially binds to the site as an activator and is then replaced by ATF3, which serves as a repressor (14). Since those initial studies, many C/EBP-ATF composite sequences have been identified, which has led to a consensus of 5′-TGATGXAAX-3′. Given that ATF4 synthesis is increased in response to a wide array stresses ranging from heme deficiency, double stranded RNA, unfolded protein response, hypoxia, and amino acid deprivation (8), the number and function of C/EBP-ATF4 responsive genes is quite large (12). To retain the original nomenclature of “C/EBP-ATF” and to reflect the broad functions of this collection of sequences, we will refer to them collectively as C/EBP-ATF response elements (CARE). Thus, the same CARE site within a particular gene can mediate transcriptional activation in response to many different stimuli. For example, when the activating stress is amino acid deprivation, these CARE sequences function as amino acid response elements (AARE), but the same site can be responsible for activating the gene following ER stress (15).
An expression microarray analysis of the AAR in HepG2 human hepatoma cells revealed increased mRNA abundance for a number of genes that mediate histone modifications (16). Among these was the gene Jumonji domain containing 3 (JMJD3), which specifically demethylates H3K27 (17–19). Di- and tri-methylation of histone H3K27, catalyzed by the mammalian polycomb group protein complex 2 (PRC2), was once considered a highly stable histone mark (20), associated with compacted chromatin, long-term gene repression, and specification/maintenance of cell identity (21, 22). However, it is now clear that dynamic and rapid regulation of JMJD3 activity and thus, H3K27 methylation, plays a critical role in the cellular response to extracellular stimuli (23–27). However, De Santa et al. (28) performed ChIP-sequencing for JMJD3 in lipopolysaccharide (LPS)-activated macrophages and discovered that although JMJD3 was recruited to more than 70% of LPS-inducible genes, only a small portion of these targets were enriched in H3K27me3 at their promoter. Furthermore, Jmjd3 deficiency does not always affect the expression of genes enriched in the H3K27me3 mark, suggesting that there are JMJD3 functions independent of the demethylase activity (28, 29). Consistent with this hypothesis, Miller et al. (30) reported that Jmjd3 can function as a bridging molecule between a transcription factor and a chromatin remodeling complex, independent of demethylase activity. There are only a few reports that address the molecular mechanisms by which the Jmjd3 gene itself is regulated in response to extracellular stimuli. De Santa et al. (24) proposed that in response to LPS treatment transcription from the Jmjd3 gene occurs from two different promoters in a cell-specific manner. While the first transcriptional start site was active in mouse embryonic stem cells (hereafter called Promoter-1), a second transcription start site, located 4.9 kb downstream in a region proximal to the next annotated exon, was the major promoter for LPS-induced Jmjd3 expression in macrophages (hereafter called Promoter-2). Agger et al. (26) demonstrated that Promoter-2 is the major promoter during senescence in fibroblasts and proposed that MEK-ERK signaling may mediate JMJD3 expression through two AP-1 sites about 2 kb upstream of Promoter-2 (26). The translation start site is in exon 4, so in absence of any evidence for alternative splicing, both promoters would generate proteins of identical composition. The mRNA from Promoter 2 would lack exon 1, which codes for 121 bp.
Understanding the regulation of the JMJD3 gene by AA deprivation may provide insight into how protein limitation in vivo leads to long-term epigenetic changes in gene expression. In this report, we have documented the AAR-induced transcriptional activation of the JMJD3 gene in several cell types and tissues. Studies indicated that Promoter-2 functions as the major basal promoter for JMJD3 in both HepG2 hepatoma cells and mouse embryonic fibroblasts (MEF), but Promoter-1, albeit less active than Promoter-2, also exhibits AA-dependent activity. Induction of Jmjd3 expression was abolished in Atf4-deficient MEF cells and chromatin immunoprecipitation (ChIP) analysis of the JMJD3 gene during the AAR demonstrated that ATF4 is recruited to a CARE sequence about 1 kb upstream of Promoter-1. Acetylation of histone H3 at lysines 9 and 14 (H3Ac) and histone H4 at lysines 5, 8, 12, and 16 (H4Ac) near the transcription start site of genes often correlates with active transcription (31). Interestingly, treating Atf4 knock-out cells with a histone deacetylase inhibitor resulted in increased histone H4 acetylation of the Jmjd3 gene and rescue of the AAR-dependent induction. These results indicate that one of the functions of ATF4 may be to alter chromatin structure by increasing histone acetylation.
MATERIALS AND METHODS
Cell Culture and Animal Studies
Human hepatoma cells (HepG2), mouse embryonic fibroblasts (MEF), and human embryonic kidney HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM), pH 7.4 (Mediatech, Herndon, VA) supplemented with 1× non-essential amino acids, 2 mm glutamine, 100 mg/ml streptomycin sulfate, 100 units/ml penicillin G, 0.25 mg/ml amphotericin B, and 10% fetal bovine serum (FBS). The cells were maintained at 37 °C in an atmosphere of 5% CO2 and 95% air. The medium for the Atf4 wild type and knock-out MEF (kindly provided by Dr. Steve Abcouwer, Pennsylvania State University) also contained 0.1% β-mercaptoethanol (12). The cells were cultured to 60–70% confluence so that they were still in a growth phase during the experimental treatment. The freshly isolated primary human hepatocytes (kindly provided by Dr. Chen Liu, Department of Pathology, University of Florida) were plated in collagen-coated 6-well plates at 0.5 × 106 cells per well for 16 h before use. For all cells, at 12 h before initiation of treatment, cells were given fresh medium and serum to ensure that no nutrient deprivation took place before the start of experimental incubations. To activate AAR signaling, cells were incubated in control DMEM or DMEM supplemented with 2 or 5 mm histidinol (HisOH) for the period of time indicated. HisOH blocks charging of histidine onto the corresponding tRNA and thus, mimics histidine deprivation and triggers activation of the AAR (32). To activate the hepatic AAR in vivo, mice were injected with asparaginase (3 IU/g body weight) which depletes cellular asparagine and glutamine, as described by Bunpo et al. (33) or fed a low protein diet for 10 days (34) prior to isolating liver RNA for mRNA analysis.
RNA Isolation and Real-time Quantitative PCR
Total RNA was isolated with the TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. A 2-μg aliquot of total RNA was used to synthesize first-strand cDNA with the SuperScript III First-Strand Synthesis Kit (Invitrogen). For real-time quantitative PCR (qPCR), synthesized cDNA was diluted 10× with TE buffer (10 mm Tris, 1 mm EDTA, pH 8.0) and then 2 μl was mixed with 10 μl of SYBR Green master mixture (Applied Biosystems, Warrington, UK) and 5 pmol of forward and reverse primers in a total volume of 20 μl. The mixture was subjected to 40 cycles at 95 °C for 30 s, 60 °C for 30 s, and 68 °C for 60 s and qPCR was performed with a DNA Engine Opticon 3 system (Bio-Rad). The primers used are listed in supplemental Table S1. After qPCR, melting curves were acquired by a stepwise increase of the temperature from 55 to 95 °C to ensure that a single product was amplified in the reaction. Either glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or ribosomal protein L7a mRNA content was used as an internal control. All calculations were based on the difference of Ct (the threshold cycle) of the analyzed gene relative to the internal control mRNA content in the same sample.
Immunoblotting
For the nuclear extract, cells were resuspended in hypotonic lysis buffer (10 mm Hepes, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 1 mm dithiothreitol) containing 1× protease inhibitor mixture (Roche Diagnostics, Mannheim, Germany) and incubated on ice for 15 min. To the swollen cells, 10% (W/V) Nonidet P-40 was added to a final concentration of 0.3%, vortexed vigorously for 10 s, and then centrifuged for 5 min at 2500 × g at 4 °C. After discarding the supernatant, the pellet was washed two more times with the same solution. The nuclear pellet was resuspended in 100 μl high salt nuclear extraction buffer (10 mm Hepes, pH 7.9, 1.5 mm MgCl2, 0.2 mm EDTA, 450 mm NaCl, 1 mm dithiothreitol) containing 1× protease inhibitor mixture. After vortexing vigorously for 30 s, the pellets were collected at 20,000 × g for 30 min at 4 °C. An aliquot of 100 μg nuclear extract was loaded per lane on an 8% Tris-HCl polyacrylamide gel (Bio-Rad) and after separation, electrotransferred to a Trans-Blot nitrocellulose membrane (0.2 μm) (Bio-Rad). The membrane was stained with Fast Green to check for equal loading and then incubated with blocking solution consisting of 5% (w/v) Carnation nonfat dry milk in TBST (30 mm Tris-base pH 7.5, 200 mm NaCl, and 0.1% (v/v) Tween-20) for 1 h at room temperature with rotating. Mouse anti-JMJD3 monoclonal antibody (kindly provided by Dr. Gioacchino Natoli at the European Institute of Oncology, Milan, Italy) was used at a concentration of 0.5 μg/ml in blocking solution and incubated overnight at 4 °C. The blots were washed 5 × 5 min in TBST and then incubated with the appropriate peroxidase-conjugated secondary antibody (Kirkegaard & Perry Laboratories, Gaithersburg, MD) for 1 h at room temperature. The blots were then washed for 5 × 5 min in TBST. The bound secondary antibody was detected using an enhanced chemiluminescence kit (Pierce) and exposing the blot to Biomax MR film (Kodak, Rochester, NY).
