Background: Thyroid hormone receptor (TR) isoforms α and β have distinct biological and physiological roles in development and in adult tissues.
Results: TRα and β are posttranslationally modified by different SUMO isoforms and require specific SUMO E3 ligases. TR sumoylation influences corepressor and coactivator recruitment.
Conclusion: TR-SUMO conjugation is important for thyroid hormone action.
Significance: Identifying a mechanism contributing to TR isoform-specific action.
Keywords: E3 Ubiquitin Ligase, Gene Regulation, Posttranslational Modification, Sumoylation, Thyroid Hormone, Coactivators, Corepressors, Thyroid Hormone Receptor
Abstract
Thyroid hormone receptor (TR) α and β mediate thyroid hormone action at target tissues. TR isoforms have specific roles in development and in adult tissues. The mechanisms underlying TR isoform-specific action, however, are not well understood. We demonstrate that posttranslational modification of TR by conjugation of small SUMO to TRα and TRβ plays an important role in triiodothyronine (T3) action and TR isoform specificity. TRα was sumoylated at lysines 283 and 389, and TRβ at lysines 50, 146, and 443. Sumoylation of TRβ was ligand-dependent, and sumoylation of TRα was ligand-independent. TRα-SUMO conjugation utilized the E3 ligase PIASxβ and TRβ-SUMO conjugation utilized predominantly PIAS1. SUMO1 and SUMO3 conjugation to TR was important for T3-dependent gene regulation, as demonstrated in transient transfection assay and studies of endogenous gene regulation. The functional role of SUMO1 and SUMO3 in T3 induction in transient expression assays was closely matched to the pattern of TR and cofactor recruitment to thyroid hormone response elements (TREs) as determined by ChIP assays. SUMO1 was required for the T3-induced recruitment of the co-activator CREB-binding protein (CBP) and release of nuclear receptor co-repressor (NCoR) on a TRE but had no significant effect on TR DNA binding. SUMO1 was required for T3-mediated recruitment of NCoR and release of CBP from the TSHβ-negative TRE. SUMO3 was required for T3-stimulated TR binding to the TSHβ-negative TRE and recruitment of NCoR. These findings demonstrate that conjugation of SUMO to TR has a TR-isoform preference and is important for T3-dependent gene induction and repression.
Introduction
Thyroid hormone action is mediated by thyroid hormone receptor (TR)3 α and β. T3-dependent gene regulation requires TR interaction with ligand, a thyroid hormone response element (TRE), coactivators, and corepressors (1, 2). TR gene “knock-out” and “knock-in” mutant mouse models and studies with TR isoform selective agonists demonstrate that TR isoforms (TRα and TRβ) play specific developmental roles and have specific actions in T3 target tissues in adults. TRα is important for normal brain and testicular development and growth (3). In the adult, TRα mediates T3 actions in the heart, brain, and potentiates sympathetic action in bone, white fat, and brown adipose tissue (4–8). TRβ is required for normal cochlear and retinal development, TSH regulation, cholesterol metabolism in the liver, and stimulation of uncoupling protein gene expression in brown fat (6, 9–12). The mechanisms underlying TR isoform-specific action in T3-mediated gene regulation, however, are not well understood.
Posttranslational modification of nuclear receptors is being increasingly recognized as an important mechanism of gene regulation. The majority of proteins modified by SUMO are transcription factors and nuclear hormone receptors (13). Sumoylation requires conjugation of SUMO to a lysine contained in the consensus recognition motif ψ-Lys-X-Glu (ψ is a large hydrophobic amino acid) of a protein and has been reported to occur in all cellular compartments and tissues (14). Sumoylation rapidly and dynamically modifies proteins involved in cellular process, including regulation of gene transcription, mRNA stability, nuclear localization, protein stability, and protein-protein interactions (15–18). Four mammalian SUMOs (SUMO1, SUMO2, SUMO3, and SUMO4) have been identified, three of which, SUMO1, SUMO2, and SUMO 3, are functionally characterized. SUMO2 and SUMO3 share 95% sequence identity, and both are >50% homologous with SUMO1. SUMO conjugation requires activating enzyme E1, conjugating enzyme UBC9, and E3 ligases, which have substrate specificity (19, 20). Sumoylation can influence the function of a transcription factor by modifying protein conformation, the interface of protein-cofactor interaction, DNA binding, and ligand binding (13).
In this study, we demonstrate that TRα and TRβ are modified by specific SUMOs. TRα and β have distinct patterns of SUMO and E3 ligase preferences and differ in their response to the ligand. SUMO conjugation of TR modulates T3-mediated gene regulation mediated by both positive and negative TREs. Sumoylation is important for the normal patterns of T3-dependent gene expression and repression. The influence of sumoylation on T3-dependent gene regulation in transient functional assays is closely paralleled by the binding of TR, corepressors, and coactivators, to response elements in endogenous T3-regulated genes.
EXPERIMENTAL PROCEDURES
Animals
Animal studies were approved by the Animal Research Committee (Veterans Affairs Greater Los Angeles Health Care System). Male BL/C57 mice were fed with normal chow and tap water ad libitum at 23 °C and light cycles of 12 h. All mice were 70–80-days-old at the time of the experiments and weighed 22–26 g. Animals were euthanized, and tissues were removed for analysis following a standard protocol.
Cell Culture, Transient Transfections, and Luciferase Assays
HepG2 cells were maintained in Eagle's Minimal Essential Medium with 10% fetal bovine serum (FBS). Rat pituitary tumor cells (GH3) were maintained in F-12K medium supplemented with 15% horse serum and 2.5% FBS. For transient transfection studies, cells were plated on 24-well dishes and grown for 24 h in serum-free medium. The test plasmid (0.1 μg) was added to each transfection, and the empty vector pcDNA3.1 was used to maintain a constant DNA concentration. Twenty-four hours after transfection, cells were treated with T3 (50 nm) for 6 h prior to the luciferase reporter assay.
Construction of Reporter Plasmids
Rat TSHβ nTRE (21, 22) (accession no. M13897) and rat GH TRE (23) (accession no. X12967) were cloned into a pGL-3 promoter vector at BglII and MluI sites. DNA oligonucleotide sequences are listed in Table 1.
TABLE 1.
Primer Sequences
| Name of primer/DNA oligonucleotide | DNA sequence |
|---|---|
| Rat TSHβ nTRE (21, 22) (M13897) | |
| Forward | 5′-agagtctgggtcatcacagcattaactcgccagtgcaaagtaaggtaggtctctacccgg-3′ |
| Rat GH TRE (23) (X12967) −198 to −158 | |
| Forward | 5′-cgcgtaaaggtaagatcagggacgtgaccgaga-3′ |
| hSUMO1 (NM_001005782.1) | |
| Forward | 5′-ccggagcgaggttctgctta-3′ |
| Reverse | 5′-aagtaatgtcactgtatc-3′ |
| hSUMO2 (NM_006937.3) | |
| Forward | 5′-gcaggatggttctgtggtg-3′ |
| Reverse | 5′-ctgcctcattgacaatccct-3′ |
| hSUMO3 (NM_006936.2) | |
| Forward | 5′-tggaggacgaggacaccat-3′ |
| Reverse | 5′-agcatgcgaggtaggacg-3′ |
| Rat TSHβ nTRE ChIP (80-bp PCR product) (M13897.1) | |
| Forward | 5′-cgaagggtataaaatgaa-3′ |
| Reverse | 5′-gcacttcattttacaggtcctg-3′ |
| Rat GH TRE ChIP (90-bp PCR product) (X1296.7) | |
| Forward | 5′-caaaaggacacattgggtgg-3′ |
| Reverse | 5′-ttgcccaggtttatgggc-3′ |
| TSHβ GSP2 (21, 22) (M13897.1) | |
| Forward | 5′-agcattaactcgccagtgca-3′ |
| TSHβ GSP3 (22) (M13897.1) | |
| Reverse | 5′-atcttatcttatcttgattattttat-3′ |
| TSHβ GSP-reverse (M13897.1) | |
| Reverse | 5′-cgtgtcatacaatacccagc-3′ |
| Rat GH (mRNA) (NM_001034848) | |
| Forward | 5′-ttgttaggtgtcagcagccag-3′ |
| Reverse | 5′-atggctgcagactctcagac-3′ |
| Consensus DR4 TRE sequence | 5′-tagctcgaggtcacaggaggtcattgcc-3′ |
| rME TRE sequence (36) −281 to −261 (M35258.1) | 5′-aaaggtaagatcagggacgtgaccgcac-3′ |
In Vitro Sumoylation
TRα and TRβ proteins were in vitro synthesized utilizing Quick T7-kit (Promega, Inc.) in the presence of [35S]methionine. In vitro sumoylation was performed using a sumoylation kit (LAE Biotech Intl., Inc.) following the manufacturer's instructions. Briefly, each sumoylation reaction (20 μl) was composed of 2 μl of synthesized protein, activating enzyme (E1, 7.5 μg/ml), UBC9 (E2 SUMO conjugating enzyme, 50 μg/ml), SUMO1 (50 μg/ml), buffer (20 mm Hepes, pH 7.5, 5 mm MgCl2, and 2 mm ATP), and extra fresh 20 mm ATP. The reaction was incubated at 37 °C for 1 h. The reaction was terminated by the addition of 5 μl of 4× SDS buffer, heated at 90 °C for 5 min, and loaded on an 8–10% SDS-PAGE. The gel was dried and imaged on a PhosphorImager.
