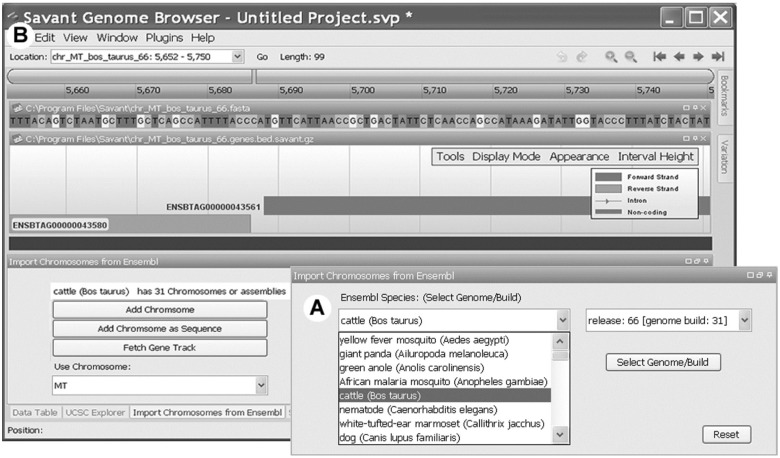
Fig. 5.
JEnsembl plug-in for Savant genome browser. (A) The user selects the desired species and release version from those available at the selected datasource (Ensembl, EnsemblGenomes or EnsemblGenomes-Bacterial). (B) A single chromosome/assembly is selected from those available for the chosen species/release. The chromosome is imported either as a simple coordinate skeleton or with the associated colour-coded genomic sequence. Currently, the only feature annotation that can be imported from the datasource is the gene track, which Savant shows aligned with the DNA Sequence