Chromatin Immunoprecipitation (ChIP)
HepG2 cells were seeded at 1.5 × 107 per 150 mm dish and ChIP analysis was performed according to our previously published protocol (35). For ChIP assay with MEF cells, the protocol was essentially the same as with HepG2 cells, except that the cells were seeded at 1.5 × 106 per 150 mm dish, 3 dishes were used for each treatment. The ATF4 rabbit polyclonal antibody was described previously (36). Commercial antibodies used were as follows: rabbit anti-total RNA Pol II polyclonal antibody (sc-899); rabbit anti-ATF3 polyclonal antibody (sc-188), rabbit anti-C/EBPβ polyclonal antibody (sc-150), and normal rabbit IgG (sc-2027) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antibodies against anti-total histone H3 polyclonal antibody (ab1791) and mouse anti-histone H3 lysine 4 monomethyl (ab8895) were purchased from Abcam. Rabbit anti-histone H3 lysine 4 trimethyl (04–745), and rabbit anti-histone H4 acetylation (06–866) polyclonal antibodies were purchased from Upstate Millipore. DNA enrichment was analyzed with qPCR and the reaction mixtures were incubated at 95 °C for 15 min, followed by amplification at 95 °C for 15 s and 60 °C for 60 s for 35 cycles. All experiments were performed in triplicate and were repeated to ensure reproducibility. The results are presented as the ratio to input DNA, and the primers used for analysis are listed in supplemental Table S1.
RESULTS
AAR-induced mRNA Expression for JMJD3 in Cells and Tissues
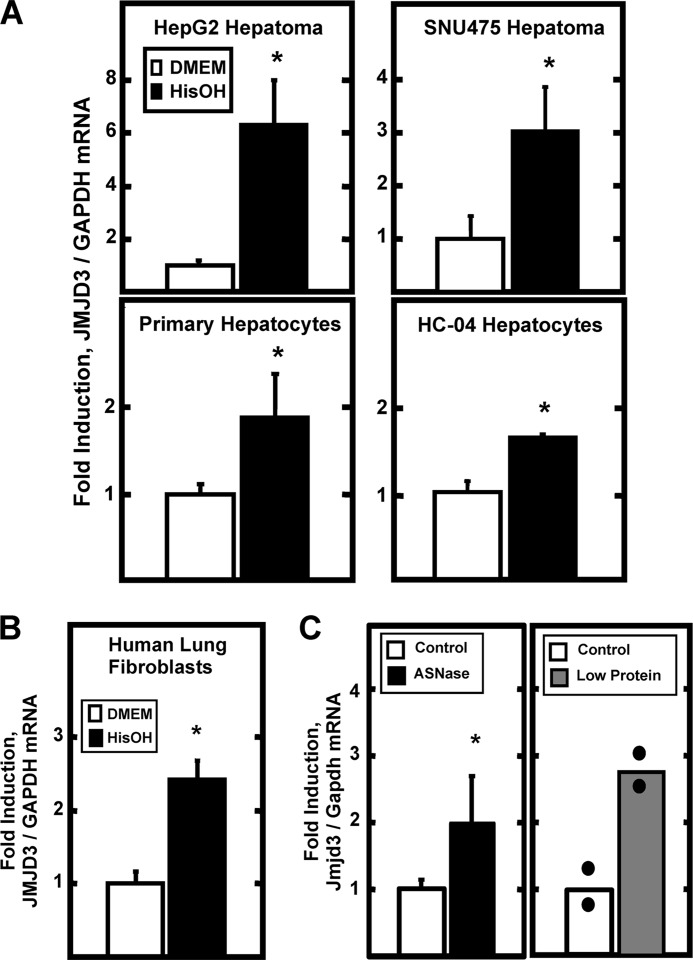
Expression profiling of HepG2 human hepatoma cells indicated that activation of the AAR caused increased expression of numerous enzymes that function as histone modification “writers” and “erasers” (supplemental Table S2) (16). Among those genes, a high level of induction was observed for the JMJD3 gene, a protein with H3K27 demethylase activity (18). To extend the microarray observations, the AAR was triggered by HisOH treatment of several human liver-derived cell lines and the JMJD3 mRNA content was measured (Fig. 1A). The AAR increased JMJD3 expression by 3–6-fold in HepG2 and SNU475 human hepatoma cells. Although the magnitude of the induction was less, a reproducibly significant increase was also observed in an immortalized human hepatocyte cell line (HC-04) and in freshly isolated primary cultures of human hepatocytes. To determine if the induction was tissue specific, human lung fibroblasts were tested and shown to exhibit increased JMJD3 expression as well (Fig. 1B). To establish whether or not the JMJD3 gene is responsive to AA deprivation in vivo, mice were treated with the drug asparaginase, which depletes circulating asparagine and glutamine levels and activates the AAR (33). Analysis of mouse liver tissue showed that asparaginase treatment resulted in about a 2-fold increase in Jmjd3 mRNA (Fig. 1C), similar to the degree observed in the non-transformed hepatocytes in culture. The asparaginase treatment result was independently confirmed by determining that hepatic Jmjd3 mRNA was observed to be about 3-fold greater in the mice fed a low protein diet (8% protein) compared with a control diet (19% protein) for 10 days (Fig. 1C).
FIGURE 1.
AAR-induced expression of JMJD3 mRNA in cells and tissue. Panel A, JMJD3 mRNA content was measured in human hepatoma cell lines (HepG2 or SNU475), primary cultures of freshly isolated human hepatocytes, or in a human non-transformed hepatocyte cell line (HC-04). The AAR was triggered by incubating the cells in DMEM ± HisOH for 8 h prior to isolation of RNA and analysis by qPCR. The data are represented as the averages ± S.D. for at least three samples and are expressed relative to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA as an internal control. Panel B, human lung fibroblasts in culture were incubated in DMEM ± HisOH for 8 h prior to analysis of JMJD3 and GAPDH mRNA by qPCR. The data are represented as the averages ± S.D. for at least three samples. The asterisks denote a significant difference of p ≤ 0.05 relative to the DMEM control. Panel C, as described in “Materials and Methods,” mice were either injected I.V. with asparaginase (ASNase) (left panel) or fed a low protein diet (right panel) to induce the AAR in vivo. Liver RNA was analyzed for Jmjd3 and Gapdh mRNA by qPCR. For the ASNase study, the data are the averages ± S.D. for at least three animals. For the low protein diet study, the bars represent the average of two animals and the actual value of each animal is shown as block dots. For all panels, the asterisks denote a significant difference of p ≤ 0.05 relative to the control.