Detection of TR Sumoylation in Mouse Tissues by Immunoprecipitation (IP) and Western Blotting (WB)
Mouse liver, heart and adipose tissues (n = 4) were freshly harvested (100 mg/tissue/mouse) and immediately homogenized in 2 ml of ice-cold IRP buffer (Millipore, Inc.) containing protease mixture (Roche Applied Science). After centrifugation at 14,000 rpm for 15 min at 4 °C, the clear supernatant (∼1.0 ml) was collected. After the lysate was pre-cleared, protein concentration of the lysate was determined and adjusted uniformly among the samples. The lysate was then incubated with the following antibodies: anti-TRα (1:100) or anti-TRβ (1:100) (Thermo Fisher Scientific, Inc.), or anti-HA tag (1:100) (Millipore) and rabbit anti-mouse IgG (1:250) as control in IPA buffer for 6 h to overnight. The immunocomplexes were captured by addition of 50 μl of protein A-agarose. After one wash with RIPA buffer and three washes with cold PBS, the immunocomplexes were resuspended in 4× SDS sample buffer (40% glycerol, 240 mm Tris-HCl, pH 6.4, 8% SDS, 0.04% Bromophenol blue and 5% β-mercaptoethanol), heated at 90 °C for 5 min, and separated by 8% or 10% SDS-PAGE. For Western blot, antibodies including full-length anti-SUMO1 antibody (1:300) (Santa Cruz Biotechnology), anti-TRα (1:250), anti-TRβ (IgG, 1:500), or anti-HA tag (1:500) (Millipore) were used to detect sumoylated TRs. Immunoreactive bands were detected by chemiluminescence (GE Life Sciences) and imaged using Kodak Imager. When the membrane was reprobed with a different antibody, the membrane was stripped with stripping reagent (Bio-Rad) for 15–30 min at 30 °C, followed by chemiluminescent exposure and imaging for residual activity, and then the membrane was incubated with blocking buffer for 1 h before incubating with antibodies.
Detection of TR Sumoylation in Cells by Immunoprecipitation, Co-immunoprecipitation, and WB
HepG2 cells were plated on a 10-cm dish, transfected with vectors expressing TRα or TRβ and hSUMO-HA, and treated with or without T3 for 4 h. Briefly, cells were lysed in 1 ml of IP buffer with protease mixture. The rest of the procedure followed the standard IP protocols. The antibody used and dilutions are the same as described above. For IP input control, the same cell lysate after IP with anti-TRs were used for IP with anti-GAPDH antibody (1:250), followed by WB detection of GAPDH. For co-immunoprecipitation, HepG2 cells were cotransfected with NCoR-FLAG TRβ and/or siRNA SUMO1. Co-immunoprecipitation was performed with anti-FLAG M2-agarose affinity resin in lysis buffer (50 mm Tris-HCl, pH 7.4, with 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100) and incubated at 4 °C with rotation for 4 h. The resin was washed three times in the wash buffer composed of 50 mm Tris-HCl, pH 7.4, 150 mm NaCl. The resin was then heated in SDS sample buffer (20 μl) at 90 °C for 5 min. The co-immunoprecipitation complexes were detected by anti-FLAG and anti-TRβ antibodies. The identity of TR-SUMO bands was confirmed by stripping the membrane after hybridization and reprobing as described previously.
Reverse Transcription, PCR and/or q-PCR
RNA (1 μg) was reverse-transcribed using the Superscript III kit (Invitrogen). For PCR and q-PCR studies, 1 μl of cDNA or DNA was used to detect mRNA or ChIP assay (primers are listed in Table 1). All q-PCR experiments were performed with triplicates.
Detection of TSHβ Transcripts
TSHβ has two transcription start sites (TSS)1 and 2. The transcript initiated from TSS2, mRNA2, is repressed in response to T3 (24). To detect distinct transcripts of TSHβ, RNA (100 ng) from GH3 cells was reverse-transcribed utilizing OminScript (Qiagen) with gene-specific forwarding primers GSP2 for TTS2 and reverse primer GSP3 containing poly(A). For q-PCR, 2 μl of cDNA was amplified using GSP2 with GSP reverse primer.
Gene Silencing, Reporter Assay, and ChIP Assay
For reporter assays, cells were transfected with 100 nm (final concentration) On-TargetTM SMARTpool (Dharmacon/Thermofisher Scientific) siRNA SUMOs or siRNA control using nucleofactor V, program 28, and then plated in 24-well plates at 2 × 104 cells per well. Knockdown efficiency was examined by PCR 48 h after knockdown. Reporter construct and plasmids expression vectors were then transfected, and cells were grown for 16–24 h in serum-replaced medium with Serum Replacement (Invitrogen). Cells were then treated with T3 (50 nm) for 6 h prior to luciferase assay. For ChIP assay, rat pituitary cells (GH3) were cotransfected with siRNA for SUMOs and TRβ. Knockdown efficiency was examined by WB 72 h after transfection. Cells were then grown in serum-replaced medium for 16 h and treated with or without T3 (50 nm) for 4 h. The ChIP assay method was as described previously (24). The antibodies used in the ChIP assay were anti-TRβ (Thermo Fisher Scientific), anti-NCoR (Millipore), and anti-CBP (Santa Cruz Biotechnology). Immunoprecipitated DNA was purified and utilized for q-PCR amplification with specific primers (Table 1).
Statistics
Statistical analysis was performed with an average of triplicates. A two-tailed Student's t test was performed to compare means. p value <0.05 was considered to be statistically significant.
RESULTS
TRα and TRβ Are Sumoylated in Vitro and in Vivo
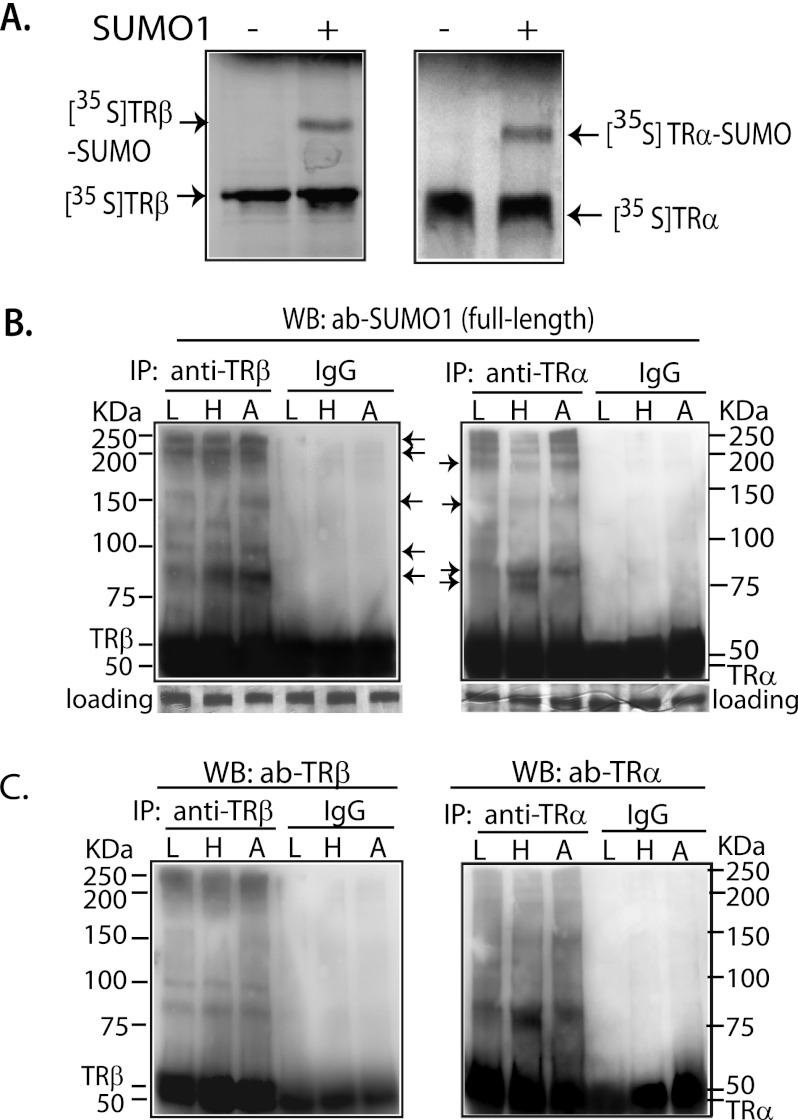
We performed in vitro sumoylation of [35S]Met-labeled TRα and TRβ to determine whether these proteins are SUMO substrates. In the presence of SUMO1, a single band migrating above the TRα and TRβ proteins was seen, indicating that both TRα and TRβ are sumoylated (Fig. 1A). We investigated in vivo sumoylation of TRs in mouse liver, heart, and adipose tissue. To detect endogenous sumoylation by any SUMO, full-length SUMO1 antibody was used, which recognizes SUMO1, SUMO2, and SUMO3 (Fig. 1B). In vivo conjugation of SUMO to TRα and TRβ was demonstrated in all tissues studied. To confirm the presence of TR in the putative TR-SUMO complex, the membranes were stripped and then incubated with TRα and TRβ antibodies (Fig. 1C). The bands detected by TR antibodies generally matched the position of the identified SUMO-TR complexes, although not all of the bands identified by the SUMO antibody contained TR. Even considering only those bands shown to contain both SUMO and TR, multiple bands were observed, indicating that TRα and TRβ have multiple sumoylation sites. There were variations in the extent of TR sumoylation among the various tissues, as well as TRα and TRβ isoform differences (Fig. 1, B and C). The molecular mass of SUMO conjugated protein species was as high as 250 kDa. SUMO usually runs at a significantly higher position in SDS-PAGE than the actual size. The high molecular weight protein species may represent both multiple TR sumoylation sites as well as polysumoylation at a single site.
FIGURE 1.
In vitro and in vivo sumoylation of TR. A, [35S]TRα and -TRβ were incubated with E1, UBC9, and SUMO1 in sumoylation buffer at 37 °C for 1 h and then separated by 10% SDS-PAGE. B, mouse liver (L), heart (H), and white adipose (A) tissues were utilized for IP with anti-TRα (rabbit IgG) and anti-TRβ (mouse IgG) antibodies. In WB, anti-full-length SUMO1 was used to detect sumoylation. The membranes shown in B were then stripped, checked by chemiluminescent exposure to confirm the absence of residual activity, and then incubated with anti-TRβ (rabbit IgG) and anti-TRα (rabbit IgG) antibodies (ab; C) to confirm that the identified TR-SUMO bands contained TR. IgG was used as nonspecific control. Arrows indicate the location of TR-SUMO complexes.
TR Isoform-specific Requirement for Ligand, SUMO Specificity, and E3 Ligase
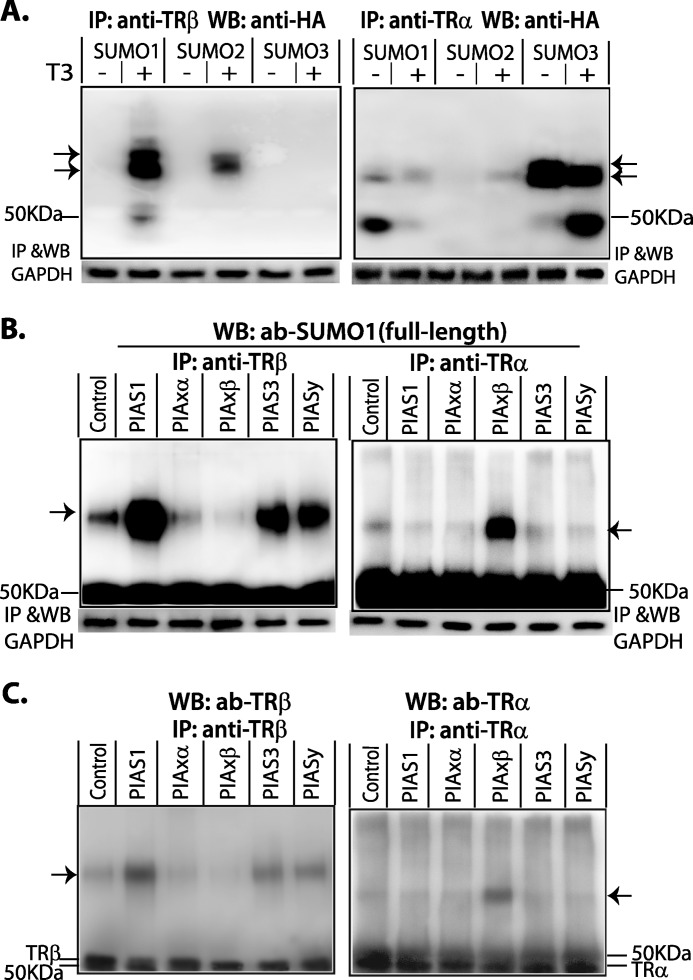
TRα and TRβ are ligand-activated receptors. We investigated the influence of ligand on TR-SUMO conjugation in HepG2 cells with transfected vectors expressing TRα, TRβ, and SUMO-HA. Cells were treated with or without T3 (50 nm) for 4 h. The cell lysate was immunoprecipitated with anti-TR antibody and detected in WB with anti-HA antibody. In the absence of T3, TRβ was not sumoylated, but after addition of T3, TRβ was preferentially sumoylated by SUMO1 (Fig. 2A, left panel). In contrast to the findings with TRβ, sumoylation of TRα was not influenced by ligand, and it was preferentially sumoylated by SUMO3 (Fig. 2A, right panel). A similar TR isoform sumoylation pattern was seen in mouse myoblast (C2C12) cells but the addition of T3 was associated with some reduction of sumoylation in TRα, suggesting some cell type variation (data not shown).
FIGURE 2.
Sumoylation of TR isoforms. A, ligand effects and ligase preference for sumoylation HepG2 cells were transfected with human SUMO (SUMO1, 2, or 3)-HA, and human TRα or human TRβ expression vectors and grown in serum-free medium for 16 h prior to treatment with or without T3 (50 nm) for 4 h. Cell lysate was used for IP with anti-TRα or anti-TRβ antibodies (ab). Immunoprecipitated protein complexes were subject to 7.5% SDS-PAGE. For detection of TR-SUMO conjugation in WB, anti-HA was used to detect SUMO(1, 2, or 3)-HA. B, E3 ligase requirement. HepG2 cells were transfected with PIAS family members. Control cells were not transfected with E3 ligase. Cells, including control cells, were treated with T3 (50 nm) for 4 h for the detection of TRβ-SUMO conjugation, but without T3 for detection of TRα-SUMO. Anti-TRα or anti-TRβ antibodies were used in IP and anti-SUMO1 (full-length) in WB. After IP with anti-TRs, the cell lysates were then used for IP with anti-GAPDH and WB GAPDH, which was used as a protein input control. C, verification of TR in SUMO-conjugated bands in B. The membrane in B was stripped, checked with chemiluminescent exposure to confirm the absence of residual activity, and then incubated with anti-TR antibodies as shown. Arrows show SUMO-TR complexes.
E1 (SUMO activation enzyme 1) and E2 (UBC9) are required enzymes for sumoylation, and E3 ligases are substrate-specific. We determined which E3 ligase was utilized for sumoylation of TRα and TRβ. HepG2 cells were transfected with vectors expressing human PIAS family members (hPIAS1, hPIASxα, hPIASβ, hPIAS3, and hPIASxy). Control cells were not transfected with E3 ligase. Cells, including the control cells, were treated with T3 (50 nm) for 4 h for detection of sumoylated TRβ and were grown without T3 treatment for detection of sumoylated TRα. The anti-SUMO1 (full-length) antibody was used to detect endogenous sumoylated TR by Western blot, following immunoprecipitation. TRβ-SUMO conjugation was primarily facilitated by PIAS1 (Fig. 2B, left panel), although PIAS3 and PIASxy also facilitated some conjugation. PIASxβ was the E3 ligase that promoted TRα-SUMO conjugation (Fig. 2B, right panel). The sumoylation of endogenous TR was stimulated by the addition of exogenous E3 ligase, compared with control without exogenous E3 ligase. This enhancement indicates that in some cells, sumoylation of TR may be limited by the availability of E3 ligase.