The AAR Induces Transcription from the JMJD3 Gene
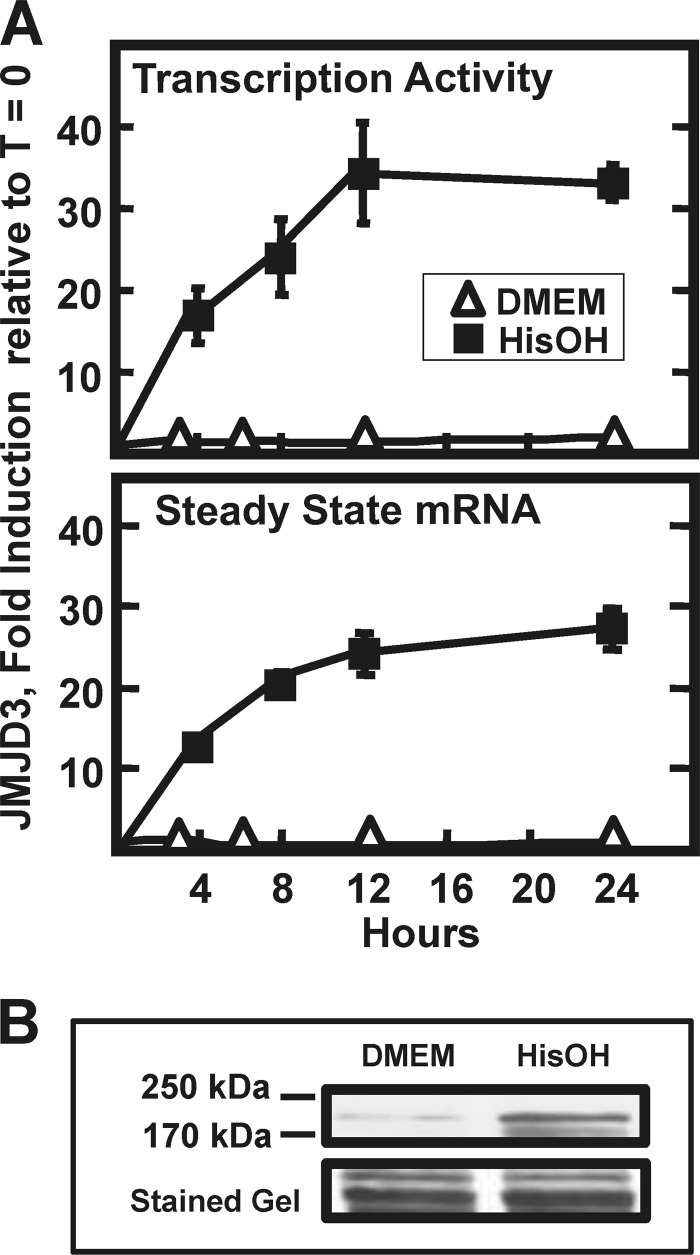
To determine the mechanism by which the AAR increased JMJD3 mRNA levels, HepG2 cells were treated with HisOH for 0–24 h and both transcription activity and steady state mRNA content were measured (Fig. 2A). The transcription activity increased rapidly during the first 4 h and then reached a plateau by 12 h and the steady state JMJD3 mRNA abundance was similar in profile. Immunoblot analysis illustrated that the JMJD3 protein content was also increased (Fig. 2B). There are no published reports describing the turnover rate for the JMJD3 mRNA. To investigate whether or not mRNA stabilization contributed to the elevation in JMJD3 mRNA levels, HepG2 cells were incubated in HisOH to activate the AAR and then transferred to control (DMEM only) or HisOH-containing media, both supplemented with actinomycin D to prevent further RNA synthesis. The results showed that in the basal state the JMJD3 mRNA half-life is ∼7 h, whereas after activation of the AAR, the half-life was increased beyond the limits of actinomycin treatment (data not shown). These results suggest that both increased transcription and mRNA stabilization contribute to increased JMJD3 expression.
FIGURE 2.
Increased transcription contributes to the induction of JMJD3 expression. Panel A, in HepG2 hepatoma cells, the time course of transcription activity and steady state mRNA content for JMJD3 was measured by qPCR, as described in “Materials and Methods.” The cells were incubated in DMEM ± HisOH for the time indicated and the data are represented as the averages ± S.D. for at least three samples. Panel B, HepG2 protein content for JMJD3 was measured by immunoblotting nuclear extract samples collected 24 h after activating the AAR as described for panel A. To illustrate equal loading, the appropriate section of the blot stained with Fast Green is shown.
Activation of the JMJD3 Transcription Is ATF4-dependent
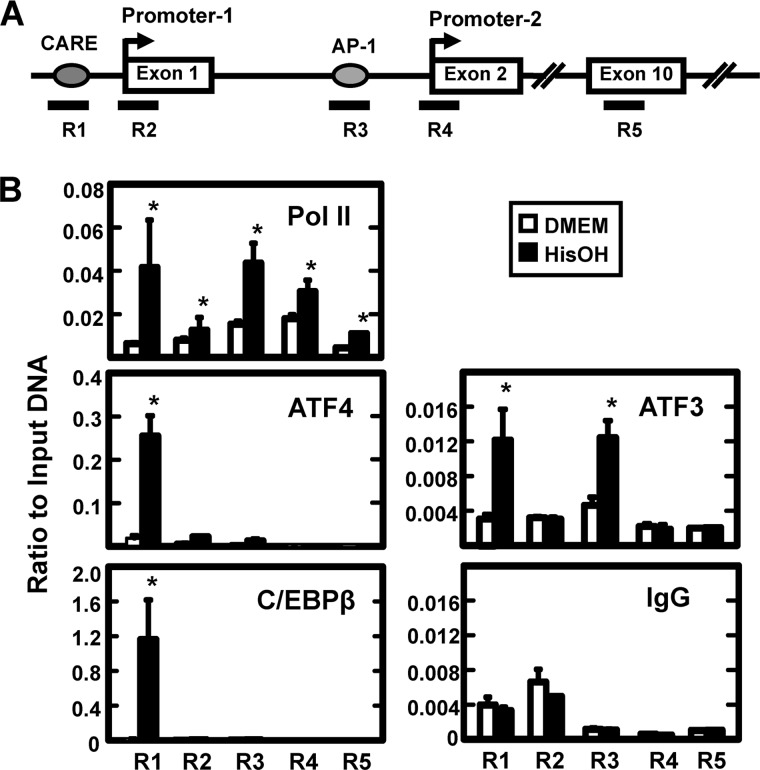
To determine if the transcriptional induction of the JMJD3 gene was dependent on ATF4, Atf4 wild type and deficient MEF cells were subjected to HisOH treatment (supplemental Fig. S1). As a positive control, Asns mRNA expression was monitored because the AAR induction of this gene is known to be highly dependent on ATF4 (37). Incubation of the wild type fibroblasts in 5 mm HisOH for 8 h enhanced Jmjd3 mRNA expression by 3–4-fold, whereas in the Atf4-deficient cells, the basal level was slightly reduced relative to the wild type cells and the AAR induction was completely blocked. As described above, AA deprivation leads to increased synthesis of ATF4, which then activates AA-responsive genes by binding to CARE sequences (5′-TGATGXAAX-3′). By computer analysis, a consensus CARE sequence was located ∼1 kb upstream from Promoter-1 (Fig. 3A). To analyze transcription factor binding, we used primers specific for the JMJD3 genomic regions containing: the CARE sequence (R1), Promoter-1 (R2), two closely localized AP-1 sequences (R3), Promoter-2 (R4), and within Exon 10 (R5). The analysis of binding within the body of the gene (exon 10) was used as a negative control. Consistent with AA-regulated transcription, there was increased association of RNA polymerase II (Pol II) observed at both promoters and within the body of the gene after activation of the AAR. Consistent with recent reports showing association of RNA Pol II with regulatory enhancers (38), the data also revealed a significantly increased recruitment of Pol II to the CARE and AP-1 regions (Fig. 3B). ATF4 binding at the CARE region in the control cells (DMEM alone) was above the “background,” taken to be the value at exon 10 with the ATF4 antibody or the value obtained with the CARE region primers after enrichment with the nonspecific IgG (Fig. 3B). Consistent with the hypothesis that the JMJD3 CARE sequence functions as an AARE, the ATF4 association was enhanced by more than 10-fold after HisOH treatment. Although primers specific for the Promoter-1 and AP-1 regions exhibited some ATF4 binding, the levels were closer to the background value for exon 10 and substantially below that for the CARE region. A program of self-limitation for ATF4 has been described whereby ATF4-mediated transcriptional activation of the C/EBPβ and ATF3 genes leads to recruitment of the corresponding proteins to the AARE where they serve as suppressors to feedback inhibit the ATF4 signal (35, 39, 40). Thus, the association of ATF3 and C/EBPβ with the JMJD3 ATF4-binding CARE site would be expected, if it is a functional AARE. ChIP analysis of the JMJD3 gene for AAR-induced C/EBPβ recruitment revealed association with the CARE region, but minimal binding to all other locations tested (Fig. 3B). For ATF3, AAR-increased association was observed not only for the CARE region, but also for the AP-1 region upstream of Promoter-2. The two sites present in this region are classic palindromic AP1 sequences (5′-TGAC/GTCA-3′). Dimers of cJUN and ATF3 have been documented to bind to AP-1 sites (41, L. Fu and M.S. Kilberg, unpublished results), but when Hsu et al. (42) transiently co-expressed ATF3 and cJUN along with AP1-driven luciferase reporter constructs, the ATF3-cJUN combination had only a weak activating capability and ATF3-JUN-B heterodimers caused inhibition. The binding partner(s) and function of ATF3 at these JMJD3 AP1 sites will require further investigation.
FIGURE 3.
The enrichment of binding for transcription factors at the JMJD3 gene as detected by ChIP. Panel A, drawing depicts the gene structure of JMJD3 and the location of five primer sets used to analyze specific gene regions (R1-R5). Primer sequences are listed in supplemental Table 1. Exon numbers are designated within the boxes. Panel B, HepG2 cells were incubated in DMEM ± HisOH for 6 h prior to ChIP analysis for the enrichment of RNA Pol II, ATF4, ATF3, and C/EBPβ. A nonspecific IgG was used as one negative control and the binding at Exon 10 (primer set R5) was taken as “background” for each antibody. The data are represented as the averages ± S.D. for at least three samples, and the asterisks denote a significant difference of p ≤ 0.05 relative to the DMEM control.