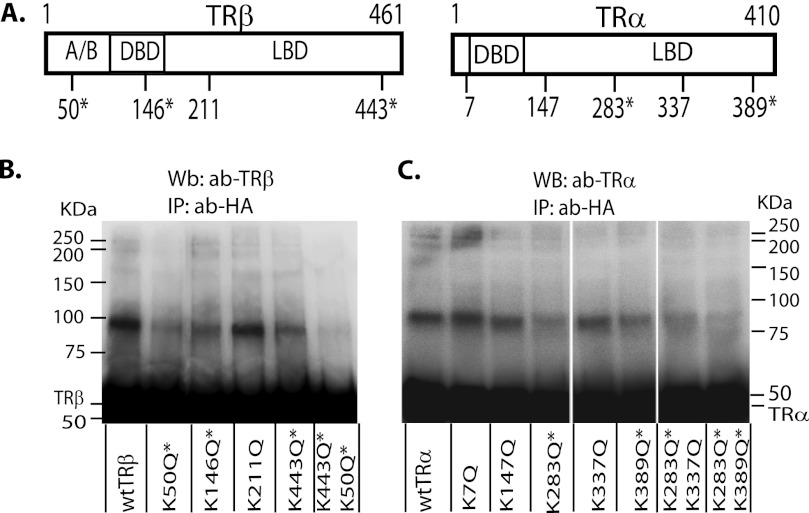
Identification of Sumoylation Sites in TRα and TRβ
Based on inspection of the amino acid sequence, we identified five potential sumoylation sites in TRα and four in TRβ (Fig. 3A). To determine the active sumoylation sites, we mutated each putative site and examined sumoylation in HepG2 cells cotransfected with the vectors expressing SUMO1-HA or SUMO3-HA. To locate the TRβ sumoylation site, cells were treated with T3 (50 nm) for 4 h prior to immunoprecipitation. To determine TRα sumoylation sites, cells were grown without T3. Active sites were identified based on loss of sumoylation as a consequence of a mutation. There were three active sites in TRβ, lysine 50, lysine 146, and lysine 443 (Figs. 3B). We confirmed two active sites in TRα, lysine 283 and lysine 389 (Figs. 3C). Mutations at these sites markedly reduced TR sumoylation.
FIGURE 3.
Identification of the sumoylation sites in TRα and TRβ. A, predicted sumoylation sites based on amino acid sequence in TRα and TRβ, with an asterisk showing confirmed sites based on mutation analysis. HepG2 cells were transfected with vectors expressing SUMO1-HA (B; for TRβ) or SUMO3-HA (C; for TRα) and wild type or mutant TRs as shown. Cells were treated with T3 (50 nm) for 4 h for TRβ sumoylation, but without T3 for TRα sumoylation. The cell lysate was immunoprecipitated with an anti-HA antibody (ab) to detect SUMO1- or SUMO3-HA, and Western blot was performed with TRβ (B) or TRα (C) antibody. *, confirmed sumoylation sites in TRs. A/B, activation domain; DBD, DNA binding domain; LDB, ligand binding domain.
The Effects of Sumoylation on T3-dependent Regulation of a Positive TRE
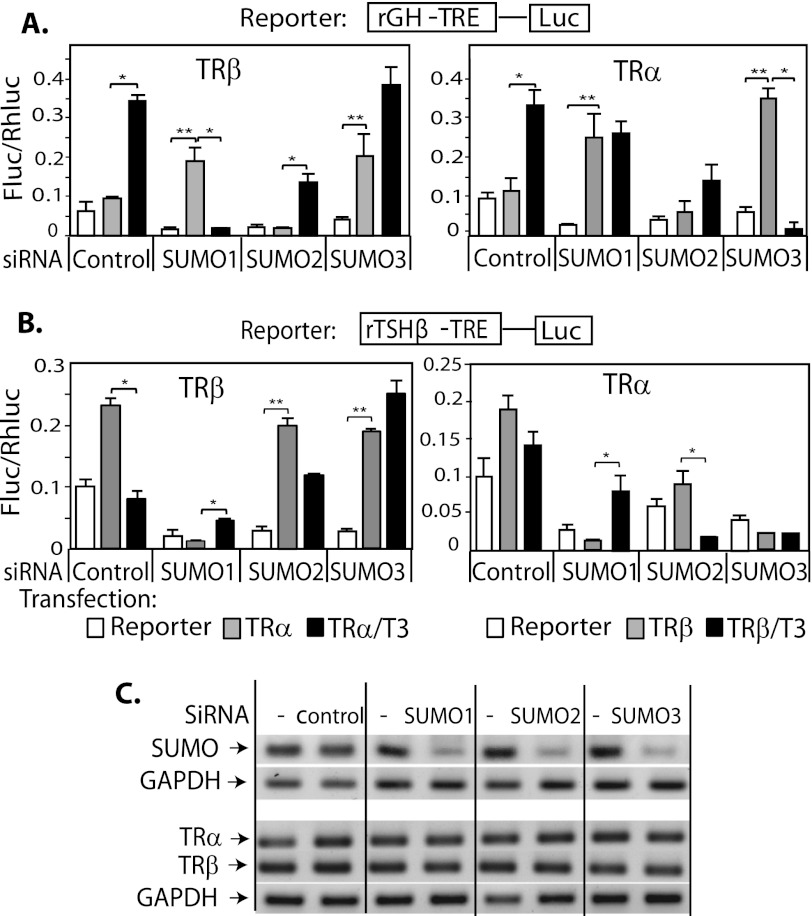
We have demonstrated that TRα and β are both sumoylated but differ with respect to the influence of ligand, preferences for SUMO isoforms and E3 ligases. We wanted to determine how these differences influenced T3-dependent gene expression. We analyzed expression of a reporter vector with a native positive TRE from the rat growth hormone gene (rGH). The rGH TRE is composed of three half-sites (A, B, and C) arranged in a combination of a DR4 and an inverted repeat (23). GH3 (rat pituitary) cells were utilized after selective SUMO knockdown using On-TargetTM SMARTpool, composed of four sets of siRNAs. We also performed siRNA knockdown using four individual siRNA for each SUMO target, which yield similar results to that using SMARTpool (supplemental Fig. S1).
GH3 cells were then transfected with the rGH TRE reporter construct and TRα or TRβ expression vectors. Reporter expression was evaluated with and without T3 (50 nm) (Fig. 4A). In the control cells, as expected, unliganded TR repressed transcription and liganded TR stimulated transcription via the rGH-positive TRE. In TRβ-transfected cells, however, T3 induction was abolished by SUMO1 knockdown and not affected by SUMO3 knockdown (Fig. 4A, left panel). Knockdown of SUMO1 or SUMO3 dramatically increased basal activity in the presence of unliganded TRα and completely abolished induction in response to Τ3. Knockdown of SUMO2 blunted TRα/T3-mediated stimulation but did not affect the pattern of T3 induction (Fig. 4A, right panel). Basal gene expression in the presence of TRs (either TRα or TRβ) and absence of ligand were significantly enhanced after SUMO1 or SUMO3 knockdown. Taken together, these results from reporter assays indicate that SUMO1 and -3 but not SUMO2 are important for TRα/TRβ T3-mediated regulation of the rGH-TRE reporter. Although SUMO3-TRβ conjugation could not be demonstrated in our in vitro system (Fig. 2A), there were functional effects of SUMO3 knockdown on basal expression by unliganded TRβ. These effects may be due to SUMO3 actions on TR cofactors. To verify the specificity of the SUMO effects for the TRE, we performed transfection studies using a reporter construct carrying a mutant rGH TRE that is no longer induced by T3 (supplemental Fig. S2). SUMO knockdown had no influence on activity of the mutant rGH TRE with or without ligand, indicating that SUMO knockdown specifically affected TR function on the TRE.
FIGURE 4.
Sumoylation is important for T3-dependent induction and repression of gene expression. GH3 cells were transfected with siRNA SUMO (1, 2, or 3) or siRNA control using Nucleofactor V, program T-28 (Lonza, Inc.) and plated onto 24-well plates. After 48 h, cells were tested for knockdown efficiency and then cotransfected with expression vectors (TRα or TRβ) and reporter construct rGH-TRE-Luc (A) or rTHSβ-TRE (B) using Effectene transfection reagent (Qiagen). T3 (50 nm) was added, and a reporter assay performed after 6 h. C, RT-PCR analysis of SUMO mRNA after siRNA SUMO knockdown and TR mRNA expression in transfected cells. *, T3 induction compared with basal (p < 0.05); **, basal compared with expression without transfected TR and T3 (p < 0.05). C, confirmation of specificity of siRNA is shown with mRNA detection by q-PCR for each SUMO condition and an absence of effect of siRNA on other SUMOs, TRs, or GAPDH.
The Effects of Sumoylation on T3-dependent Regulation of a Negative TRE
We next investigated the importance of sumoylation for T3-dependent repression mediated by the well characterized TSHβ-negative TRE (nTRE). The reporter construct TSHβ nTRE-luc was cotransfected with expression vectors for TRα and TRβ in GH3 cells, after selective knockdown of SUMO. In the control cells, unliganded TR enhanced reporter expression and addition of T3 repressed it (Fig. 4B). TRβ was more effective mediating T3 repression, compared with TRα, in agreement with the predominant role of TRβ in regulation of TSH. After SUMO1 knockdown, unliganded-TRα or -TRβ reduced rather than the expected enhancement of basal expression. Compared with control, T3 induced reporter expression 8-fold in TRα-transfected cells and 2.7-fold in TRβ-transfected cells, rather than the usual repression (Fig. 4B). The TSHβ nTRE did not confer T3-dependent repression in the absence of SUMO1. SUMO2 knockdown had minimal effects on TRα or TRβ -mediated T3-dependent repression. In TRβ-transfected cells, SUMO3 knockdown substantially enhanced reporter expression in the presence or absence of T3 (Fig. 4B, left panel). This is consistent with TRβ recruiting coactivator, but not corepressor, to the TSHβ nTRE in the absence of SUMO3. After SUMO3 knockdown, neither TRα nor Τ3 influenced reporter expression (Fig. 4B, right panel), indicating that SUMO3 is required for TRα-mediated T3 induction.
Although SUMO1 and SUMO3 are similar, their effects on transcription are distinct, as shown in the reporter assay with the rGH TRE. To confirm our findings, we studied an additional TRE, the consensus DR4TRE in a reporter assay (supplemental Fig. S3). The results are similar to those utilizing the rGHTRE regarding siRNA SUMO2 effects, although the magnitude of the effect on transcription activity varied.
Influence of TRα and β Sumoylation Mutations on T3 Regulation of a TRE and nTRE
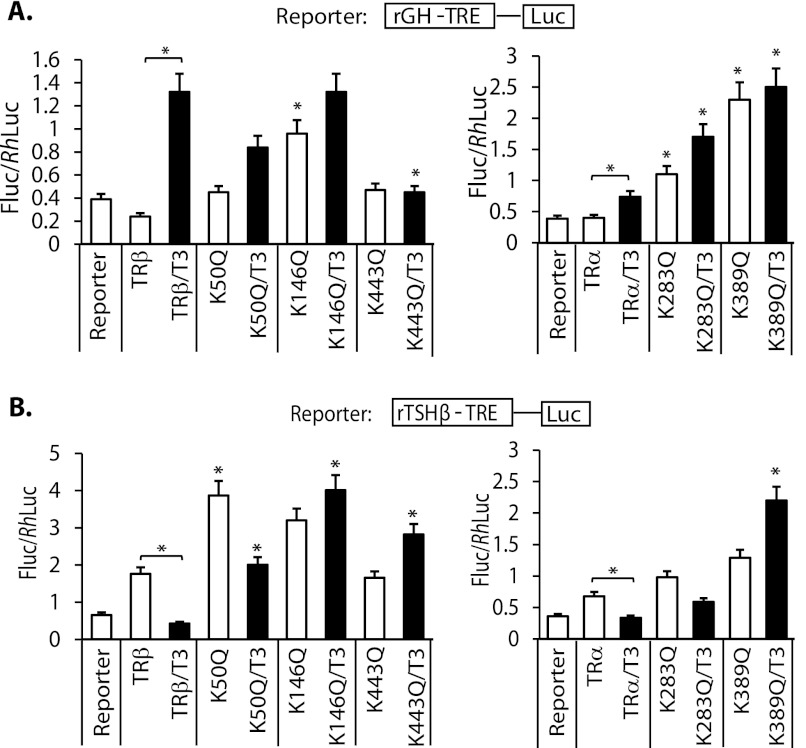
We wanted to determine the specificity of the influence of SUMO knockdown on T3 induction and repression to TR sumoylation. We, therefore, performed transient transfection assays with the rGH TRE and β-TSH nTRE and transfected wild-type TR α and β compared with TRs with introduced sumoylation-site mutations in HepG2 cells (Fig. 5). The TRβ mutations K146Q and K443Q were associated with loss of T3-dependent induction and repression, consistent with the findings with SUMO1 knockdown. The K50Q mutation retained T3 induction and repression, but with significant reduction in the magnitude. The TRα mutation K389Q was associated with complete loss of T3 induction and repression. The K283Q mutation retained some T3 induction and repression, but with reduced magnitude. This shows that TR sumoylation is important for the observed functional effects after SUMO knockdown.
FIGURE 5.
Effects of sumoylation site TRα and β mutations on T3-mediated gene transcription. Wild-type TRs and TR mutants cotransfected with reporter constructs rGH TRE-luc (A) and TSHβ TRE-luc (B) in HepG2 cells. Cells were grown in serum-replaced medium for 16 h and then treated with T3 (50 nm; black bars) or without (white bars) for 4 h prior to luciferase assay. *, with linking bar shows significant effect, p < 0.05 of T3 treatment compared with control; *, compares mutant TR with or without ligand to control, p < 0.05.
Sumoylation Modulates TR Binding and Recruitment of Cofactors to an Endogenous TRE
Our data demonstrate that SUMO1 and SUMO3 are important for T3-dependent gene induction and repression and influenced by the specific TRE and TR isoforms. We next determined the influence of SUMO knockdown on endogenous T3-regulated gene expression of a positively (GH) and negatively (TSHβ) regulated gene in GH3 cells. We also determined the influence of SUMO on TR binding and the recruitment of endogenous corepressor (NCoR) and coactivator (CBP), in the presence and absence of T3.
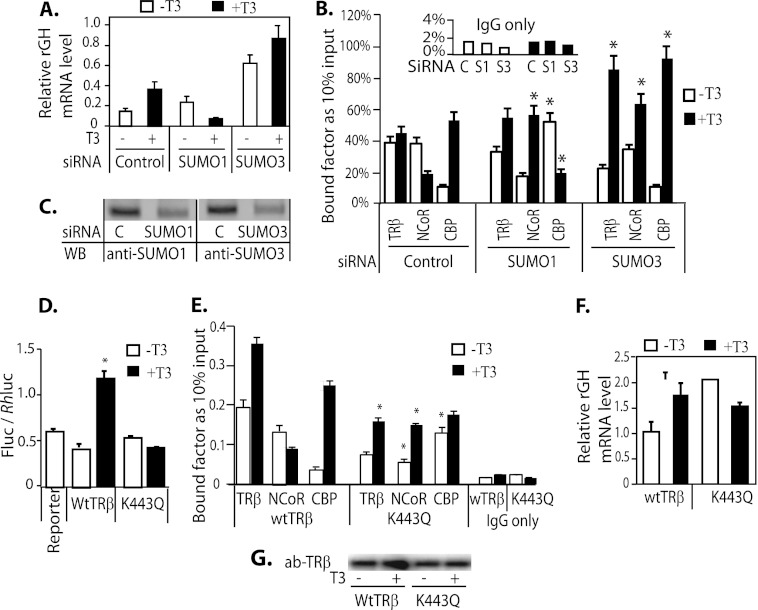
The net effect of SUMO1 knockdown on endogenous growth hormone gene regulation, shown by the level of GH mRNA expression in GH3 cells, was an increase in basal expression and a 3-fold T3 repression, compared with the normal T3 induction in controls (Fig. 6A). After SUMO3 knockdown, GH mRNA expression at base line and after T3 treatment was markedly increased. In the control, NCoR was recruited to the rGH TRE in the absence of ligand, and the addition of ligand resulted in loss of NCoR binding and recruitment of CBP (Fig. 6B). This is consistent with functional repression in the absence of ligand and induction with the addition of ligand. After SUMO1 knockdown, the patterns of NCoR and CBP recruitment to the rGH TRE, however, were reversed from that of controls, with a net effect of T3 repression (Fig. 6B). After SUMO3 knockdown and treatment with T3, TRβ DNA-binding, NCoR, and CBP recruitment were enhanced compared with control group. Our data from the reporter assay in transient transfection, endogenous GH mRNA, and ChIP assay showed that SUMO1 had the most influence on TRβ/T3-mediated rGH gene stimulation in GH3 cells.