Activation of JMJD3 Transcription Is MEK-dependent
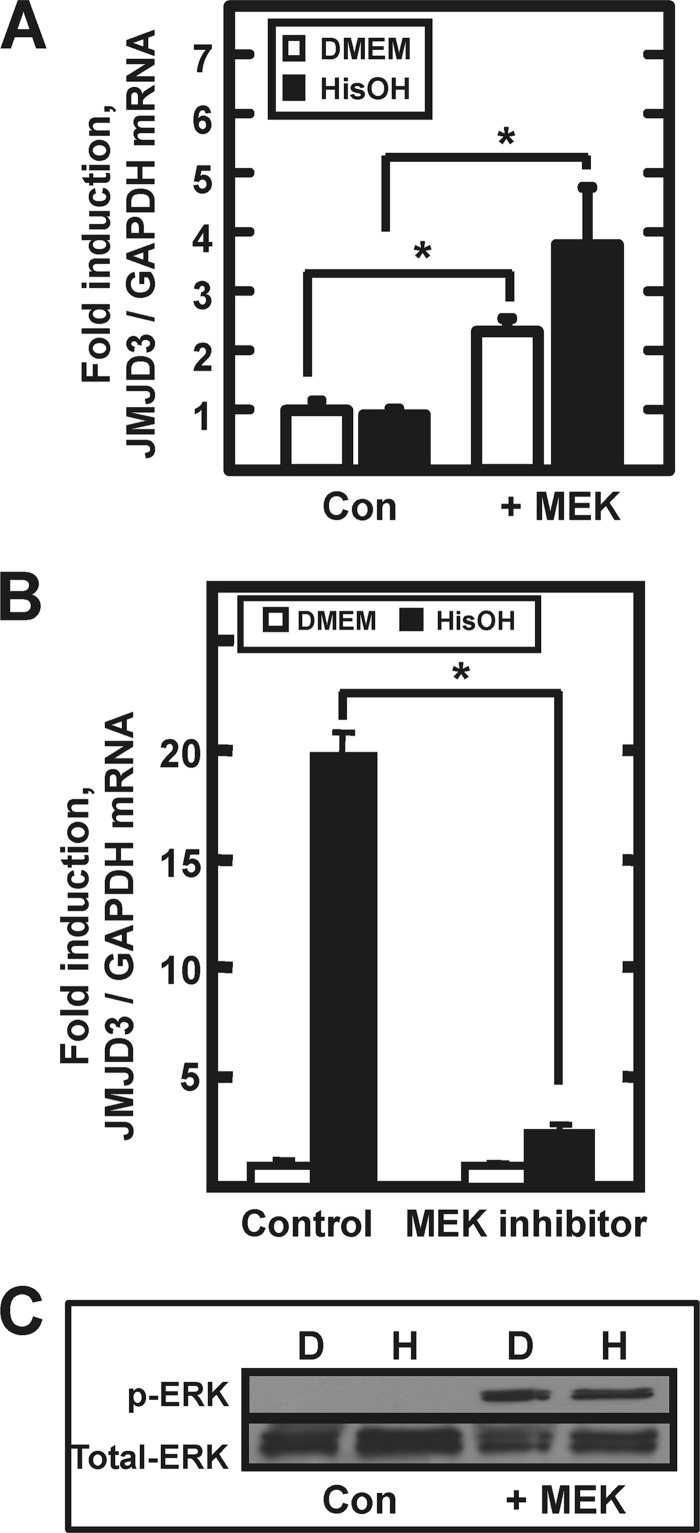
One of the cell lines that was surveyed for induction of JMJD3 expression was human embryonic kidney 293 cells (HEK293), but in repeated experiments, little or no AAR-induced increase in JMJD3 mRNA was detected (Fig. 4A, Con). We have documented that in HepG2 hepatoma cells increased ATF4 synthesis after activation of the AAR is dependent on MEK-ERK signaling (43) and accordingly, the induction of JMJD3 mRNA expression in HepG2 cells is blocked by inhibition of MEK activity (Fig. 4B). However, the HEK293 cells do not exhibit MEK-dependent ERK phosphorylation (Fig. 4C). To test for a role of MEK-ERK in JMJD3 expression, HEK293 cells were transiently transfected with a construct encoding a constitutively active MEK (44) and to confirm MEK expression, phosphorylated ERK was measured by immunoblot (Fig. 4C). Consistent with the hypothesis that ERK activity is necessary for ATF4 production, qPCR analysis showed that basal (DMEM medium alone) JMJD3 mRNA levels were increased significantly by the exogenous MEK activity and the treatment with HisOH resulted in significant induction (Fig. 4A, +MEK).
FIGURE 4.
Induction of JMJD3 mRNA requires MEK-ERK signaling. Panel A, HEK293T cells were transfected with a control vector (Con) or a vector encoding a constitutively active MEK (+MEK) construct. After 36 h post-transfection, the cells were incubated in DMEM ± HisOH for 8 h prior to analysis of JMJD3 and GAPDH mRNA by qPCR. The asterisks denote a significant difference of p ≤ 0.05. Panel B, HepG2 cells ere incubated for 8 h with or without HisOH in the absence (Control) or presence (MEK inhibitor) of 50 μm PD98059. Panel C, for the HEK293 cells described in panel A, analysis of total and p-ERK by immunoblotting was used as a control to demonstrate MEK signaling after transfection.
Histone Marks on the JMJD3 Gene
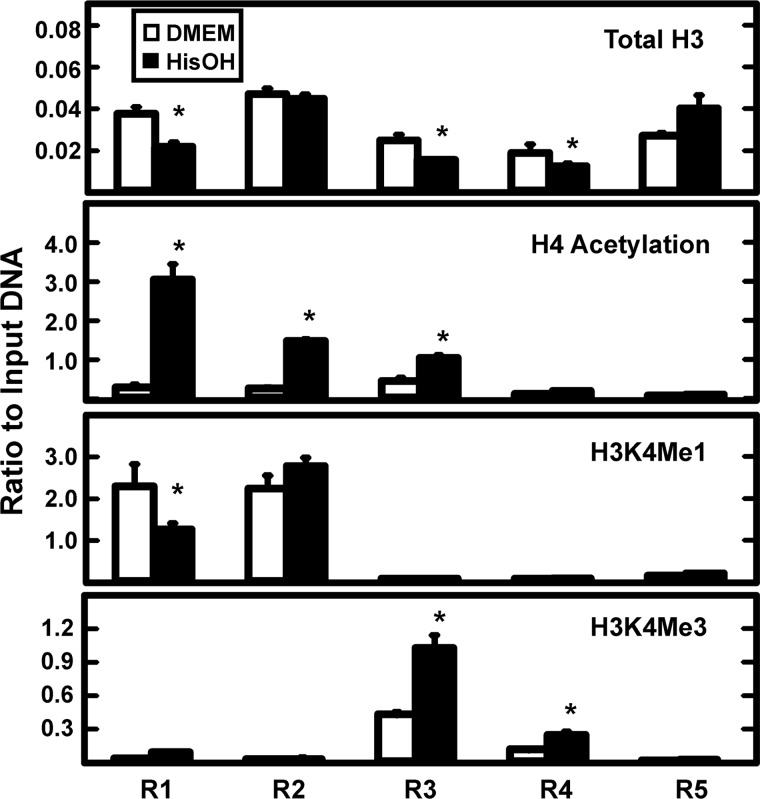
To further investigate the mechanisms by which the AAR regulates transcriptional activation of the JMJD3 gene, histone modifications at specific locations within the gene locus (see Fig. 3A) were monitored after HisOH treatment (Fig. 5). The loss of total histone H3 protein can be taken as indirect evidence for nucleosome remodeling (45, 46) and therefore, H3 content was analyzed by ChIP analysis. The results showed that activation of the AAR caused a decrease in the H3 content at the CARE (R1), AP-1 (R3), and Promoter-2 (R4) regions. There was a high level of H3 near Promoter-1 (R2), but no change after HisOH treatment and the H3 association was actually increased at exon 10 (R5). Histone H4 acetylation near the transcription start site of genes often correlates with active transcription (31, 47). Following AAR activation, this histone mark was significantly increased at the CARE, Promoter-1, and AP-1 regions, but to a negligible extent at Promoter-2 and in the body of the gene (Fig. 5). Histone H3 lysine 4 mono-methylation (H3K4me1), a mark often associated with active enhancer regions, was only present to a large extent at the CARE and Promoter-1 sites and HisOH treatment caused a 50% decline at the CARE site with no change at Promoter-1. The H3K4me3 mark is associated with active promoters (47, 48). There was a minimal amount of H3K4me3 at either the CARE or Promoter-1 regions (Fig. 5). In contrast, the level of H3K4Me3 at the AP-1 and Promoter-2 regions was much greater and the amount was clearly induced after activation of the AAR.