FIGURE 6.
The influence of SUMO1 and SUMO3 expression on endogenous rGH mRNA expression and recruitment of TRβ and cofactors to the TRE. A, GH3 cells were transfected with siRNA SUMO1 or SUMO3. Three days after transfection, the medium was changed to serum-replaced medium, and cells were allowed to grow for 16 h. Cells were treated with or without T3 (50 nm) for 4 h prior to isolating RNA. The endogenous rGH mRNA expression in GH3 cells was detected by q-PCR. C, control. B, ChIP assays were performed using GH3 cells transfected with TRβ and siRNA SUMO1 or siRNA SUMO3 and antibodies (ab) anti-TRβ, anti-NCoR, and anti-CBP. C, WB analysis of SUMO1 and -3 knockdown. D, the rGH TRE-luc reporter activity was analyzed in TRβ- and TRβ K443Q-transfected GH3 cells with or without addition of 50 nm T3. E, ChIP assay of TR binding to rGH TRE and interaction with NCoR and CBP. GH3 cells were transfected with TRβ and TRβ K443Q and treated with or without T3 as described in A. Antibodies used in ChIP assay were anti-TRβ, anti-NCoR, and anti-CBP. The specific region of the rGH TRE was q-PCR quantified (see Table 1 for primers). F, endogenous rGH mRNA expression in the presence of transfected TRβ or TRβ K443Q. T3 treatment is the same as described in A. G, WB shows the protein level of TRβ and TRβ K443Q in transfected cells from the experiment shown in E. *, indicate p < 0.05 compared with controls.
The observed impact of SUMO knockdown on T3-dependent gene expression could be the direct result of TR sumoylation but could also be indirectly related to sumoylation of other cofactors. We wanted to determine the influence of specific disruption of TR sumoylation. We utilized the TRβ sumoylation site mutant, K443Q, to determine the influence of SUMO on TR/T3-mediated transcription and recruitment of cofactors in GH3 cells. In a reporter assay, T3 induction was significantly diminished with transfection of TRβ K443Q, compared with wild-type TRβ (Fig. 6D). In the presence of TRβ K443Q, T3 enhanced rather than disrupted NCoR binding to the rGH TRE (Fig. 6E). With transfection of TRβ K443Q, the GH mRNA level was increased 2.2-fold in the absence of T3 and reduced 0.6-fold with T3 treatment, compared with the actions of WT TRβ (Fig. 6F). These data indicate that the functional effects of SUMO knockdown are reproduced with a TR sumoylation mutant, indicating that sumoylation of TR is sufficient to explain the effects of sumoylation on T3 signaling. Sumoylation of cofactors may additionally influence T3 action but are not the primary sumoylation targets that explain the functional results.
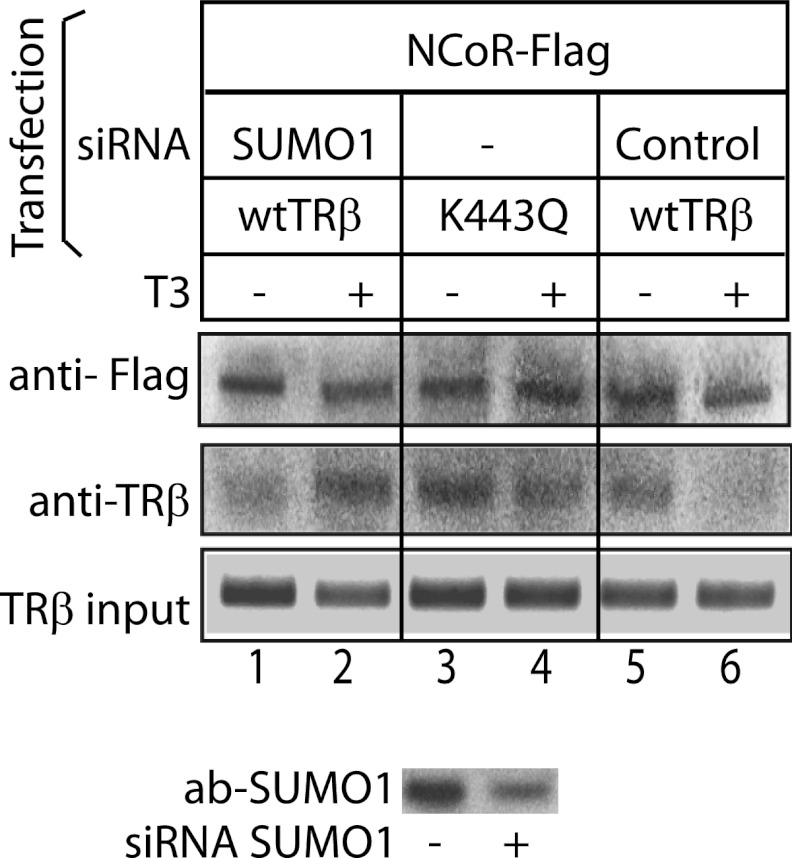
The ChIP assay demonstrated that SUMO knockdown resulted in impairment of ligand-dependent release of NCoR. We performed co-immunoprecipitation using HepG2 cell cotransfected with NCoR-FLAG and TRβ or TRβ K443Q with or without siRNA SUMO1 (Fig. 7). In the control condition, NCoR and TRβ were co-immunopreciptated in the absence of T3, but TRβ did not bind NCoR in the presence of T3 (Fig. 7, lanes 5 and 6). After either SUMO1 knockdown (Fig. 7, lanes 1 and 2) or in the presence of the TRβ K443Q (Fig. 7, lanes 3 and 4), addition of T3 did not disrupt the NCoR-TRβ complex. Sumoylation of TRβ, therefore, is important for ligand-dependent release of NCoR from TRβ.
FIGURE 7.
Ligand-dependent release of corepressor from TRβ was disrupted by TRβ K443Q mutation. HepG2 cells were cotransfected with NCoR-FLAG and TRβ or TRβ K443Q. Cells were cotransfected with siRNAs and expression vectors as indicated in the figure. Coimmunoprecipitation was performed using affinity anti-FLAG resin in coimmunoprecipitation buffer (see “Experimental Procedures”). The immunoprecipitated complexes were detected in WB using anti-FLAG antibody (ab) to detect NCoR and anti-TRβ antibody. SUMO1 knockdown efficiency is shown in the WB in the lower panel.
Previously, we observed the differential effects of SUMO2 and SUMO3 in reporter expression, suggesting differential effects of SUMO2/3 on interaction of TR with cofactors. We performed a ChIP assay using GH3 cells with transfected siRNA SUMO2 and TRβ with or without T3 treatment (supplemental Fig. S4). After siRNA SUMO2 knockdown, the protein interaction pattern on the rGH TRE was similar to control cells, as NCoR was dissociated from TR after T3 treatment. In contrast, after knockdown of SUMO3 NCoR association with TR was not disrupted by T3 (Fig. 6). The differential effects of SUMO2 and SUMO3 has been reported by others (25, 26).