FIGURE 5.
The AAR-induced change in histone modifications across the JMJD3 gene locus. The five primer sets (R1-R5), shown in Fig. 3A, were used to scan the JMJD3 gene for changes in histone modifications during the AAR. HepG2 cells were incubated in DMEM ± HisOH for 6 h prior to ChIP analysis for the indicated histone form. The data are represented as the averages ± S.D. for at least three samples. The asterisks denote a significant difference of p ≤ 0.05 relative to the DMEM control.
Increased Acetylation Can Rescue JMJD3 Gene Activation in Atf4-deficient Cells
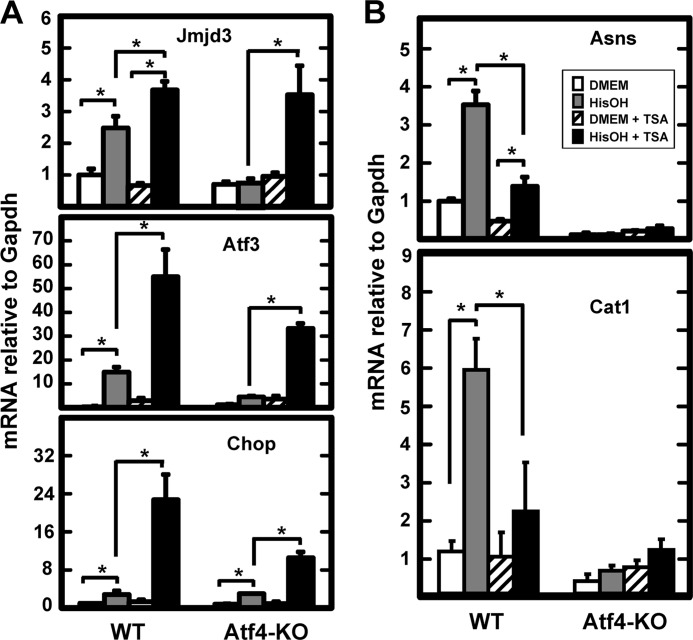
Within 30 min after activation of the AAR, there is recruitment of ATF4 to the CARE sites of AA-responsive genes, which is associated with increased acetylation of histones H3 and/or H4 (35, 39, 49). For the CHOP (49) and ATF3 (50) genes, the histone acetyltransferase (HAT) has been identified, but for all other cases the acetyltransferase identity is unknown. Given the strong increase in H4 acetylation at the CARE region of the JMJD3 gene (Fig. 5), the association between increased acetylation and ATF4 action on the JMJD3 gene was investigated. Cycling of histone acetylation-deacetylation has been linked to gene regulation with hyper-acetylated histones considered a hallmark of active transcription. Histone deacetylase (HDAC) inhibitors block the cycling and therefore, cause hyper-acetylation across the genome (51). To determine the relationship between ATF4 action and increased histone acetylation for the JMJD3 gene, wild type and Atf4 knock-out MEF cells were incubated in DMEM ± HisOH, with or without trichostatin A (TSA), a HDAC inhibitor. The effectiveness of the TSA was demonstrated by immunoblotting for acetylated H4 (supplemental Fig. S2). In the wild type MEF cells, TSA treatment alone had a slightly inhibitory effect on the expression of Jmjd3 mRNA (Fig. 6A). Activation of the AAR by HisOH increased Jmjd3 mRNA content as expected and the presence of TSA further enhanced the induction by about 30%. Consistent with the results shown in supplemental Fig. S1, there was no induction of Jmjd3 mRNA in the HisOH-treated Atf4-deficient cells. Surprisingly, when the MEF cells lacking Atf4 were treated with the combination of HisOH and TSA, the induction of Jmjd3 expression was equal to that in the wild type cells (Fig. 6A). The data suggest that in the absence of Atf4, induction of the Jmjd3 gene can be rescued by hyper-acetylation of histones. These results lead to the hypothesis that one of the functions of Atf4 is to increase the accessibility of the chromatin region containing the Jmjd3 gene through recruitment of a histone acetyltransferase (HAT) activity. To determine if this function applies to all Atf4-dependent genes, a number of other AA-responsive genes were tested for the effect of TSA in the Atf4 knock-out MEF cells. Qualitatively, the Atf3 and Chop genes showed the same response to TSA treatment as the Jmjd3 gene (Fig. 6A). However, in contrast, for induction of the Asns and Cat1 genes, which are known to be highly dependent on Atf4 for AAR activation (37, 40), TSA treatment significantly inhibited the induction in the wild type MEF cells and did not lead to increased expression in the Atf4-deficient cells (Fig. 6B). To ensure that the effect of TSA on mRNA levels of the Jmjd3, Atf3, and Chop genes was transcriptional in nature, a time course of induction in Atf4 knock-out MEF cells was performed (supplemental Fig. S3). The results show that TSA treatment alone does not enhance transcription activity, but the combination of TSA+HisOH induces transcription from all three genes in the absence of Atf4. Collectively, these results reveal the observation that the absence of Atf4 can be rescued by histone hyperacetylation, presumably leading to a change in chromatin conformation that is typically triggered by Atf4 binding. Conversely, the data for the Asns and Cat1 genes also document that this function of ATF4 is not universal for all AA-responsive genes suggesting that there are significant differences in the genomic mechanisms by which ATF4 acts on AAR target genes.
FIGURE 6.
For selected genes, loss of AAR-induced expression by Atf4 deficiency is rescued by TSA treatment. Wild type (WT) and Atf4-deficient (Atf4-KO) mouse embryonic fibroblasts were incubated in medium containing the HDAC inhibitor TSA for 24 h. During the last 8 h, the cells were incubated in DMEM ± HisOH as indicated. Total RNA was isolated and analyzed by qPCR for the mRNA content of the indicated gene relative to the GAPDH internal control. For each gene, all data were normalized to the DMEM control value in the wild type cells and are shown as the averages ± S.D. for at least three samples. The asterisks denote a significant difference of p ≤ 0.05. Panel A illustrates the data fro the TSA-responsive genes, whereas panel B shows the results for two genes that were not rescued by TSA treatment.
TSA Effect on Histone Acetylation Across the Jmjd3 Gene
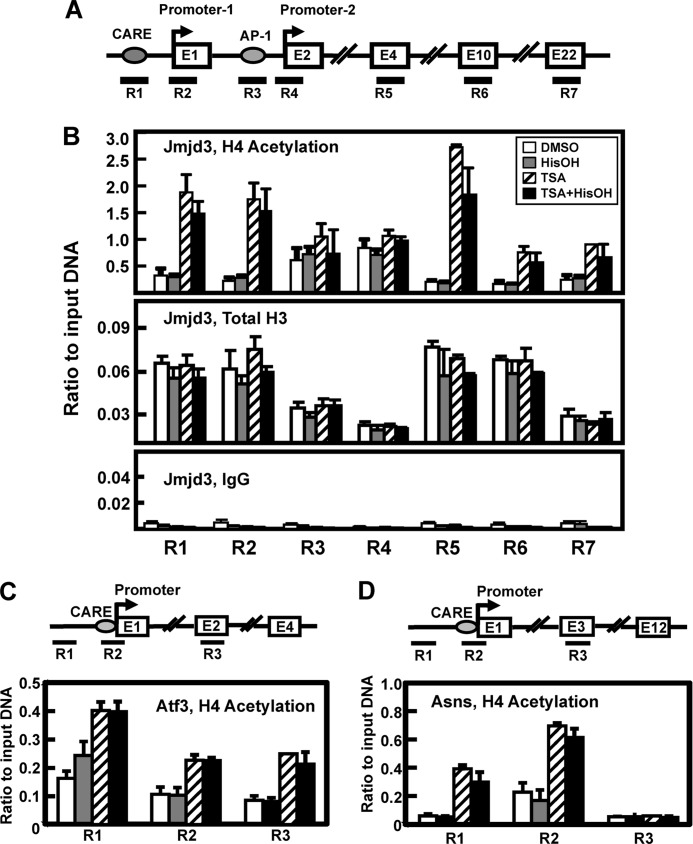
To determine the effect of TSA treatment on H4 acetylation within the Jmjd3 gene in the absence of Atf4, ChIP analysis was performed to analyze changes in histone association in Atf4 knock-out MEF cells using primer sets that spanned the Jmjd3 locus from the CARE to exon 22 (Fig. 7A). The results show that the Atf4-deficient cells exhibit no change in total H3 or H4Ac in response to HisOH treatment (Fig. 7B). This result is consistent with the lack of transcriptional induction of Jmjd3 in the absence of Atf4 (supplemental Fig. S1). In contrast, when the cells were treated with TSA alone or TSA+HisOH, H4Ac increased in abundance at the CARE (R1), Promoter-1 (R2), and the coding regions (R5-R7) of the Jmjd3 gene (Fig. 7B). Interestingly, whereas total H3 association was depleted at the AP-1 (R3) and Promoter-2 (R4) regions relative to the other Jmjd3 regions tested, histone H4 acetylation was present but unchanged at these two areas of the gene. TSA-driven transcription from the Atf3 gene responded similarly to Jmjd3 (Fig. 6A), and the TSA effect on H4Ac at this locus was also similar in that, TSA±HisOH caused hyper-acetylation at a region about 0.5 kb upstream of the promoter (R1), the CARE/promoter region (R2), and within the coding region (R3) (Fig. 7C). On the other hand, transcription from the Asns gene could not be rescued by TSA in the Atf4 knock-out MEF cells (Fig. 7B) and yet, TSA±HisOH caused an increase in H4Ac association at a site 0.5 kb upstream and at the CARE/promoter region of Asns (Fig. 7D). One feature that distinguished the Asns gene from Jmjd3 and Atf3 was that no increase in TSA-mediated H4Ac occurred within the coding region of the gene. When H4 acetylation was tested at Asns exons 2, 3, 7, and 12, there was no increase in response to TSA ± HisOH. The data for exon 3 are shown for an example (Fig. 7D). These results indicate that, like transcription activity, differences exist in the TSA-induced changes in histone acetylation within individual AA-responsive genes.