Sumoylation Modulates TR Binding and Recruitment of Cofactors to an Endogenous nTRE
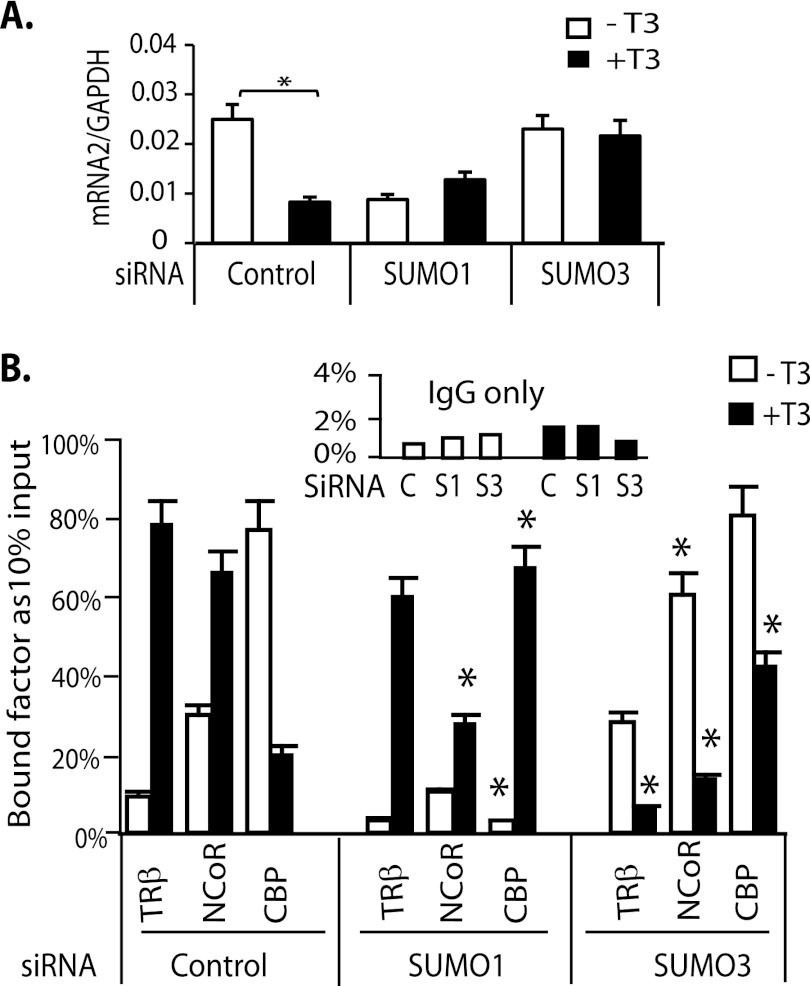
We investigated whether SUMO knockdown influenced endogenous TSHβ mRNA expression in a similar fashion to the TSHβ nTRE in transient transfection. The rat TSHβ gene has two transcription start sites (TSS1 and TSS2), which generate a longer and shorter transcript. The shorter transcript is repressed by T3 and reflects T3 regulation of TSH (27). We measured the shorter transcript as the TSHβ mRNA and determined the effects of siRNA SUMO knockdowns in GH3 cells on TSHβ mRNA expression (Fig. 8A). In control cells, TSHβ was repressed 2.9-fold by T3 (10 nm). In cells transfected with siRNA SUMO1, the TSHβ mRNA was markedly reduced in the absence of T3 compared with the control cells. In the presence of T3, mRNA was increased but not to a significant level (p < 0.066). After SUMO3 knockdown, however, the level of TSHβ mRNA expression did not change in response to T3 (Fig. 8A). These results indicate that SUMO3 is required for endogenous TSHβ gene regulation by TRβ, as it was required for regulation of the TSHβ nTRE in the transient transfection assay.
FIGURE 8.
The influence of SUMO1 and SUMO3 on endogenous TSHβ mRNA expression and recruitment of endogenous TRβ and cofactors to the TSHβ nTRE. A, the endogenous T3-responsive TSHβ mRNA was determined using gene-specific primer (GSP2 and GSP3) for cDNA synthesis with cell culture and treatment conditions as described in the legend to Fig. 6. q-PCR primers GSP2 and GSP-reverse were used (see Table 1 for primer sequences). B, ChIP assays using GH3 cells. T3 treatment and assay conditions were the same as described in the legend to Fig. 6. *, indicates significance p < 0.05 compared with control.
We demonstrated by ChIP assay that SUMO influences endogenous TR DNA binding and interaction with cofactors. In the control condition (transfected with non-targeting siRNA), T3 significantly enhanced TRβ binding to the TSHβ nTRE, resulting in an increase in recruitment of NCoR and release of CBP (Fig. 8B). This is the expected pattern of TR-DNA binding and cofactor interaction on the TSHβ nTRE in response to ligand. After SUMO1 knockdown, the magnitude of NCoR recruitment was reduced compared with the control, either in the absence or presence of T3. CBP binding was diminished in the absence of T3 but significantly increased in the presence of T3. These data support the functional data showing T3 induction after SUMO1 knockdown. SUMO3 knockdown promoted TRβ interaction with DNA and recruitment of NCoR in the absence of T3 (Fig. 8B), consistent with increased basal activity in the reporter assay. Conversely, in the presence of T3, TRβ binding was reduced 87%, NCoR recruitment was reduced 75%, and CBP was increased 47% compared with control, consistent with T3 induction seen in reporter assay. These results demonstrate that sumoylation influences TR recruitment of corepressor and coactivator to the TSHβ nTRE.
DISCUSSION
We have demonstrated that TRα and TRβ are SUMO substrates. Multiple forms of SUMO-TR were observed, with the highest molecular mass form ∼250 kDa. There were some variations in the extent and nature of TR sumoylation among the different tissues studied. Although SUMO runs larger in SDS-PAGE than its actual size, we suspect a poly-SUMO chain associated with the higher molecular weight species. Conjugation by a poly-SUMO chain usually has more repressive effects on transcription than single SUMO conjugation. Repressive and activating actions of TR vary by the ligand status and interaction with a TRE or nTRE, so it is unlikely that TR-sumoylation alone is the sole determining factor for repression or activation of transcription.
There were distinct roles for SUMO1 and SUMO3 in sumoylation of TRβ and TRα and in T3-dependent gene regulation, with little apparent role for SUMO2. The influence of sumoylation of TR on positive and negative gene regulation had variable effects that were a function of the individual SUMO, the TRE/nTRE, and TR isoforms. SUMO1 and SUMO3 were critical for T3-induced gene expression. The rGH TRE showed distinct patterns of the influence of SUMO1 and SUMO3 on gene expression with TR isoform preference. For example, SUMO1 and SUMO3 were important for TRβ/T3-induction and SUMO3 for TRα/T3 induction.
SUMO modification influences protein-protein interaction, which has been demonstrated in androgen receptor, glucocorticoid receptor, Smad4, LRH-1 (28–31), and cofactors NCoR and RIP140 (32, 33). We demonstrated by ChIP assay that SUMO1 influences the recruitment of CBP to both positive and negative TREs. SUMO had diverse effects on TR binding to DNA and recruitment of NCoR and CBP. SUMO conjugation to TR may act by modifying TR interactions with cofactors or may also protect TR from proteolytic targeting by ubiquitin (34, 35). The most potent effect of TR sumoylation, however, is likely on its interaction with transcriptional cofactors.
The role of sumoylation in T3-mediated induction and repression was shown in functional studies and TR and cofactor binding to the response elements by ChIP assays after SUMO knockdown. Although we demonstrated that TR is sumoylated, SUMO knockdown may secondarily influence TR by the action on other SUMO substrates, especially nuclear receptor coactivators and corepressors. Both NCoR and CBP are sumoylated by SUMO1 (30, 33). SUMO1-NCoR conjugation increases NCoR interaction with Sin3 and histone deacetylase and promotes repression. Desumoylation of CBP by SENP1 enhances CBP interaction with SRC and histone acetyltransferase and potentiates transcriptional activity. The observed effects, therefore, are likely the result of both TR and cofactor sumoylation.
Several findings, however, support a significant role for TR sumoylation, especially with respect to TR isoform specificity. Although both TR isoforms required sumoylation for T3 induction and repression, the influence of ligand, SUMO type, and E3 ligase preference varied between TRα and β. Isoform specificity, therefore, may be mediated by variations in sumoylation.
To isolate the effects of TR sumoylation, from cofactors that are SUMO substrates and essential for transcription, we studied a TRβ sumoylation mutant, K443Q. The SUMO mutant had a similar profile in the functional assay to the wild-type TR after SUMO knockdown. This sumoylation mutant eliminated T3 induction and prevented the release of NCoR from the endogenous rGH TRE. The TRβ K443Q mutation, however, may influence cofactor interaction as well as SUMO conjugation and will require further study. The cotransfection of the sumoylation mutant may also be influenced by endogenous TRβ, although the comparison with cotransfected wild-type TR demonstrates a clear difference with the TRβ K443Q. The other sumoylation site mutants had similar impairments in T3 induction, although the range of the magnitude of the effects suggests that the SUMO sites likely have different impact on function.
Sumoylation modulates T3-induced gene induction and repression. The specificity of SUMOs, TR isoforms, and E3 ligases support the importance of this posttranslational modification. There was concordance of functional requirements for SUMOs in transient transfection with TR and cofactor binding to endogenous genes. A systematic study of TR and cofactor SUMO mutants will ultimately be needed to determine the relative influence of sumoylation on TR and cofactors in vivo, although the current evidence supports that sumoylation of TR is important. Complimentary approaches to point mutations, however, will be needed as mutations of SUMO-interacting amino acids may secondarily influence receptor structure and other interactions important for function.
This work was supported by Veterans Affairs Merit Review Funds.

This article contains supplemental “Methods” and Figs. S1–S4.
- TR
- thyroid hormone receptor
- T3
- triiodothyronine
- SUMO
- small ubiquitin-like modifier
- PIAS
- protein inhibitor of activated STAT
- UBC9
- ubiquitin carrier protein 9 or SUMO conjugation enzyme
- TRE
- thyroid hormone response element
- CBP
- CREB-binding protein
- nTRE
- negative TRE
- q-PCR
- quantitative PCR
- luc
- luciferase
- CREB
- cAMP-responsive element-binding protein.
REFERENCES
- 1. Lazar M. A. (2003) Thyroid hormone action: a binding contract. J. Clin. Invest. 112, 497–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen J. D., Evans R. M. (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377, 454–457 [DOI] [PubMed] [Google Scholar]
- 3. Bernal J. (2007) Thyroid hormone receptors in brain development and function. Nat. Clin. Pract. Endocrinol. Metab. 3, 249–259 [DOI] [PubMed] [Google Scholar]
- 4. Liu Y. Y., Schultz J. J., Brent G. A. (2003) A thyroid hormone receptor α gene mutation (P398H) is associated with visceral adiposity and impaired catecholamine-stimulated lipolysis in mice. J. Biol. Chem. 278, 38913–38920 [DOI] [PubMed] [Google Scholar]
- 5. Morreale de Escobar G., Obregon M. J., Escobar del Rey F. (2004) Role of thyroid hormone during early brain development. Eur. J. Endocrinol. 151, U25–37 [DOI] [PubMed] [Google Scholar]
- 6. Ribeiro M. O., Carvalho S. D., Schultz J. J., Chiellini G., Scanlan T. S., Bianco A. C., Brent G. A. (2001) Thyroid hormone-sympathetic interaction and adaptive thermogenesis are thyroid hormone receptor isoform-specific. J. Clin. Invest. 108, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sjögren M., Alkemade A., Mittag J., Nordström K., Katz A., Rozell B., Westerblad H., Arner A., Vennström B. (2007) Hypermetabolism in mice caused by the central action of an unliganded thyroid hormone receptor alpha1. EMBO J. 26, 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y. Y., Brent G. A. (2010) Thyroid hormone cross-talk with nuclear receptor signaling in metabolic regulation. Trends Endocrinol. Metab. 21, 166–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rusch A., Ng L., Goodyear R., Oliver D., Lisoukov I., Vennstrom B., Richardson G., Kelley M. W., Forrest D. (2001) Retardation of cochlear maturation and impaired hair cell function caused by deletion of all known thyroid hormone receptors. J. Neurosci. 21, 9792–9800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gauthier K., Chassande O., Plateroti M., Roux J. P., Legrand C., Pain B., Rousset B., Weiss R., Trouillas J., Samarut J. (1999) Different functions for the thyroid hormone receptors TRα and TRβ in the control of thyroid hormone production and postnatal development. EMBO J. 18, 623–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Forrest D., Hanebuth E., Smeyne R. J., Everds N., Stewart C. L., Wehner J. M., Curran T. (1996) Recessive resistance to thyroid hormone in mice lacking thyroid hormone receptor β: evidence for tissue-specific modulation of receptor function. EMBO J. 15, 3006–3015 [PMC free article] [PubMed] [Google Scholar]
- 12. Gullberg H., Rudling M., Saltó C., Forrest D., Angelin B., Vennström B. (2002) Requirement for thyroid hormone receptor beta in T3 regulation of cholesterol metabolism in mice. Mol. Endocrinol. 16, 1767–1777 [DOI] [PubMed] [Google Scholar]
- 13. Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 14. Zhao J. (2007) Sumoylation regulates diverse biological processes. Cell Mol. Life Sci. 64, 3017–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheng C. H., Lo Y. H., Liang S. S., Ti S. C., Lin F. M., Yeh C. H., Huang H. Y., Wang T. F. (2006) SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 20, 2067–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gill G. (2005) Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15, 536–541 [DOI] [PubMed] [Google Scholar]
- 17. Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K. M., Jackson S. P. (2009) Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamard P. J., Boyer-Guittaut M., Camuzeaux B., Dujardin D., Hauss C., Oelgeschläger T., Vigneron M., Kedinger C., Chatton B. (2007) Sumoylation delays the ATF7 transcription factor subcellular localization and inhibits its transcriptional activity. Nucleic Acids Res. 35, 1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meulmeester E., Melchior F. (2008) Cell biology: SUMO. Nature 452, 709–711 [DOI] [PubMed] [Google Scholar]
- 20. Hay R. T. (2005) SUMO: a history of modification. Mol Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 21. Darling D. S., Burnside J., Chin W. W. (1989) Binding of thyroid hormone receptors to the rat thyrotropin-β gene. Mol. Endocrinol. 3, 1359–1368 [DOI] [PubMed] [Google Scholar]
- 22. Carr F. E., Wong N. C. (1994) Characteristics of a negative thyroid hormone response element. J. Biol. Chem. 269, 4175–4179 [PubMed] [Google Scholar]
- 23. Brent G. A., Larsen P. R., Harney J. W., Koenig R. J., Moore D. D. (1989) Functional characterization of the rat growth hormone promoter elements required for induction by thyroid hormone with and without a co-transfected β type thyroid hormone receptor. J. Biol. Chem. 264, 178–182 [PubMed] [Google Scholar]
- 24. Liu Y. Y., Nakatani T., Kogai T., Mody K., Brent G. A. (2011) Thyroid hormone and COUP-TF1 regulate kallikrein-binding protein (KBP) gene expression. Endocrinology 152, 1143–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosendorff A., Sakakibara S., Lu S., Kieff E., Xuan Y., DiBacco A., Shi Y., Shi Y., Gill G. (2006) NXP-2 association with SUMO-2 depends on lysines required for transcriptional repression. Proc. Natl. Acad. Sci. U.S.A. 103, 5308–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu G., Warbrick E. (2006) The p66 and p12 subunits of DNA polymerase delta are modified by ubiquitin and ubiquitin-like proteins. Biochem. Biophys. Res. Commun. 349, 360–366 [DOI] [PubMed] [Google Scholar]
- 27. Carr F. E., Need L. R., Chin W. W. (1987) Isolation and characterization of the rat thyrotropin β-subunit gene. Differential regulation of two transcriptional start sites by thyroid hormone. J. Biol. Chem. 262, 981–987 [PubMed] [Google Scholar]
- 28. Poukka H., Karvonen U., Janne O. A., Palvimo J. J. (2000) Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl. Acad. Sci. U.S.A. 97, 14145–14150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tian S., Poukka H., Palvimo J. J., Jänne O. A. (2002) Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem. J. 367, 907–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuo H. Y., Chang C. C., Jeng J. C., Hu H. M., Lin D. Y., Maul G. G., Kwok R. P., Shih H. M. (2005) SUMO modification negatively modulates the transcriptional activity of CREB-binding protein via the recruitment of Daxx. Proc. Natl. Acad. Sci. U.S.A. 102, 16973–16978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Venteclef N., Jakobsson T., Ehrlund A., Damdimopoulos A., Mikkonen L., Ellis E., Nilsson L. M., Parini P., Jänne O. A., Gustafsson J. A., Steffensen K. R., Treuter E. (2010) GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRbeta in the hepatic acute phase response. Genes Dev. 24, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rytinki M. M., Palvimo J. J. (2008) SUMOylation modulates the transcription repressor function of RIP140. J. Biol. Chem. 283, 11586–11595 [DOI] [PubMed] [Google Scholar]
- 33. Tiefenbach J., Novac N., Ducasse M., Eck M., Melchior F., Heinzel T. (2006) SUMOylation of the corepressor N-CoR modulates its capacity to repress transcription. Mol. Biol. Cell 17, 1643–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson E. S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 35. Uzunova K., Göttsche K., Miteva M., Weisshaar S. R., Glanemann C., Schnellhardt M., Niessen M., Scheel H., Hofmann K., Johnson E. S., Praefcke G. J., Dohmen R. J. (2007) Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282, 34167–34175 [DOI] [PubMed] [Google Scholar]
- 36. Petty K. J., Desvergne B., Mitsuhashi T., Nikodem V. M. (1990) Identification of a thyroid hormone response element in the malic enzyme gene. J. Biol. Chem. 265, 7395–7400 [PubMed] [Google Scholar]