FIGURE 7.
TSA treatment of Atf4-deficient cells results in an increase in H4 acetylation at specific regions within the AAR target genes. Panel A, drawing depicts the gene structure of JMJD3 and the location of seven primer sets used to analyze specific gene regions (R1-R7). Primer sequences are listed in supplemental Table S1. Exon numbers (E) are designated within the boxes. Panel B, MEF cells lacking Atf4 were treated with DMSO or TSA ± HisOH for 1 h and then analyzed by ChIP for H4 acetylation and total H3 protein association on the Jmjd3 gene. Panels C and D, ChIP samples used to obtain the data shown in panel B were also analyzed for three regions (R1-R3) on the Asns (panel C)and Atf3 (panel D) genes. For both genes, region R1 is located about 0.5 kb upstream of the transcription start site, region R2 includes the CARE (nt −68 for Asns and nt −23 for Atf3) and the proximal promoter, and region R3 is located downstream within the exon (E) indicated. The data are represented as the averages ± S.D. for at least three samples, and the asterisks denote a significant difference of p ≤ 0.05 relative to the DMEM control.
DISCUSSION
The present report documents the AA-dependent regulation of the enzyme JMJD3 that can function as an epigenetic “eraser” through its histone H3K27 demethylase activity. The data in this report provide the following novel insights. 1) Although a number of AA-responsive genes have been identified, JMJD3 is the first histone-modifying enzyme documented to be transcriptionally regulated by AA deprivation and thereby, providing evidence for epigenetic change in response to nutrient availability. 2) ATF4 protein is recruited to a CARE sequence upstream of Promoter-1 in an AAR-dependent manner and activation of JMJD3 transcription does not occur in Atf4-deficient MEF cells. 3) ATF3 is not only recruited to the CARE sequence, consistent with published data for other AA-responsive genes (35, 39, 40), but ATF3 also binds in an AAR-dependent manner to the AP-1 containing region upstream of the JMJD3 Promoter-2. 4) Also shown is evidence that for the JMJD3 gene at least one of the functions of ATF4 is to act as a pioneer factor to alter the chromatin structure, as evidenced by the fact that AAR-induced transcription in Atf4-deficient cells can be rescued by inhibition of histone deacetylation. This function of ATF4 also applies to the regulation of the ATF3, and CHOP genes.
The range of AAR and ATF4 target genes is broad as documented by several independent microarray studies (12, 16, 52–54). After activation of the AAR in human HepG2 cells, microarray analysis revealed the induction of a number of genes associated with histone modification and chromatin remodeling, one of which was the H3K27me3 demethylase JMJD3 (supplemental Table S2) (16). Given that H3K27 demethylation is associated with “opening” of chromatin structure and activation of gene expression (51), understanding the regulation of the JMJD3 gene by AA deprivation is likely to provide insight into how limiting dietary protein leads to long-term epigenetic changes. Consistent with this proposal, it is known that dietary protein deprivation during embryo development not only has immediate effects on gene expression, but also has long-term effects through epigenetic mechanisms (3–6). A common theme of such studies is that epigenetic changes resulting from a poor maternal protein diet results in altered expression of key transcription factors in the offspring. These changes can ultimately cause significant alterations in metabolism and other cellular functions, consistent with the fetal origins of adult disease (FOAD) hypothesis (7). Methylation of H3K27 is generally associated with repression of transcription, in particular, for genes involved in differentiation of specific cell types (20). However, it has become clear that the methylation status of H3K27 is complex and is rapidly altered during short-term gene regulation. For example, Agger et al. (26) have demonstrated that increased transcription from the JMJD3 gene occurs within 2 h of activating the RAS-RAF-MEK-ERK pathway, possibly mediated through two AP-1 sequences about 2 kb upstream of Promoter 2. We have demonstrated that in HepG2 cells AA deprivation leads to activation of the MEK-ERK pathway (43) and that AP-1 sequences can function as AARE (55). In this report, we document that in HepG2 cells the AAR-dependent induction of JMJD3 expression is also dependent MEK-ERK signaling. Furthermore, in HEK293 cells that lack endogenous MEK activity and do not exhibit induction of JMJD3 transcription, transiently expressing exogenous MEK activity in the cells results in AAR-mediated induction of the JMJD3 gene, presumably by promoting ATF4 synthesis.
The majority of the genes induced by AA deprivation are dependent on the rapid translational increase in ATF4 protein, which then binds to CARE sites that act as enhancer sequences within the activated genes (2). The actual function of ATF4 within the chromatin has not been described, but we have reported previously that along with ATF4 binding there are concurrent changes in histone modifications and increased recruitment of the general transcription machinery (35, 56). Characterization of the JMJD3 gene as an AAR target has led to new and valuable insight by revealing that a primary function of ATF4 for this and other genes may be to act as a pioneer factor that leads to increased histone acetylation at many regions across a gene locus. Remarkably, in the absence of ATF4 protein, increasing that acetylation by HDAC inhibition with TSA treatment is sufficient to rescue the AAR-dependent activation of the Jmjd3, Atf3, and Chop genes. In this instance, the function of ATF4 most likely is to recruit/activate a HAT or expel/inactivate a HDAC. While the data reveal significant changes in histone modification that correlate well with the presence of ATF4 or TSA+HisOH treatment, there is a possibility that the target protein(s) rescued by the TSA treatment may be a non-chromatin-associated protein, perhaps a signaling protein or a transcription factor, that upon acetylation then leads to increased histone acetylation. Regardless of these possible upstream steps, it is clear that ultimately ATF4 causes altered chromatin structure and thus, can be added to the list of proteins that act as pioneer transcription factors for which the function is to trigger an open chromatin state. Other known examples of such pioneer factors include FOXA1 (57) and C/EBPβ (58). In contrast, despite the fact that AAR-induced transcription from the Asns and Cat1 genes are largely dependent on Atf4, TSA treatment inhibited their expression in wild type MEF cells and had no positive effect in the Atf4-deficient cells. These results clearly demonstrate that for some AAR-inducible genes ATF4 has required functions other than, or in addition to, its action leading to increased histone acetylation.
Based on the data collected to date, the ability of TSA to rescue transcription in Atf4-deficient cells appears to correlate with a change in H4 acetylation associated with the target gene, both upstream of the transcription start site and within the body of the gene. Following activation of the AAR, increased acetylation of both H3 and H4, in parallel with a rapid increase in ATF4 binding, has been observed at the ASNS and ATF3 proximal promoters (35, 39). Carraro et al. observed that AAR-induction led to an increase in H2B, H3, and H4 acetylation levels at the TRB3 CARE site (59). The sodium-dependent neutral amino acid transporter 2 (SNAT2) gene has an intronic CARE enhancer (60). For the SNAT2 gene, AA deprivation induces H3 acetylation at both the promoter and the downstream CARE, but there is no change in H4 acetylation at either location (61). The lack of a change in H4Ac for SNAT2 is interesting because TSA+HisOH treatment of Atf4 knock-out MEFs caused no induction of SNAT2 transcription (data not shown). Histone acetyltransferase activity has been found be associated with elongating RNA polymerase II complexes (62), perhaps a necessary step to allow the transcriptional machinery to overcome the impediment imposed by compacted chromatin within the body of the gene. Other than the elongating transcriptional machinery, histone acetylation in the body of the gene is also promoted by nucleosome bound proteins, such as a high mobility group nucleosome-binding (HMGN) family members. For example, HMGN3a/b stimulates H3K14 acetylation within the Glyt1 gene which leads to increased transcription (63). Thus, enrichment of H4 acetylation within the body of the Jmjd3 and Atf3 genes, but not for Asns may contribute to the expression difference in response to the TSA+HisOH. One possibility is that the coding regions of these two classes of genes are associated with different TSA-responsive HDAC activities. Further study will be necessary to definitively identify these mechanisms.
Bruhat et al. observed an AAR-induced increase in H4 and H2B acetylation at the CHOP promoter, but no change in H3 acetylation (49). Additionally, those authors showed that p-ATF2, known to encode HAT activity (64), mediates the AAR-induced histone acetylation at the CHOP and ATF3 promoters, but not at the ASNS promoter. Consequently, p-ATF2 could be considered as a possible mediator of the TSA+HisOH induction in the absence of ATF4, but data for the Chop and Atf3 genes from the Fafournoux laboratory indicates that p-Atf2 cannot activate transcription in the absence of Atf4 (49, 50). Using knock-out MEF cells, they showed that loss of Atf4 largely blocks AAR induction of transcription from these genes, but does not prevent the increase in p-Atf2 abundance, p-Atf2 recruitment to the gene, and histone acetylation. We have confirmed this result in our Atf4-deficient cells (data not shown). Thus, AAR-driven p-Atf2 association and its corresponding histone modification are not sufficient for transcriptional activation. Therefore, in the absence of Atf4, TSA+HisOH must activate transcription through an AAR-associated activity other than p-Atf2.
Collectively, the results show that AA deprivation can regulate the expression of specific enzymes that are critical mediators of the histone code. The present data are consistent with previously published work showing that dietary protein limitation during pregnancy has profound effects not only on immediate fetal development, but also on disease occurrence during adulthood of the resulting offspring, likely through epigenetic mechanisms. A decrease or increase in JMJD3 protein expression does not cause global changes in H3K27 methylation and some of its functions are independent of its demethylase activity (28). Indeed, an immunoblot of HepG2 extracts after HisOH treatment confirmed no change in total H3K27 trimethylation (data not shown) and so, one must also consider that the role of JMJD3 within the AAR is something other than H3K27 demethylation. Based on ChIP-sequencing data, it is proposed that there are a limited number of genes regulated by JMJD3 (28, 29). Thus, the emerging picture for the role that JMJD3 plays in gene regulation is one of selective gene action rather than a broad function across the genome. Future studies will focus on identifying the target genes for JMJD3 action during activation of the AAR, but the present observations already reveal epigenetics as a new avenue by which AA availability impacts gene expression, and consequently, cell function. The observation that drug-induced hyper-acetylation can replace the function of ATF4 in regulating certain genes also represents an important advance in our mechanistic knowledge of how this critical stress factor regulates gene expression. Conversely, the fact that not all ATF4-dependent genes function in this manner is also intriguing because it demonstrates that ATF4 controls stress-induced transcription through multiple mechanisms.
Acknowledgments
We thank other members of the laboratories for technical advice and helpful discussion. We also acknowledge with appreciation the gifts of the Atf4 knock-out MEF cells and the JMJD3 antibody from Dr. Steve Abcouwer (Pennsylvania State University, Hershey) and Dr. Gioacchino Natoli (European Institute of Oncology, Milan, Italy), respectively. We thank Dr. Chen Liu, Department of Pathology, University of Florida, for the primary human a hepatocytes.
This research was supported, in whole or in part, by Grants DK092062 and DK094729 (to M. S. K.) and HD070487 (to T. G. A.) from the National Institutes of Health.

This article contains supplemental Tables S1 and S2 and Figs. S1–S3.
- AA
- amino acid
- AAR
- amino acid response
- AARE
- amino acid response element
- ASNS
- asparagine synthetase
- ATF
- activating transcription factor
- C/EBP
- CCAAT-enhancer binding protein
- CARE
- C/EBP-ATF response element
- DMSO
- dimethylsufoxide
- ERK
- extracellular-regulated kinase
- GCN2
- general control non-derepressible 2
- HAT
- histone acetyltransferase
- HDAC
- histone deacetylase
- HisOH
- histidinol
- JMJD
- jumonji domain-containing
- MEK
- MAPK/ERK kinase
- qPCR
- real time quantitative PCR
- SNAT2
- system A neutral amino acid transporter 2
- TSA
- trichostatin A.
REFERENCES
- 1. Kilberg M. S., Balasubramanian M., Fu L., Shan J. (2012) The transcription factor network associated with the amino acid response in mammalian cells. Adv. Nutr. 3, 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kilberg M. S., Shan J., Su N. (2009) ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 20, 436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lillycrop K. A., Phillips E. S., Jackson A. A., Hanson M. A., Burdge G. C. (2005) Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J. Nutr. 135, 1382–1386 [DOI] [PubMed] [Google Scholar]
- 4. Burdge G. C., Slater-Jefferies J., Torrens C., Phillips E. S., Hanson M. A., Lillycrop K. A. (2007) Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br. J. Nutr. 97, 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zheng S., Rollet M., Pan Y. X. (2011) Maternal protein restriction during pregnancy induces CCAAT/enhancer-binding protein (C/EBPβ) expression through the regulation of histone modification at its promoter region in female offspring rat skeletal muscle. Epigenetics 6, 161–170 [DOI] [PubMed] [Google Scholar]
- 6. Zheng S., Rollet M., Pan Y. X. (2012) Protein restriction during gestation alters histone modifications at the glucose transporter 4 (GLUT4) promoter region and induces GLUT4 expression in skeletal muscle of female rat offspring. J. Nutr. Biochem., 23, 1064–1071 [DOI] [PubMed] [Google Scholar]
- 7. Barker D. J. (2007) The origins of the developmental origins theory. J. Intern. Med. 261, 412–417 [DOI] [PubMed] [Google Scholar]
- 8. Wek R. C., Jiang H. Y., Anthony T. G. (2006) Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11 [DOI] [PubMed] [Google Scholar]
- 9. Lu P. D., Harding H. P., Ron D. (2004) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vattem K. M., Wek R. C. (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ameri K., Lewis C. E., Raida M., Sowter H., Hai T., Harris A. L. (2004) Anoxic induction of ATF-4 through HIF-1-independent pathways of protein stabilization in human cancer cells. Blood 103, 1876–1882 [DOI] [PubMed] [Google Scholar]
- 12. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 13. Wolfgang C. D., Chen B. P., Martindale J. L., Holbrook N. J., Hai T. (1997) gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol. Cell Biol. 17, 6700–6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fawcett T. W., Martindale J. L., Guyton K. Z., Hai T., Holbrook N. J. (1999) Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress respone. Biochem. J. 339, 135–141 [PMC free article] [PubMed] [Google Scholar]
- 15. Barbosa-Tessmann I. P., Chen C., Zhong C., Siu F., Schuster S. M., Nick H. S., Kilberg M. S. (2000) Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. J. Biol. Chem. 275, 26976–26985 [DOI] [PubMed] [Google Scholar]
- 16. Shan J., Lopez M. C., Baker H. V., Kilberg M. S. (2010) Expression profiling after activation of the amino acid deprivation response in HepG2 human hepatoma cells. Physiol. Genomics 41, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burgold T., Spreafico F., De S. F., Totaro M. G., Prosperini E., Natoli G., Testa G. (2008) The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS One. 3, e3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hong S., Cho Y. W., Yu L. R., Yu H., Veenstra T. D., Ge K. (2007) Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. 104, 18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Agger K., Cloos P. A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A. E., Helin K. (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734 [DOI] [PubMed] [Google Scholar]
- 20. Jones R. S. (2007) Epigenetics: reversing the 'irreversible'. Nature 450, 357–359 [DOI] [PubMed] [Google Scholar]
- 21. Trojer P., Reinberg D. (2006) Histone lysine demethylases and their impact on epigenetics. Cell 125, 213–217 [DOI] [PubMed] [Google Scholar]
- 22. Lan F., Bayliss P. E., Rinn J. L., Whetstine J. R., Wang J. K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E., Roberts T. M., Chang H. Y., Shi Y. (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449, 689–694 [DOI] [PubMed] [Google Scholar]
- 23. Shaw T., Martin P. (2009) Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 10, 881–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Santa F., Totaro M. G., Prosperini E., Notarbartolo S., Testa G., Natoli G. (2007) The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094 [DOI] [PubMed] [Google Scholar]
- 25. Ishii M., Wen H., Corsa C. A., Liu T., Coelho A. L., Allen R. M., Carson W. F., 4th, Cavassani K. A., Li X., Lukacs N. W., Hogaboam C. M., Dou Y., Kunkel S. L. (2009) Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114, 3244–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agger K., Cloos P. A., Rudkjaer L., Williams K., Andersen G., Christensen J., Helin K. (2009) The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 23, 1171–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barradas M., Anderton E., Acosta J. C., Li S., Banito A., Rodriguez-Niedenführ M., Maertens G., Banck M., Zhou M. M., Walsh M. J., Peters G., Gil J. (2009) Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 23, 1177–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De Santa F., Narang V., Yap Z. H., Tusi B. K., Burgold T., Austenaa L., Bucci G., Caganova M., Notarbartolo S., Casola S., Testa G., Sung W. K., Wei C. L., Natoli G. (2009) Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 28, 3341–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Satoh T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y., Miyake T., Matsushita K., Okazaki T., Saitoh T., Honma K., Matsuyama T., Yui K., Tsujimura T., Standley D. M., Nakanishi K., Nakai K., Akira S. (2010) The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11, 936–944 [DOI] [PubMed] [Google Scholar]
- 30. Miller S. A., Mohn S. E., Weinmann A. S. (2010) Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 40, 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 32. Thiaville M. M., Dudenhausen E. E., Zhong C., Pan Y. X., Kilberg M. S. (2008) Deprivation of protein or amino acid induces C/EBPβ synthesis and binding to amino acid response elements, but its action is not an absolute requirement for enhanced transcription. Biochem. J. 410, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bunpo P., Dudley A., Cundiff J. K., Cavener D. R., Wek R. C., Anthony T. G. (2009) GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent l-asparaginase. J. Biol. Chem. 284, 32742–32749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Su N., Thiaville M. M., Awad K., Gjymishka A., Brant J. O., Yang T. P., Kilberg M. S. (2009) Protein or amino acid deprivation differentially regulates the hepatic forkhead box protein A (FOXA) genes through an activating transcription factor-4-independent pathway. Hepatology 50, 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen H., Pan Y. X., Dudenhausen E. E., Kilberg M. S. (2004) Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive basic region/leucine zipper transcription factors as well as localized histone acetylation. J. Biol. Chem. 279, 50829–50839 [DOI] [PubMed] [Google Scholar]
- 36. Su N., Kilberg M. S. (2008) C/EBP homology protein (CHOP) interacts with activating transcription factor 4 (ATF4) and negatively regulates the stress-dependent induction of the asparagine synthetase gene. J. Biol. Chem. 283, 35106–35117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Siu F., Bain P. J., LeBlanc-Chaffin R., Chen H., Kilberg M. S. (2002) ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 277, 24120–24127 [DOI] [PubMed] [Google Scholar]
- 38. Kim T. K., Hemberg M., Gray J. M., Costa A. M., Bear D. M., Wu J., Harmin D. A., Laptewicz M., Barbara-Haley K., Kuersten S., Markenscoff-Papadimitriou E., Kuhl D., Bito H., Worley P. F., Kreiman G., Greenberg M. E. (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pan Y. X., Chen H., Thiaville M. M., Kilberg M. S. (2007) Activation of the ATF3 gene through a coordinated amino acid-sensing response program that controls transcriptional regulation of responsive genes following amino acid limitation. Biochem. J. 401, 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lopez A. B., Wang C., Huang C. C., Yaman I., Li Y., Chakravarty K., Johnson P. F., Chiang C. M., Snider M. D., Wek R. C., Hatzoglou M. (2007) A feedback transcriptional mechanism controls the level of the arginine/lysine transporter cat-1 during amino acid starvation. Biochem. J. 402, 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hai T., Curran T. (1991) Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. 88, 3720–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hsu J. C., Bravo R., Taub R. (1992) Interactions among LRF-1, JunB, c-Jun, and c-Fos define a regulatory program in the G1 phase of liver regeneration. Mol. Cell Biol. 12, 4654–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thiaville M. M., Pan Y. X., Gjymishka A., Zhong C., Kaufman R. J., Kilberg M. S. (2008) MEK signaling is required for phosphorylation of eIF2α following amino acid limitation of HepG2 human hepatoma cells. J. Biol. Chem. 283, 10848–10857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mansour S. J., Matten W. T., Hermann A. S., Candia J. M., Rong S., Fukasawa K., Vande Woude G. F., Ahn N. G. (1994) Transformation of mammalian cells by constitutively active MAP kinase kinase. Science 265, 966–970 [DOI] [PubMed] [Google Scholar]
- 45. Lee C. K., Shibata Y., Rao B., Strahl B. D., Lieb J. D. (2004) Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36, 900–905 [DOI] [PubMed] [Google Scholar]
- 46. Li B., Carey M., Workman J. L. (2007) The role of chromatin during transcription. Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 47. Bhaumik S. R., Smith E., Shilatifard A. (2007) Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14, 1008–1016 [DOI] [PubMed] [Google Scholar]
- 48. Bernstein B. E., Meissner A., Lander E. S. (2007) The mammalian epigenome. Cell 128, 669–681 [DOI] [PubMed] [Google Scholar]
- 49. Bruhat A., Chérasse Y., Maurin A. C., Breitwieser W., Parry L., Deval C., Jones N., Jousse C., Fafournoux P. (2007) ATF2 is required for amino acid-regulated transcription by orchestrating specific histone acetylation. Nucleic Acids Res. 35, 1312–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chaveroux C., Jousse C., Cherasse Y., Maurin A. C., Parry L., Carraro V., Derijard B., Bruhat A., Fafournoux P. (2009) Identification of a novel amino acid response pathway triggering ATF2 phosphorylation in mammals. Mol. Cell Biol. 29, 6515–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Z., Zang C., Cui K., Schones D. E., Barski A., Peng W., Zhao K. (2009) Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Endo Y., Fu Z., Abe K., Arai S., Kato H. (2002) Dietary protein quantity and quality affect rat hepatic gene expression. J. Nutr. 132, 3632–3637 [DOI] [PubMed] [Google Scholar]
- 53. Lee J. I., Dominy J. E., Jr., Sikalidis A. K., Hirschberger L. L., Wang W., Stipanuk M. H. (2008) HepG2/C3A cells respond to cysteine deprivation by induction of the amino acid deprivation/integrated stress response pathway. Physiol. Genomics 33, 218–229 [DOI] [PubMed] [Google Scholar]
- 54. Deval C., Chaveroux C., Maurin A. C., Cherasse Y., Parry L., Carraro V., Milenkovic D., Ferrara M., Bruhat A., Jousse C., Fafournoux P. (2009) Amino acid limitation regulates the expression of genes involved in several specific biological processes through GCN2-dependent and GCN2-independent pathways. FEBS J. 276, 707–718 [DOI] [PubMed] [Google Scholar]
- 55. Fu L., Balasubramanian M., Shan J., Dudenhausen E. E., Kilberg M. S. (2011) Auto-activation of c-JUN gene by amino acid deprivation of hepatocellular carcinoma cells reveals a novel c-JUN-mediated signaling pathway. J. Biol. Chem. 286, 36724–36738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Palii S. S., Thiaville M. M., Pan Y. X., Zhong C., Kilberg M. S. (2006) Characterization of the amino acid response element within the human sodium-coupled neutral amino acid transporter 2 (SNAT2) System A transporter gene. Biochem. J. 395, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sérandour A. A., Avner S., Percevault F., Demay F., Bizot M., Lucchetti-Miganeh C., Barloy-Hubler F., Brown M., Lupien M., Métivier R., Salbert G., Eeckhoute J. (2011) Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 21, 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Plachetka A., Chayka O., Wilczek C., Melnik S., Bonifer C., Klempnauer K. H. (2008) Mol. Cell Biol. 28, 2102–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carraro V., Maurin A. C., Lambert-Langlais S., Averous J., Chaveroux C., Parry L., Jousse C., Ord D., Ord T., Fafournoux P., Bruhat A. (2010) Amino acid availability controls TRB3 transcription in liver through the GCN2/eIF2α/ATF4 pathway. PloS One 5, e15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Palii S. S., Chen H., Kilberg M. S. (2004) Transcriptional control of the human sodium-coupled neutral amino acid transporter system A gene by amino acid availability is mediated by an intronic element. J. Biol. Chem. 279, 3463–3471 [DOI] [PubMed] [Google Scholar]
- 61. Gjymishka A., Palii S. S., Shan J., Kilberg M. S. (2008) Despite increased ATF4 binding at the C/EBP-ATF composite site following activation of the unfolded protein response, system A transporter 2 (SNAT2) transcription activity is repressed in HepG2 cells. J. Biol. Chem. 283, 27736–27747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Winkler G. S., Kristjuhan A., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2002) Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. 99, 3517–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Barkess G., Postnikov Y., Campos C. D., Mishra S., Mohan G., Verma S., Bustin M., West K. L. (2012) The chromatin-binding protein HMGN3 stimulates histone acetylation and transcription across the Glyt1 gene. Biochem. J. 442, 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kawasaki H., Schiltz L., Chiu R., Itakura K., Taira K., Nakatani Y., Yokoyama K. K. (2000) ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature 405, 195–200 [DOI] [PubMed] [Google Scholar]